Summary
The immortal strand hypothesis proposes that stem cells retain a template copy of genomic DNA (i.e. an ‘immortal strand’) to avoid replication-induced mutations. An alternative hypothesis suggests that certain cells segregate sister chromatids non-randomly to transmit distinct epigenetic information. However, this area of research has been highly controversial, with conflicting data even from the same cell types. Moreover, historically, the same term of ‘non-random sister chromatid segregation’ or ‘biased sister chromatid segregation’ has been used to indicate distinct biological processes, generating a confusion in the biological significance and potential mechanism of each phenomenon. Here, we discuss the models of non-random sister chromatid segregation, and we explore the strengths and limitations of the various techniques and experimental model systems used to study this question. We also describe our recent study on Drosophila male germline stem cells, where sister chromatids of X and Y chromosomes are segregated non-randomly during cell division. We aim to integrate the existing evidence to speculate on the underlying mechanisms and biological relevance of this long-standing observation on non-random sister chromatid segregation.
Key words: Drosophila, Asymmetric cell division, Immortal strand hypothesis, Niche, Stem cells
Introduction
Many adult stem cells are known to undergo asymmetric cell division to generate daughters with distinct cell fates: a daughter cell that self-renews the stem cell identity and one that gives rise to differentiated cells that comprise mature tissue (Morrison and Spradling, 2008). Asymmetric stem cell division can be achieved by asymmetric segregation of intrinsic fate determinants and/or asymmetric placement of daughter cells into different microenvironments (i.e. inside versus outside the stem cell niche) (Knoblich, 2008). In addition to these asymmetries, which are clearly related to their function and fate, a series of studies have suggested that other types of asymmetries occur during stem cell division, including a non-random segregation of sister chromatids (Tajbakhsh, 2008), of the midbody (Ettinger et al., 2011; Kuo et al., 2011) or of protein aggregates (Rujano et al., 2006). However, the relevance of these asymmetries is yet to be established.
Among these, non-random sister chromatid segregation is undoubtedly the one that has been most extensively studied (reviewed by Charville and Rando, 2013; Tajbakhsh, 2008). We begin this Hypothesis with a description of the different modes and models for non-random sister chromatid segregation, then present the various techniques that are used to study sister chromatid segregation. Finally, we discuss our recent study in Drosophila germline stem cells that provides new clues into the molecular mechanisms by which sister chromatids are distinguished and segregated non-randomly and the biological relevance of non-random sister chromatid segregation. We also attempt to speculate how distinct types of non-random sister chromatid segregation may be mechanistically and/or biologically related.
Distinct types of non-random sister chromatid segregation
Although being classified as ‘non-random’ sister chromatid segregation, various types of non-random pattern with potentially different biological meaning have been observed or proposed. The most extensively studied type of non-random sister chromatid segregation is the one in which sister chromatids are coordinated among all chromosomes: one cell inherits all the sister chromatids that contain the older strand as a template, whereas the other cell inherits those that contain the newer strand as a template (extensively reviewed by Yennek and Tajbakhsh, 2013). In the case of the immortal strand hypothesis (ISH), it is speculated that the cell that inherits all of the older stands is the one that has to protect its genome from replication-induced mutations, such as a stem cell (Cairns, 1975). However, in many reports, the correlation between cell fate and strand segregation was not tested or unambiguously determined.
In another type of biased segregation, sister chromatids of paternal and maternal chromosomes coordinate their segregation; for example, it was reported that mouse maternal and paternal chromosomes 7 coordinate sister chromatid segregation in such a way that the maternal sister chromatid containing the Watson strand as a template always co-segregates with the paternal sister chromatid that also contains the Watson strand as a template (Armakolas and Klar, 2006). In this case, the relationship between the sister chromatids of chromosome 7 and other chromosomes, or the relationship between the segregation pattern of chromosome 7 and cell fates, have not been addressed. Thus, it is unclear how this phenomenon relates to the immortal strand hypothesis. Furthermore, more recent work using the chromosome-oriented fluorescence in situ hybridization (CO-FISH) method (see below) did not detect a non-randomness of sister chromatid segregation of chromosome 7 (Sauer et al., 2013), implying that the previously observed non-random segregation (Armakolas and Klar, 2006) could be the result of mitotic recombination that was used here as a method to examine sister chromatid segregation pattern.
However, in another type of biased sister chromatid segregation that is observed in fission yeast, the difference between sister chromatids being synthesized as a leading strand or lagging strand during S phase determines whether the daughter cells switch mating type or not (Klar, 1987a; Klar, 1987b). This is a clear example of where the difference between sister chromatids correlates with fate determination. However, it is not known whether sister chromatids of chromosome II [on which the mating type (mat) locus is located] are coordinated with other chromosomes (i.e. chromosome I or III). Therefore, it remains unclear whether the observed ‘biased segregation’ of the mat locus during the division of fission yeast is functionally or mechanistically related to other types of biased sister chromatid segregation, such as the immortal strand hypothesis or the coordination of maternal and paternal chromosomes 7 in mouse cells.
