Summary
Four members of the structural maintenance of chromosome (SMC) protein family have essential functions in chromosome condensation (SMC2/4) and sister-chromatid cohesion (SMC1/3). The SMC5/6 complex has been implicated in chromosome replication, DNA repair and chromosome segregation in somatic cells, but its possible functions during mammalian meiosis are unknown. Here, we show in mouse spermatocytes that SMC5 and SMC6 are located at the central region of the synaptonemal complex from zygotene until diplotene. During late diplotene both proteins load to the chromocenters, where they colocalize with DNA Topoisomerase IIα, and then accumulate at the inner domain of the centromeres during the first and second meiotic divisions. Interestingly, SMC6 and DNA Topoisomerase IIα colocalize at stretched strands that join kinetochores during the metaphase II to anaphase II transition, and both are observed on stretched lagging chromosomes at anaphase II following treatment with Etoposide. During mitosis, SMC6 and DNA Topoisomerase IIα colocalize at the centromeres and chromatid axes. Our results are consistent with the participation of SMC5 and SMC6 in homologous chromosome synapsis during prophase I, chromosome and centromere structure during meiosis I and mitosis and, with DNA Topoisomerase IIα, in regulating centromere cohesion during meiosis II.
Key words: Centromere, Meiosis, Mitosis, Mouse, SMC5, SMC6, Synaptonemal complex, DNA Topoisomerase IIα
Introduction
Structural maintenance of chromosomes (SMC) proteins are conserved chromosomal ATPases that regulate nearly all aspects of chromosome biology and are critical for genomic stability. In eukaryotes, SMC proteins form heterodimers which associate with different non-SMC proteins to form holocomplexes that regulate different aspects of chromosome biology during both mitosis and meiosis (Carter and Sjögren, 2012). There are three distinct SMC complexes in eukaryotes. SMC1–SMC3-based cohesin complexes play important roles not only in sister-chromatid cohesion, but also in repair of damaged DNA, regulation of gene expression and development (Nasmyth and Haering, 2009). SMC2/4-based condensin complexes play a major role in chromosome condensation, but also, similar to cohesin complexes, in DNA recombination and repair, and in gene regulation (Hirano, 2012). The third SMC complex, the SMC5/6 complex, which does not yet have a functionally descriptive name, has been implicated mainly in DNA repair, but also in chromosome replication and segregation (Murray and Carr, 2008; De Piccoli et al., 2009; Kegel and Sjögren, 2010; Wu and Yu, 2012). In yeast the Smc5/6 complex is composed of Smc5, Smc6 and six non-SMC elements (Nse), including Nse1-6 (Duan et al., 2009). The kleisin Nse4 bridges the ATPase head domains of Smc5 and Smc6. Nse1 forms an ubiquitin ligase complex together with Nse3, and both bind to Nse4. Nse2, which has a SUMO ligase activity, interacts with the coiled-coil region of Smc5. In mammals, interactions between corresponding orthologs have been identified for SMC5, SMC6 and NSE/NSMCE1-4 (Taylor et al., 2001; Potts and Yu, 2005; Taylor et al., 2008).
Recent data indicate that the SMC5/6 complex might be involved in sister-chromatid cohesion and dissolution of aberrant DNA-mediated sister-chromatid linkages after DNA replication (Behlke-Steinert et al., 2009; Outwin et al., 2009; Bermúdez-López et al., 2010; Stephan et al., 2011). Exactly how the three SMC complexes carry out different but related tasks in molecular terms is still not known. Likewise, the function of SMC5/6 complexes during meiosis is unclear (Firooznia et al., 2005). The analysis of fission yeast mutants for different non-SMC subunits of the SMC5/6 complex suggests that this complex is crucial for the timely resolution of joint molecules, in precursor intermediates for cross-over formation during homologous recombination, and for chromosome segregation during meiotic divisions (Pebernard et al., 2004; Wehrkamp-Richter et al., 2012). By contrast, recent analyses of mutants for Smc5 and Smc6 during Caenorhabditis elegans meiosis indicate that the Smc5/6 complexes are required for successful completion of meiotic homologous recombination repair, but are dispensable for accurate chromosome segregation (Bickel et al., 2010). Yet another study has shown that budding yeast Smc5 and Smc6 mutants do not exhibit altered homologous recombination, but do show aberrant meiotic divisions (Farmer et al., 2011).
The function of SMC5/6 complexes during mammalian meiosis is presently unknown. The only existing data indicate that SMC5 and SMC6 are highly expressed in mouse testis and associate with sex chromosomes during the pachytene stage (Taylor et al., 2001), and there is a testis-specific kleisin subunit, NSMCE4B (Taylor et al., 2008). In this study we have analyzed the pattern of expression of all the SMC5/6 subunits during the first wave of spermatogenesis in mouse. We have also analyzed by immunofluorescence the subcellular distribution of SMC5, SMC6 and the non-SMC subunit NSMCE1 on surface-spread spermatocyte chromatin from wild-type and various mutant mice, and for comparison, in somatic cells, including spermatogonia and 3T3 cells. Our results show that the SMC5/6 complex proteins are present at the central region of the synaptonemal complex (SC), the heterochromatic chromocenters and the sex body during prophase I stages. We also found that SMC5/6 components localize to the centromeres and along the chromatid axes, colocalizing with DNA Topoisomerase IIα, during both meiotic divisions and during mitosis. However, our immunoprecipitation data showed that SMC6 does not form a stable complex with DNA Topoisomerase IIα. Additionally, we asked whether the distribution of SMC5 and SMC6 at the central region of the SC was compromised in knockout mice lacking components of the SC and cohesin subunits. We found that the loading of SMC6 at the central region depends on the SC transverse filament protein SYCP1, but not on SC central element protein SYCE3, or REC8- and SMC1β-containing cohesin complexes. However, SMC5 and SMC6 do not localize to γH2AX-rich chromatin during pachytene when the sex body fails to form in the SMC1β mutant. We also found that the loading of SMC5, SMC6 and NSMCE1 at the heterochromatic chromocenters during prophase I, and its redistribution at the centromeres during the diplotene to metaphase I transition, are independent of the shugoshin SGOL2. Together, these findings portray dynamic relationships of the SMC5/6 complexes and associated proteins in the assembly of the SC during prophase I, chromosome and centromere structure during mammalian meiosis I and mitosis, and sister-chromatid centromere cohesion during meiosis II.
