Abstract
The molecular mechanisms involved in the development of type 2 diabetes are poorly understood. Starting from genome-wide genotype data for 1,924 diabetic cases and 2,938 population controls generated by the Wellcome Trust Case Control Consortium, we set out to detect replicated diabetes association signals through analysis of 3,757 additional cases and 5,346 controls, and by integration of our findings with equivalent data from other international consortia. We detected diabetes susceptibility loci in and around the genes CDKAL1, CDKN2A/CDKN2B and IGF2BP2 and confirmed the recently described associations at HHEX/IDE and SLC30A8. Our findings provide insights into the genetic architecture of type 2 diabetes, emphasizing the contribution of multiple variants of modest effect. The regions identified underscore the importance of pathways influencing pancreatic beta cell development and function in the etiology of type 2 diabetes.
The pathophysiological basis of type 2 diabetes (T2D) remains unclear despite its growing global importance (1). Candidate gene and positional cloning efforts have suggested many putative susceptibility variants, but unequivocal replications are so far limited to variants in just three genes: PPARG, KCNJ11 and TCF7L2 (2-4).
Improved understanding of the correlation between genetic variants (linkage disequilibrium [LD]), allied to advances in genotyping technology, have enabled systematic searches for disease-associated common variants on a genome-wide scale. The Wellcome Trust Case Control Consortium (WTCCC) recently completed such a genome-wide association (GWA) scan in 1,924 T2D cases and 2,938 population controls from the UK, using the Affymetrix GeneChip Human Mapping 500k Array Set (5). The strongest association signals genome-wide were observed for single nucleotide polymorphisms (SNPs) in TCF7L2 (e.g. rs7901695 OR 1.37 [95% CI 1.25-1.49], p=6.7×10−13). The other known T2D susceptibility variants were detected with effect sizes consistent with previous reports (2, 3).
Here, we describe how integration of data from the WTCCC scan and our own replication studies with similar information generated by the Diabetes Genetics Initiative [DGI] (6) and the Finland-United States Investigation of NIDDM Genetics [FUSION] (7) has identified several additional susceptibility variants for T2D.
In the WTCCC study, analysis of 490,032 autosomal SNPs in 16,179 samples yielded 459,448 SNPs that passed initial quality control (5). We considered only the 393,453 autosomal SNPs with minor allele frequency (MAF) exceeding 1% in both cases and controls and no extreme departure from Hardy Weinberg equilibrium (HWE) (p<10−4 in cases or controls) (8). This T2D-specific dataset shows no evidence of substantial confounding from population substructure and genotyping biases (8).
To distinguish true associations from those reflecting fluctuations under the null or residual errors arising from aberrant allele calling, we first submitted putative signals from the WTCCC study to additional quality control including cluster plot visualization and validation genotyping on a second platform (8). Next, we attempted replication of selected signals in up to 3,757 additional cases and 5,346 controls (replication sets RS1-RS3). RS1 comprised 2,022 cases and 2,037 controls from the UK Type 2 Diabetes Genetics Consortium collection (UKT2DGC: all from Tayside, Scotland). RS2 included 632 additional T2D cases and 1,750 population controls from the Exeter Family Study of Child Health (EFSOCH). A subset of SNPs were typed in RS3, comprising a further 1,103 cases and 1,559 controls from the UKT2DGC (Table S1).
The first wave of validated SNPs sent for replication was selected from the 30 SNPs, in 9 distinct chromosomal regions (excluding TCF7L2), which had, in the WTCCC scan alone, attained the most extreme (p<10−5) significance values on Cochran-Armitage tests of association. Genotyping of 21 representative SNPs generated evidence of replication (p<0.05) for three of these 9 regions (Tables 1, S2).
Table 1. Confirmed T2D susceptibility variants.
Representative SNPs are shown for each signal with ORs and 95% CIs reported (for the Cochran-Armitage 1df test) with respect to the risk allele (denoted in bold, with the ancestral allele underlined where known). SNPs selected for inclusion are those with the strongest evidence for association in the UK datasets (except in the case of TCF7L2, where, to maximize consistency across the datasets rs7901695 is presented). In the case of HHEX, the UK meta-analysis combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU). Since DGI and FUSION had not typed the identical SNPs in all samples, results shown for those studies feature the SNP generating the strongest association: in all cases these were SNPs in strong LD (minimum r2 0.95, except TCF7L2) and with consistent direction of effect with the SNP reported in the UK data (see Table S3 for details). The use of different SNPs may result in slightly different estimates of p values and OR between the three studies. Combined estimates of the ORs were calculated by weighting the logORs of each study by the inverse of their variance.