Models of non-random sister chromatid segregation during stem cell division
Now, it is clear that a term ‘non-random sister chromatid segregation’ does not necessarily describe a single biological phenomenon. As such, each type of non-random sister chromatid segregation can have a distinct biological meaning, and be carried out through distinct molecular or cellular mechanisms. Non-random sister chromatid segregation during stem cell division has been intensively studied in recent years in a broad range of stem cell populations (Tajbakhsh, 2008). There are two major models for the biological relevance of non-random sister chromatid segregation. The first model proposes that stem cells retain older template DNA strands of all the chromosomes to limit replication-induced errors (Cairns, 1975). This idea is termed the immortal strand hypothesis or ISH, because stem cells would inherit the template strand for many cell cycles (essentially forever), making the template strand ‘immortal’. However, the validity of this hypothetical meaning remains untested, because there is only limited data that compare the number of mutations in stem cells and differentiated cells (Rossi et al., 2007). Moreover, retaining older template strands would not prevent mutations caused by other naturally occurring DNA-damaging events (environmental factors and cellular stresses). Although this hypothesis has been intensively studied, the interpretation from some of these studies remains controversial, owing to the differences in techniques used and complexities of the different model systems employed (Yennek and Tajbakhsh, 2013).
The second model proposes that stem cells might non-randomly segregate sister chromatids of all or only a subset of chromosomes, perhaps to retain epigenetic memories (Klar, 1994; Lansdorp, 2007). However, it remains a mystery how exactly distinct epigenetic information is placed on two sister chromatids during replication (or perhaps during the subsequent G2 phase) and how information is segregated during division in such a way that instructs distinct cell fates.
Techniques to study sister chromatid segregation
Addressing the sister chromatid segregation pattern in a stem cell population can be very challenging as a result of multiple factors. First, the ambiguity with regard to the characteristics of stem cells (e.g. identity and mode of cell division as either symmetric or asymmetric) has hampered a clear-cut interpretation of the experimental results (Yamashita, 2013). Second, the insufficient resolution of sister chromatid detection methods has added to the complexity of interpreting the results. Below, we briefly describe the various techniques used to address this question, and discuss their strengths and limitations.
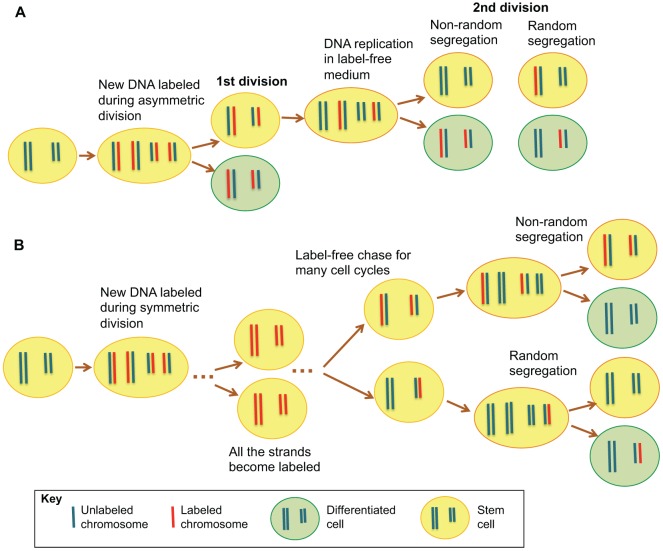
In the majority of studies, nucleotide analogs such as 5-bromo-2′-deoxyuridine (BrdU) have been used to label and distinguish two sister chromatids. In a pulse–chase experiment (Fig. 1A), nucleotide analogs are used to label the newly synthesized DNA strands during asymmetric division of stem cells, and the segregation of BrdU-labeled chromatids is monitored during the chase period. If stem cells are indeed segregating all the template DNA strands, the differentiated cell would inherit all the BrdU-labeled chromatids. By contrast, if sister chromatids are randomly segregated, both the resulting stem cell and the differentiating daughter cell randomly inherit BrdU-labeled chromatids. Alternatively, BrdU can also be administered for extended periods to label the nascent DNA when stem cells or their precursors are dividing symmetrically, so that immortal template strands, if they exist, will be labeled. If cells follow the ISH model, the labeled DNA will continue to be inherited by the stem cells despite undergoing many rounds of cell divisions during the label-free chase period (Fig. 1B). Such DNA-labeling experiments have been performed for small and large intestinal cells (Potten et al., 2002; Quyn et al., 2010), neural stem cells (Karpowicz et al., 2005), skeletal muscle satellite cells (Conboy et al., 2007; Shinin et al., 2006), mammary epithelial cells (Smith, 2005) and others, and support the ISH model. However, experiments using the same technique that involved for instance mouse hematopoietic stem cells (Kiel et al., 2007), epidermal basal cells (Sotiropoulou et al., 2008), hair follicle stem cells (Waghmare et al., 2008), Drosophila male germline stem cells (Yadlapalli et al., 2011) and neocortical precursor cells (Fei and Huttner, 2009) showed that stem cells do not inherit the immortal strands. It should be noted that because BrdU is incorporated into the DNA strand of all the chromosomes, one can only address non-random sister chromatid segregation that applies to the whole set of the chromosomes. The non-random segregation of a small subset of chromosomes would not be detected by this method.
Fig. 1.
Pulse–chase assay to study sister chromatid segregation during stem cell division. (A) In an asymmetrically dividing stem cell, new DNA strands are labeled with a nucleotide analog during the S phase of one cell cycle. Inheritance of all the label by the differentiating cell in the second cell cycle would indicate that stem cells inherit ‘immortal’ strands. If the label is inherited symmetrically, it would suggest the cells do not follow the ISH model. (B) All stem cell DNA strands are labeled through administration of nucleotide analogs over multiple generations when the stem cells or their precursors are dividing symmetrically. If immortal template strands exist, they would become labeled. During the following label-free chase period, stem cells would retain the labeled strands even after many cell divisions if they follow the ISH model. However, if they do not, the label would be quickly diluted and lost from the stem cells.