Results
SMC5/6 complex subunits are expressed in meiotic prophase I
To set a framework for protein expression and localization, we analyzed the expression of the mRNAs encoding each subunit of the SMC5/6 complex, in comparison to expression of the transcript for the SC transverse filament component SYCP1, during the first wave of spermatogenesis. We found that the mRNAs for each SMC5/6 complex subunit – Smc5, Smc6, Nsmce1, Nsmce2, Nsmce3, Nsmce4A and Nsmce4B – were expressed during the first wave of spermatogenesis in a pattern similar to that of Sycp1 (Fig. 1A). To best visualize subtle differences in expression throughout the first wave of spermatogenesis, Fig. 1A illustrates the product from a 25-cycle PCR reaction. However, in a 35-cycle PCR reaction, product was evident for SMC5/6 components as early as 7 d.p.p., reflecting spermatogonial expression (data not shown). We next determined the protein level of the SMC5/6 complex subunits SMC5, SMC6, NSMCE1, NSMCE4A, NSMCE4B, as well as SYCP1, during the first wave of spermatogenesis. Each protein was expressed at significantly higher levels between 16 d.p.p. to 22 d.p.p., when pachytene spermatocytes are abundant, than at 7 d.p.p. to 13 d.p.p., when spermatogonia and early meiotic prophase spermatocytes are most abundant, reflecting transcript data (Fig. 1B). The SMC6 antibodies from Abcam (SMC6 Ab) and from the Lehmann group (SMC6 Le) detected SMC6 bands at the same mobilities (Fig. 1B), a point relevant to differences detected in localization with these antibodies (see below). Altogether, these data indicate that all SMC5/6 complex subunits are similarly expressed during meiotic prophase I, coincident with the expression of SYCP1.
Fig. 1.
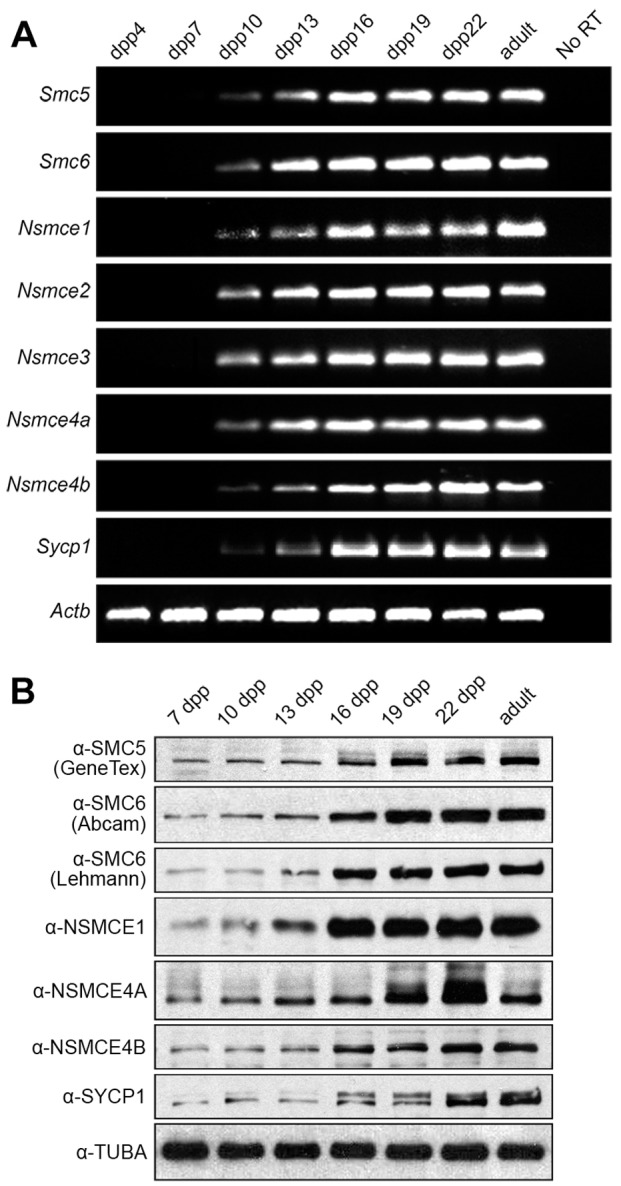
The subunits of the SMC5/6 complex are expressed in the first wave of spermatogenesis, beginning when zygotene spermatocytes appear. (A) PCR analysis of mRNA expression of SMC5/6 complex subunits Smc5, Smc6, Nsmce1-4b during the first wave of spermatogenesis, from mice aged 4, 7, 10, 13, 16, 19, 22 and 56 (adult) d.p.p. Sycp1 was used as a control for progression of the first wave of spermatogenesis and Actb was used as an mRNA loading control. (B) Western blot analysis of protein expression of SMC5/6 complex subunits SMC5 (GeneTex), SMC6 (Abcam), SMC6 (Lehmann), NSMCE1, NSMCE4A and NSMCE4B during the first wave of spermatogenesis, from mice aged 7, 10, 13, 16, 19, 22 and 56 (adult) d.p.p. SYCP1 was used as a control for progression of the first wave of spermatogenesis and α-tubulin (TUBA) was used as a protein loading control.