| WTCCC | Replication meta-analysis |
All UK sample meta-analysis |
DGI | FUSION | All Combined | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1924 cases | 3757 cases | 5681 cases | 6529 cases | 2376 cases | 14586 cases | ||||||||||||
| 2938 controls | 5346 controls | 8284 controls | 7252 controls | 2432 controls | 17968 controls | ||||||||||||
| rs | chr | position | A1 | A2 | Region | OR (95%Cis) |
Padd | OR (95%Cis) |
Padd | OR (95%Cis) |
Padd | OR (95%Cis) |
Padd | OR (95%Cis) |
Padd | OR (95% Cis) |
Padd |
| rs8050136 | 16 | 52373776 | A | C | FTO | 1.27 (1.16-1.37) |
2.0×10−8 | 1.22 (1.12-1.32) |
5.4×10−7 | 1.23 (1.18-1.32) |
7.3×10−14 | 1.03 (0.91-1.17) |
0.25 | 1.11 (1.02-1.20) |
0.017 | 1.17 (1.12-1.22) |
1.3×10−12 |
| rs10946398 | 6 | 20769013 | A | C | CDKAL1 | 1.20 (1.10-1.31) |
2.5×10−5 | 1.14 (1.07-1.22) |
8.4×10−5 | 1.16 (1.10-1.22) |
1.3×10−8 | 1.08 (1.03-1.14) |
2.4×10−3 | 1.12 (1.03-1.22) |
9.5×10−3 | 1.12 (1.08,1.16) |
4.1×10−11 |
| rs5015480 | 10 | 94455539 | C | T | HHEX | 1.22 (1.12-1.33) |
5.4×10−6 | - | - | ||||||||
| rs1111875 | 10 | 94452862 | C | T | HHEX | - | - | 1.08 (1.01-1.15) |
0.020 | 1.13 (1.07-1.19) |
4.6×10−6 | 1.14 (1.06-1.22) |
1.7×10−4 | 1.10 (1.01-1.19) |
0.025 | 1.13 (1.08-1.17) |
5.7×10−10 |
| rs10811661 | 9 | 22124094 | C | T | CDKN2B | 1.22 (1.09-1.37) |
7.6×10−4 | 1.18 (1.08-1.28) |
1.7×10−4 | 1.19 (1.11-1.28) |
4.9×10−7 | 1.20 (1.12-1.28) |
5.4×10−8 | 1.20 (1.07-1.36) |
2.2×10−3 | 1.20 (1.14-1.25) |
7.8−10−15 |
| rs564398 | 9 | 22019547 | C | T | CDKN2B | 1.16 (1.07-1.27) |
3.2×10−4 | 1.12 (1.05-1.19) |
8.6×10−4 | 1.13 (1.08-1.19) |
1.3×10−6 | 1.05 (0.94-1.17) |
0.5 | 1.13 (1.01-1.27) |
0.039 | 1.12 (1.07-1.17) |
1.2×10−7 |
| rs4402960 | 3 | 186994389 | G | T | IGF2BP2 | 1.15 (1.05-1.25) |
1.7×10−3 | 1.09 (1.01-1.16) |
0.018 | 1.11 (1.05-1.16) |
1.6×10−4 | 1.17 (1.11-1.23) |
1.7×10−9 | 1.18 (1.08-1.28) |
2.4×10−4 | 1.14 (1.11-1.18) |
8.6×10−16 |
| rs13266634 | 8 | 118253964 | C | T | SLC30A8 | 1.12 (1.02-1.23) |
0.020 | 1.12 (1.04-1.19) |
1.2×10−3 | 1.12 (1.05-1.18) |
7.0×10−5 | 1.07 (1.00-1.16) |
0.047 | 1.18 (1.09-1.29) |
7.0×10−5 | 1.12 (1.07-1.16) |
5.3×10−8 |
| rs7901695 | 10 | 114744078 | C | T | TCF7L2 | 1.37 (1.25-1.49) |
6.7×10−13 | - | - | - | - | 1.38 (1.31-1.46) |
2.3×10−31 | 1.34 (1.21-1.49) |
1.4×10−8 | 1.37 (1.31-1.43) |
1.0×10−48 |
| rs5215 | 11 | 17365206 | C | T | KCNJ11 | 1.15 (1.05-1.25) |
1.3×10−3 | - | - | - | - | 1.15 (1.09-1.21) |
1.0−10−7 | 1.11 (1.02-1.20) |
0.014 | 1.14 (1.10-1.19) |
5.0×10−11 |
| rs1801282 | 3 | 12368125 | C | G | PPARG | 1.23 (1.09-1.41) |
1.3×10−3 | - | - | - | - | 1.09 (1.01-1.16) |
0.019 | 1.20 (1.07-1.33) |
1.4×10−3 | 1.14 (1.08-1.20) |
1.7×10−6 |
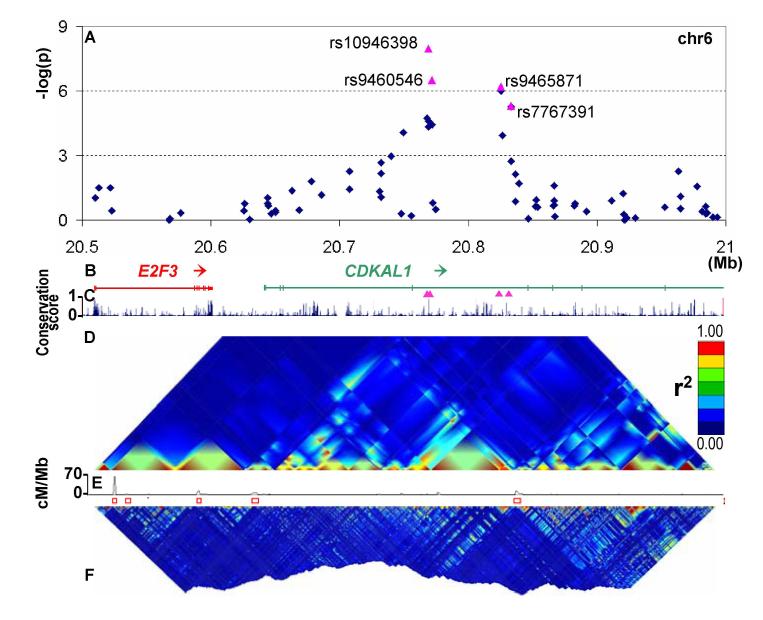
Rs8050136 (mapping to the FTO [fat mass associated with obesity] gene region on chr16) was among a cluster of SNPs generating the strongest evidence for association outside TCF7L2 in the original scan (risk allele OR 1.27 [1.16-1.37] p=2.0×10−8) (Figure S1). This SNP showed strong replication (OR 1.22 [1.12-1.32], p=5.4×10−7). As we recently reported (9), this effect on T2D risk is mediated through a primary effect on adiposity, and adjustment for BMI abolishes the T2D association. Replication was also obtained for SNPs within the CDKAL1 locus on chromosome 6, including rs9465871 and rs10946398. Although rs9465871 generated the stronger signal in the WTCCC scan, replication