Multi-isotope imaging mass spectrometry (MIMS) is a novel technique to image stable isotopes in cells with a new type of secondary ion mass spectrometer (Steinhauser et al., 2012). This method is essentially the same as the use of nucleotide analogs in that the isotope would label all the chromosomes. However, this has a strong advantage over the use of nucleotide analogs, because isotopes are non-toxic and their concentration within the cell can be precisely measured by mass spectrometry. Using this technique, Steinhauser and colleagues showed that stem cells in the mouse small intestine do not follow the ISH model (Steinhauser et al., 2012).
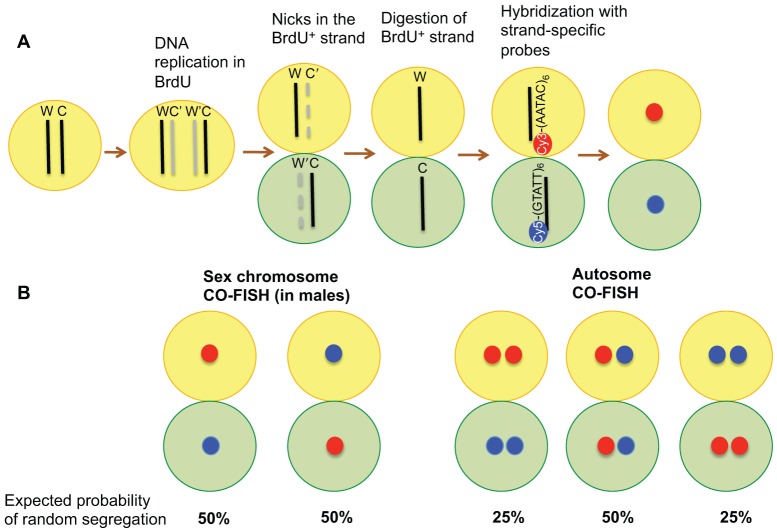
CO-FISH allows the distinction of the sister chromatids that contain the Watson strand as a template from those that contain the Crick strand as a template (Bailey et al., 2004; Falconer et al., 2010). In the CO-FISH protocol, cells are allowed to replicate once in the presence of BrdU. Following BrdU incorporation, cells complete mitosis in BrdU-free medium, such that the sister chromatids are segregated into stem cell and daughter pairs. The BrdU-containing strands are removed by treatment with ultraviolet irradiation and exonuclease III. The remaining template strands can be identified using differentially labeled strand-specific probes (Fig. 2A). This technique has been used to follow the segregation patterns of sister chromatids in mouse colon epithelial cells in vivo (Falconer et al., 2010). The authors observed significant non-random sister chromatid segregation in the differentiating cells (i.e. transit-amplifying cells), but not in the Lgr5+ stem cells in the colon crypts. Another study applied CO-FISH to mouse muscle satellite stem cells and reported that stem cells retain the template strands with a strong bias, thus supporting the ISH model (Rocheteau et al., 2012). It should be noted that these studies used probes that are targeted to centromeric and telomeric repeats of all chromosomes, and thus it is impossible to detect any non-random sister chromatid segregation that applies to only a small subset of the chromosomes. However, if probes are designed to distinguish each chromosome (for example, using repeat sequences that exist only on a particular chromosome), the two sister chromatids of a particular chromosome can be distinguished. Using such a single-chromosome probe, we recently found that sister chromatids of sex chromosomes are non-randomly segregated during Drosophila male germline stem cell divisions (Yadlapalli and Yamashita, 2013) (see below).
Fig. 2.
CO-FISH to identify sister chromatids of individual chromosomes. (A) Schematic illustration of the CO-FISH protocol. Cy3–(AATAC)6 and Cy5–(GTATT)6 probes used to identify the sister chromatids of Drosophila Y chromosome are shown as an example. Upon DNA replication in the presence of BrdU, only newly synthesized strands will contain BrdU. After fixation and irradiation with UV, the BrdU-containing strands specifically will be nicked. Treatment with exonuclease III is used to remove the nicked strand, leaving the template strand intact. Upon hybridization of the CO-FISH probe (Y chromosome probes are shown), sister chromatids that contain (AATAC)6 as a template versus (GTATT)6 as a template can be distinguished. C, Crick strand; W, Watson strand; C′ and W′ indicate newly synthesized strands after replication. Newly synthesized BrdU strands are shown in gray, template strands are shown in black. Green, differentiated cells; yellow, stem cells. (B) Expected probabilities of CO-FISH signal pattern based on random segregation. In the case of sex chromosomes, if there is no bias in sister chromatid segregation, we expect that a stem cell inherits a red (Cy3-based) signal and a differentiated cell a blue (Cy5-based) signal in 50% of the cases, and the opposite, i.e. a blue signal in the stem cell and a red signal in the differentiated cell, in the other 50%. For autosomes, two signals per cell (originating from paternal and maternal chromosomes) are expected. If there is no bias or coordination between homologous (i.e. maternal and paternal chromosomes), stem cells would inherit either two red signals, a blue and a red signal or two blue signals with the probability of 25%, 50%, 25%, respectively. A skewed pattern would suggest the presence of biased segregation (see Fig. 4B).
