Background: DNA damage-induced ubiquitylation is important in regulating the DNA damage response.
Results: PRT4165 inhibits histone H2A ubiquitylation and the accumulation of ubiquitin at the DNA double-strand break (DSB) sites.
Conclusion: PRT4165 is a novel drug for studying DSB response.
Significance: PRT4165 may constitute a novel approach for studying DSB response and for development of new cancer therapy.
Keywords: Chromatin, DNA Damage Response, DNA Repair, Polycomb, Ubiquitylation
Abstract
Polycomb-repressive complex 1 (PRC1)-mediated histone ubiquitylation plays an important role in aberrant gene silencing in human cancers and is a potential target for cancer therapy. Here we show that 2-pyridine-3-yl-methylene-indan-1,3-dione (PRT4165) is a potent inhibitor of PRC1-mediated H2A ubiquitylation in vivo and in vitro. The drug also inhibits the accumulation of all detectable ubiquitin at sites of DNA double-strand breaks (DSBs), the retention of several DNA damage response proteins in foci that form around DSBs, and the repair of the DSBs. In vitro E3 ubiquitin ligase activity assays revealed that PRT4165 inhibits both RNF2 and RING 1A, which are partially redundant paralogues that together account for the E3 ubiquitin ligase activity found in PRC1 complexes, but not RNF8 nor RNF168. Because ubiquitylation is completely inhibited despite the efficient recruitment of RNF8 to DSBs, our results suggest that PRC1-mediated monoubiquitylation is required for subsequent RNF8- and/or RNF168-mediated polyubiquitylation. Our results demonstrate the unique feature of PRT4165 as a novel chromatin-remodeling compound and provide a new tool for the inhibition of ubiquitylation signaling at DNA double-strand breaks.
Introduction
Ubiquitylation is a critical post-translational modification taking place in response to DNA double-strand breaks (DSBs)3 (reviewed in Ref. 1). The canonical pathway is dependent on the phosphorylation of histone H2AX and MDC1 (1). When phosphorylated, MDC1 recruits the E3 ubiquitin ligase, RNF8. RNF8 is then thought to initiate ubiquitylation and this involves, at least in part, the monoubiquitylation of histone H2A and H2AX at lysine 119. After the initiation of the ubiquitylation cascade, RNF168 and the E2 enzyme, UBC13, conjugate K63-linked polyubiquitin chains onto histones H2A/H2AX. Recently, RNF8 was found to be unable to ubiquitylate nucleosomal H2A, and evidence was presented that RNF168 initiated signaling through novel ubiquitylation sites in the N terminus of histone H2A (2). Nonetheless, RNF8 E3 ubiquitin ligase activity is necessary for the retention of RNF168 at sites of DSBs (3, 4).
Although there are good inhibitors to inhibit the phosphorylation cascade taking place near DSBs, it has only been possible to date to inhibit ubiquitylation indirectly by inhibiting the proteasome (3, 5). This inhibits ubiquitylation at DSBs by sequestering ubiquitin in the cytoplasm in lysine 48-linked polyubiquitin chains (6). Recently, an inhibitor of BMI1/RING1-mediated polyubiquitylation of topoisomerase II was identified (7). BMI1/RING1 are subunits of the polycomb repressive complex 1 (PRC1) (8, 9). PRC1 is a histone H2A/H2AX E3 ubiquitin ligase and has a well established role in the regulation of differentiation and the maintenance of stem cell populations (10). Not surprisingly, overexpression of PRC1 components is commonly associated with poor prognosis across a range of human cancer types (11–13). The role of PRC1 in the maintenance of a less differentiated phenotype is thought to be important in human cancer and may explain this relationship. However, we and others have recently demonstrated that PRC1 is rapidly recruited to DSBs, where it can monoubiquitylate histones H2A and H2AX (DSBs) (14–18). This raises the possibility that PRC1 may be involved in the initiation of the ubiquitylation cascade by providing a monoubiquitylated substrate for RNF8 and RNF168.
There is a consensus that the PRC1 components BMI1 and RNF2 recruit to DSBs, where they contribute to DSB repair (14–17, 19). However, placement of PRC1 within the DDR signaling cascade is controversial. For example, we initially observed that knockdown of BMI1 diminished downstream DDR functions at DSBs, including the focal accumulation of ubiquitin and of DDR factors such as 53BP1 and BRCA1 (14). Upstream DDR signaling events including the function of the MRE11-RAD50-NBS1 complex, activation of the ATM kinase by phosphorylation (p-ATM), and the phosphorylation-dependent recruitment of MDC1 each remained unaffected by BMI1 knockdown. In contrast, a study examining glioblastoma multiforme and neural stem cells found that knockdown of BMI1 also inhibited one of the earliest DDR processes, focal accumulation of upstream DDR components MRE11, γ-H2AX, and MDC1 (16). Focal accumulation of downstream factors RNF8 and 53BP1 were also reduced, as was DSB repair. At the extremes, mutation of histone H2AX lysines 119 and 120 to arginine and knockdown of RNF2 were both shown to inhibit histone H2AX phosphorylation (18, 19), whereas at the other extreme, defects in signaling were not observed, but homologous recombination was impaired (15). The differences between these studies may reflect compensation by paralogues (10), differences in knockdown efficiency, and the potential reprogramming of gene expression that could accompany the lengthy incubation periods required in knockdown experiments. These sources of experimental variation may be overcome with a broadly acting inhibitor of PRC1 ligase. This prompted us to further investigate the specificity of the potential PRC1 inhibitor, PRT4165.
We find that PRT4165 inhibits the in vitro histone H2A E3 ubiquitin ligase activity of RING1, its more active paralogue, RNF2, and a BMI1/RNF2 complex. Treatment of cells with this drug resulted in the near complete loss of ubiquitylated H2A within the first hour of drug treatment. We also found that the drug was an effective inhibitor of H2AX ubiquitylation taking place in response to DNA double-strand breaks. This allowed us to test whether or not transient inhibition of PRC1 could reproduce any of the results obtained using knockdown and knock-out approaches. We find that many of the signaling defects observed in knockdown and knock-out experiments are reproduced by transient inhibition of PRC1. We also find that inhibition of PRC1 dramatically reduces the accumulation of ubiquitin at sites of DSBs and strongly inhibits the repair of DSBs.
EXPERIMENTAL PROCEDURES
Cell Culture, Vector Construct, and Transfections
U2OS cells were cultured in DMEM containing 10% FCS at 37 °C and 5% CO2. NLS-VX3-EGFP plasmid was obtained from Addgene (plasmid identification no. 35529). Unless otherwise stated, cells were irradiated in ambient air using a model CS-600 137Cs irradiator (Picker, Glendale, CA) at a dose rate of 2 Gy/min. PRC1 inhibitor (PRT4165) was purchased from TimTec and was dissolved in Me2SO at concentration of 21 mm, aliquoted, and kept at −20 °C. When added to the cells, the inhibitor was added in serum-free medium.
Immunofluorescence Microscopy
Immunofluorescence staining was performed as described previously (14) with some modifications. Briefly, cells were permeabilized with cytoskeleton buffer (100 mm NaCl, 300 mm sucrose, 10 mm Pipes, pH 6.8, 3 mm MgCl2,0.5% Triton X-100) for 2 min before being fixed at the indicated time points after exposing cells to radiation (IR). The cells were fixed with 4.0% paraformaldehyde in PBS, pH 7.5, for 5 min at room temperature. Next, the cells were washed twice with PBS, inverted onto 50-μl aliquots of an appropriate primary antibody, and incubated at room temperature for 30 min. Coverslips were rinsed with PBS containing 0.1% Triton X-100 and washed twice with PBS before a 30-min incubation with an appropriate secondary antibody conjugated to a fluorophore. The cells were rinsed with PBS containing 0.1% Triton X-100 and washed twice with PBS. Coverslips were mounted onto slides containing ∼10 μl of a 90% glycerol-PBS-based medium containing 1 mg of paraphenylenediamine/ml and 0.5 μg DAPI/ml. A panel of commercially available primary antibodies, directed against various DNA damage proteins and the polycomb group proteins BMI1 (see below).