at this SNP was modest (p=0.023). The replication signal at rs10946398 was more striking (OR 1.14 [1.07-1.22], p=8.4×10−5) (Tables 1, S2). Consistent evidence of association is provided by the DGI (p=4.1×10−4 at rs7754840) and FUSION groups (p=9.5×10−3 at rs471253) (Tables 1, S3) (6, 7), both SNPs being strong (r2>0.99) proxies for rs10946398. Across all studies, combined evidence for association at CDKAL1 is compelling (p~4.1×10−11). All associated SNPs map to a large (90kb) intron within CDKAL1 (Figure 1). Flanking recombination hotspots define a 200kb interval likely to contain the etiological variant(s). CDKAL1 (cyclin-dependent kinase 5 [CDK5] regulatory subunit associated protein 1-like 1) encodes a 579-residue, 65kD protein of unknown function. We have detected expression of CDKAL1 mRNA in human pancreatic islet and skeletal muscle (Figure S2). CDKAL1 shares considerable protein domain and amino acid homology with CDK5 regulatory subunit associated protein 1 (CDK5RAP1), a known inhibitor of CDK5 activation. CDK5 has been implicated in the regulation of pancreatic beta cell function, through formation of p35/CDK5 complexes that downregulate insulin expression (11, 12).
Figure 1. Overview of CDKAL1 signal region.
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (25) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (26).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
The third replicated association maps to the HHEX (homeobox, hematopoietically expressed) gene region on chromosome 10. This gene showed both strong association in the WTCCC scan (rs5015480: risk allele OR 1.22 [1.12-1.33], p=5.4×10−6) and is a powerful biological candidate (13, 14). We could not optimize a replication assay for rs5015480, but observed evidence for replication at a perfect proxy, rs1111875 (risk allele OR 1.08 [1.01-1.15], p=0.02) (Tables 1, S2, S3). Both DGI and FUSION studies showed modest, but consistent association signals generating strong combined evidence (p~5.7×10−10) for a role in T2D susceptibility (Tables 1, S3). A fourth genome-wide association scan, in French subjects, recently reported independent evidence for a T2D signal in this region (10). The signal resides within an extended (295kb) region of LD containing not only HHEX (highly expressed in fetal and adult pancreas [Figure S2]) but also the genes encoding kinesin-interacting factor (KIF11) and insulin degrading enzyme (IDE) (Figure S3). IDE represents a second strong biological candidate given postulated effects on both insulin signalling and islet function, and data from rodent models (15-17).
Of the remaining regions selected in the first wave, none showed any evidence of replication in UK samples (Table S2), and for none was there strong support from the DGI and FUSION scans.
The relatively strict thresholds imposed for SNP selection in the first wave (i.e. point-wise p<10−5) help to limit false discovery, but many genuine susceptibility variants will fail to reach them. We initiated a second wave of replication based around SNPs for which the WTCCC scan generated more modest evidence for association (Cochran-Armitage p ~10−2 to 10−5). We prioritized the 5367 SNPs in this range, using additional criteria: (a) evidence of association in DGI and FUSION (6, 7); (b) presence of multiple, independent (r2<0.4) associations within the same locus; and (c) biological candidacy (8, 18).