A novel sequencing technique, called strand-seq, which sequences only the parental template strands of all chromosomes from single cells has recently been developed to study sister chromatid segregation during cell division (Falconer et al., 2012). Briefly, newly synthesized strands are labeled with BrdU, the genome is fragmented and a paired-end library is constructed. Prior to PCR amplification, the nascent BrdU-labeled strand is nicked so that it is not amplified during the subsequent PCR step. As a result, only the original intact template strand is selectively amplified, resulting in directional library fragments. The library fragments can then be sequenced and the resulting paired short sequencing reads can be used to identify the original template strands that have been inherited from the parental cell. This technique allows the detection of non-random sister chromatid segregation at a single chromosome resolution, without being limited by the availability of suitable probe sequences. Using this technique, it has been shown that in mouse embryonic stem (ES) cells, chromosome 7 is randomly segregated (Falconer et al., 2012); this is in contrast to the earlier study mentioned above that found a biased segregation of chromosome 7 sister chromatids (Armakolas and Klar, 2006), but in agreement with another recent report that used the CO-FISH technique (Sauer et al., 2013).
Taken together, the development of these new techniques has been instrumental in moving the field forward and addressing the question of sister chromatid segregation with greater clarity.
Drosophila male germline stem cells as a model to study non-random sister chromatid segregation
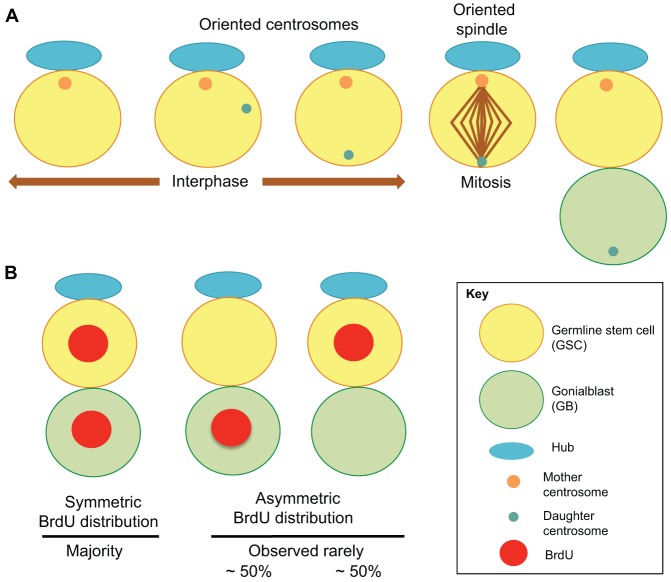
Drosophila male germline stem cells (GSCs) provide an ideal model system to study non-random sister chromatid segregation. The well-characterized behavior of GSCs provides the precision that is required for unambiguously testing non-random sister chromatid segregation. At the apical tip of the testis, approximately nine GSCs physically attach to a cluster of somatic cells called the hub, which is the major component of the stem cell niche and thus instructs stem cell self-renewal (Yamashita et al., 2010). It is known that GSCs divide asymmetrically by orienting the spindle perpendicular to the hub, so that the daughter cell that is in contact with the hub retains stem cell identity, whereas the daughter cell that is located away from the hub becomes a gonialblast (GB) and initiates differentiation (Yamashita et al., 2003). This stereotypical orientation of the spindle in GSCs is set up by the precisely controlled positioning of the centrosomes during interphase; the mother centrosome normally remains adjacent to the hub and is inherited by the GSC, whereas the daughter centrosome migrates to the opposite side of the cell and is inherited by the gonialblast (Yamashita et al., 2007) (Fig. 3A). Centrosome asymmetry has also been demonstrated in other stem cell populations, such as radial glial progenitor cells (Wang et al., 2009) and Drosophila neuroblasts (Conduit and Raff, 2010; Januschke et al., 2011). It has been proposed that the mother centrosome anchors the immortal DNA strands during repeated cell divisions, thereby retaining the immortal strand within stem cells (Tajbakhsh, 2008; Tajbakhsh and Gonzalez, 2009).
Fig. 3.
Drosophila male GSCs do not follow the ISH model despite an asymmetric centrosome segregation. (A) Schematic illustration of asymmetric Drosophila male GSC division. The mitotic spindle is oriented perpendicularly to the hub cells. This specific orientation of the spindle is established by the movement of the centrosomes during interphase. The mother centrosome (orange) normally remains close to the hub cells and is inherited by the GSC, whereas the daughter centrosome (green) migrates to the opposite side of the cell and is inherited by the gonialblast (GB). (B) GSCs do not follow the ISH model. The pulse–chase labeling method in which the newly synthesized strands are labeled with BrdU and their segregation pattern is followed (see Fig. 1A). During the chase period, the majority of the GSC–GB pairs show a symmetric distribution of the BrdU label (left). In a few cases, an apparent asymmetric BrdU segregation is observed. This probably reflects those cases in which all BrdU-containing strands are inherited by one cell by chance (based on random segregation). Consistently, even in the cases of such apparent asymmetry, GSCs and GBs inherit all the BrdU label at equal frequency (i.e. randomly).
