Background: Salivary biomarkers for systemic diseases have been undermined due to lack of mechanistic and biological rationale.
Results: Suppression of exosome biogenesis leads to ablation of salivary biomarkers.
Conclusion: Tumor-derived exosomes provide a mechanism for discriminatory biomarkers in saliva.
Significance: Tumor-derived exosomes provide the scientific rationale that connects pancreatic tumors and the oral cavity leading to salivary biomarkers.
Keywords: Biomarkers, Cancer, Exosomes, mRNA, Pancreas, Saliva
Abstract
Recent studies have demonstrated that discriminatory salivary biomarkers can be readily detected upon the development of systemic diseases such as pancreatic cancer, breast cancer, lung cancer, and ovarian cancer. However, the utility of salivary biomarkers for the detection of systemic diseases has been undermined due to the absence of the biological and mechanistic rationale as to why distal diseases from the oral cavity would lead to the development of discriminatory biomarkers in saliva. Here, we examine the hypothesis that pancreatic tumor-derived exosomes are mechanistically involved in the development of pancreatic cancer-discriminatory salivary transcriptomic biomarkers. We first developed a pancreatic cancer mouse model that yielded discriminatory salivary biomarkers by implanting the mouse pancreatic cancer cell line Panc02 into the pancreas of the syngeneic host C57BL/6. The role of pancreatic cancer-derived exosomes in the development of discriminatory salivary biomarkers was then tested by engineering a Panc02 cell line that is suppressed for exosome biogenesis, implanting into the C56BL/6 mouse, and examining whether the discriminatory salivary biomarker profile was ablated or disrupted. Suppression of exosome biogenesis results in the ablation of discriminatory salivary biomarker development. This study supports that tumor-derived exosomes provide a mechanism in the development of discriminatory biomarkers in saliva and distal systemic diseases.
Introduction
Discriminatory salivary biomarkers for the detection of systemic diseases have emerged in the last decade with the availability of high throughput omics technologies and systems biology tools. Because collection is noninvasive and easily accessible, using saliva for the early detection of systemic diseases is ideal. To date, transcriptomic and proteomic salivary biomarkers discriminatory for systemic diseases such as breast, ovarian, lung, and pancreatic cancer have been successfully discovered and pre-validated (1–3). However, with the development of these disease-specific biomarkers arises the pertinent question of how discriminatory profiles of biomarkers for diseases developing distally from the oral cavity would appear in saliva. Presently, the answer to this puzzling question remains elusive.
In 2010, Palanisamy et al. (4) showed that saliva contained exosomes where these transferable saliva-derived microvesicles were found to contain proteins and functional mRNA. Moreover, we recently showed that breast cancer-derived exosome-like microvesicles are capable of activating transcription in salivary gland cells and ultimately altering the salivary gland cell-derived exosome-like microvesicles both proteomically and transcriptomically (5). These findings suggested that tumor-derived exosomes could function as the shuttle between the distal tumor and the oral cavity leading to the development of discriminatory salivary biomarkers.
Exosomes are durable, cell-specific lipid microvesicles (30–100 nm) that are able to migrate systemically through the vasculature of the body promoting intercellular communication (6). They reside in a multitude of biofluids, including urine, blood, breast milk, bronchial lavage fluid, cerebral spinal fluids, and saliva (4, 7–11). Even though specific physiological implications of exosomes have yet been elucidated, the literature suggests that they are involved in a variety of functions, ranging from immune response regulators to tumor invasion promoters (12, 13).
This study provides an in vivo examination of the mechanism for salivary biomarker development in a systemic disease. We hypothesized that tumor-derived exosomes play a pivotal role in the development of pancreatic cancer-specific salivary transcriptomic biomarkers. We tested our hypothesis by using an orthotopic, syngeneic pancreatic cancer mouse model. First, we developed a panel of salivary transcriptomic biomarkers discriminatory for pancreatic cancer in the rodent model. Second, we demonstrated that inhibiting the biogenesis and secretion of exosomes at the tumor source disrupted the panel's pancreatic cancer-specific transcriptomic salivary biomarkers. The findings of this in vivo study illustrated that the inhibition of pancreatic cancer-derived exosomes in mice altered the disease-specific salivary transcriptomic biomarker profile, supporting that tumor-derived exosomes play a role in the development of disease-specific discriminatory biomarkers in saliva.
EXPERIMENTAL PROCEDURES
Reagents
The following reagents were used: McCoy's 5A medium (DMEM, Invitrogen); fetal bovine serum (FBS, Cellgro); 100× penicillin/streptomycin (5000 μg/ml, Cellgro), phosphate-buffered saline (PBS, Invitrogen); Lipofectamine (Invitrogen); paraformaldehyde (Sigma); green fluorescence protein (GFP) antibody (Cell Signaling); β-actin antibody (Sigma); horseradish peroxidase-coupled secondary antibody (Invitrogen); ketamine (60 mg/ml; Phoenix Scientific, St. Joseph, MO); xylazine (8 mg/ml; Phoenix Scientific); pilocarpine (0.05 mg pilocarpine/100g body weight; Sigma); geneticin (G418, 400 μg/ml; Sigma); Lipofectamine 2000 (Invitrogen); DNase (Ambion); RiboAmp RNA amplification kit (Molecular Devices); and GeneChip Expression 3′-amplification reagents (Affymetrix).
Animals
Six- to 8-week-old C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed in the Division of Laboratory Medicine at UCLA. The Chancellor's Animal Research Committee at UCLA approved the experimental protocols.
In Vivo Pancreatic Tumor Induction
The pancreatic cancer mouse model was induced by orthotopic injection of 1.0 × 106 Panc02 cells in 30 μl of PBS into the mouse pancreas. After 4 weeks, mice were sacrificed, and pancreatic tumors were observed histologically.
Collection of Mouse Saliva, Blood, and Tumor Tissue
Saliva, blood, and tumor tissue from mice were collected as described previously (26). Upon the development of the pancreatic tumor (4 weeks), saliva was collected, and then the mouse was sacrificed. Mild anesthesia was induced by intramuscular of 1 μl/kg body weight of a solution containing 60 mg/ml ketamine and 8 mg/ml xylazine. Salivation was induced by subcutaneous injection of pilocarpine (0.05 mg pilocarpine/100 g body weight) between the ears of the mice; saliva was obtained from the oral cavity by micropipette and immediately placed on pre-chilled 1.5-ml microcentrifuge tubes. The samples were stored in −80 °C until analyzed.