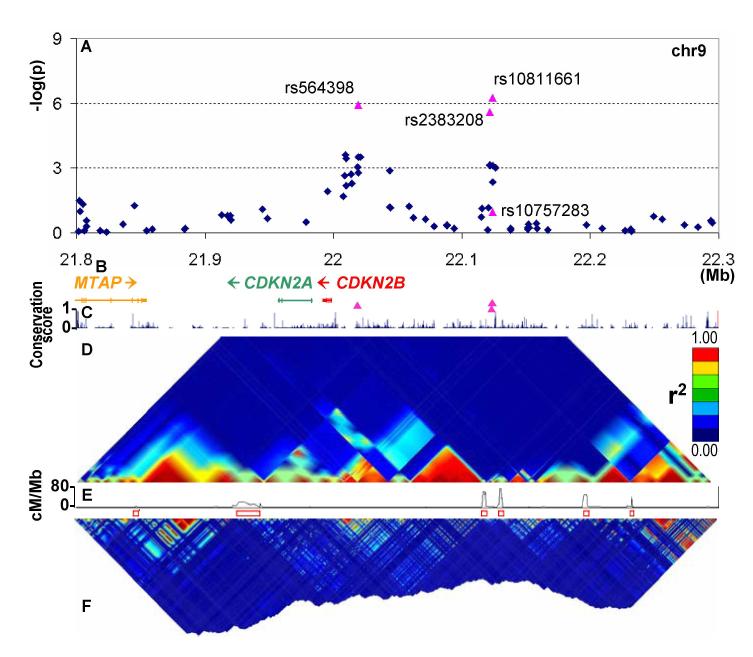
Analysis of the 56 SNPs, representing 49 putative signals, selected for this “second wave” of replication (Table S4) yielded two further regions implicated in T2D-susceptibility. A cluster of SNPs on chromosome 9 (represented by rs10811661) generated a promising signal in all three scans. Replication was observed in UK samples (rs10811661:OR 1.18 [1.08-1.28], p=1.7×10−4), as well as DGI (p=2.2×10−5) and FUSION follow-up studies (rs2383208, p=9.7×10−3). A second signal from the WTCCC scan located ~100kb 5′ (rs564398, OR 1.16 [1.07-1.27], p=3.2×10−4) was weakly supported in the FUSION, but not the DGI scan (Tables 1, S3) and replicated in the UK RS samples (OR 1.12 [1.05-1.19], p=8.6×10−4) (Tables 1, S3).
These two association signals are separated by a recombination hotspot (D′ between rs10811661 and rs564398 is 0.057, r2<0.001) (Figure 2). Across all studies, the combined evidence for association is stronger for the 3′ (p~7.8×10−15) than the 5′ (p~1.2×10−7) peak (Table 1). The 3′ signal maps to sequence with no characterized genes, while the recombination interval enclosing the 5′ signal includes the full coding sequences of CDKN2B and CDKN2A (encoding p15INK4b and p16INK4a respectively). CDKN2A is a known tumour suppressor and its product, p16INK4a, inhibits CDK4 (cyclin-dependent kinase 4), a powerful regulator of pancreatic beta cell replication (19-21). Overexpression of Cdkn2a leads to decreased islet proliferation in ageing mice (22). Cdkn2b overexpression is also causally related to islet hypoplasia and diabetes in murine models (23). Both CDKN2B and CDKN2A display high levels of expression in pancreatic islets and pituitary (Figure S2).
Figure 2. Overview of chr9 signal region.
Panel layout as per Figure 1
A fifth replicated association lies within the IGF2BP2 gene on chromosome 3. We observed some evidence of association for SNPs in this region in the WTCCC scan (5) (e.g. rs4402960: OR 1.15 [1.05-1.25], p=1.7×10−3). Consistent associations in the DGI and FUSION scans (6, 7) and the biological candidacy of the gene (a known regulator of insulin-like growth factor 2 [IGF2] translation), prompted replication. We obtained only modest evidence for replication at rs4402960 (OR 1.09 [1.01-1.16], p=0.018) (Tables 1, S4), but combined evidence across all studies (p~8.6×10−16) establishes this as a genuine T2D signal (Tables 1, S3). The associated SNPs map to a 57kb region spanning the promoter and first 2 exons of IGF2BP2 (Figure S4).
Most of the remaining 50 “second wave” SNPs can be discounted as susceptibility variants based on their failure to replicate (Table S4), though some merit further consideration. One such example is rs9369425, located 57kb downstream of the VEGFA (vascular endothelial growth factor A) gene on chromosome 6 (Figure S5). Evidence for association in the WTCCC scan (OR 1.16 [1.06-1.27], p=8.6×10−4) is supported by nominal replication in UK samples (1.08 [1.01-1.15], p=0.03) and by DGI scan results (1.17 [1.04-1.32], p=4.4×10−3). While no signal is apparent in the FUSION study, this does not allow us to reject this association. For 80% power to detect an OR of 1.11 (α=0.05), over 3,000 case-control pairs are needed.
In the French genome-wide scan (10), variants in both the HHEX and SLC30A8 genes were implicated in T2D susceptibility. As the associated SNPs in SLC30A8 are poorly captured on the Affymetrix chip (r2<0.01), the WTCCC scan was not informative for this locus. However, we genotyped rs13266634 independently and obtained replication of the finding (risk allele OR 1.12 [1.05-1.18], p=7.0×10−5 in all UK data) and across all three studies (p~5.3×10−8, Tables 1, S4).
The present analysis has contributed to identification of several confirmed T2D susceptibility loci. One of these (FTO) exerts its primary effect on T2D risk through an impact on adiposity (9): none of the other signals was attenuated by adjustment for BMI or waist circumference (Tables S5-S7). One of the remaining four loci (HHEX/IDE) represents a strong replication of findings recently reported (10). The other three loci (near CDKAL1, IGF2BP2 and CDKN2A), all showing extensive replication across the three studies represent novel T2D susceptibility loci.