Immunoblotting and Acid Extraction of Histones
U2OS cells were subcultured the previous day and used at 70–80% confluency on the day of the experiment. Cells, when necessary, were exposed to drugs or IR, then harvested using 0.53 mm EDTA in PBS, and washed once with cold PBS. Nuclear extracts were prepared as per the procedure described previously (14). Histones extraction was prepared using 0.4 n sulfuric acid from cells exposed to radiation and incubated for 60 min at 37 °C. After washing with cold PBS, the cells were scraped off the plates, and H2AX phosphorylation was measured as described previously (14). In brief, nuclear extracts were separated on 6–18% SDS-PAGE (depending on the molecular weight of the protein) and transferred to 0.2-μm nitrocellulose blotting membrane (Bio-Rad) according to standard protocols. The distribution of various nuclear proteins was examined by immunoblotting according to standard procedures using 5% BSA in TBST (TBS with 0.05% Tween) as the blocking buffer and antibody incubation buffer. The primary antibodies used were: γ-H2AX (Millipore), H2AX, p-ATM (active motif), H2A (Millipore), p-SMC1-S966 (Bethyl Laboratories), p-NBS1 S343 (Bethyl Laboratories), 53BP-1 (Lake Placid), and MDC1 (Abcam). Secondary antibodies used were conjugated with infrared-specific dyes (Alexa Fluor 680, Alexa Fluor 750, or IRDye 800), and all immunoblots were imaged on the Odyssey infrared imaging scanner imaging system (LI-COR Biosciences).
In Vitro Ubiquitylation Assay
GST-RNF2, GST-BMI1, ubiquitin, UBCH13, UBCH5c, E1, GST-RING1, and GST-RNF8 were either GST purified or purchased from Boston Biochem or Abnova. Reactions were performed in 30 μl of ubiquitylation buffer (50 mm Tris, pH 7.5, 2.5 mm MgCl2, 0.5 mm DTT) containing ubiquitin-activating enzyme E1, UBCH5c, or UBC13, ubiquitin, 0.2 mm ATP, H2A, and 1 μg of indicated GST fusion protein. The reactions were thoroughly mixed and incubated at 37 °C for 60 min. The reaction was then stopped by the addition of Laemmli sample buffer, and proteins were resolved by SDS-PAGE and immunoblotted using H2A antibody (Abcam).
G2/M Checkpoint Analysis
U2OS cells were treated with different concentrations of the PRT4165 60 min prior to irradiation (2 Gy). The cells were harvested 2 h after IR. The cells were washed with PBS twice and then fixed with 1% paraformaldehyde at 37 °C for 10 min. After cooling on ice for 1 min, the cells were permeabilized with 90% methanol and stored at −20 °C overnight. Fixed cells were washed with PBS twice and blocked with FACS incubation buffer (0.5% BSA in PBS) for 10 min. The cells were then stained with anti-phosphohistone H3 (serine 10) antibody at 1:500 dilution in FACS incubation buffer for 1 h at room temperature. All washes were carried out in FACS incubation buffer. For secondary antibody staining, FITC-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch) diluted 1:500 was used, and cells were incubated for 1 h at room temperature. After secondary antibody staining, the cells were washed and then incubated with 5 μg/ml propidium iodide (Sigma-Aldrich) and 250 μg/ml RNase A in PBS. More than 10,000 cells were analyzed by flow cytometry (Becton Dickinson). The data were analyzed by Cell Quest Pro (Becton Dickinson).
Image Processing and Figure Construction
Where necessary, images were subjected to a 3 × 3 median filter to reduce pixel noise. MetaMorph images were exported as 16-bit TIFF files. These were subsequently rescaled over an 8-bit data range in Photoshop CS4. In most cases, the background fluorescence of the medium and the base signal from the detector were subtracted to better represent the dynamic range of the data content in the image. In some instances, three-dimensional image sets were imported into Imaris 7 (BitPlane), and three-dimensional image sets were generated. In this instance, the image was scaled to map the data over the range of the display, and the screen capture function in Imaris 7 was used to capture the image used in the figure. In general, images were scaled to span the 8-bit data depth, reducing background in the process, and then pasted into a composite canvas that was either 8-bit grayscale or 24-bit RGB color. If necessary, images were interpolated to 300 dpi using Photoshop.
RESULTS
PRT4165 Inhibits the E3 Ubiquitin Ligase Activity of PRC1
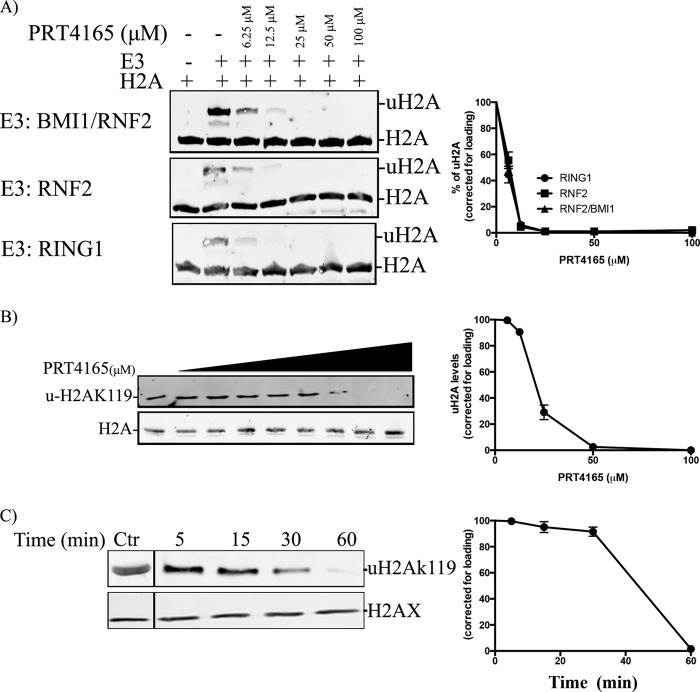
A recent report identified an active inhibitor of the E3 ubiquitin ligase complex BMI1/RING1, PRT4165 (7). We wished to determine whether or not this inhibitor also acted on the more common and more active BMI1/RNF2 complex, which is required if the drug is to be useful as a broadly acting inhibitor of PRC1-mediated histone H2A ubiquitylation. When RING1, RNF2, or a BMI1/RNF2 complex was tested for in vitro ubiquitylation activity toward histone H2A, we found that all three were able to ubiquitylate histone H2A, with the BMI1/RNF2 complex being most efficient (Fig. 1A), as expected. In the presence of PRT4165, however, H2A ubiquitylation could be completely inhibited regardless of whether RING1 or RNF2 contributed the E3 ubiquitin ligase activity (Fig. 1A). RNF2 is thought to be responsible for most of the H2A E3 ubiquitin ligase activity, but RING1 also contributes (8). The direct inhibition of these two E3 ubiquitin ligases suggested that this inhibitor had the potential to effectively inhibit H2A ubiquitylation in cells. To test whether or not PRT4165 can broadly inhibit PRC1 H2A E3 ubiquitin ligase activity in vivo, we titrated the concentration of the inhibitor (Fig. 1B) and performed time course experiments where the ubiquitylation of H2A was monitored by immunoblotting. We noted that PRT4165 efficiently inhibited H2A ubiquitylation at lysine 119 (uH2AK119) and that this inhibition was dependent on both the concentration of inhibitor and the incubation time with the inhibitor (Fig. 1C). Treatment of cells for 60 min with 50 μm PRT4165 resulted in a dramatic reduction in total ubiquitylated histone H2A (Fig. 1C). This result reveals that H2A ubiquitylation turns over quite rapidly, particularly in comparison with lysine 27 methylation mediated by the polycomb repressive complex 2 (data not shown), and is consistent with the established role of the PRC1 complex as the major histone H2A E3 mono-ubiquitin ligase in the cell.