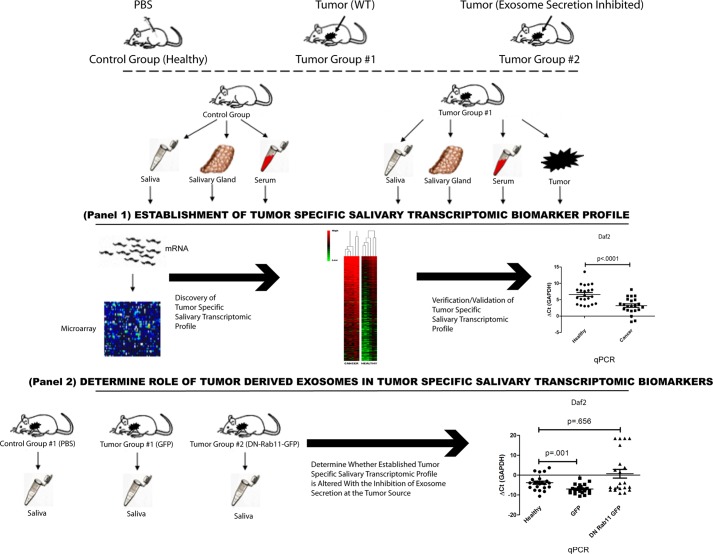
For the validation of pancreatic cancer-specific salivary transcriptomic biomarkers, C57BL/6 mice were randomly assigned to a control group and a tumor group (22 animals per group). To determine whether inhibition of tumor-derived exosomes altered pancreatic cancer-specific salivary transcriptomic biomarkers, C57BL/6 mice were randomly assigned to a control group, tumor group 1 (with Panc02-expressing GFP) and tumor group 2 (with Panc02-expressing DN-Rab11-GFP; 22 animals per group). Fig. 1 provides the schematic summarizing the complete procedure.
FIGURE 1.
Summary of study. Illustration depicting the study conducted for the examination of whether tumor-derived exosomes are involved in pancreatic cancer-discriminatory salivary transcriptomic profile. The study was conducted in a bi-fold fashion, first by establishing a mouse model that yielded pancreatic cancer-discriminatory transcriptomic biomarkers (Panel 1). Then, using the established model, we used the Panc02 cell line that is manipulated with defective ability to secrete exosomes and determine whether the established salivary transcriptomic profile is affected (Panel 2).
Blood was collected in a Serum Gel Z/1.1 tube, and serum was extracted by centrifugation at 14,000 × g for 10 min after mice were sacrificed. Salivary glands and pancreatic tumor tissues were removed from mice, snap-frozen in liquid, and stored in −80 °C until analyzed.
RNA Extraction from Mouse Salivary Glands and Pancreases
On ice, tissue sample was submerged in TRIzol (Invitrogen; 1 ml of TRIzol/50–100 mg of tissue). Tissues were homogenized in the TRIzol solution until no visible particles remained. Addition of chloroform (Sigma) and subsequent centrifugation at 14,000 × g separated the mixture into three phases (lower red phenol/chloroform phase, interphase, and upper aqueous phase). The upper aqueous phase was then extracted and mixed with the 100% isopropyl alcohol (Sigma) and centrifuged at 12,000 × g at 4 °C. RNA pellet was washed with 75% ethanol, centrifuged at 7500 × g, air dried, resuspended in RNase-free water, and analyzed.
Handling, Fixing, and Staining of Mouse Pancreases for Microscopy
Tissues were fixed in 4% paraformaldehyde (Sigma) for 6 h, washed with PBS, and then submerged in 30% EtOH overnight at 4 °C. Tissues were submerged in O.C.T. compound (Tissue-Tek) and snap-frozen in liquid nitrogen. The tissues were subsequently sectioned at ∼5 μm slices and stained with hematoxylin and eosin solutions at the Pathology and Laboratory Medicine Microscopic Techniques Core Facility at the UCLA Brain Research Institute.
Maintenance and Stable Transfection of Panc02
Murine pancreatic cell line Panc02 was obtained from Dr. Guido Eibl, David Geffen School of Medicine at UCLA. Panc02 is a methylcholanthrene-induced pancreatic carcinoma that is syngeneic to C57BL/6 mice (27). Cells were cultured in McCoy's 5A medium with 10% FCS and penicillin/streptomycin. All cells were maintained in an atmosphere of 5% CO2 at 37 °C.
Plasmids pEGFP-C1 (GFP) and pEGFP-C1-Rab11-DN (Addgene plasmid 12678; 4 μg; DN-Rab11-GFP) were transfected into Panc02 cultured in 6-well plates at ∼85% confluency using Lipofectamine 2000 via the manufacturer's protocol. Panc02 cells were passaged at 1:10 into a 12-well plate 24 h post-transfection and into fresh McCoy's 5A medium with 10% FCS and penicillin/streptomycin. 48 h after transfection, 400 μg/ml of geneticin were added; after 3 weeks in selection, the surviving colonies were isolated to a 24-well plate and expanded.
Discovery of Pancreatic Cancer-discriminatory Candidate Genes in Saliva
For the discovery of candidate pancreatic cancer-discriminatory salivary transcriptomic biomarkers, saliva RNA was isolated using the RNeasy mini kit (Qiagen) according to manufacturer's protocol. There were 30 mice each in the control and tumor group. Samples derived from five mice in each group were pooled for RNA extraction. Pooling was necessary to ensure that sufficient salivary mRNA was obtained for microarray analyses. Isolated total RNA was treated with recombinant DNase. For microarray analysis, mRNA from mouse saliva was linearly amplified using the RiboAmp RNA amplification kit (Molecular Devices). After purification, cDNAs were transcribed in vitro and biotinylated using GeneChip Expression 3′-amplification reagents for labeling of transcripts (Affymetrix). The labeled RNAs (15 μg each) were subsequently fragmented and sent to the UCLA Microarray Core Facility for array hybridization and scanning. The GeneChip Mouse Genome 430 2.0 array, which represents >39,000 transcripts and variants, was used for profiling analysis. The microarray data have been uploaded to www.ncbi. nlm.gov. The accession number is GSE47811.
Arrays were analyzed using R 2.7.0. The probe logarithmic intensity error estimation (PLIER) expression measures were computed after background correction and quantile normalization. Quantile normalization was performed across all samples to make the distributions similar and comparable among all probe sets. For every probe set, the two-sample t test was applied to identify differential expression between saliva samples obtained from pancreatic cancer-bearing mice and healthy mice. After obtaining the estimates and p values for each probe set, we corrected the p values for the false discovery rate. A score was then generated based on the corrected p values and differential expression levels. The heat map dendrogram was constructed using agglomerative hierarchical clustering of the 209 significantly up-regulated genes.