Across the four T2D scans completed (5, 6, 7, 10), TCF7L2 clearly emerges as the largest association signal. On current evidence, all other confirmed loci display more modest effect sizes (between 1.10 and 1.25 per allele). Extensive resequencing and fine-mapping will be required to define the full spectrum of etiological variation at each locus and these may yet identify variants with greater impact. Our findings offer clear lessons for the design of future studies. Robust identification of variants with such effect sizes is only feasible with large-scale sample sets (13,965 individuals were typed in the present study). Further, the exchange of data between groups (providing data on up to 32,554 samples) was key to the rapid and unequivocal identification of the signals we report.
As a result of the four GWA studies reported to date (5, 6, 7, 10), the number of genuine, replicated T2D susceptibility signals has climbed from 3 to 9 (adding HHEX/IDE, SLC30A8, CDKAL1, CDKN2A, IGF2BP2 and FTO). However, these loci explain only a small proportion of the observed familiality (the sibling relative risk, λs, attributable to all loci in the UK samples is only ~1.07). We expect additional loci to be revealed by further rounds of replication initiated by more systematic meta-analysis of these and other scans. Our study provides an important validation of the genome-wide indirect association mapping approach and a demonstration of the value of aggressive data sharing efforts. It also generates insights into T2D pathogenesis emphasizing the likely importance of pathways involved in pancreatic beta cell development, regeneration and function. In-depth physiological and functional studies are now needed to establish the precise mechanisms involved.
Supplementary Material
Figure S1: Overview of FTO signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S2: Expression patterns of CDKAL1, CDKN2A, CDKN2B, HHEX, IGF2BP2. Messenger RNA expression profiles are shown for the genes listed for a range of human tissues, as determined by RT-PCR. Figures on the y axes refer to the test transcript levels relative to two separate endogenous control genes (beta glucuronidase [BGUS] and beta 2 microglobulin [B2M]). Each test:control ratio was then normalized to that of adult human pancreas for tissue to tissue comparison.
Figure S3: Overview of HHEX signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples. Note that rs5015480 was typed in the WTCCC scan and rs1111875 in the replication set, so the meta-analysis result is based on a combined analysis of the two (r2=1 in HapMap CEU) and the position of this signal denoted at both locations.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S4: Overview of IGF2BP2 signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S5: Overview of VEGFA signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S6: Quantile-Quantile plots for the 393,453 SNPs passing T2D-specific quality control. The plot compares the observed and expected distributions for the 1df chi-square statistics generated from single-point Cochran-Armitage tests.
Figure S7: Correlation plot of association statistics for the WTCCC scan genotypes before and after adjusting for population structure. The plot compares the 1df χ2 (single-point Cochran-Armitage) values obtained from a naïve trend test for the 393.453 SNPs passing T2D-specific quality control, with the equivalent statistics generated after correcting for population substructure using Eigenstrat (S8). The high correlation overall (r2>0.99), and in particular the strong linearity for high χ2 values (top right) indicates minimal confounding from population substructure after implementation of the various QC measures described.
Figure S8: Single-point and haplotype-based analysis results for the chr9 signal region using GENEBPM. Circles denote single-point analysis results and the continuous line represents 5 SNP sliding window haplotype-based analyses using GENEBPM (S15). The two peaks of association are separated by a recombination hotspot (Figure 2). Multipoint analyses reveal much stronger evidence for association at the 3′ peak in this region. Common (>0.01) high risk haplotypes in the 3′ signal share alleles T and T at SNPs rs10811661 and rs10757283 respectively. In the WTCCC scan (S1), Cochran-Armitage p values for rs2891169 were similar to those of SNPs rs10811661 and rs10757283, which were selected for replication. In contrast, GENEBPM analysis indicated stronger evidence for single-point association for rs2891169.
Table S1: Clinical characteristics of UK samples. WTCCC controls came from two sources. No data on age, waist circumference or BMI are available for the UK Blood Service controls. Control individuals from the 1958 Birth Cohort were last reviewed at age 41. Under the terms of access, waist circumference and BMI values from these controls are not available to WTCCC researchers. Only 46% (all male) of the RS2 control individuals had available waist circumference measures.
Table S2: Replication results for SNPs selected for Cochran-Armitage p<10−5 on WTCCC scan. Alleles in this table are named alphabetically (as per the forward strand) with the ancestral allele underlined (where known). A2F denotes allele 2 frequency. For consistency, all ORs in this table are reported for allele 2 (and may therefore be the reciprocal of the ORs reported in the text and Table 1). It was not possible to design a working replication assay for rs5015480, and the UK meta-analysis for this signal combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU).
Table S3: Confirmed T2D susceptibility signals: SNPs reported for the DGI and FUSION studies. As DGI and FUSION did not always type the same SNPs as the UK study in all their samples, results in Table 1 include data from the SNPs generating the strongest association in their respective studies. Table S3 gives details of the SNPs reported for DGI and FUSION, and their LD relationships (based on HapMap CEU and/or genome-wide or imputation data as available) with the UK index SNP. In all cases these proxies were SNPs in strong LD (r2>0.95, except TCF7L2) and showed consistent direction of effect with the SNP reported in the UK data.