Using BrdU pulse–chase experiments, in which newly synthesized strands were labeled, we have shown that male GSCs do not follow the ISH model (Yadlapalli et al., 2011). Throughout the label-free chase period in which we followed the segregation patterns of BrdU-labeled chromatids, we observed that in the majority of cases, the BrdU label was equally distributed to GSC–GB pairs until BrdU is finally diluted to undetectable levels (Fig. 3B), suggesting that GSCs do not maintain the immortal strands for the entire genome.
Although male GSCs do not retain the immortal strands for their entire genome, our recent study that used the above-described single chromosome-resolution CO-FISH (Fig. 2A) showed that X and Y chromosomes show non-random sister chromatid segregation (Yadlapalli and Yamashita, 2013) (Fig. 4A). For instance, we used a Cy3-labeled (AATAC)6 probe (red) and the complementary Cy5-labeled (GTATT)6 probe (blue) to examine the sister chromatid segregation pattern of the Y chromosome, because these sequences are repeated uniquely on the Y chromosome (Bonaccorsi and Lohe, 1991). Using this method, we found that in 85% of GSC divisions, GSCs inherited the sister chromatid of Y chromosome that contains (GTATT)6 repeats as a template [and thus hybridize to the Cy3–(AATAC)6 probes] (Fig. 4A) (Yadlapalli and Yamashita, 2013). A similar trend (of ∼85∶15) was observed for X chromosome segregation. Despite the comparable segregation bias for both X and Y chromosomes, X and Y chromosomes are not co-segregated, suggesting that sister chromatids of X and Y chromosomes are segregated independently of each other. Interestingly, in spite of the strong bias, the X and Y chromosome template strands in GSCs are not ‘immortal’; instead, each GSC appears to switch the template strands once in approximately seven cell divisions on average (∼15%). This type of non-random sister chromatid segregation is novel in that both chromosomes (X and Y) show a bias in sister chromatid segregation with respect to the cell fate (i.e. stem cell and differentiating cell). Therefore, this is distinct from the other types of biased sister chromatid segregation that we described above. Furthermore, this provides a clear example of biased segregation that does not lead to retention of the immortal strand, suggesting that the data obtained by the use of BrdU labeling need to be interpreted with caution. In many studies that supported the ISH model, the segregation pattern following BrdU treatment was assessed after two cell cycles only, but not in subsequent cell cycles, and the bias observed in these experiments might therefore not necessarily support the immortality of the strand.
Fig. 4.

Non-random sister chromatid segregation of X and Y chromosomes during GSC division. (A) Sister chromatid segregation pattern of X and Y chromosomes examined by CO-FISH. GSCs inherit the red (Cy3-based) signal in a majority of the cases, suggesting that GSCs inherit particular sister chromatids of X and Y chromosomes with a striking bias (85%∶15%) (compare with Fig. 2B). (B) Sister chromatid segregation pattern of the autosomes observed in GSCs. GSCs inherit two Cy3 signals or two Cy5 signals with equal probability, but never a Cy3 and a Cy5 signal, suggesting the existence of a certain type of bias (compare with Fig. 2B). The CO-FISH experiments using the chromosome II probe and chromosome III probe showed the same trend. (C) Model of non-random sister chromatid segregation of X and Y chromosomes. Sister chromatids might be distinctly recognized by the SUN–KASH components of the LINC complex, resulting in the anchorage of particular DNA strands to the mother centrosome that is mediated by microtubule–LINC interactions (see text for further details). Adaptor protein(s) might link SUN–KASH proteins and the sister chromatid.
Using CO-FISH with autosome probes, we noticed that GSCs always inherited either two Cy3 signals or two Cy5 signals, but never a Cy3 and a Cy5 signal. If the paternal and maternal chromosomes behave independently, one would expect to observe a distribution of Cy3–Cy3∶Cy3–Cy5∶Cy5–Cy5 equal to 25%∶50%∶25% (1∶2∶1) (Fig. 2B). However, we observed a distribution pattern of 50%∶0%∶50% (or 1∶0∶1) (Fig. 4B). Although it was random with regard to which signals (either Cy3–Cy3 or Cy5–Cy5) are inherited by GSCs, this pattern is clearly distinct from numbers that would be expected from a ‘truly random’ segregation pattern, suggesting the existence of certain bias. This is similar to the sister chromatid segregation pattern previously reported in Drosophila (Beumer et al., 1998) and mouse ES cells (Armakolas and Klar, 2006). Future investigation is required to determine whether these coordinated segregations are related with regard to their biological significance and/or underlying molecular mechanisms.
Molecular mechanisms of non-random sister chromatid segregation
There has been much speculation and many hypotheses regarding how and for what reasons a cell might non-randomly segregate sister chromatids (Lew et al., 2008; Tajbakhsh and Gonzalez, 2009). However, the cellular machinery responsible for non-random sister chromatid segregation remains elusive. Our recent study showed that the centrosomal component cnn, nuclear envelope components (i.e. SUN–KASH-domain proteins) and Dnmt2 are required for the non-random segregation of the X and Y chromosomes. This provides the first insights into how cells might mechanistically carry out non-random sister chromatid segregation (Yadlapalli and Yamashita, 2013) (Fig. 4C).