Direct Saliva Transcriptomic Analysis
DSTA2 was conducted as described previously (20). Saliva collected from mice was analyzed by a two-step RT-PCR followed by quantitative PCR (qPCR), operated separately. The PCR primers for the 20 genes (supplemental Table S1) were designed using Primer Express 3.0 software (Applied Biosystems). Multiplex RT-PCR preamplification of the 20 mRNAs was performed by using SuperScript III platinum qRT-PCR system (Invitrogen) with a pool of outer primer sets (100 μmol/ml for each), and conducted by a GeneAmp PCR-System 9700 (Applied Biosystems) with a fixed thermal cycling program.
SDS-PAGE and Protein Staining
Cell lysates were resuspended in Laemmli Sample Buffer (62.5 mm Tris-HCl, pH 6.8, 2% w/v SDS, 50 mm dithiothreitol, 0.01% bromphenol blue, w/v) and analyzed by SDS-PAGE followed by staining according to the manufacturer's instructions.
Western Blotting
The membrane was blocked with 5% milk solution and incubated with GFP (Cell Signaling) or β-actin antibody (Abcam), followed by incubation with the appropriate horseradish peroxidase-coupled secondary antibody. The proteins were detected using the ECL Western blotting detection system (GE Healthcare).
Microscopy
Panc02 cells or pancreas images were taken on Olympus IX80 using ×20 or ×40 objectives.
Isolation of Exosomes
Panc02 cells were grown to 80% confluency and incubated in DMEM + 10% exosome-free FBS for 48 h. The culture supernatant was centrifuged at 300 × g for 10 min to remove suspended cells. The cell pellet was discarded, and the supernatant was centrifuged at 2000 × g for 10 min to remove dead cells and then 10,000 × g for 30 min to remove cell debris. Next, the supernatant was centrifuged at 100,000 × g for 70 min, the supernatant removed, the pellet washed with PBS, and centrifuged at 100,000 × g for 70 min, and then filtered using a 0.22-μm PVDF syringe filter (Millipore), resulting in purified exosome-like microvesicles.
Acetylcholine Esterase Assay
The amount of released exosomes was quantified by using the Amplex® red acetylcholine/acetylcholinesterase assay kit (Molecular Probes) according to the manufacturer's protocol. Acetylcholinesterase (AchE) is an enzyme that is specifically directed to exosomes (15), and the data represent AchE enzymatic activity (nanounits/microliter/cell) after 30 min of incubation.
Magnetic Bead-based Exosome Extraction
For rapid, effective extraction, magnetic bead-based technology was applied to isolate and accumulate exosomes. The magnetic beads were ∼1–2 μm in diameter, as measured by TEM. Briefly, the streptavidin-coated magnetic beads (Invitrogen) were mixed with biotinylated lectin specific to α-2,6-linked sialic acid (28) (Vector Laboratories) on a mixer for 30 min at room temperature. Then, 10 μl of Panc02 cell medium, serum, or saliva were incubated with the lectin-conjugated magnetic beads in casein-PBS (Invitrogen). Samples were mixed for 2 h at room temperature to form exosome-magnetic bead complexes. Next, exosome-magnetic bead complexes were attracted onto the electrochemical sensor by applying an array of magnets. The unattached species were removed by washing.
Exosomal mRNA Release and Measurement
Seven exosomal mRNAs were measured by EFIRM. For mRNA detection, the electrodes were coated with a surface matrix of conducting polymer pyrrole. Then the surface of the electrode was pre-coated with oligonucleotide capture probes with sequences specific for the selected seven genes in addition to GAPDH as a reference gene and a biotin label at the 5′ end. The immobilizations of the capture probes were carried out by applying a csw-E field for 20 cycles of 9 s at −300 mV followed by 1 s at +200 mV (200 s total) (29).
After the exosome extraction step, the exosome-magnetic bead complex was collected onto the capture probe-coated electrode with an array of magnets placed underneath the electrochemical sensor. When loading the sample onto the electrode, 10 nm of detector probes (with a fluorescein-labeled 3′ end) were mixed in with the exosome-magnetic bead complexes. The csw-E field was then applied to release the exosomal mRNA from the exosomes. The csw-E field was 20 cycles of 9 s at −300 mV followed by 1 s at +200 mV (200 s total) for the hybridization with the oligonucleotide capture and detector probes. Then, we added 150 units/ml of anti-fluorescein antibody conjugated to horseradish peroxidase (HRP; 1:1000 dilution; Roche Applied Science). Finally, the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate for horseradish peroxidase was loaded, and an amperometric signal was read out.
TEM Measurement of Exosomes
The serum and saliva samples were loaded onto carbon-coated grids. Because of interference between the magnetic beads and the electric field of the electron microscope, specific grids were used to hold the magnetic beads. In addition, we used support film for TEM with a wide dynamic range to accommodate the size differences between the magnetic beads (1–2 μm diameter) and the exosomes (60–100 nm diameter). We selected Lacey Formvar/Carbon with 200-mesh copper (Ted-Peller) for the TEM support film. The samples were examined with a JEOL 100CX transmission electron microscope (JEOL USA, Inc., Peabody, MA).
Electrochemical Sensor
The electrochemical sensor was an array of 16 bare gold electrode chips (Gene Fluidics). Each unit of the array had a working electrode, a counter electrode, and a reference electrode. The species to be measured were immobilized on the working electrode. The array of 16 plastic wells separated each three-electrode set, which avoided cross-contamination between different sensors. A conducting polymer was deposited on the working electrodes as supporting film. The 16-channel electrochemical reader (Gene Fluidics Inc.) controlled the electrical field applied to the 16 array sensors and simultaneously recorded the amperometric current (29, 30).
All the electrical potentials in the following steps were relative to the gold reference electrode, which was determined to be +218 mV. In contrast, saturated calomel electrode was determined by measuring cyclic voltammetric curves of 0.1 mm [Fe(CN)6]3−/4−. For the experiments, solutions were loaded onto the whole area of the three-electrode region contained within the plastic wells. For the release and reaction steps, the csw-E field was applied.
The signal readout was the electrochemical current from the HRP reaction. The TMB substrate (TMB/H2O2, low activity) (Neogen) was loaded, and amperometric detection was carried out by applying −200 mV to each electrode unit, followed by a parallel amperometric signal readout after 60 s of equilibration. TMB acts as a mediator; it is reduced at −200 mV, and in turn, the reduced TMB reduces the oxidized form of HRP. HRP then reduces H2O2 to two H2O molecules, and the HRP is re-oxidized.
Statistical Analysis
All graphs were made and statistical analyses performed using R 2.7.0, GraphPad Prism, SAS 9.3, or Microsoft Excel 2007. The heat map dendrogram was constructed using agglomerative hierarchical clustering. Groups were formed based on the average linkage function. One-way analysis of variance, two-sample t tests were used for testing the difference between expression values. Wilcoxon rank-sum tests were used to determine whether the adjusted ΔCt values differed between groups (p values <0.05 were considered statistically significant). Data are expressed as mean ± S.E.