Table S4: Replication results for SNPs selected for the “second wave” of replication. Alleles in this table are named alphabetically (as per the forward strand) with the ancestral allele underlined (where known). A2F denotes allele 2 frequency. For consistency, all ORs in this table are reported for allele 2 (and may therefore be the reciprocal of the ORs reported in the text). These signals were (with the exception of rs13266634 in SLC30A8) selected on the basis of Cochran-Armitage test p values between 10−2 and 10−5 on the WTCCC scan, prioritized on the basis of biological candidacy, multiple independent associations and/or support from DGI and/or FUSION scans. It was not possible to design a working replication assay for rs11140802, and the UK meta-analysis for this signal combines data from rs11140802 and rs12346884 (r2=1 in HapMap CEU). Rs13266634 in SLC30A8 was not captured on the Affymetrix chip but was selected for replication on the basis of the associations in French (S18) and FUSION (S17) subjects.
Table S5: Associations between T2D susceptibility variants and (a) BMI, (b) waist circumference in cases and controls. Analyses were performed using linear regression on (a) log10-transformed BMI values and (b) on waist circumference values (in cm) using gender as a covariate. Beta values, 95% CIs and asymptotic p values (t statistic) are reported. Fixed-effects meta-analyses are shown. BMI and waist circumference information was not available for WTCCC controls. In the case of HHEX, the UK meta-analysis combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU). Rs8050136 was not typed in RS3. At rs13266634, in the WTCCC cases, the common allele (C; T2D risk allele) homozygotes have a waist circumference that is on average 3.2 cm and a BMI that is 2.0 kg/m2 less than the rare allele (TT) homozygotes.
Table S6: Effects of adjusting T2D associations for BMI and waist circumference. In this table, ORs and 95% CIs are reported with respect to the risk allele (denoted in bold, with the ancestral allele underlined where known). Analyses report ORs and CIs before and after adjustment for log10BMI or waist circumference and gender, by logistic regression, and fixed-effects meta-analysis. These analyses are only possible for the replication sets, since BMI and waist circumference values were not available in the WTCCC controls. Only the T2D associations at FTO are attenuated by adjustment for BMI and waist circumference.
Table S7: Associations between T2D susceptibility variants and age of diagnosis in cases. Analyses were performed using linear regression on square root-transformed age of diagnosis values using gender as a covariate. Beta values, 95% CIs and asymptotic p values (t statistic) are reported. Fixed-effects meta-analyses are shown. In the case of HHEX, the UK meta-analysis combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU). Rs8050136 were not typed in RS3. T2D- and adiposity-predisposing variants at FTO are associated with earlier age of diagnosis. At rs8050136, rare allele (A; T2D risk allele) homozygotes have an age of diagnosis that is on average 1.7 years earlier than the common allele (CC) homozygotes.
Table S8: Genotype counts, association p values under different genetic models and test of departure from additivity for robustly replicating signals. The most significant model for each SNP is shown in bold. In the case of HHEX, the UK meta-analysis combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU).
Table S9: Selection criteria used for “second wave” SNPs.
Table S10: Pairwise interaction analyses of replicating SNPs, and known T2D susceptibility variants in TCF7L2, KCNJ11 and PPARG. Odds ratios (for interactions under a log additive model) are reported to the risk allele at each SNP, as defined in Table 1. In the replication sets, the analyses were adjusted for the three strata. The results are ordered by interaction p value. Rs1801282, rs5215 and rs7901695 were not typed in the replication sets. Rs1111875 was typed as an r2=1 proxy (HapMap CEU) for rs5015480 in the replication sets.
Acknowledgments
We are grateful to all the study participants. We acknowledge the support of Diabetes UK, BDA Research, the UK Medical Research Council, the Wellcome Trust, European Commission (EURODIA LSHG-CT-2004-518153) and Peninsula Medical School. Personal funding comes from the Wellcome Trust (EZ, ATH, LRC); UK Medical Research Council (JRBP); Diabetes UK (RMF); Throne-Holst Foundation (CML); Leverhulme Trust (APM); Research Councils UK (LWH); and the Scottish Executive Generations Scotland Initiative (CNAP, ADM). MNW is Vandervell Foundation Research Fellow at the Peninsula Medical School. We are grateful to members of the DGI and FUSION teams for sharing data.
Footnotes
Supporting Online Material
Materials and Methods
Replication set-only combined effect size estimation
Haplotype-based analysis results
Overlap between association and linkage signals
Quantitative trait analysis results
Departures from additivity
Data access details
Membership of WTCCC
Detailed acknowledgements
References
Figs. S1, S2, S3, S4, S5, S6, S7, S8
Tables S1, S2, S3, S4, S5, S6, S7, S8, S9, S10
REFERENCES
- 1.Stumvoll M, Goldstein BJ, van Haeften TW. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.Altshuler D, et al. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 3.Gloyn AL, et al. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 4.Grant SF, et al. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 5.Donnelly P, the WTCCC, personal communication Data from the Wellcome Trust Case Control Consortium scan.
- 6.Diabetes Genetics Initiative Science. (this issue) [Google Scholar]
- 7.Scott LJ, et al. Science. (this issue) [Google Scholar]
- 8.Materials and methods are available as supporting material on Science Online
- 9.Frayling TM, et al. Science. Published online April 12 2007.