Although much has yet to be learnt to fully elucidate the mechanisms that allow non-random sister chromatid segregation of X and Y chromosomes, the genes that are required for non-random sister chromatid segregation allow us to propose the following model (Fig. 4C). First, the mother centrosome is anchored to the adherens junctions at the hub–GSC interface throughout the cell cycle (Fig. 3A) (Yamashita et al., 2007). Through its association with microtubules, the mother centrosome is linked to the SUN–KASH-domain proteins located on the nuclear envelope that form the linker of nucleoskeleton and cytoskeleton (LINC) complex (Razafsky and Hodzic, 2009). The LINC complex might associate only with a particular sister chromatid to allow biased segregation (Hiraoka and Dernburg, 2009; Razafsky and Hodzic, 2009). Two sister chromatids could carry distinct epigenetic information that allows their distinction and biased segregation. It is tempting to speculate that chromosomal components (such as centromeres or other regions) and associated proteins (such as kinetochore proteins or other chromatin-associated proteins) are distinct between the sister chromatids (Thorpe et al., 2009), thereby allowing for the selective capture of a particular sister chromatid by the mother centrosome.
Little is known about the nature of epigenetic information that could allow non-random sister chromatid segregation. Dnmt2 appears to confer epigenetic (non-genetic) information to the X and Y chromosomes, which starts during the gametogenesis of parents. Non-random sister chromatid segregation of the Y chromosome specifically relies on the function of the Dnmt2 gene in the father (who provides the original Y chromosome to the individual) and zygotic expression of Dnmt2. Similarly, non-random segregation of the X chromosome is dependent on the Dnmt2 gene function in the mother (Yadlapalli and Yamashita, 2013). These results reveal the striking possibility that the original X and Y chromosome from the parents must carry the appropriate epigenetic information to achieve a non-random segregation of the X and Y chromosomes in the GSCs of their progeny.
Despite the clear involvement of Dnmt2 in non-random sister chromatid segregation, the molecular function of Dnmt2 remains unclear. Although Dnmt2 is the sole gene in the Drosophila genome that encodes a potential DNA methyltransferase (Kunert et al., 2003), the function of Dnmt2 in Drosophila is highly controversial; some studies suggest that it functions as a DNA methyltransferase (Phalke et al., 2009), whereas others have suggested that it functions as a tRNA methyltransferase (Goll et al., 2006; Schaefer et al., 2010). Therefore, future investigations are needed to elucidate how GSCs distinguish between sister chromatids.
Biological relevance of non-random sister chromatid segregation
Non-random sister chromatid segregation is compromised in tumor-like overproliferation of GSCs, suggesting that the stem cell program regulates the sister chromatid segregation pattern (Yadlapalli and Yamashita, 2013). Yet, it is unlikely that non-random sister chromatid segregation determines GSC identity, because the Drosophila mutants that are defective in non-random segregation (cnn, koi, klar, dnmt2) do not exhibit GSC overproliferation or depletion (Yadlapalli and Yamashita, 2013). Therefore, the question of how stem cell identity affects sister chromatid segregation needs further investigation.
We have also shown that dedifferentiated GSCs do not maintain non-random sister chromatid segregation. Dedifferentiation is a well-described process by which partially differentiated cells move into the niche and revert back to stem cell identity (e.g. GSCs) (Brawley and Matunis, 2004). Such dedifferentiated GSCs are believed to have the same functionality as bona fide GSCs, because they are capable of producing differentiated cells and reconstitute spermatogenesis. However, randomized sister chromatid segregation in dedifferentiated GSCs demonstrates that dedifferentiated GSCs are indeed somewhat different from bona fide GSCs. A careful characterization of dedifferentiated GSCs will therefore be necessary to determine whether there are any functional differences between the dedifferentiated and original GSCs.
Interestingly, we found that non-random sister chromatid segregation of X and Y chromosomes might be involved in the suppression of stellate and its corresponding piRNA expression in Drosophila testis (Yadlapalli and Yamashita, 2013). stellate, which encodes polypeptides and whose derepression is known to reduce fertility, is located on the X chromosome, and the piRNAs that suppress stellate expression [Su(ste)] are located on the Y chromosome (Aravin et al., 2001; Tulin et al., 1997). We found that stellate is derepressed in all the mutants that are defective in non-random sister chromatid segregation. Although our current data do not provide direct evidence that randomized sister chromatid segregation is the underlying molecular reason for derepressed stellate expression, they raise the intriguing possibility that non-random sister chromatid segregation might serve to transmit an epigenetically modified copy of Su(ste) and/or stellate, thereby contributing to suppression of stellate. To obtain a definitive answer to this question, it is important to determine whether the stellate or Su(ste) loci have any distinct epigenetic marks that are segregated asymmetrically during GSC division.
Conclusions
Here, we discuss the non-random segregation of sister chromatids during stem cell division and other cell types. Accumulating evidence highlights that cells are indeed capable of distinguishing and segregating sister chromatids that are genetically identical. With distinct types of non-randomness observed in various cell types, it remains unclear whether they are related with respect to the biological relevance and/or underlying molecular mechanisms, or whether there are multiple mechanisms and purposes. To answer the question we raise in the title of this Hypothesis – whether DNA asymmetry in stem cells is mortal or immortal – we do not know yet in how many of the cell types that exhibit non-random segregation this leads to immortality of the DNA strands. At least in Drosophila male GSCs, sister chromatids of specific chromosomes are segregated non-randomly but in a non-immortal and thus mortal manner. The development of new techniques as outlined here will help to unambiguously answer this question in the near future.