RESULTS
Induction of Pancreatic Cancer in Mice and Discovery of Pancreatic Cancer-specific Salivary Transcriptomic Candidate Markers
The Panc02/C57BL/6 orthotopic, syngeneic mouse pancreatic cancer model is schematically shown in Fig. 1. Orthotopic injections of Panc02 cells into mouse pancreases induced pancreatic cancer in ∼4 weeks (supplemental Fig. S1, A–H).
Expression microarray analysis of the saliva collected from both the cancer-bearing and noncancer control mice revealed different saliva profiles between the two groups. The supplemental Fig. 5 shows a heat map with two distinct groups formed by the cluster analysis of 209 up-regulated genes differentially expressed in the saliva of pancreatic cancer mice versus noncancer control mice (p value<0.05; fold change ≥2). Statistical analysis selected the 20 top-scoring candidate genes based on p values and fold change used for validation, listed in Table 1 and supplemental Table S4.
TABLE 1.
Discovery of pancreatic cancer-discriminatory transcriptomic biomarkers in mouse saliva
Array analysis of saliva extracted from pancreatic cancer-bearing mice and healthy mice revealed differential transcriptomic saliva profile between the two populations. Listed here are the top scoring 20 up-regulated transcripts, their respective functions, p values, and fold differences between control and cancer saliva. A list of the top 20 up-regulated transcripts was determined using R 2.7.0 according to their p values and fold change. Two-sample t tests were used to determine significance (p values <0.05).
| Gene | Function | p value | -Fold change |
|---|---|---|---|
| Sema 6d | Immune response and gastric cancer | 0.03 | 12.77 |
| Daf2 | Associated with insulin receptor | 0.04 | 11.97 |
| Slc6a1 | GABA transporter | 0.03 | 11.84 |
| Usp12 | Deubiquitinase | 0.03 | 11.6 |
| Rhobtb3 | Small GTPase for actin filamentation | 0.04 | 11.5 |
| Zfp551 | Zinc finger protein | 0.03 | 11.32 |
| Aspn | Binds collagen and calcium | 0.03 | 11.1 |
| Apbb1ip | Actin reorganization | 0.03 | 10.64 |
| Gng2 | G protein signaling | 0.03 | 9.79 |
| BC031781 | Unknown | 0.03 | 9.68 |
| Tchhl1 | Unknown | 0.03 | 9.47 |
| Tardbp | Transcription repressor | 0.03 | 8.52 |
| Apex2 | Endonuclease; excision repair | 0.03 | 8.18 |
| Cog5 | Golgi morphology and function | 0.04 | 8.09 |
| Prl3c1 | Regulator of postimplantation intrauterine events | 0.03 | 7.34 |
| Foxp1 | Tumor suppressor | 0.03 | 7.62 |
| Abcg1 | Macrophage cholesterol and phospholipid transport | 0.03 | 7.5 |
| Ttll9 | Ligase | 0.04 | 7.48 |
| Zic4 | Zinc finger protein for embryo development | 0.04 | 7.33 |
| Incenp | Required for mitosis | 0.03 | 7.31 |
Validation of Pancreatic Cancer-specific Salivary Transcriptomic Biomarkers
The 20 top-ranking candidate salivary transcriptomic biomarkers discriminatory for mouse pancreatic cancer were examined for their biomarker performance using qPCR in an independent group of Panc02/C57BL/6 (n = 22) and noncancer control mice (n = 22) (Fig. 2; Table 2). Seven out of the 20 candidate transcriptomic markers were validated and significantly elevated (p < 0.05) in pancreatic cancer-bearing mouse saliva when compared with noncancer control saliva based on their ΔCt differences (Apbb1ip, Aspn, BCO31781, Daf2, Foxp1, Gng2, and Incenp). The validation of pancreatic cancer-specific salivary biomarkers in this rodent model parallels the translational studies in humans where discriminatory salivary transcriptomic markers have been developed (1).
FIGURE 2.
Scatterplot of the seven validated pancreatic cancer-specific transcripts in mice saliva. Seven out of the 20 discovered genes with discriminatory salivary biomarkers for pancreatic cancer in mice were validated by DSTA.
TABLE 2.
Summary of the seven validated pancreatic cancer-specific transcripts in mouse saliva
| Gene | p value | -Fold change |
|---|---|---|
| Apbb1ip | 0.006 | 4.11 up-regulated |
| Aspn | 0.039 | 2.55 up-regulated |
| BC031781 | <0.0001 | 11.79 up-regulated |
| Daf2 | <0.0001 | 10.63 up-regulated |
| Foxp1 | <0.0001 | 114.56 up-regulated |
| Gng2 | 0.018 | 17.38 up-regulated |
| Incenp | <0.0001 | 11.55 up-regulated |
Detection of the Seven Validated Pancreatic Cancer-specific Salivary Transcriptomic Markers in Pancreatic Cancer Tissues, Salivary Gland, and Saliva Exosomes
We examined the potential origins of the seven validated pancreatic cancer-specific salivary transcriptomic markers by examining whole pancreases and salivary glands of tumor-bearing mice and controls. All seven transcriptomic markers were found to be significantly up-regulated in the pancreases bearing tumors in comparison with noncancer control pancreases (supplemental Table S2). Of the seven validated markers, Daf2 and Gng2 were significantly down-regulated in the tissues derived from submandibular salivary glands of pancreatic cancer-bearing mice in comparison with noncancer controls, whereas the expression of the remaining five genes did not show significant differences upon development of pancreatic cancer (supplemental Table S3).
We then examined the biofluid compartments that connect the distal tumor (pancreatic cancer), blood, and saliva as well as conditioned media from Panc02 cells by selectively capturing exosomes from these bio-fluid compartments based on our working model that exosomes mediate the systemic dissemination of the tumor-derived biological functions. We have developed an exosome-specific detection technology, “Electric Field-induced Release and Measurement (EFIRM),” to selectively capture CD63 (or other exosomal membrane protein markers)-positive exosomes and concurrently perform on chip real time detection for nucleic acids and/or proteins. Using the EFIRM technology, we were able to capture exosomes from saliva, serum, and Panc02 culture media and observed that all seven validated pancreatic cancer-specific transcriptomic markers and GAPDH were detectable in the exosomes derived from saliva, serum, and Panc02 cells (supplemental Fig. S3). Moreover, of the seven markers, six (Apbb1ip (A), Daf2 (C), Foxp1 (D), Incenp (E), BCO31781 (F), and Gng2 (G); see supplemental Fig. S3) were found to be up-regulated in both serum- and saliva-derived exosomes of tumor-bearing mice, although Aspn (supplemental Fig. S3B) was up-regulated in saliva but not in the serum-derived exosomes.