- 10.Sladek R, et al. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 11.Ubeda M, Rukstalis JM, Habener JF. J. Biol. Chem. 2006;281:28858–28864. doi: 10.1074/jbc.M604690200. [DOI] [PubMed] [Google Scholar]
- 12.Wei FY, et al. Nat. Med. 2005;11:1104–1108. doi: 10.1038/nm1299. [DOI] [PubMed] [Google Scholar]
- 13.Foley AC, Mercola M. Genes Dev. 2005;19:387–396. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bort R, Martinez-Barbera JP, Beddington RS, Zaret KS. Development. 2004;131:797–806. doi: 10.1242/dev.00965. [DOI] [PubMed] [Google Scholar]
- 15.Fakhrai-Rad H, et al. Hum. Mol. Genet. 2000;9:2149–2158. doi: 10.1093/hmg/9.14.2149. [DOI] [PubMed] [Google Scholar]
- 16.Seta KA, Roth RA. Biochem. Biophys. Res. Commun. 1997;231:167–71. doi: 10.1006/bbrc.1997.6066. [DOI] [PubMed] [Google Scholar]
- 17.Farris W, et al. Am. J. Pathol. 2004;164:1425–1434. doi: 10.1016/s0002-9440(10)63229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GeneSniffer (www.genesniffer.org) was used to prioritize genes for biological candidacy by integrating information from diverse online databases (including Entrez Gene, OMIM, PubMed and MGI).
- 19.Rane SG, et al. Nat. Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 20.Mettus RV, Rane SG. Oncogene. 2003;22:8413–8421. doi: 10.1038/sj.onc.1206888. [DOI] [PubMed] [Google Scholar]
- 21.Marzo N, et al. Diabetologia. 2004;47:686–694. doi: 10.1007/s00125-004-1372-0. [DOI] [PubMed] [Google Scholar]
- 22.Krishnamurthy J, et al. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 23.Moritani M, et al. Mol. Cell Endocrinol. 2005;229:175–184. doi: 10.1016/j.mce.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Blanchette M, et al. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersson F, Jonsson O, Cardon LR. Bioinformatics. 2004;20:3241–3243. doi: 10.1093/bioinformatics/bth341. [DOI] [PubMed] [Google Scholar]
- 26.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. Science. 2005;310:321. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Overview of FTO signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S2: Expression patterns of CDKAL1, CDKN2A, CDKN2B, HHEX, IGF2BP2. Messenger RNA expression profiles are shown for the genes listed for a range of human tissues, as determined by RT-PCR. Figures on the y axes refer to the test transcript levels relative to two separate endogenous control genes (beta glucuronidase [BGUS] and beta 2 microglobulin [B2M]). Each test:control ratio was then normalized to that of adult human pancreas for tissue to tissue comparison.
Figure S3: Overview of HHEX signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples. Note that rs5015480 was typed in the WTCCC scan and rs1111875 in the replication set, so the meta-analysis result is based on a combined analysis of the two (r2=1 in HapMap CEU) and the position of this signal denoted at both locations.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S4: Overview of IGF2BP2 signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S5: Overview of VEGFA signal region
A Plot of −log(p values) for T2D (Cochran-Armitage test for trend) against chromosome position in Mb. Blue diamonds represent primary scan results and pink triangles denote meta-analysis results across all UK samples.
B Genomic location of genes showing intron and exon structure (NCBI Build 35). Pink triangles show position of replication SNPs relative to gene structure.
C MULTIZ (S24) vertebrate alignment of 17 species showing evolutionary conservation.
D GoldSurfer2 (S19) plot of linkage disequilibrium (r2) for SNPs genotyped in WTCCC scan (passing T2D-specific quality control) in WTCCC T2D cases.
E Recombination rate given as cM/MB. Red boxes represent recombination hotspots (S25).
F GoldSurfer2 plot of linkage disequilibrium (r2) for all HapMap SNPs across the region (HapMap CEU data).
Figure S6: Quantile-Quantile plots for the 393,453 SNPs passing T2D-specific quality control. The plot compares the observed and expected distributions for the 1df chi-square statistics generated from single-point Cochran-Armitage tests.
Figure S7: Correlation plot of association statistics for the WTCCC scan genotypes before and after adjusting for population structure. The plot compares the 1df χ2 (single-point Cochran-Armitage) values obtained from a naïve trend test for the 393.453 SNPs passing T2D-specific quality control, with the equivalent statistics generated after correcting for population substructure using Eigenstrat (S8). The high correlation overall (r2>0.99), and in particular the strong linearity for high χ2 values (top right) indicates minimal confounding from population substructure after implementation of the various QC measures described.
Figure S8: Single-point and haplotype-based analysis results for the chr9 signal region using GENEBPM. Circles denote single-point analysis results and the continuous line represents 5 SNP sliding window haplotype-based analyses using GENEBPM (S15). The two peaks of association are separated by a recombination hotspot (Figure 2). Multipoint analyses reveal much stronger evidence for association at the 3′ peak in this region. Common (>0.01) high risk haplotypes in the 3′ signal share alleles T and T at SNPs rs10811661 and rs10757283 respectively. In the WTCCC scan (S1), Cochran-Armitage p values for rs2891169 were similar to those of SNPs rs10811661 and rs10757283, which were selected for replication. In contrast, GENEBPM analysis indicated stronger evidence for single-point association for rs2891169.