FIGURE 1.
PRT4165 inhibits the PRC1 E3 monoubiquitin ligase activity toward H2A. A, RING1, RNF2, or BMI-1/RNF2 complex exhibits ubiquitin ligase activity in the presence of the E2 enzyme, UbcH5c, and E1 enzyme. Ubiquitin E3 ligase activity was investigated by incubating histone H2A substrate with members of the PRC1 complex in the presence of E1 enzyme and E2 enzyme (as indicated). Reactions were carried out in the presence of different concentrations of the PRT4165 with a 30-min preincubation step as described under “Experimental Procedures.” H2A ubiquitylation products were analyzed by SDS-PAGE and visualized using a H2A antibody. B, effect of PRT4165 on H2A ubiquitylation in cells. The cells were treated with different concentrations of PRT4165 as in A, and acid extracts were prepared and immunoblotted using H2A and uH2AK119 antibodies. Detection of H2A ubiquitylation in vivo. C, U2OS cells were either mock treated or incubated with 50 μm of the PRT4165 for different time points. Histone extracts were prepared, and samples were run on SDS-PAGE. H2A ubiquitylation was detected using antibody against the uH2AK119, and H2AX was used as a loading control (Ctr). The dividing line indicates that the intervening lanes were spliced out. Quantifications of the immunoblots from two independent experiments were done using Odyssey software and were plotted on the right using Prism Software.
PRT4165 Inhibits H2AX Ubiquitylation at Sites of DNA Damage
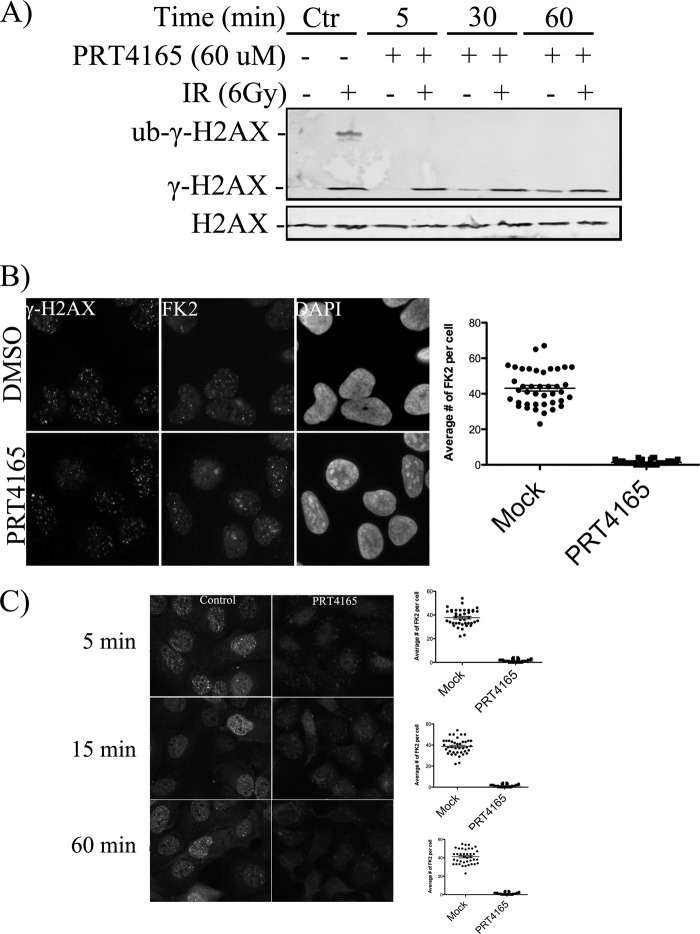
We next wished to determine whether or not this inhibitor would also inhibit H2AX ubiquitylation that was taking place in response to DSBs. Because H2AX is rapidly phosphorylated in response to DSBs but is otherwise phosphorylated at very low abundance during interphase, this could be directly tested by examining the ubiquitylation of phosphorylated H2AX (γ-H2AX). The inhibition of H2AX ubiquitylation at sites of DNA damage, revealed by immunoblotting with γ-H2AX antibodies, was complete (Fig. 2A). This inhibition did not reflect the turnover of steady-state ubiquitylation because we were able to detect inhibition as early as 5 min after the addition of the PRT4165, which was insufficient to turn over steady-state H2A ubiquitylation (Fig. 1C). Consistent with a role for PRT4165 in maintaining genomic stability, we found that longer exposure of the cells with the PRT4165 (30 and 60 min) led to increased levels of γ-H2AX in unirradiated cells (Fig. 2A). We have previously observed increased basal γ-H2AX in BMI1 knock-outœnockdown cells (14). These results are consistent with increased endogenous DNA damage arising from inhibition of PRC1 E3 ubiquitin ligase activity.
FIGURE 2.
PRT4165 inhibits the PRC1 E3 monoubiquitin ligase activity toward γ-H2AX in vivo. A, U2OS cells were either mock treated or treated with the PRT4165 (50 μm) before they were exposed to radiation (6 Gy). The cells were allowed to recover for 5 min. Histone extracts were prepared, and samples were run on SDS-PAGE. γ-H2AX ubiquitylation was detected using antibody against the phosphorylated form of H2AX, which detects both the phosphorylated and ubiquitylated form of H2AX. B, immunofluorescence staining of the effect of PRT4165 on the formation of mono- and polyubiquitin conjugates at the sites of DNA damage. U2OS cells were either mock treated or irradiated (2 Gy) in the presence of 50 μm of the PRT4165. The cells were allowed to recover for 5 after irradiation. The cells were then fixed and stained as indicated. C, time course of the PRT4165 inhibition of FK2 IRIF. U2OS cells were pretreated with PRT4165 for 5 min and allowed to recover for different time points as indicated after irradiation (2 Gy). The average number of FK2 and BRCA1 IRIF per cell was quantified and plotted as indicated. The scale bars represent 10 μm. Ctr, control.
We next tested the effect of PRT4165 on DNA damage-induced ubiquitin conjugation occurring directly at the sites of DSBs. This was accomplished by immunofluorescence using the FK2 antibody. FK2 recognizes both mono- and polyubiquitylated proteins and, hence, reports on total ubiquitylation (21). The cells were irradiated with 2 Gy in the presence of 50 μm PRT4165, allowed to recover for 5 min, and then stained using the monoclonal antibody FK2. Remarkably, we found that the accumulation of ubiquitin conjugates at the sites of DNA damage was inhibited upon treatment with PRT4165, similar to the proteasome inhibitor, MG132, which was used as a positive control (Fig. 2B and data not shown). MG132 inhibits ubiquitylation at sites of DNA damage through sequestration of ubiquitin in polyubiquitin chains in the cytoplasm (6). Similar results were obtained when cells were pretreated with 50 μm PRT4165 for 5 min before irradiation and allowed to recover for 5, 30, and 60 min (Fig. 2C). Thus, PRT4165 inhibits ongoing ubiquitylation at the sites of DNA damage.