Dominant Negative Rab-11 Transfection Inhibits Exosome Secretion by Panc02 Cells
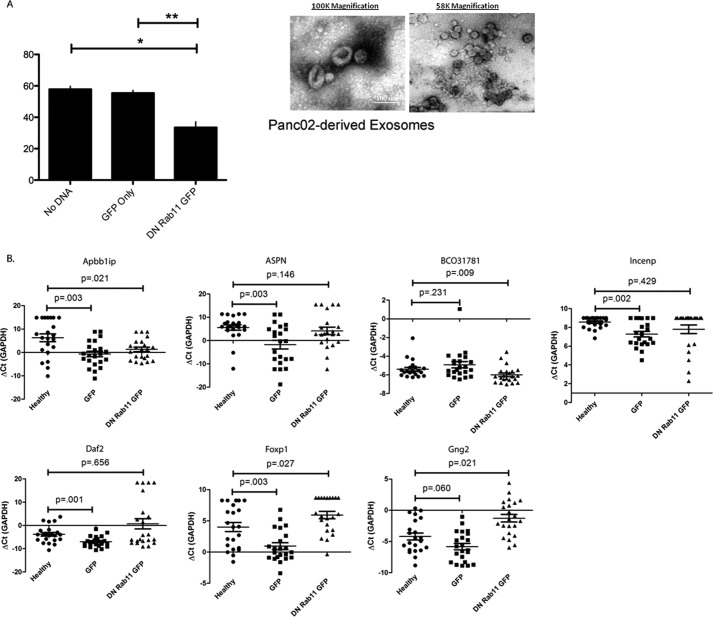
Sevina et al. (14, 15) showed that the expression of the dominant negative form of the GTPase Rab11 (DN-Rab11) could effectively inhibit the biogenesis of exosomes in chronic myelogenous leukemic cell line K562. To examine whether DN-Rab11 expression can inhibit the biogenesis and exosome secretion in Panc02 cells, the mouse pancreatic cancer cell line was stably transfected with either GFP or DN-Rab11-GFP, as confirmed by confocal microscopy and Western blot (supplemental Fig. S2, A and B). To demonstrate that the DN-Rab11 expression can alter exosome biogenesis, the acetylcholine esterase assay was used (14). AchE is an enzyme that is specifically directed to exosomes, and the AchE assay revealed that Panc02 cells stably expressing GFP-Rab11-DN exhibited a significant decrease in exosome secretion/AchE activity in comparison with Panc02 cells expressing GFP only (Fig. 3A). Exosomes extracted from the culture media of untransfected Panc02 cells exhibited AchE activity of 57.81 ± 1.94 nanounits/μl per cell, from Panc02 cells stably transfected with the GFP plasmid exhibited AchE activity of 55.45 ± 1.71 nanounits/μl per cell, and Panc02 cells stably transfected with DN-Rab11-GFP exhibited a significant decrease in AchE activity at 33.53 ± 3.65 nanounits/μl per cell (p < 0.05).
FIGURE 3.
Secretion of exosomes is inhibited by expression of DN-Rab11-GFP in Panc02 cells and altered the pancreatic cancer-discriminatory biomarker profile in mice saliva. DN-Rab11-GFP expression significantly inhibited AchE activity from the exosomes isolated from Panc02 cells when compared with Panc02 cells that are untransfected or expressing only GFP (A). Orthotopic injections of Panc02 cells developing tumors that are expressing either GFP or DN-Rab11-GFP altered the previously established salivary transcriptomic profile discriminatory for pancreatic cancer (B). Five out of the seven salivary biomarkers discriminatory for pancreatic cancer (Apbb1ip, Aspn, Incenp, Daf2, and Foxp1) were significantly up-regulated in the saliva of mice bearing GFP-expressing tumors in comparison with control. Only one out of the seven pancreatic cancer-specific salivary biomarkers (Apbb1ip) was still significantly up-regulated in the saliva of mice bearing DN-Rab11-GFP-expressing tumors in comparison with control.
Dominant Negative Rab11 Expression in Mouse Pancreatic Tumor Significantly Disrupted the Pancreatic Cancer-discriminatory Saliva Biomarker Profile
To determine whether inhibition of tumor-derived exosome biogenesis and secretion will disrupt the profile of pancreatic cancer-specific biomarkers established as seen in the first mouse cohort, Panc02 cells that were stably expressing with GFP, DN-Rab11-GFP, or noncancer control were orthotopically injected into individual mouse pancreases. Confocal microscopy analysis of the tumor-bearing pancreases confirmed that there is expression of GFP or DN-Rab11-GFP by the resulting tumors 4 weeks post-injection (supplemental Fig. S1, I–P). Furthermore, we did not see significant differences in tumor weight and volume between tumors expressing GFP or DN-Rab11-GFP. The average weight of tumors expressing GFP was 289.2 ± 9.97 mg and had an average volume of 576.8 ± 44.66 mm3. Tumors expressing DN-Rab11-GFP had an average weight of 266.9 ± 13.27 mg with an average volume of 523.1 ± 46.16 mm3 (supplemental Fig. S2, C and D).
Independent qPCR analysis of the saliva collected from the second mouse cohort of Panc02-implanted mice (three groups: noncancer control (n = 22), GFP Panc02 control (n = 22), and DN-Rab11-GFP (n = 22)) revealed that five (Apbb1ip, Aspn, Daf2, Foxp1, and Incenp) out of the seven pancreatic cancer-specific saliva transcriptomic biomarkers established in the first mouse cohort were significantly up-regulated between the noncancer and GFP-Panc02 groups, although two markers (BCO31781 and Gng2) did not change based on their ΔCt values. Three mice died in each group so the saliva from 22 mice were analyzed. Expression of DN-Rab11-GFP in Panc02 cells inhibited exosome secretion at the tumor source and disrupted the established panel of pancreatic cancer-specific salivary biomarkers where only one of the seven saliva biomarkers (Apbb1ip) remained significantly validated as shown in Fig. 3B, Table 3, and supplemental Table S5.
TABLE 3.