SMC6 shows dynamic relationships with the SC and centromeres throughout prophase I and during meiotic divisions
We used immunofluorescence to determine the distribution of SMC6 on surface-spread spermatocytes. To differentiate between different meiotic stages we performed a double-immunolabeling of SMC6 with SYCP3, a structural protein of the axial elements (AEs) and lateral elements (LEs) of SCs (Parra et al., 2004). We used three antibodies against different regions of human SMC6: one kindly provided by A. Lehmann (SMC6 Le) raised against N-terminal amino acids; one from Santa Cruz Biotechnology (SMC6 Sc), raised against central portion amino acids, and one from Abcam (SMC6 Ab) raised against C-terminal amino acids. We found varying degrees of accessibility of these three different epitopes dependent on localization, as described below.
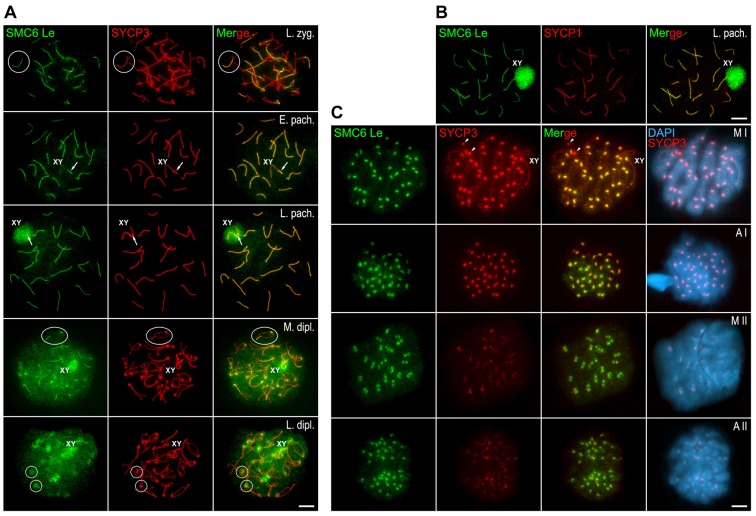
During early prophase I, the SMC6 Le and SMC6 Sc antibodies revealed SMC6 first at early zygotene, as short lines that colocalized with regions where AEs, as detected by SYCP3, appeared thickened and thus paired. With progression of homologous pairing, the SMC6 signals extended in length and colocalized with the SYCP3-labeled LEs involved in SC assembly (Fig. 2A). At early pachytene, SMC6 colocalized with fully assembled autosomal SCs (Fig. 2A). At this stage, SMC6 also appeared at the pseudoautosomal region (PAR) of homology between the sex chromosomes, where their AEs appeared synapsed (Fig. 2A). In more advanced pachytene nuclei, SMC6 was also detected as a cloud on the chromatin of the sex body (Fig. 2A). Because we detected SMC6 at synapsed chromosome regions during zygotene and pachytene stages, we performed a double-immunolabeling with SYCP1 to corroborate the presence of SMC6 at the SC central region. Our results showed that, indeed, SMC6 and SYCP1 colocalized at the central region (Fig. 2B). By diplotene, SMC6 was observed as lines at the remaining paired chromosomal regions. These SMC6 lines became progressively shorter and fainter with ongoing desynapsis (Fig. 2A). Likewise, SMC6 labeling at the sex body decreased gradually throughout diplotene (Fig. 2A). Interestingly, by late diplotene, SMC6 was detected only at the centromeres (Fig. 2A).
Fig. 2.
Distribution of SMC6 Le during meiosis. (A,C) Double-immunolabeling of SMC6 Le (green) and SYCP3 (red) on spread mouse spermatocytes at different meiotic stages: late zygotene (L. zyg.), early pachytene (E. pach.), late pachytene (L. pach.), mid diplotene (M. dipl.), late diplotene (L. dipl.), metaphase I (M I), anaphase I (A I), metaphase II (M II) and anaphase II (A II). Chromatin is stained with DAPI (blue) in C. Selected bivalents in late zygotene and mid diplotene panels, and centromere regions in the late diplotene panel, are encircled. The sex bivalent (XY) is indicated. Arrows in early pachytene and late pachytene panels indicate the PAR region between the sex chromosomes. Arrowheads in the metaphase I panel indicate large SYCP3 agglomerates in the cytoplasm. (B) Double-immunolabeling of SMC6 Le (green) and SYCP1 (red) on a spread late pachytene spermatocyte. The sex body (XY) is indicated. Scale bars: 10 µm.
In metaphase I and anaphase I chromosomes, SMC6 colocalized with SYCP3 at the inner centromere domain as defined previously (Parra et al., 2004; Gómez et al., 2007) (Fig. 2C). This SMC6 centromere labeling was absent by late telophase I and reappeared at chromocenters in late interkinesis nuclei (data not shown). By metaphase II, a pair of separated SMC6 signals was observed at each centromere (Fig. 2C). Single SMC6 signals were detected at the centromere of each segregating chromatid during anaphase II (Fig. 2C).