PRT4165 Does Not Inhibit RNF8 Ubiquitin Ligase Activity in Vitro nor Its Recruitment to Sites of DNA Damage
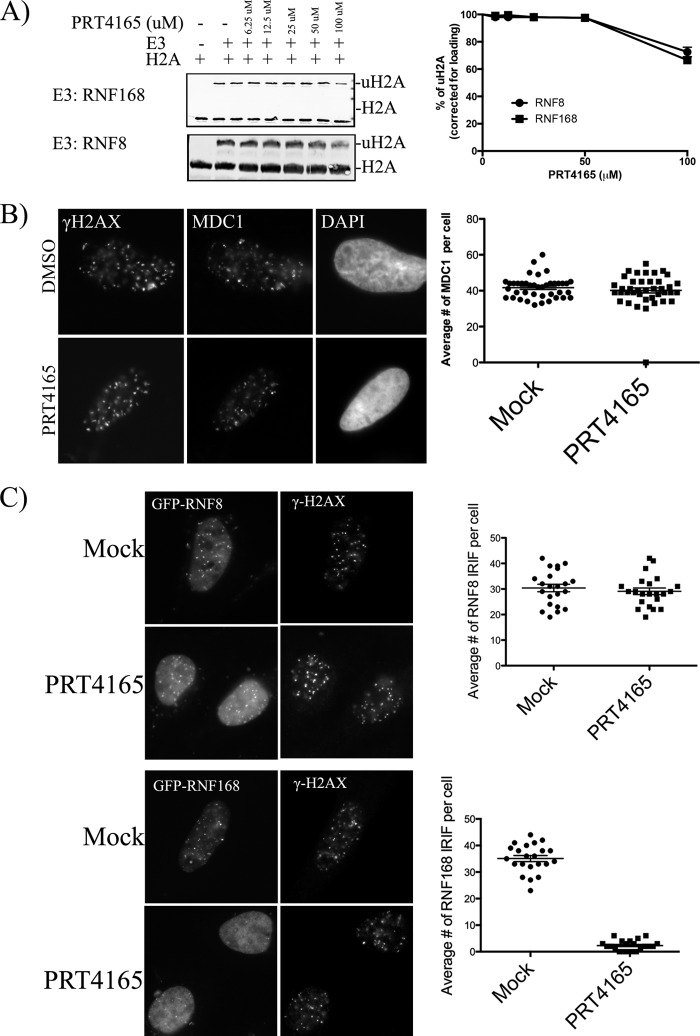
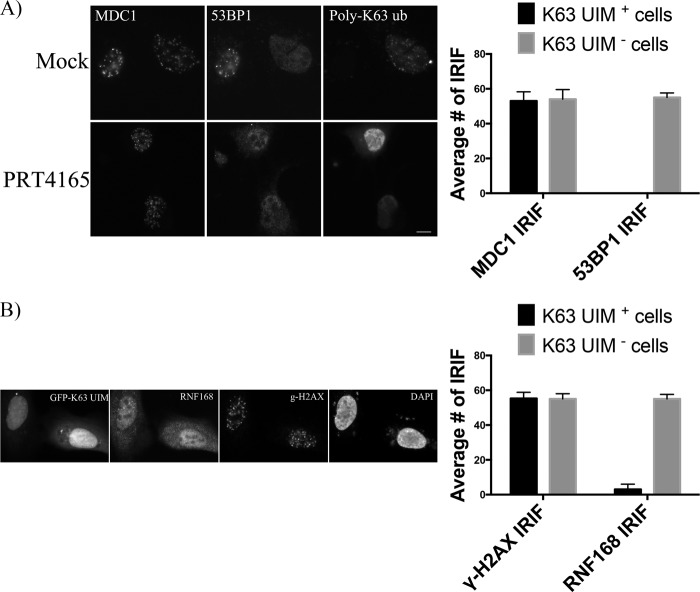
The loss of all detectable ubiquitylation was surprising given that histone H2AX was efficiently phosphorylated and, thus, would be expected to recruit RNF8, an established DDR histone H2AX E3 ubiquitin ligase (5, 22). Because RNF8 is the upstream E3 ubiquitin ligase in the canonical ubiquitin signaling cascade at DSBs, the failure to ubiquitylate proteins at IRIF could reflect the inhibition of either or both of the two additional E3 ubiquitin ligases implicated in this signaling, RNF8 and RNF168. We thus tested whether or not PRT4165 inhibited RNF8 or RNF168 in an in vitro ubiquitylation assay. Only at the highest concentrations of the drug (e.g., 100 μm) could we observe a modest inhibition of RNF8 and RNF168 (Fig. 3A). Although RNF8 and RNF168 were not significantly inhibited at 50 μm, it may be that PRT4165 is able to prevent their recruitment to DSBs. Thus, we determined whether or not signaling immediately downstream of H2AX phosphorylation was altered. Fig. 3B shows that MDC1, which is required for the recruitment of RNF8, was efficiently recruited to IR-induced foci enriched in γ-H2AX. We next tested whether or not the E3 ubiquitin ligases RNF8 and RNF168 are recruited to IRIF. RNF8 and RNF168 were shown to facilitate the accumulation of ubiquitin conjugates at the sites of DNA damage (3–5, 21, 22). Unfortunately, antibodies to RNF8 or RNF168 failed to recognize endogenous proteins in IRIF under control conditions. Consequently, we used transiently transfected GFP fusion proteins and examined their retention in IRIF in the presence and absence of PRT4165. To minimize any artifacts caused by overexpression of RNF8/RNF168 proteins, only low and medium GFP-expressing cells were selected. We found that RNF168 accumulation at DSBs was compromised in cells treated with PRT4165, whereas RNF8 IRIFs were not noticeably affected (Fig. 3C). Thus, the absence of ubiquitylation at sites of DSBs upon treatment with PRT4165 cannot be explained by a failure to recruit RNF8, nor can it be explained by inhibiting the RNF8 or RNF168 E3 ubiquitin ligases. Rather, the results are most consistent with PRC1-mediated monoubiquitylation being required for subsequent chain extension by RNF8 and RNF168. Consistent with this interpretation, we and others have found that monoubiquitylation of γ-H2AX remains robust upon RNF8 knockdown, whereas di- and polyubiquitylation are strongly reduced (14, 21). In contrast, BMI1 knockdown resulted in a suppression of both mono- and polyubiquitylated species of γ-H2AX (14). To further confirm this hypothesis, we tested the ability of a PRT4165 to inhibit DNA damage-induced K63 ubiquitin chains. For this experiment, we employed a recently developed synthetic GFP-tagged high affinity K63-linked polyubiquitin-binding protein (NLS-Vx3-EGFP) to label the K63-linked polyubiquitin chains in U2OS cells (23). We found that in response to IR, K63-linked polyubiquitin enriched in foci that co-localize with MDC1 IRIF (Fig. 4A). In contrast, we found that PRT4165 abrogates K63-linked polyubiquitin formation in IRIF (Fig. 4A). As a positive control, we establish that these K63 ubiquitin-binding domains function in a dominant-negative manner (23) and prevent the accumulation of 53BP1 and RNF168 in IRIFs (Fig. 4, A and B). In contrast, MDC1, whose recruitment does not require ubiquitylation, was not inhibited.
FIGURE 3.
PRT4165 does not inhibit the E3 ubiquitin ligase activity of RNF8 in vitro. A, effect of PRT4165 on the E3 ubiquitin ligase activity of RNF8/RNF168 in vitro. Ubiquitin E3 ligase activity was investigated by incubating histone H2A substrate with RNF8/RNF168 in the presence of E1 and E2 enzymes. Reactions were carried out in the presence of different concentrations of the PRT4165 inhibitor with a 30-min preincubation step as described under “Experimental Procedures.” H2A ubiquitylation products were analyzed by SDS-PAGE and visualized using an H2A antibody. Quantifications of the immunoblots from two independent experiments were done using Odyssey software and were plotted on the right using Prism Software. B, effect of PRT4165 on MDC1 IRIF. U2OS cells were either mock treated or treated with 50 μm of the PRT4165 for 5 min before they were exposed to radiation (2 Gy). The cells were allowed to recover for 30 min in the presence of the PRT4165. The cells were then fixed and stained as indicated. The average number of MDC1 IRIF per cell was quantified and plotted as indicated. C, effect of PRT4165 on RNF8/RNF168 IRIF. U2OS cells transiently transfected with either GFP-RNF8 or GFP-RNF168. Medium and low GFP-expressing cells were selected by G418. The cells were either mock treated or treated with 50 μm of the PRT4165 for 5 min before they were exposed to radiation (2 Gy). The cells were allowed to recover for 30 min in the presence of the PRT4165. The cells were then fixed and stained as indicated. The GFP signal was used to monitor the RNF8/RNF168 IRIF. The average number of GFP-RNF8/GFP-RNF168 IRIF per cell was quantified and plotted as indicated. The scale bars represent 10 μm.