DN-Rab11-GFP expression altered the established pancreatic cancer-specific transcriptomic biomarker profile in mice saliva
| Gene | GFP |
DN-Rab11-GFP |
||
|---|---|---|---|---|
| p value | -Fold change | p value | -Fold change | |
| Apbb1ip | 0.003 | 124.5 up-regulated | 0.021 | 31.34 up-regulated |
| Aspn | 0.003 | 167.73 up-regulated | 0.148 | 2.77 up-regulated |
| Incenp | 0.002 | 2.39 up-regulated | 0.429 | 6.68 up-regulated |
| Daf2 | 0.001 | 9.13 up-regulated | 0.056 | 23.1 down-regulated |
| Foxp1 | 0.003 | 8.34 up-regulated | 0.027 | 3.78 down-regulated |
| BCO31781 | 0.231 | 1.39 down-regulated | 0.009 | 1.52 up-regulated |
| Gng2 | 0.06 | 3.07 up-regulated | 0.021 | 7.57 down-regulated |
Pancreatic Cancer Transcriptomic Biomarkers Were Detected in Panc02, Saliva, and Serum-derived Exosomes
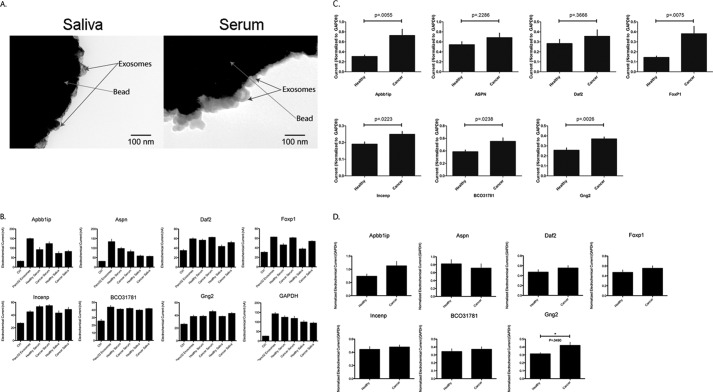
EFIRM shows that the seven validated pancreatic cancer-specific salivary transcripts and GAPDH are found in exosomes derived from Panc02 cells, saliva, and serum (Fig. 4B and supplemental Fig. S4). Beads coated with mCD63 antibody captured exosomes from both saliva and serum (Fig. 4A). Six out of the seven genes were up-regulated in both saliva (Fig. 4C) and serum-derived exosomes (Fig. 4D) of tumor-bearing mice. Moreover, five out of the seven cancer-specific salivary biomarkers were significantly up-regulated in the exosomes found in the saliva of tumor-bearing mice.
FIGURE 4.
Beads coated with mCD63 antibody captured exosomes from both saliva and serum (A). EFIRM shows that the seven validated pancreatic cancer-specific salivary transcripts and GAPDH are found in exosomes derived from Panc02 cells, saliva, and serum (B). All seven genes were found to be up-regulated in both saliva (C) and serum-derived exosomes (D) of tumor-bearing mice; however, Aspn was only up-regulated in saliva-derived exosomes but down-regulated in the serum-derived exosomes of tumor-bearing mice. Five of the seven genes were found to be significantly up-regulated in saliva-derived exosomes, although Gng2 was found to be significantly up-regulated in serum-derived exosomes of tumor-bearing mice.
DISCUSSION
Our previous studies have demonstrated that systemic diseases can be detected by discriminatory salivary biomarkers (1, 2). Despite successfully discovering, verifying, and validating discriminatory salivary biomarkers for cancers, including pancreatic, ovarian, breast, and lung (1–3, 16), how a disease developing distally from the oral cavity could exert its profile into the saliva is not yet known. This study demonstrates that a discriminatory panel of transcriptomic salivary biomarkers presents itself upon the development of pancreatic cancer in mice. Moreover, the results of this study support that tumor-derived exosomes provide the scientific and mechanistic rationale that systemically connect between pancreatic tumors and the oral cavity in the development of discriminatory salivary biomarkers.
To investigate whether pancreatic tumor-derived exosomes play a role in the presence of disease-specific salivary biomarkers in mice, we first established a panel of pancreatic cancer-specific salivary transcriptomic biomarkers. Orthotopic injections of Panc02 cells into the mouse pancreases successfully induced pancreatic cancer. We collected saliva from each mouse four weeks post-injection, and we pooled the saliva as indicated previously so that there were sufficient amounts of RNA for microarray analysis. To note, we did not observe any metastasis to distal organs at 4 weeks post-injection; however, few mice did exhibit moribund behavior prior to the saliva collection period and were immediately sacrificed. For control, we injected sterile PBS into the mouse pancreases to ensure that the salivary transcriptomic biomarkers discovered are due to the development of pancreatic cancer, rather than the inflammation from the surgery procedure.
Microarray analyses of the saliva collected from the pancreatic cancer-bearing mice and noncancer mice exhibited different gene profiles. The top-scoring 20 up-regulated candidate salivary biomarkers discriminatory for pancreatic cancer were picked based upon statistical evaluation, as indicated under “Experimental Procedures.” Literature research shows that Daf2 is a gene that encodes an insulin-like receptor in Caenorhabditis elegans (17). Moreover, Sema 6d and Foxp1 have been shown to be associated with the development of cancer, where expression of the Sema 6d gene (Semaphorin 6D) has been linked to gastric cancer (18), and the Foxp1 gene (Forkhead box protein P1 gene) has been implicated as a tumor suppressor that is absent in various cancers, including breast, lung, colon renal cell carcinoma, and head and neck carcinoma (19).
Validation of the 20 candidate genes discriminatory for pancreatic cancer was conducted by DSTA as described previously (20). To better assess the potential utility of the prospective markers, saliva collected from 22 noncancer and 22 pancreatic cancer-bearing mice was individually analyzed, without pooling. Of the 20 candidate genes, seven genes were found as significantly up-regulated in the saliva of pancreatic cancer-bearing mice in comparison with noncancer control. This validation showed that within a controlled environment, the development of pancreatic cancer in mice induced a discerning profile of salivary transcriptomic biomarkers, despite developing distally from the oral cavity. Moreover, it is interesting to note that all seven of the validated salivary biomarkers were significantly up-regulated in tumor-bearing pancreases in comparison with noncancer healthy pancreases, suggesting a strong correlation between the development of pancreatic cancer and the presence of discriminating salivary biomarkers. Only one of the seven pancreatic cancer-specific salivary biomarkers that was found in the saliva was significantly up-regulated in the serum of cancer-bearing mice in comparison with control (data not shown), although two of the seven salivary biomarkers were found to be significantly down-regulated in the submandibular gland. We attribute this lack of linearity between the salivary glands and serum with the saliva to our discovery process. Our objective was not to search for a panel of pancreatic cancer-specific transcriptomic biomarkers that exhibit linearity between saliva and serum or salivary gland, rather our discovery is a de novo search within saliva for discriminatory biomarkers. However, our findings here in mouse saliva concurred with our previous findings in human saliva that upon the development of pancreatic cancer a discriminatory salivary transcriptomic biomarker profile can be discovered and validated (1). Moreover, by validating seven salivary biomarkers discriminatory for pancreatic cancer in mice, a model was established to examine the role of exosomes mechanistically responsible for the presence of salivary biomarkers specific for pancreatic cancer and other systemic diseases.