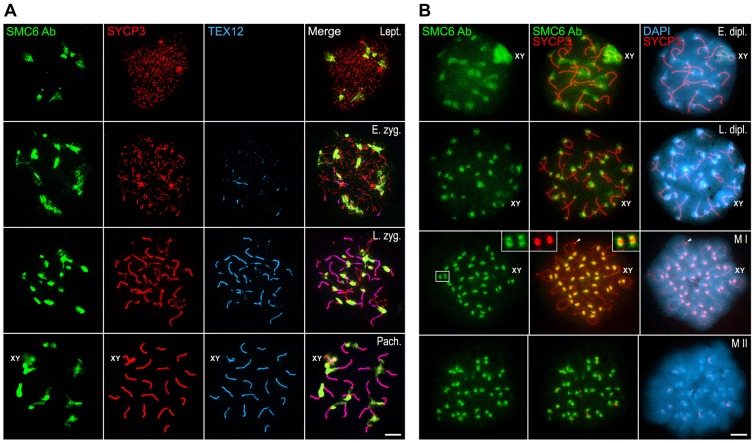
In contrast to this pattern, when the SMC6 Ab antibody, which recognizes the C-terminal region of the protein, was used, we observed SMC6 at heterochromatin-rich chromocenters, which represent clustered centromere heterochromatic regions of different autosomes, from leptotene up to pachytene, when it also became evident at the sex body (Fig. 3A). Unlike localization with antibodies against the N-terminal and central domains, the SMC6 Ab antibody did not localize to the SC central region, detected by antibodies against TEX12, a structural protein present at the SC central element (Hamer et al., 2006) (Fig. 3A). During early diplotene, SMC6 Ab label was still evident at the chromocenters and at the sex body (Fig. 3B). Similar to the pattern with SMC6 Le and SMC6 Sc antibodies, the SMC6 labeling at the sex body detected by SMC6 Ab disappeared gradually throughout middle and late diplotene (Fig. 3B). By metaphase I, the SMC6 Ab antibody, similar to SMC6 Le and SMC6 Sc, detected the centromeres as two bright and close signals that partially colocalized with the SYCP3 signals at the inner centromeric domains (Fig. 3B). This same SMC6 Ab labeling was also detected at anaphase I centromeres (data not shown). By metaphase II, two signals were detected per centromere (Fig. 3B). Taken together, these results with a diverse array of antibodies recognizing different functional domains of SMC6 illustrate a dynamic relationship of this subunit of the SMC5/6 complex with the central region of SCs and centromeres during prophase I and the subsequent divisions.
Fig. 3.
Distribution of SMC6 Ab during meiosis. (A) Triple-immunolabeling of SMC6 Ab (green), SYCP3 (red) and TEX12 (light blue) on spread leptotene (Lept.), early zygotene (E. zyg.), late zygotene (L. zyg.) and pachytene (Pach.) spermatocytes. The sex body (XY) is indicated in the pachytene panel. (B) Double-immunolabeling of SMC6 Ab (green) and SYCP3 (red) and counterstaining of the chromatin with DAPI (blue) on spread early diplotene (E. dipl.), late diplotene (L. dipl.), metaphase I (M I) and metaphase II (M II) spermatocytes. The sex body and the sex bivalent (XY) are indicated in the diplotene and metaphase I panels, respectively. The boxed area in the metaphase I panel is enlarged in the corresponding insets. Scale bars: 10 µm.
SMC6 and Topo IIα are present at the centromeres and chromatid axes in both meiotic and mitotic chromosomes
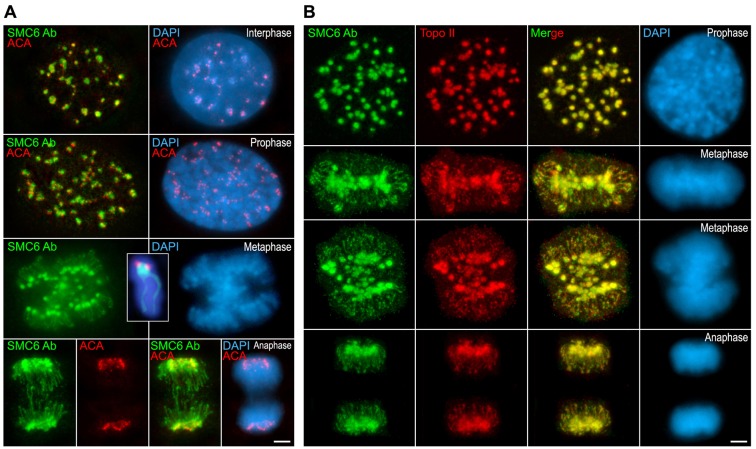
Because the SMC6 signals observed at metaphase I and metaphase II centromeres were reminiscent of those found for DNA Topoisomerase IIα (Topo IIα) (Cobb et al., 1999), we double-immunolabeled SMC6 with Topo IIα on surface-spread spermatocytes to analyze their relative localization (Fig. 4A). Our results showed that both proteins colocalized at the heterochromatic chromocenters during prophase I stages (Fig. 4Aa), and at the centromeres in metaphase I bivalents (Fig. 4Ab). In enlarged metaphase I autosomal bivalents, SMC6 and Topo IIα were detected as two masses per centromere (Fig. 4Ac,d), and as faint lines along the chromatid axes (Fig. 4Ac). This same labeling was observed at anaphase I chromosomes (Fig. 4Af). In metaphase II chromosomes, SMC6 and Topo IIα also colocalized at centromeres (Fig. 4Ag,h). These proteins appeared as faint and small signals at the centromere of the Y chromosome in metaphase I (Fig. 4Ae) and metaphase II (Fig. 4Ag). Interestingly, when these proteins were immunolabeled on metaphase II chromosomes from squashed spermatocytes we found that both proteins were detected as two masses, one per chromatid, and at an additional strand between them (Fig. 4Ai,j). This strand appeared surprisingly stretched at early anaphase II (Fig. 4Ak). Together, these data indicate that SMC6 colocalizes with Topo IIα at the centromeric heterochromatin during all meiotic stages, at the chromatid axes in metaphase I and anaphase I chromosomes, and at a strand connecting the chromatids in meiosis II.
Fig. 4.