FIGURE 4.
PRT 4165 inhibits K63 polyubiquitin chains IRIF in cells. U2OS cells transiently transfected with plasmid DNA encoding NLS-Vx3-EGFP were either left untreated or treated with PRT4165 for 5 min before they were exposed to ionizing radiation. The cells were allowed to recover for 30 min before they were fixed and stained with the indicated antibodies. A, 53BP1 and MDC1. B, RNF168 and γ-H2AX. The average number of IRIF per cell was quantified and plotted as indicated. The scale bars represent 10 μm.
PRT4165 Abrogates the Retention of Several DDR Proteins in IRIF
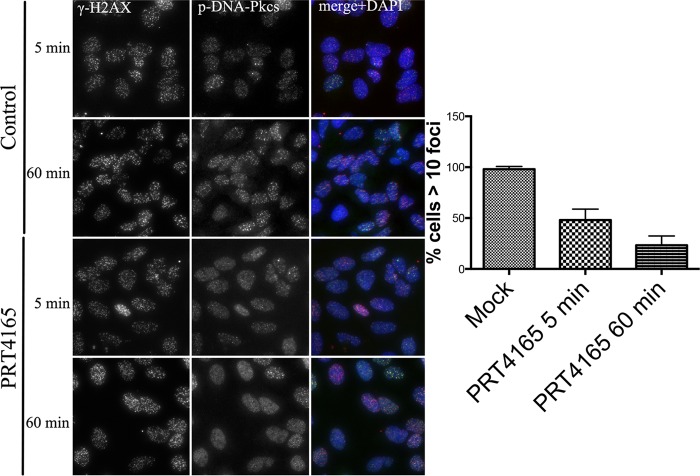
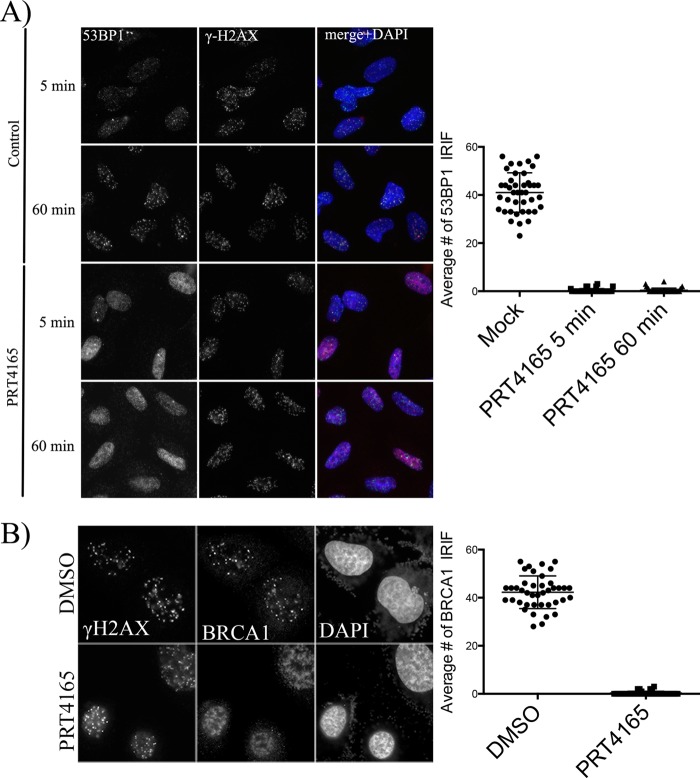
There have been considerable differences in the reported consequences of BMI1 knockdown on the recruitment of DDR proteins to IRIFs (14, 16, 18, 19, 24, 25). The recruitment of phosphorylated ATM to IRIFs, for example, differed between laboratories. We therefore sought to determine the consequences of inhibition with PRT4165 in the assembly of IRIFs at DSBs following IR treatment. U2OS cells were either mock treated or treated with 50 μm PRT4165 for 5 min before exposing them to radiation (2 Gy). This short incubation time was chosen to minimize any indirect effects of the PRT4165 that may arise from derepressing genes upon loss of H2A ubiquitylation Surprisingly, the phosphorylated form of DNA-PKcs failed to efficiently accumulate in IRIFs when PRC1 was inhibited (Fig. 5). The fact that γ-H2AX accumulates in IRIF in the presence of PRC1 inhibitor indicates that it is the retention, rather than the recruitment of these proteins that is altered under conditions of PRC1 inhibition. Consistent with the previously described requirement for ubiquitylation in the recruitment of 53BP1 and BRCA1, both proteins failed to recruit to IRIFs when PRC1 was inhibited (5, 14, 21, 22) (Fig. 6, A and B).
FIGURE 5.
Effect of PRT5165 on DNA-PKcs recruitment to sites of DNA damage. U2OS cells were either mock treated or treated with 50 μm of the PRT4165 for 5 min before they were exposed to radiation (2 Gy). The cells were allowed to recover for different time points (5 and 60 min). Cells were then fixed and stained as indicated. p-DNA-PKcs and γ-H2AX. The average number of p-DNA-PKcs IRIF per cell was quantified and plotted as indicated. The scale bars represent 10 μm.
FIGURE 6.
Effect of PRT5165 on the localization of 53BP1 and BRAC1 into sites of DNA damage. A, cells were treated as in Fig. 5 and stained with 53BP1 and γ-H2AX. B, γ-H2AX and BRCA1. The average number of p-DNA-PKcs and BRCA1 IRIF per cell was quantified and plotted as indicated. The scale bars represent 10 μm.
PRT4165 Treatment Delays Recovery from Genotoxic Stress
To test the role of PRT4165-sensitive events in the DDR, we treated U2OS cells with PRT4165 and then exposed these cells to a moderate dose of IR (2 Gy). We next followed the formation of MDC1 IRIF in individual nuclei over time. Because MDC1 accumulation at sites of DNA damage is independent of PRC1-dependent ubiquitylation (Fig. 3C), we used the dissolution of MDC1 foci as a measure of DSB repair. The number of nuclei with MDC1 foci detected early after IR (5, 15, 30, and 60 min) was very similar in control and PRT4165-treated cells. However, although MDC1 foci were progressively lost over time in control cells, inhibition of PRC1 activity caused sustained persistence of MDC1 foci even at time points when the control cells had repaired most of their DSBs (Fig. 7A). This suggests delayed DSB repair in PRT4165 inhibited cells, a notion that was further supported by prolonged phosphorylation of two genome caretakers targeted by ATM after DNA damage, NBS1 and SMC1 (Fig. 7B). Based on these results, we conclude that PRC1 E3 ubiquitin ligase activity is required for efficient recovery from genotoxic stress.
FIGURE 7.