Table S1: Clinical characteristics of UK samples. WTCCC controls came from two sources. No data on age, waist circumference or BMI are available for the UK Blood Service controls. Control individuals from the 1958 Birth Cohort were last reviewed at age 41. Under the terms of access, waist circumference and BMI values from these controls are not available to WTCCC researchers. Only 46% (all male) of the RS2 control individuals had available waist circumference measures.
Table S2: Replication results for SNPs selected for Cochran-Armitage p<10−5 on WTCCC scan. Alleles in this table are named alphabetically (as per the forward strand) with the ancestral allele underlined (where known). A2F denotes allele 2 frequency. For consistency, all ORs in this table are reported for allele 2 (and may therefore be the reciprocal of the ORs reported in the text and Table 1). It was not possible to design a working replication assay for rs5015480, and the UK meta-analysis for this signal combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU).
Table S3: Confirmed T2D susceptibility signals: SNPs reported for the DGI and FUSION studies. As DGI and FUSION did not always type the same SNPs as the UK study in all their samples, results in Table 1 include data from the SNPs generating the strongest association in their respective studies. Table S3 gives details of the SNPs reported for DGI and FUSION, and their LD relationships (based on HapMap CEU and/or genome-wide or imputation data as available) with the UK index SNP. In all cases these proxies were SNPs in strong LD (r2>0.95, except TCF7L2) and showed consistent direction of effect with the SNP reported in the UK data.
Table S4: Replication results for SNPs selected for the “second wave” of replication. Alleles in this table are named alphabetically (as per the forward strand) with the ancestral allele underlined (where known). A2F denotes allele 2 frequency. For consistency, all ORs in this table are reported for allele 2 (and may therefore be the reciprocal of the ORs reported in the text). These signals were (with the exception of rs13266634 in SLC30A8) selected on the basis of Cochran-Armitage test p values between 10−2 and 10−5 on the WTCCC scan, prioritized on the basis of biological candidacy, multiple independent associations and/or support from DGI and/or FUSION scans. It was not possible to design a working replication assay for rs11140802, and the UK meta-analysis for this signal combines data from rs11140802 and rs12346884 (r2=1 in HapMap CEU). Rs13266634 in SLC30A8 was not captured on the Affymetrix chip but was selected for replication on the basis of the associations in French (S18) and FUSION (S17) subjects.
Table S5: Associations between T2D susceptibility variants and (a) BMI, (b) waist circumference in cases and controls. Analyses were performed using linear regression on (a) log10-transformed BMI values and (b) on waist circumference values (in cm) using gender as a covariate. Beta values, 95% CIs and asymptotic p values (t statistic) are reported. Fixed-effects meta-analyses are shown. BMI and waist circumference information was not available for WTCCC controls. In the case of HHEX, the UK meta-analysis combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU). Rs8050136 was not typed in RS3. At rs13266634, in the WTCCC cases, the common allele (C; T2D risk allele) homozygotes have a waist circumference that is on average 3.2 cm and a BMI that is 2.0 kg/m2 less than the rare allele (TT) homozygotes.
Table S6: Effects of adjusting T2D associations for BMI and waist circumference. In this table, ORs and 95% CIs are reported with respect to the risk allele (denoted in bold, with the ancestral allele underlined where known). Analyses report ORs and CIs before and after adjustment for log10BMI or waist circumference and gender, by logistic regression, and fixed-effects meta-analysis. These analyses are only possible for the replication sets, since BMI and waist circumference values were not available in the WTCCC controls. Only the T2D associations at FTO are attenuated by adjustment for BMI and waist circumference.
Table S7: Associations between T2D susceptibility variants and age of diagnosis in cases. Analyses were performed using linear regression on square root-transformed age of diagnosis values using gender as a covariate. Beta values, 95% CIs and asymptotic p values (t statistic) are reported. Fixed-effects meta-analyses are shown. In the case of HHEX, the UK meta-analysis combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU). Rs8050136 were not typed in RS3. T2D- and adiposity-predisposing variants at FTO are associated with earlier age of diagnosis. At rs8050136, rare allele (A; T2D risk allele) homozygotes have an age of diagnosis that is on average 1.7 years earlier than the common allele (CC) homozygotes.
Table S8: Genotype counts, association p values under different genetic models and test of departure from additivity for robustly replicating signals. The most significant model for each SNP is shown in bold. In the case of HHEX, the UK meta-analysis combines data from rs5015480 and rs1111875 (r2=1 in HapMap CEU).
Table S9: Selection criteria used for “second wave” SNPs.
Table S10: Pairwise interaction analyses of replicating SNPs, and known T2D susceptibility variants in TCF7L2, KCNJ11 and PPARG. Odds ratios (for interactions under a log additive model) are reported to the risk allele at each SNP, as defined in Table 1. In the replication sets, the analyses were adjusted for the three strata. The results are ordered by interaction p value. Rs1801282, rs5215 and rs7901695 were not typed in the replication sets. Rs1111875 was typed as an r2=1 proxy (HapMap CEU) for rs5015480 in the replication sets.