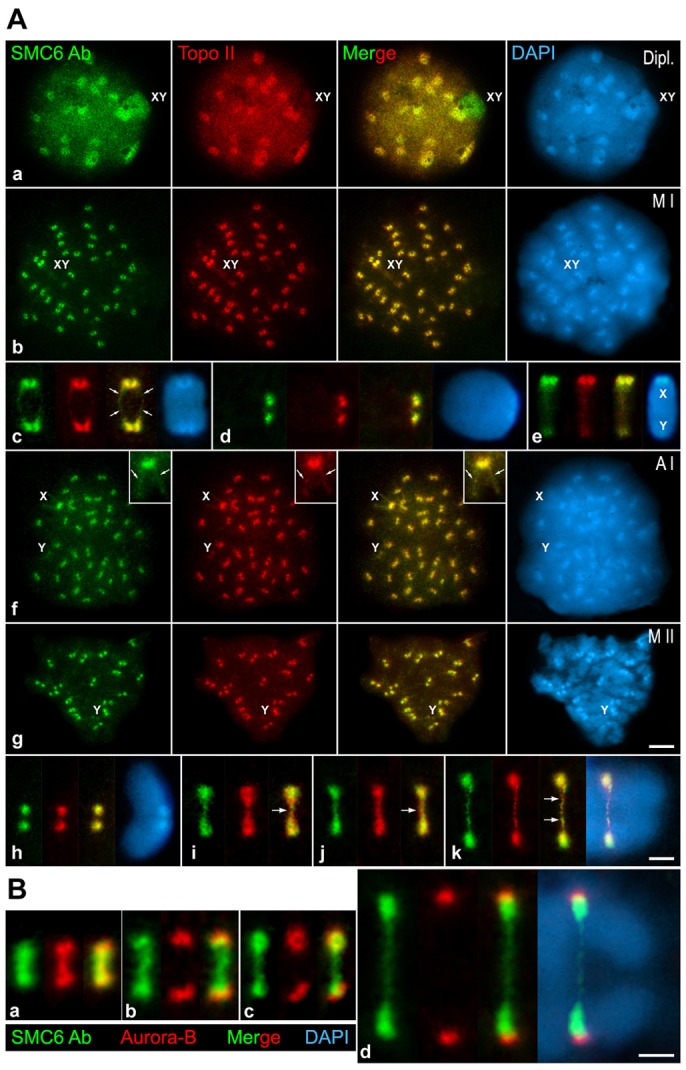
SMC6 Ab colocalizes with Topo IIα at prophase I chromocenters and at the centromeres during both meiotic divisions. (A) Double-immunolabeling of SMC6 Ab (green) and Topo IIα (red) and counterstaining of the chromatin with DAPI (blue) on spread diplotene (Dipl.), metaphase I (M I), anaphase I (A I) and metaphase II (M II) spermatocytes. Enlarged spread metaphase I autosomal (c,d) and sex bivalents (e), spread (h) and squashed (i, j) metaphase II chromosome, and squashed early anaphase II chromosome (k). The sex body (XY) in a, the sex bivalent (XY) in b and the sex chromosomes (X,Y) are indicated. The arrows in c and in the insets in f mark the chromatid axes, whereas in i–k, the arrows indicate a thin strand between the Topo IIα masses at the kinetochore regions. (B) Double-immunolabeling of SMC6 Ab (green) and Aurora-B (red) and counterstaining of the chromatin with DAPI (blue) on squashed metaphase II (a–c), and early anaphase II (d) spermatocytes. Scale bars: 10 µm (Aa,b,f,g); 5 µm (Ac–e,h–k and B).
To further define the centromeric localization of SMC6, we double-immunolabeled SMC6 Ab with an ACA serum that reveals the kinetochores (supplementary material Fig. S1). We observed that at metaphase I and anaphase I centromeres SMC6 appeared as two masses distal to the sister kinetochores (supplementary material Fig. S1A,B), but only as one signal below the kinetochores at metaphase II and anaphase II centromeres (supplementary material Fig. S1C,G). Interestingly, on squashed metaphase II spermatocytes, when their centromeres are subjected to tension towards opposite poles, SMC6 was also detected as a strand that connects the sister kinetochores and traverses the inner centromere domain in all chromosomes (supplementary material Fig. S1D,E). This SMC6 strand appeared stretched during early anaphase II (supplementary material Fig. S1F).
Because SMC6 was unexpectedly detected at a strand that stretched between sister kinetochores during the metaphase II to anaphase II transition, we investigated the behavior of SMC6 relative to Aurora-B, a chromosomal passenger complex kinase that redistributes at the inner centromere domain during meiosis II (Parra et al., 2006). Our results showed that SMC6 and Aurora-B colocalized at the inner domain of 41.7% (15 out of 36) metaphase II centromeres (Fig. 4Ba), whereas in the remaining 58.3% (21 out of 36) SMC6 persisted as a strand but Aurora-B had redistributed to the kinetochore regions (Fig. 4Bb,c). Accordingly, SMC6 was detected at a stretched strand between segregating early anaphase II chromatids when Aurora-B was found at the kinetochores (Fig. 4Bd).
We also examined the distribution of SMC6 in mitotic chromosomes. We found that in metaphase spermatogonia (data not shown) and in mouse 3T3 metaphase and anaphase chromosomes SMC6 labeled the centromeres and the chromatid axes (Fig. 5A). By performing double-immunolabeling of SMC6 and Topo IIα we found that both proteins colocalized at interphase chromocenters, and at the centromeres and chromatid axes during all mitotic stages (Fig. 5B). Together, these results suggest that SMC6 and Topo IIα functionally interact at centromeres and chromatid axes during both meiosis and mitosis.
Fig. 5.
SMC6 Ab colocalizes with Topo IIα at the centromeres and chromatid axes in mitotic chromosomes. (A) Double-immunolabeling of SMC6 Ab (green) and an ACA serum revealing kinetochores (red) and counterstaining of the chromatin with DAPI (blue) on interphase, prophase, metaphase and anaphase 3T3 cultured cells. The inset in the metaphase panel shows an enlarged metaphase chromosome. (B) Double-immunolabeling of SMC6 Ab (green) and Topo IIα (red) and counterstaining of the chromatin with DAPI (blue) on prophase, metaphase and anaphase 3T3 cultured cells. Scale bars: 2.5 µm.