Using the EFIRM technology, we determined that all seven of the validated genes were found in exosomes derived from saliva, serum, and also Panc02 cells (Fig. 4). Moreover, six of the seven genes were found to be up-regulated in both saliva- and serum-derived exosomes of tumor-bearing mice when compared with control, although five of the seven genes were found to be significantly up-regulated in saliva-derived exosomes, and Gng2 was found to be significantly up-regulated in serum-derived exosomes of tumor-bearing mice. These results imply that there is compositional linearity between serum- and saliva-derived exosomes transcriptomically, although we did not see biomarker profile linearity between the saliva supernatant and the serum. Moreover, this demonstrated linearity between saliva and saliva exosomes, although serum and serum exosomes did not exhibit the same linearity. Hence, aside from whole serum, tumor, tumor-derived exosomes, serum-derived exosomes, saliva, and saliva-derived exosomes, all exhibited up-regulation of most, if not all, of the seven validated pancreatic cancer-specific salivary transcriptomic biomarkers. This finding potentially explains the difficulties of developing serum biomarkers for disease detection and reinforces the utilization of saliva for tumor-specific biomarkers. Because of the high concentration of various molecular constituents in blood, transcriptomic biomarkers that arise upon the development of pancreatic cancer and released into the blood as exosomes can become muted and indiscoverable. Although in saliva, a more diluted, lower concentration of molecular constituents means disease-specific biomarkers can be more readily discovered. Additionally, even though these results are suggestive that tumor-derived exosomes may travel through the blood and directly into the saliva, these results do not clarify the path of tumor-derived exosomes. Whether tumor-derived exosomes travel through the blood and directly into the saliva through the gingival crevice or whether the tumor-derived exosomes encounter and interact with the salivary glands remains unclear.
The Rab family of proteins is a member of the Ras superfamily of monomeric G proteins (21, 22). This family of GTPases has been implicated in many stages of cellular trafficking, including vesicle formation, vesicle movement, and membrane fusion (23). Recent studies have strongly suggested that the Rab family of proteins is correlated with the biogenesis of exosomes, where Rab2 and Rab7 proteins are commonly found in exosomes (24), although Rab27a and Rab27b have been implicated with the release of exosomes (25). Moreover, Sevina et al. (14, 15) reported that the Rab11 protein regulates exosome release in K562 cells. Sevina et al. (15) demonstrated that by overexpressing a dominant negative mutant form of Rab11, secretion of exosomes was significantly inhibited in the chronic myelogenous leukemia cell line. Hence, we examined whether the overexpression mutant form of Rab11 can inhibit secretion of exosomes by Panc02 cells. Because AchE activity is proportional to the amount of exosomes present (14, 15), AchE assay revealed that stable overexpression of the dominant negative form of Rab11 significantly inhibited the secretion of exosomes by Panc02 cells. This finding allowed us to generate a mouse pancreatic cancer cell line that is defective in its ability to fully secrete exosomes and induce mouse pancreatic tumors with hindered exosome secretion.
Orthotopic injection of Panc02 cells stably expressing GFP or DN-Rab11-GFP into mouse pancreases induced pancreatic tumors after 4 weeks. Hematoxylin and eosin stain and confocal microscopy confirmed the successful development of pancreatic tumors with expression of either GFP or DN-Rab11-GFP. Moreover, we observed that pancreatic tumors expressing either GFP or DN-Rab11-GFP did not significantly differ in weight or volume, hence indicating that the expression of DN-Rab11-GFP did not affect the development of the tumors. Analysis of the salivary RNA via DSTA showed that five out of the seven pancreatic cancer-discriminatory salivary biomarkers established from the initial cohort of mice were found to be significantly up-regulated in the saliva of mice bearing pancreatic tumors that expressed GFP, in comparison with saliva derived from healthy noncancer-bearing mice. Notably, in the mice bearing pancreatic tumors that expressed DN-Rab11-GFP, only one out of the seven established pancreatic cancer-specific salivary transcriptomic biomarkers (Apbb1ip) was found to be significantly up-regulated, in comparison with the saliva from healthy mice. Even though Apbb1ip was still significantly up-regulated in the saliva of pancreatic cancer mouse saliva, inhibition of exosomes drastically decreased the fold difference between cancer and noncancer saliva. Thus, these results suggested that tumor-derived exosomes might be involved in the connectivity between the pancreatic tumors and the oral cavity leading to pancreatic cancer-specific transcriptomic biomarkers in saliva.
Utilizing saliva to detect the early onset of pancreatic cancer and other systemic diseases is ideal, as it is noninvasive and easily accessible. Here, we examined whether tumor-derived exosomes are involved in the presence of pancreatic cancer-specific transcriptomic biomarkers in the saliva. By using a mouse model, we showed that pancreatic cancer-specific salivary transcriptomic biomarkers were present. Moreover, we demonstrated that inhibiting exosome secretion by pancreatic tumors disrupted the panel of discriminating salivary biomarkers. Hence, through these findings we developed a working model and a central hypothesis as to how tumor-derived exosomes relay information from a distal site of the body to the oral cavity to generate discriminatory salivary biomarkers. We reason that tumor-derived exosomes can induce discriminatory salivary biomarkers by altering transcription in the salivary glands by direct contact with the salivary glands. We think that this interaction between the tumor-derived exosomes can alter transcription of the salivary glands and consequently change the contents of the released salivary exosomes with discriminatory biomarkers.
Future studies must be conducted to decipher the path of the tumor-derived exosomes as they are secreted because this study does not provide the complete mechanistic explanation as to why and how communication occurs between a tumor distal to the oral cavity and saliva leading to salivary biomarkers for systemic diseases. Whether the interplay between tumor-derived exosomes and the salivary glands triggers disease-specific biomarkers will be the next focus. We will aim to examine whether tumor-derived exosomes are the shuttles that travel through the body's vasculature and into the saliva delivering disease-specific messages or whether they are messengers that trigger the release of secondary messengers (i.e. cytokines and hormones) inducing transcriptional modification of the salivary glands releasing biomarkers into the saliva.
Acknowledgments
We express our appreciation for the scientific discussions with Dr. Gordon Proctor and Dr. James Melvin during the planning of this project.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DE17170, UH2/UH3 TR000923, and T32DE07296 from USPHS. This work was also supported by SOD Faculty Seed Grants 441901–69749-FWEI-FY11DR from the UCLA School of Dentistry. D. Wong is co-founder of RNAmeTRIX Inc., a molecular diagnostic company. He holds equity in RNAmeTRIX. Intellectual property that David Wong invented and that was patented by University of California has been licensed to RNAmeTRIX. Additionally, he is a paid consultant to PeriRx.