We have only just started to understand how cells might be able to distinguish sister chromatids, and segregate them in a non-random manner. It will be interesting to see whether the molecular mechanisms involved in the non-random sister chromatid segregation of X and Y chromosomes in Drosophila male GSCs (involving cnn, SUN–KASH proteins and methyl transferase) have similar roles in other systems. What purpose the non-random segregation of sister chromatids might be serving remains very much an open question. Our recent study provides a tantalizing clue that non-random sister chromatid segregation might be involved in regulating repression of repetitive elements. We foresee exciting research in the future that will help us to improve our understanding of how and why stem cells non-randomly segregate their sister chromatids.
Acknowledgments
We thank the Yamashita laboratory members for discussions.
Footnotes
Funding
This work was supported by the American Heart Association [grant number 12PRE9630000]; the National Institutes of Health [grant number 1F31HD071727-01] to S.Y. and Keck Foundation to Y.M.Y. Y.M.Y. is supported by the MacArthur Foundation. Deposited in PMC for release after 12 months.
References
- Aravin A. A., Naumova N. M., Tulin A. V., Vagin V. V., Rozovsky Y. M., Gvozdev V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027 10.1016/S0960-9822(01)00299-8 [DOI] [PubMed] [Google Scholar]
- Armakolas A., Klar A. J. S. (2006). Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science 311, 1146–1149 10.1126/science.1120519 [DOI] [PubMed] [Google Scholar]
- Bailey S. M., Goodwin E. H., Cornforth M. N. (2004). Strand-specific fluorescence in situ hybridization: the CO-FISH family. Cytogenet. Genome Res. 107, 14–17 10.1159/000079565 [DOI] [PubMed] [Google Scholar]
- Beumer K. J., Pimpinelli S., Golic K. G. (1998). Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila. Genetics 150, 173–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Lohe A. (1991). Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: relationships between satellite sequences and fertility factors. Genetics 129, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley C., Matunis E. (2004). Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 304, 1331–1334 10.1126/science.1097676 [DOI] [PubMed] [Google Scholar]
- Cairns J. (1975). Mutation selection and the natural history of cancer. Nature 255, 197–200 10.1038/255197a0 [DOI] [PubMed] [Google Scholar]
- Charville G. W., Rando T. A. (2013). The mortal strand hypothesis: Non-random chromosome inheritance and the biased segregation of damaged DNA. Semin. Cell Dev. Biol doi: 10.1016/j.semcdb.2013.05.006; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy M. J., Karasov A. O., Rando T. A. (2007). High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 5, e102 10.1371/journal.pbio.0050102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P. T., Raff J. W. (2010). Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20, 2187–2192 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- Ettinger A. W., Wilsch-Bräuninger M., Marzesco A. M., Bickle M., Lohmann A., Maliga Z., Karbanová J., Corbeil D., Hyman A. A., Huttner W. B. (2011). Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat. Commun 2, 503 10.1038/ncomms1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E., Chavez E. A., Henderson A., Poon S. S., McKinney S., Brown L., Huntsman D. G., Lansdorp P. M. (2010). Identification of sister chromatids by DNA template strand sequences. Nature 463, 93–97 10.1038/nature08644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E., Hills M., Naumann U., Poon S. S. S., Chavez E. A., Sanders A. D., Zhao Y. J., Hirst M., Lansdorp P. M. (2012). DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat. Methods 9, 1107–1112 10.1038/nmeth.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J. F., Huttner W. B. (2009). Nonselective sister chromatid segregation in mouse embryonic neocortical precursor cells. Cereb. Cortex 19, Suppl. 1i49–i54 10.1093/cercor/bhp043 [DOI] [PubMed] [Google Scholar]
- Goll M. G., Kirpekar F., Maggert K. A., Yoder J. A., Hsieh C. L., Zhang X. Y., Golic K. G., Jacobsen S. E., Bestor T. H. (2006). Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311, 395–398 10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Dernburg A. F. (2009). The SUN rises on meiotic chromosome dynamics. Dev. Cell 17, 598–605 10.1016/j.devcel.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Januschke J., Llamazares S., Reina J., Gonzalez C. (2011). Drosophila neuroblasts retain the daughter centrosome. Nat. Commun 2, 243 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P., Morshead C., Kam A., Jervis E., Ramunas J., Cheng V., van der Kooy D. (2005). Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically in vitro. J. Cell Biol. 170, 721–732 10.1083/jcb.200502073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M. J., He S. H., Ashkenazi R., Gentry S. N., Teta M., Kushner J. A., Jackson T. L., Morrison S. J. (2007). Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature 449, 238–242 10.1038/nature06115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J. S. (1987a). Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature 326, 466–470 10.1038/326466a0 [DOI] [PubMed] [Google Scholar]
- Klar A. J. S. (1987b). The mother-daughter mating type switching asymmetry of budding yeast is not conferred by the segregation of parental HO gene DNA strands. Genes Dev. 1, 1059–1064 10.1101/gad.1.10.1059 [DOI] [PubMed] [Google Scholar]
- Klar A. J. S. (1994). A model for specification of the left-right axis in vertebrates. Trends Genet. 10, 392–396 10.1016/0168-9525(94)90055-8 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583–597 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Kunert N., Marhold J., Stanke J., Stach D., Lyko F. (2003). A Dnmt2-like protein mediates DNA methylation in Drosophila. Development 130, 5083–5090 10.1242/dev.00716 [DOI] [PubMed] [Google Scholar]