SMC6 and Topo IIα colocalize at stretched strands joining the chromatids of lagging chromosomes induced with Etoposide
Since the previous results pointed to a possible functional interaction between SMC6 and Topo IIα at centromeres, we next analyzed the behavior of SMC6 after the inhibition of the enzymatic activity of Topo IIα by Etoposide. This is a potent Topo IIα inhibitor whose effect lies in blocking the activity of Topo IIα once it has cut the concatenated DNA molecules, but prior to the occurrence of the religation (Nitiss, 2009). Our previous data demonstrate that Etoposide induces the appearance of lagging chromosomes during the second meiotic division, and that the chromatids remain associated by Topo IIα strands joining their kinetochores (Gómez et al., 2013). Indeed, Etoposide does not disturb the chromosomal Topo IIα distribution, but only its enzymatic activity (Nitiss, 2009). We performed double-immunolabeling of SMC6 and Topo IIα to determine whether these proteins colocalized at such strands in lagging chromosomes in Etoposide-treated individuals. Our results showed that both proteins colocalized at stretched strands present along chromatin bridges connecting the chromatids of lagging chromosomes (Fig. 6A). The double-immunolabeling of SMC6 and the kinetochores demonstrated that these chromatin bridges lay between the sister kinetochores of the lagging chromosomes (Fig. 6B). These results support the functional interaction between SMC5/6 complexes and Topo IIα at the inner centromere domain.
Fig. 6.
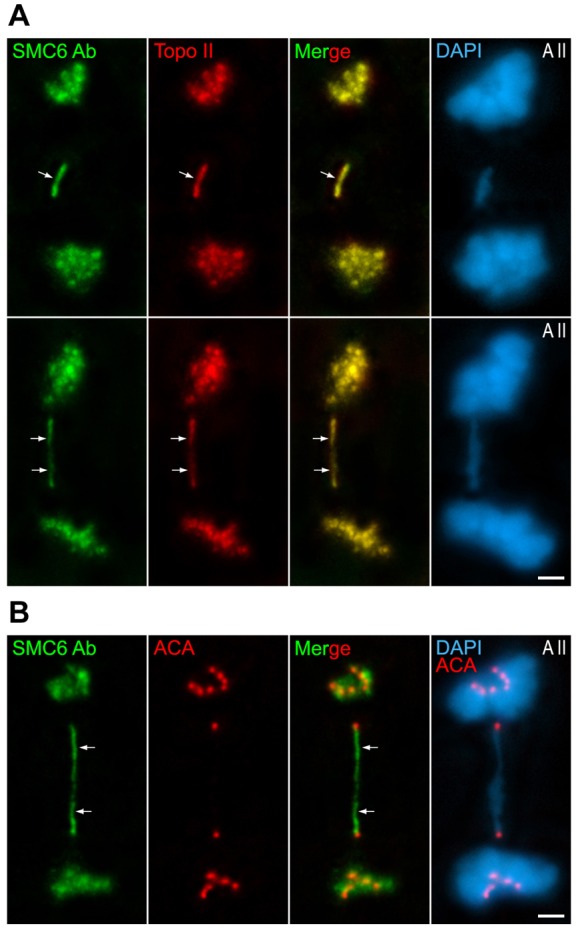
SMC6 Ab colocalizes with Topo IIα at the strands present in lagging chromosomes induced by Etoposide. (A) Double-immunolabeling of SMC6 Ab (green) and Topo IIα (red) and counterstaining of the chromatin with DAPI (blue) on two squashed representative anaphase II (A II) spermatocytes. The arrows indicate thin strands between the lagging chromatids. (B) Double-immunolabeling of SMC6 Ab (green) and an ACA serum revealing kinetochores (red) and counterstaining of the chromatin with DAPI (blue) on a squashed anaphase II (A II) spermatocyte. The arrows indicate a thin strand between the kinetochores of lagging chromatids. Scale bars: 10 µm.
In order to determine whether SMC6 and Topo IIα form a complex in testis nuclear extracts, we performed an immunoprecipitation (IP) assay (supplementary material Fig. S2). By IP using SMC6 antibody, we were able to confirm that SMC5 co-immunoprecipitates with SMC6 (supplementary material Fig. S2B). However, we were unable to detect a stable interaction between TopoIIα and SMC6 (supplementary material Fig. S2C).
Distribution of SMC5 and NSMCE1 during male meiosis
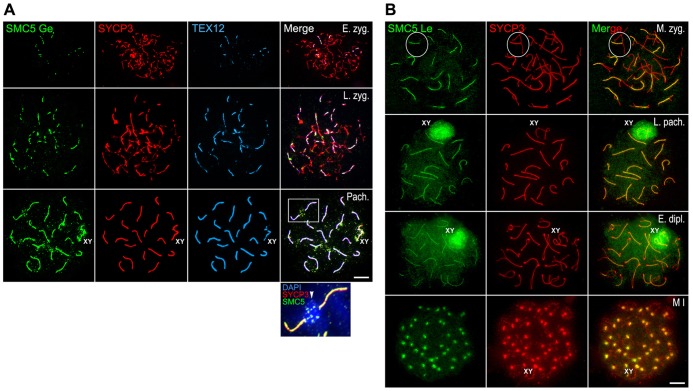
We analyzed the pattern of distribution of SMC5 on mouse spread spermatocytes, using two different antibodies raised against human SMC5, one from Dr Lehmann's group (SMC5 Le) raised against amino acids 181–305, and one from GeneTex (SMC5 Ge) raised against amino acids 257–493. Both antibodies exhibited the same localization pattern in mouse spermatocytes, one similar to that of SMC6 Le and SMC6 Sc. Thus, SMC5 appeared at the central regions of SCs colocalizing with TEX12 during prophase I stages (Fig. 7), at the sex body during pachytene and diplotene (Fig. 7A,B), and at chromocenters during pachytene (inset in Fig. 7A). In metaphase I bivalents, SMC5 was observed at the inner centromere domains colocalizing with SYCP3 (Fig. 7B).