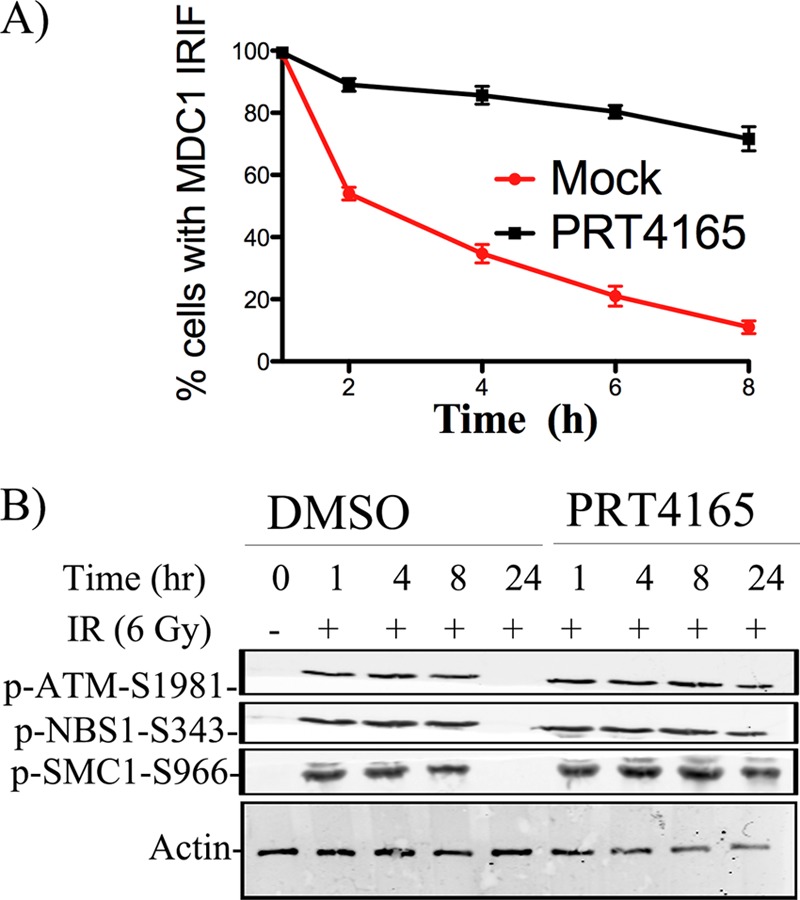
Inhibition of PRC1 activity in cells delays recovery from genotoxic stress. A, U2OS cells were either mock treated or treated with 50 μm of the PRT5164 for 5 min before they were exposed to radiation (2 Gy). The cells were allowed to recover for different amounts of time before they were fixed and stained with MDC1 antibody. The proportion of cells with MDC1 IRIF were quantified and plotted as indicated. B, effect of the PRT4165 on the activation of the DDR. U2OS cells were either mock treated or treated with the PRT4165 (50 μm) for different amounts of time. Nuclear extracts were prepared and immunoblotted with the indicated phospho-specific antibodies.
PRT4165 Inhibits DSB Repair
The above results imply that there is a DSB repair defect in cells treated with PRT4165. Previous studies, including our own, have knocked down individual PRC1 components and reported an increase in radiation sensitivity of ∼2–3-fold. Although significant, this defect in DSB repair is not as dramatic as, for example, the inhibition of DNA PKcs signaling. The delayed dissolution of MDC1 foci suggested a more severe phenotype when PRC1 E3 ubiquitin ligase activity is inhibited. Thus, we tested the effect of PRT4165 treatment on DSB repair. U2OS cells were either left untreated or treated with 50 μm PRT4165 for 5 min before exposing them to 2 Gy of IR. The cells were then left to recover either in the presence or absence of PRT4165 for different time points. The cells were stained for γ-H2AX, and the number of γ-H2AX IRIF was counted. We found that, consistent with the previous reports, PRT4165 inhibits DSB repair at the 8-h time point compared with mock treated cells. This defect becomes even more pronounced after the 24-h time point (Fig. 8A). Interestingly, this repair defect is similar in magnitude to the inhibition of DNA-PKcs, a core component of the nonhomologous end joining DSB repair pathway.
FIGURE 8.
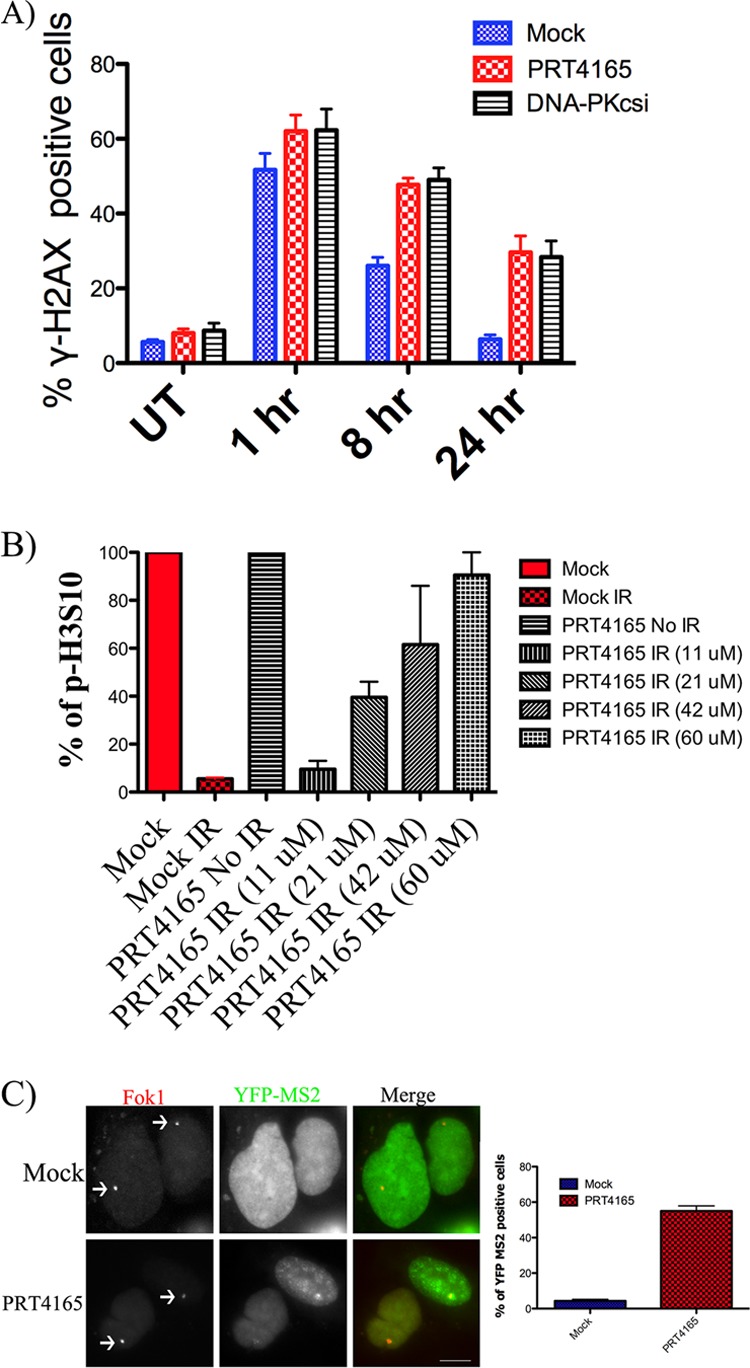
Effect of PRT5165 on G2/M DNA damage checkpoints and DSB repair. A, U2OS cells were either mock treated or treated with 50 μm of the PRT4165 for 5 min before they were exposed to radiation (2 Gy). The cells were allowed to recover for different amounts of time before they were fixed and stained with γ-H2AX. γ-H2AX IRIF were counted and plotted versus time. B, U2OS cells were either mock treated or treated with 50 μm of the PRT4165 for 2 h before they were exposed to radiation (2 Gy). The fraction of cells in G2 phase was determined by flow cytometry using phospho-specific antibody against H3 at serine 10. The graph is a summary from two experiments. The error bars represent standard error. C, U2OS cells containing the stably integrated reporter transiently transfected with plasmid DNA encoding the mCherry tagged wild-type FokI nuclease fusion (FokI WT) were either left untreated or treated with PRT4165 for 60 min before the initiation of transcription by doxycycline. Representative images of mCherry-FokI and YFP-MS2 loci are shown. Quantification of YFP-MS2 relative mean fluorescence intensity from C was done using ImageJ software (National Institutes of Health). The data were derived from three independent experiments. The scale bars represent 10 μm.