- Kuo T. C., Chen C. T., Baron D., Onder T. T., Loewer S., Almeida S., Weismann C. M., Xu P., Houghton J. M., Gao F. B. et al. (2011). Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 13, 1214–1223 10.1038/ncb2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp P. M. (2007). Immortal strands? Give me a break. Cell 129, 1244–1247 10.1016/j.cell.2007.06.017 [DOI] [PubMed] [Google Scholar]
- Lew D. J., Burke D. J., Dutta A. (2008). The immortal strand hypothesis: how could it work? Cell 133, 21–23 10.1016/j.cell.2008.03.016 [DOI] [PubMed] [Google Scholar]
- Morrison S. J., Spradling A. C. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalke S., Nickel O., Walluscheck D., Hortig F., Onorati M. C., Reuter G. (2009). Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 41, 696–702 10.1038/ng.360 [DOI] [PubMed] [Google Scholar]
- Potten C. S., Owen G., Booth D. (2002). Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 115, 2381–2388 [DOI] [PubMed] [Google Scholar]
- Quyn A. J., Appleton P. L., Carey F. A., Steele R. J. C., Barker N., Clevers H., Ridgway R. A., Sansom O. J., Näthke I. S. (2010). Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 6, 175–181 10.1016/j.stem.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Razafsky D., Hodzic D. (2009). Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 186, 461–472 10.1083/jcb.200906068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheteau P., Gayraud-Morel B., Siegl-Cachedenier I., Blasco M. A., Tajbakhsh S. (2012). A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125 10.1016/j.cell.2011.11.049 [DOI] [PubMed] [Google Scholar]
- Rossi D. J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., Weissman I. L. (2007). Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 10.1038/nature05862 [DOI] [PubMed] [Google Scholar]
- Rujano M. A., Bosveld F., Salomons F. A., Dijk F., van Waarde M. A. W. H., van der Want J. J. L., de Vos R. A. I., Brunt E. R., Sibon O. C. M., Kampinga H. H. (2006). Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 4, e417 10.1371/journal.pbio.0040417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer S., Burkett S. S., Lewandoski M., Klar A. J. (2013). A CO-FISH assay to assess sister chromatid segregation patterns in mitosis of mouse embryonic stem cells. Chromosome Res. 21, 311–328 10.1007/s10577-013-9358-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., Lyko F. (2010). RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24, 1590–1595 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinin V., Gayraud-Morel B., Gomès D., Tajbakhsh S. (2006). Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat. Cell Biol. 8, 677–682 10.1038/ncb1425 [DOI] [PubMed] [Google Scholar]
- Smith G. H. (2005). Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development 132, 681–687 10.1242/dev.01609 [DOI] [PubMed] [Google Scholar]
- Sotiropoulou P. A., Candi A., Blanpain C. (2008). The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells 26, 2964–2973 10.1634/stemcells.2008-0634 [DOI] [PubMed] [Google Scholar]
- Steinhauser M. L., Bailey A. P., Senyo S. E., Guillermier C., Perlstein T. S., Gould A. P., Lee R. T., Lechene C. P. (2012). Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature 481, 516–519 10.1038/nature10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S. (2008). Stem cell identity and template DNA strand segregation. Curr. Opin. Cell Biol. 20, 716–722 10.1016/j.ceb.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Gonzalez C. (2009). Biased segregation of DNA and centrosomes: moving together or drifting apart? Nat. Rev. Mol. Cell Biol. 10, 804–810 10.1038/nrm2784 [DOI] [PubMed] [Google Scholar]
- Thorpe P. H., Bruno J., Rothstein R. (2009). Kinetochore asymmetry defines a single yeast lineage. Proc. Natl. Acad. Sci. USA 106, 6673–6678 10.1073/pnas.0811248106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A. V., Kogan G. L., Filipp D., Balakireva M. D., Gvozdev V. A. (1997). Heterochromatic Stellate gene cluster in Drosophila melanogaster: structure and molecular evolution. Genetics 146, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare S. K., Bansal R., Lee J., Zhang Y. V., McDermitt D. J., Tumbar T. (2008). Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 27, 1309–1320 10.1038/emboj.2008.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Q., Tsai J. W., Imai J. H., Lian W. N., Vallee R. B., Shi S. H. (2009). Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947–955 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadlapalli S., Yamashita Y. M. (2013). Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature 498, 251–254 10.1038/nature12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadlapalli S., Cheng J., Yamashita Y. M. (2011). Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J. Cell Sci. 124, 933–939 10.1242/jcs.079798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y. M. (2013). Biased DNA segregation in Drosophila male germline stem cells. Semin. Cell Dev. Biol. doi: 10.1016/j.semcdb.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Yamashita Y. M., Jones D. L., Fuller M. T. (2003). Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 10.1126/science.1087795 [DOI] [PubMed] [Google Scholar]
- Yamashita Y. M., Mahowald A. P., Perlin J. R., Fuller M. T. (2007). Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315, 518–521 10.1126/science.1134910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y. M., Yuan H. B., Cheng J., Hunt A. J. (2010). Polarity in stem cell division: asymmetric stem cell division in tissue homeostasis. Cold Spring Harb Perspect Biol. 2, a001313 10.1101/cshperspect.a001313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yennek S., Tajbakhsh S. (2013). DNA asymmetry and cell fate regulation in stem cells. Semin. Cell Dev. Biol. doi: 10.1016/j.semcdb.2013.05.008 [DOI] [PubMed] [Google Scholar]