Fig. 7.
SMC5 localizes to the synaptonemal complex central region and at the sex body during prophase I, and at centromeres in metaphase I. (A) Triple-immunolabeling of SMC5 Ge (green), SYCP3 (red) and TEX12 (light blue) on spread mouse spermatocytes at early zygotene (E. zyg.), late zygotene (L. zyg.) and pachytene (Pach.). The sex body (XY) is indicated. The boxed area in the pachytene merged image is enlarged in the inset to show the presence of SMC5 Ge at the heterochromatin-rich chromocenter that is intensely labeled with DAPI. (B) Double-immunolabeling of SMC5 Le (green) and SYCP3 (red) on spread mouse spermatocytes at mid zygotene (M. zyg.), late pachytene (L. pach.), early diplotene (E. dipl.), and metaphase I (M I). A bivalent is encircled in the mid zygotene panel. The sex body and the sex bivalent (XY) are indicated. Scale bars: 10 µm.
We also analyzed the distribution of the SMC5/6 complex subunit NSMCE1, and found that NSMCE1 was present at the chromocenters during leptotene and zygotene (data not shown). During prophase I NSMCE1 remained at the chromocenters and also localized to the sex body (supplementary material Fig. S3). NSMCE1 localized to the centromeres during metaphase I (supplementary material Fig. S3). This pattern of distribution is identical to that found for SMC6 Ab.
The loading of SMC6 at the synaptonemal complex central region depends on SYCP1
The distribution of proteins detected by SMC6 Le, SMC6 Sc, SMC5 Ge and SMC5 Le at paired chromosome regions colocalizing with the central regions during prophase I stages prompted us to test whether the SC distribution of SMC5/6 complexes was dependent on the presence of transverse filaments or a central element at the synaptonemal complex central region. We first analyzed the distribution of SMC6 Le in knockout spermatocytes for the central element protein SYCE3 (Schramm et al., 2011). In these mutants, spermatocytes arrest at a pachytene-like stage with normally assembled AEs/LEs that appear paired and aligned homologously along their entire lengths, but never completely synapse. The homologous AEs/LEs can appear clearly separated or converging at sites called axial associations that are not connected by transverse filaments, and do not show a central element between them (Schramm et al., 2011). We found that SMC6 appeared as foci along aligned AEs/LEs in pachytene-like spermatocytes (Fig. 8A,B). Careful inspection demonstrated three patterns of SMC6 foci (Fig. 8C–E): as single foci on or protruding from the AEs/LEs, mainly at separated AEs (white arrowheads); as pairs of facing foci, one per AE, at close AEs (blue arrowheads), or as single foci between close AEs or at axial associations (pink arrowheads). However, it is important to note that the SMC6 foci were more abundant on the AEs or LEs that were closely aligned (Fig. 8C) than on those more separated (Fig. 8D,E). These data indicate that the presence of SMC6 foci along the AEs/LEs is not dependent on the presence of the synaptonemal complex central element, because in the absence of SYCE3, other central element proteins required downstream for its assembly such as SYCE1, SYCE2 and TEX12, are not present and do not allow the formation of the central element (Fraune et al., 2012).
Fig. 8.
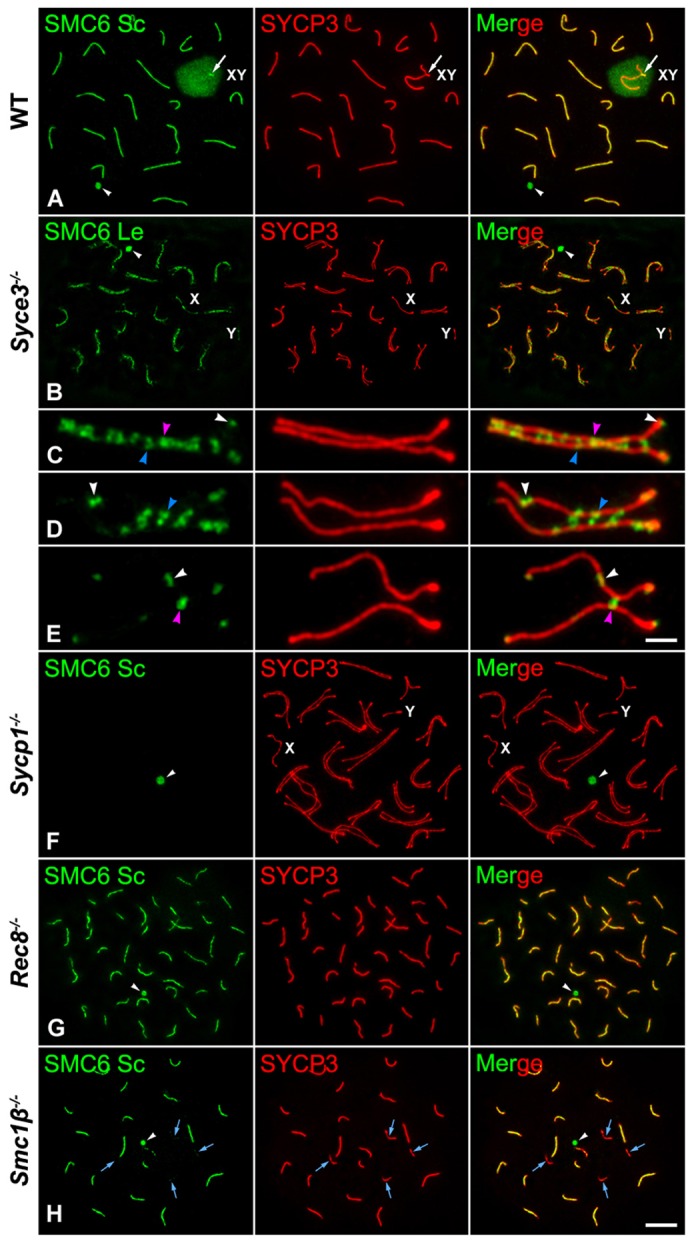
The loading of SMC6 at the synaptonemal complex central region is dependent on SYCP1, but independent of REC8 and SMC1β-containing cohesin axes. Double immunolabeling of SMC6 Sc and Le (green) and SYCP3 (red) on spread wild-type (WT) pachytene (A), and pachytene-like Syce3−/− (B–E), Sycp1−/− (F), Rec8−/− (G) and Smc1β−/− (H) mutant spermatocytes. Enlarged pachytene-like autosomal Syce3−/− ‘pseudobivalents’ (C–E). The arrows in A indicate the PAR region between the sex chromosomes. The sex body (XY) and sex univalents (X,Y) are indicated. White arrowheads in (A,B,F–H) indicate a round body in the nucleoplasm. In mutant Syce3−/− ‘pseudobivalents’ (C–E), white arrowheads indicate single SMC6 Le foci on or protruding from the AEs and LEs, blue arrowheads mark pairs of facing foci, one per AE, at close AEs, and pink arrowheads depict single foci between adjacent AEs. Blue arrows in H indicate univalents. Scale bars: 10 µm (A,B,F–H); 3 µm (C–E).