Effects of PRT4165 on G2/M DNA Damage Checkpoints
DNA damage activates cell cycle checkpoints, which arrest cell cycle progression to allow time for DNA repair. We next wanted to examine whether PRT4165 treatment alters the DNA damage G2/M checkpoints. To examine this, U2OS cells were plated in the presence of different concentrations of PRT4165. Cell cycle progression was monitored by FACS analysis using an antibody recognizing phosphorylated serine 10 of histone H3 (26) and propidium iodide 2.5 h postirradiation. The percentage of G2/M cells was 20% in the untreated cell population (Fig. 8B). Cells treated with increasing concentrations of PRT4165 showed increasing numbers of cells in G2/M (Fig. 8B). These data suggest that inhibition of PRC1 results in G2/M checkpoint failure.
PRT4165 Inhibits DSB-induced Transcriptional Silencing
Upon introduction of a DSB, nearby genes are transcriptionally repressed. RNF8 and BMI1 knockdown have both been found to reduce transcriptional silencing at DNA damage sites, although only the former has been demonstrated directly in response to DSBs (27). Because transcriptional repression through H2A monoubiquitylation is the principal mechanism whereby PRC1 is thought to function in the developmental regulation of gene expression, we tested whether or not inhibition by PRT4165 would inhibit DNA damage-associated transcriptional repression. We utilized a transcriptional reporter system where a targeted DSB can be introduced upstream of a gene that encodes a transcript containing a MS2 binding site (28). DSBs were introduced by fusing the nuclease domain of the FokI endonuclease to mCherry-LacI, targeting nuclease activity to the lac operator repeats. Transcription can be directly detected following transfection with YFP-MS2. This experimental system was previously used to demonstrate that RNF8 and ubiquitylation of H2A were required for DNA damage induced transcriptional silencing. Using this reporter system, we found that following DSB induction there was a 10-fold reduction in transcript levels in damaged cells compared with controls. PRT4165 was administered 1 h before the initiation of transcription by doxycycline, and nascent transcript levels were assessed 3 h later. Remarkably, PRT4165 almost completely restored transcriptional activity in cells expressing FokI WT but had no effect in control cells, demonstrating that a PRT4165-sensitive E3 ubiquitin ligase activity is required for DSB-induced transcriptional silencing (Fig. 8C).
DISCUSSION
In this study, we have characterized PRT4165, a small molecule identified in a screen for inhibitors of BMI1/RING1A-mediated polyubiquitylation of topoisomerase II-α upon trapping the DSB intermediate with the topoisomerase II inhibitor etoposide (7). We demonstrate that PRT4165 inhibits the two E3 ligase paralogues, RING1A and RNF2, in vitro, and that it can be used to inhibit H2A ubiquitylation in cells. Consequently, this inhibitor will be valuable in studying the contribution of H2A ubiquitylation to gene repression. Although we did not specifically study how this drug influences PRC1-mediated gene repression, we did find that the inhibitor was able to prevent the silencing of transcription that takes place near DSBs. This was shown to be dependent on H2A ubiquitylation (28).
There have been multiple studies that have employed knockdown, knock-out, and mutation analysis to directly implicate PRC1 and H2AX ubiquitylation in the regulation of signaling at sites of DSBs (14–19, 27, 29, 30). These studies have consistently shown that PRC1 is rapidly recruited to sites of DNA damage. However, there have been some differences in the consequences of PRC1 down-regulation even when the same cell system has been used. There are two important limitations of these studies apart from technical differences, such as the efficiency of the knockdown or adaptation of knock-out cells. The most important limitation of these studies is that PRC1 is a major repressor of transcription that silences target genes through the ubiquitylation of histone H2A (8). Knockdown and knock-out experiments are therefore complicated by significant differences in gene expression profiles that are likely to take place that complicate the interpretation of any observed differences. A second important difference is that every subunit of the PRC1 complex has at least one paralogue (10), and many of these paralogues have been shown to recruit to sites of laser microirradiation-induced strand breaks (27). Consequently, we applied this drug to determine which of the observed signaling and repair defects observed in knockdown and knock-out experiments could be replicated upon transient inhibition of PRC1. Using this approach, we could show complete inhibition of ubiquitylation of phosphorylated H2AX, reflecting ubiquitylation taking place near DSBs, if we added the drug at the time of irradiation and then measured ubiquitylation 10 min later. This illustrates that we could effectively inhibit the ubiquitylation of H2A that takes place upon DNA damage.
Employing this transient inhibition of PRC1-mediated H2A ubiquitylation, we were able to replicate several observations made previously using knockdown and knock-out strategies. We found that two of the best characterized ubiquitin-dependent proteins, BRCA1 and 53BP1, failed to be retained in IRIF in cells treated with PRT4165. We also observed the inhibition of H2A ubiquitylation within the phosphorylated histone H2AX pool associated with the DSBs, as expected. In addition, we confirm that PRC1 is required for the inhibition of transcription near sites of DSBs, as reported previously in BMI1 knock-out mouse embryonic fibroblasts (17).
The inhibition of ubiquitylation at DSBs in the presence of PRT4165 is remarkably robust. Not only is H2A/H2AX ubiquitylation inhibited, the accumulation of K63-linked polyubiquitin chains is also inhibited. When using drugs in micromolar concentrations and above, it is expected that there will be off-target effects, and we do not know at this point what other E3 ubiquitin ligases or other classes of enzymes may be inhibited by this drug. K63-linked polyubiquitylation is known to require two E3 ubiquitin ligases. RNF8 is recruited by binding to phosphorylated MDC1 and is thought to initiate the ubiquitylation cascade (reviewed in Ref. 31). RNF168 is recruited by binding to the ubiquitin deposited by RNF8 and is required for the amplification K63-linked polyubiquitin signaling at DSBs (reviewed in Ref. 32). Consequently, we tested these two enzymes for potential inhibition by PRC1. Neither enzyme was significantly inhibited at the concentrations used in this study. Moreover, RNF8 was efficiently retained in IRIF. We would therefore expect that if the established pathway is correct, RNF8 should be able to recruit RNF168 to IRIFs and K63-linked polyubiquitin chains should be formed. We do not see this. The least likely explanation for this result is that PRT4165 directly inhibits RNF8 in cells despite having little activity toward RNF8 in vitro.
With respect to H2A/H2AX ubiquitylation upon DSB formation, it was recently shown that despite the robust H2A ubiquitylation activity of RNF8 using purified H2A, RNF8 could not ubiquitylate H2A when H2A was reconstituted into nucleosomes (2). These experiments identified a previously unknown ubiquitylation site in the N terminus of H2A, lysine 13 or 15, that was monoubiquitylated by RNF168 and then extended by RNF8 into polyubiquitin chains (2). It is also unlikely that PRT4165 inhibits K63-linked polyubiquitin chain formation through the direct inhibition of RNF168 in cells. It is more likely that there is at least one upstream component that is required for the initiation of this pathway, and this upstream activity is inhibited by PRT4165.
It is possible that PRT4165 inhibits an unknown upstream component of this signaling pathway other than PRC1. The simplest explanation, however, is that PRC1-mediated monoubiquitylation is required for K63-linked polyubiquitin chain extension. If that target is histone H2A/H2AX, there are two possibilities. The first is that PRC1-mediated ubiquitylation of the H2A C terminus is required for RNF168-mediated ubiquitylation of the H2A N terminus, which is then used as the substrate for K63-linked chain formation. A second possibility is that RNF168-mediated monoubiquitylation of H2A in the N terminus requires RNF8-mediated ubiquitylation to recruit RNF168 to the site of the DSB. In this case, PRC1, rather than RNF168, initiates the signaling through the monoubiquitylation of the C terminus. RNF8 then extends this ubiquitin into chains sufficiently long to recruit RNF168, which, in turn, monoubiquitylates H2A/H2AX at lysine 15. Consistent with this interpretation, lysine 15 monoubiquitylation was recently shown to be required for 53BP1 retention in IRIF (20). Durocher and co-workers (20) argue that RNF8-mediated generation of K63-linked polyubiquitin chains serves to recruit RNF168, which then monoubiquitylates H2A/H2AX on lysine 15.