This article contains supplemental Figs. S1–S5 and Tables S1–S5.
- DSTA
- direct saliva transcriptomic analysis
- AchE
- acetylcholinesterase
- TMB
- 3,3′,5,5′-tetramethylbenzidine
- qPCR
- quantitative PCR
- EFIRM
- electric field-induced release and measurement
- TEM
- transmission electron microscopy
- csw
- cyclic square wave.
REFERENCES
- 1. Zhang L., Farrell J. J., Zhou H., Elashoff D., Akin D., Park N. H., Chia D., Wong D. T. (2010) Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 138, 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang L., Xiao H., Karlan S., Zhou H., Gross J., Elashoff D., Akin D., Yan X., Chia D., Karlan B., Wong D. T. (2010) Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the noninvasive detection of breast cancer. PLoS ONE 5, e15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xiao H., Zhang L., Zhou H., Lee J. M., Garon E. B., Wong D. T. (2012) Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 11, M111.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palanisamy V., Sharma S., Deshpande A., Zhou H., Gimzewski J., Wong D. T. (2010) Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE 5, e8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lau C. S., Wong D. T. (2012) Breast cancer exosome-like microvesicles and salivary gland cells interplay alters salivary gland cell-derived exosome-like microvesicles in vitro. PLoS ONE 7, e33037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnstone R. M. (2006) Exosomes biological significance: A concise review. Blood Cells Mol. Dis. 36, 315–321 [DOI] [PubMed] [Google Scholar]
- 7. Pisitkun T., Shen R. F., Knepper M. A. (2004) Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 101, 13368–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnstone R. M., Bianchini A., Teng K. (1989) Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood 74, 1844–1851 [PubMed] [Google Scholar]
- 9. Admyre C., Johansson S. M., Qazi K. R., Filén J. J., Lahesmaa R., Norman M., Neve E. P., Scheynius A., Gabrielsson S. (2007) Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 179, 1969–1978 [DOI] [PubMed] [Google Scholar]
- 10. Qazi K. R., Torregrosa Paredes P., Dahlberg B., Grunewald J., Eklund A., Gabrielsson S. (2010) Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis. Thorax 65, 1016–1024 [DOI] [PubMed] [Google Scholar]
- 11. Vella L. J., Greenwood D. L., Cappai R., Scheerlinck J. P., Hill A. F. (2008) Enrichment of prion protein in exosomes derived from ovine cerebral spinal fluid. Vet. Immunol. Immunopathol. 124, 385–393 [DOI] [PubMed] [Google Scholar]
- 12. Somasundaram R., Herlyn M. (2012) Melanoma exosomes: messengers of metastasis. Nat. Med. 18, 853–854 [DOI] [PubMed] [Google Scholar]
- 13. Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., Garcia-Santos G., Ghajar C. M., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B. A., Callahan M. K., Yuan J., Martins V. R., Skog J., Kaplan R. N., Brady M. S., Wolchok J. D., Chapman P. B., Kang Y., Bromberg J., Lyden D. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Savina A., Fader C. M., Damiani M. T., Colombo M. I. (2005) Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 6, 131–143 [DOI] [PubMed] [Google Scholar]
- 15. Savina A., Vidal M., Colombo M. I. (2002) The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci. 115, 2505–2515 [DOI] [PubMed] [Google Scholar]
- 16. Lee Y. H., Kim J. H., Zhou H., Kim B. W., Wong D. T. (2012) Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J. Mol. Med. 90, 427–434 [DOI] [PubMed] [Google Scholar]
- 17. Iser W. B., Wolkow C. A. (2007) DAF-2/insulin-like signaling in C. elegans modifies effects of dietary restriction and nutrient stress on aging, stress and growth. PLoS ONE 2, e1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu L. X., Liu Z. H., Jiang H. C., Qu X., Zhang W. H., Wu L. F., Zhu A. L., Wang X. Q., Wu M. (2002) Profiling of differentially expressed genes in human gastric carcinoma by cDNA expression array. World J. Gastroenterol. 8, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Banham A. H., Beasley N., Campo E., Fernandez P. L., Fidler C., Gatter K., Jones M., Mason D. Y., Prime J. E., Trougouboff P., Wood K., Cordell J. L. (2001) The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 61, 8820–8829 [PubMed] [Google Scholar]
- 20. Lee Y. H., Zhou H., Reiss J. K., Yan X., Zhang L., Chia D., Wong D. T. (2011) Direct saliva transcriptome analysis. Clin. Chem. 57, 1295–1302 [DOI] [PubMed] [Google Scholar]
- 21. Zahraoui A., Touchot N., Chardin P., Tavitian A. (1989) The human Rab genes encode a family of GTP-binding proteins related to yeast YPT1 and SEC4 products involved in secretion. J. Biol. Chem. 264, 12394–12401 [PubMed] [Google Scholar]
- 22. Chardin P. (1988) The ras superfamily proteins. Biochimie 70, 865–868 [DOI] [PubMed] [Google Scholar]
- 23. Novick P., Brennwald P. (1993) Friends and family: the role of the Rab GTPases in vesicular traffic. Cell 75, 597–601 [DOI] [PubMed] [Google Scholar]
- 24. Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. (2001) Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 166, 7309–7318 [DOI] [PubMed] [Google Scholar]
- 25. Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C. F., Schauer K., Hume A. N., Freitas R. P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M. C., Darchen F., Amigorena S., Moita L. F., Thery C. (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 [DOI] [PubMed] [Google Scholar]
- 26. Gao K., Zhou H., Zhang L., Lee J. W., Zhou Q., Hu S., Wolinsky L. E., Farrell J., Eibl G., Wong D. T. (2009) Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS ONE 4, e5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morikane K., Tempero R. M., Sivinski C. L., Nomoto M., Van Lith M. L., Muto T., Hollingsworth M. A. (1999) Organ-specific pancreatic tumor growth properties and tumor immunity. Cancer Immunol. Immunother. 47, 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kesimer M., Scull M., Brighton B., DeMaria G., Burns K., O'Neal W., Pickles R. J., Sheehan J. K. (2009) Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 23, 1858–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wei F., Patel P., Liao W., Chaudhry K., Zhang L., Arellano-Garcia M., Hu S., Elashoff D., Zhou H., Shukla S., Shah F., Ho C. M., Wong D. T. (2009) Electrochemical sensor for multiplex biomarkers detection. Clin. Cancer Res. 15, 4446–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei F., Liao W., Xu Z., Yang Y., Wong D. T., Ho C. M. (2009) Bio/abiotic interface constructed from nanoscale DNA dendrimer and conducting polymer for ultrasensitive biomolecular diagnosis. SMALL 5, 1784–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]