We favor the explanation that PRC1 is required to initiate the ubiquitylation by forming monoubiquitylated substrate, which need not be histone H2A, but which is required for RNF8/UBC13 to extend into K63-linked polyubiquitin chains. We have ruled out that there is an unknown target of PRT4165 that inhibits the phosphorylation-dependent recruitment of RNF8, but it remains possible that another activity or an E3 ubiquitin ligase other than PRC1 is required for the initiation of the RNF8-dependent K63-linked polyubiquitin chain formation.
Whether or not the sole DDR-related target of PRT4165 is PRC1, PRT4165 should prove to be a very useful drug for studying the DNA damage response because it appears to only inhibit the ubiquitylation-dependent signaling that is taking place at DSBs. The ATM and related kinases responsible for the phosphorylation signaling cascade appear to be fully operative. The only other known class of drugs that is able to achieve this is the proteosome inhibitors, which inhibit ubiquitylation by sequestering ubiquitin in polyubiquitylated proteins targeted for degradation. Because this is a very indirect way of inhibiting ubiquitylation signaling at DSBs, PRT4165 may provide a better alternative for chemical inhibition of ubiquitin signaling at DSBs.
In conclusion, we have shown that PRT4165 can be used to rapidly inhibit ongoing histone H2A ubiquitylation in cells. This drug represents the first chemical inhibitor that is available to inhibit the PRC1 E3 ubiquitin ligase and should provide an important tool for gene expression studies. We also show that PRT4165 can be used to inhibit ubiquitylation signaling taking place at DSBs while not impairing the phosphorylation of histone H2AX and the assembly of ubiquitin-independent components of IRIF.
Acknowledgments
We thank Dr. Randall Gieni for a critical reading and editing of the manuscript. We thank Drs. Andre Nussenzweig, Roger Greenberg, Susanne Janicki, Daniel Durocher, Robert Cohen, and Junji Chen for providing valuable reagents. We also thank the Cellular Imaging Facility at the Cross Cancer Institute for the use of microscopes and image analysis software.
This work was supported by grants from the Alberta Cancer Foundation and the Canadian Institute of Health Research.
- DSB
- double-strand break
- PRC
- polycomb repressive complex
- K63
- lysine 63.
REFERENCES
- 1. Polo S. E., Jackson S. P. (2011) Dynamics of DNA damage response proteins at DNA breaks. A focus on protein modifications. Genes Dev. 25, 409–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mattiroli F., Vissers J. H., van Dijk W. J., Ikpa P., Citterio E., Vermeulen W., Marteijn J. A., Sixma T. K. (2012) RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 [DOI] [PubMed] [Google Scholar]
- 3. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 4. Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 5. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 6. Dantuma N. P., Groothuis T. A., Salomons F. A., Neefjes J. (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 173, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alchanati I., Teicher C., Cohen G., Shemesh V., Barr H. M., Nakache P., Ben-Avraham D., Idelevich A., Angel I., Livnah N., Tuvia S., Reiss Y., Taglicht D., Erez O. (2009) The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2α cleavage complex. A potentially new drug target. PLoS One 4, e8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 9. Cao R., Tsukada Y., Zhang Y. (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20, 845–854 [DOI] [PubMed] [Google Scholar]
- 10. Gieni R. S., Hendzel M. J. (2009) Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochem. Cell Biol. 87, 711–746 [DOI] [PubMed] [Google Scholar]
- 11. Raaphorst F. M., van Kemenade F. J., Blokzijl T., Fieret E., Hamer K. M., Satijn D. P., Otte A. P., Meijer C. J. (2000) Coexpression of BMI-1 and EZH2 polycomb group genes in Reed-Sternberg cells of Hodgkin's disease. Am. J. Pathol. 157, 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleer C. G., Cao Q., Varambally S., Shen R., Ota I., Tomlins S. A., Ghosh D., Sewalt R. G., Otte A. P., Hayes D. F., Sabel M. S., Livant D., Weiss S. J., Rubin M. A., Chinnaiyan A. M. (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 100, 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Kemenade F. J., Raaphorst F. M., Blokzijl T., Fieret E., Hamer K. M., Satijn D. P., Otte A. P., Meijer C. J. (2001) Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood 97, 3896–3901 [DOI] [PubMed] [Google Scholar]
- 14. Ismail I. H., Andrin C., McDonald D., Hendzel M. J. (2010) BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191, 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ginjala V., Nacerddine K., Kulkarni A., Oza J., Hill S. J., Yao M., Citterio E., van Lohuizen M., Ganesan S. (2011) BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell Biol. 31, 1972–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Facchino S., Abdouh M., Chatoo W., Bernier G. (2010) BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 30, 10096–10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chagraoui J., Hébert J., Girard S., Sauvageau G. (2011) An anticlastogenic function for the Polycomb Group gene Bmi1. Proc. Natl. Acad. Sci. U.S.A. 108, 5284–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pan M. R., Peng G., Hung W. C., Lin S. Y. (2011) Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol. Chem. 286, 28599–28607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu C. Y., Kang H. Y., Yang W. L., Wu J., Jeong Y. S., Wang J., Chan C. H., Lee S. W., Zhang X., Lamothe B., Campos A. D., Darnay B. G., Lin H. K. (2011) Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J. Biol. Chem. 286, 30806–30815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fradet-Turcotte A., Canny M. D., Escribano-Díaz C., Orthwein A., Leung C. C., Huang H., Landry M. C., Kitevski-LeBlanc J., Noordermeer S. M., Sicheri F., Durocher D. (2013) 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sims J. J., Scavone F., Cooper E. M., Kane L. A., Youle R. J., Boeke J. D., Cohen R. E. (2012) Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat. Methods 9, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uhrhammer N., Bay J. O., Bignon Y. J. (1998) Seventh International Workshop on Ataxia-Telangiectasia. Cancer Res. 58, 3480–3485 [PubMed] [Google Scholar]
- 25. Opeskin K., Waterston J., Nirenberg A., Hare W. S. (1998) Ataxia telangiectasia with long survival. J. Clin. Neurosci. 5, 471–473 [DOI] [PubMed] [Google Scholar]
- 26. Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 [DOI] [PubMed] [Google Scholar]
- 27. Chou D. M., Adamson B., Dephoure N. E., Tan X., Nottke A. C., Hurov K. E., Gygi S. P., Colaiácovo M. P., Elledge S. J. (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 107, 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A. (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ismail I. H., Gagné J. P., Caron M. C., McDonald D., Xu Z., Masson J. Y., Poirier G. G., Hendzel M. J. (2012) CBX4-mediated SUMO modification regulates BMI1 recruitment at sites of DNA damage. Nucleic Acids Res. 40, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nacerddine K., Beaudry J.-B., Ginjala V., Westerman B., Mattiroli F., Song J.-Y., van der Poel H., Ponz O. B., Pritchard C., Cornelissen-Steijger P., Zevenhoven J., Tanger E., Sixma T. K., Ganesan S., van Lohuizen M. (2012) Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. J. Clin. Invest. 122, 1920–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jackson S. P., Durocher D. (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 [DOI] [PubMed] [Google Scholar]
- 32. Panier S., Durocher D. (2009) Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair 8, 436–443 [DOI] [PubMed] [Google Scholar]