We next tested whether the presence of SMC6 at the synaptonemal complex central region was dependent on SYCP1, by analyzing the distribution of SMC6 Sc in Sycp1−/− knockout spermatocytes (de Vries et al., 2005; Hamer et al., 2008). These mutant spermatocytes show, similar to Syce3−/− spermatocytes, an arrest at a pachytene-like stage with normally assembled AEs/LEs that also appear paired and aligned homologously along their entire lengths. In these Sycp1−/− pachytene-like spermatocytes we were not able to detect the presence of SMC6 along aligned AEs (Fig. 8F). Consequently, the loading of SMC6 at the synaptonemal complex central region depends on SYCP1.
The loading of SMC5 and SMC6 at the synaptonemal complex central region is independent of REC8 and SMC1β-containing cohesin axes
Next, we analyzed whether the presence of SMC5 and/or SMC6 at the central region was dependent on the presence of cohesin axes that serve as a scaffold for the assembly of AEs/LEs (Pelttari et al., 2001). For this purpose we analyzed the distribution of SMC6 Sc and SMC5 Ge in mutants for the meiosis-specific cohesin subunits REC8 (Bannister et al., 2004), knockout for SMC1β (Smc1β−/−) (Revenkova et al., 2004), and ethylnitrosourea (ENU) induced mutants for SMC1β (Smc1βrepro11/repro11), hereafter referred to as Smc1βrepro11 (Handel et al., 2006).
Rec8−/− mutant spermatocytes arrest at a pachytene-like stage with chromosome cores that are not homologously synapsed, and are shorter than the LEs in wild-type pachytene spermatocytes. Consequently, 40 short chromosome cores corresponding to univalents are found, composed of two closely associated axes, one per chromatid, with SYCP1 detected between them (Bannister et al., 2004; Xu et al., 2005). Accordingly, it has been proposed that in the absence of REC8, there is an inappropriate synapsis between sister chromatids that leads to the formation of a SC-like structure between sister cores (Xu et al., 2005). We found that SMC6 appeared as continuous lines that colocalized with SYCP3 along most of the chromosome cores (Fig. 8G). Thus, these data also support that the loading of SMC5/6 complexes at the central region can be directed by SYCP1, even between sister chromatid cores, but is independent of REC8-containing cohesin complexes.
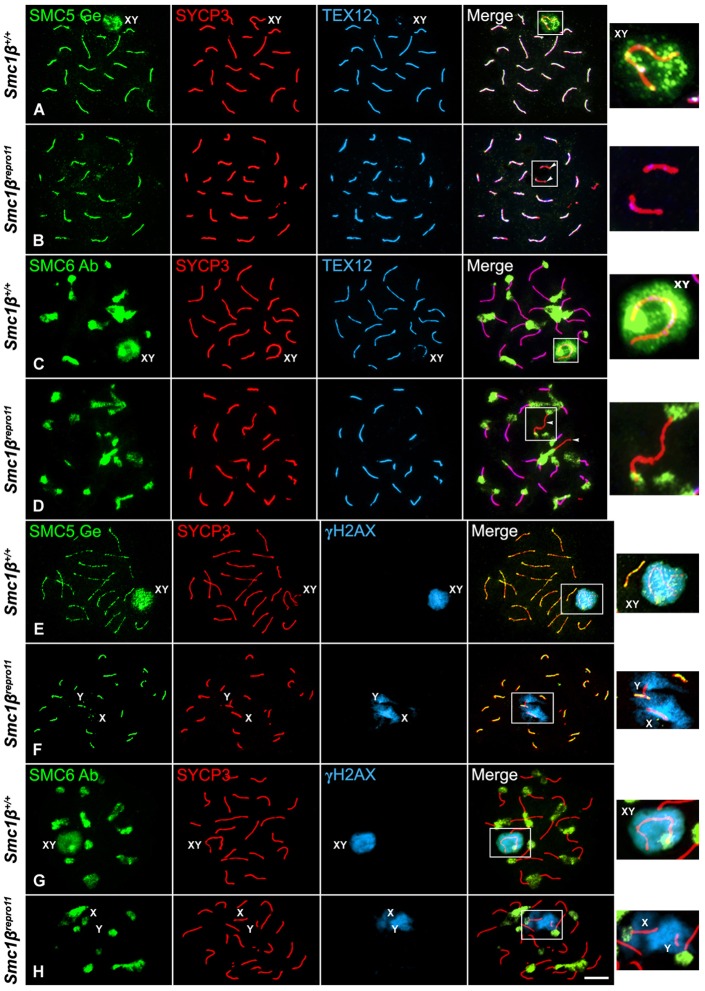
Smc1β−/− mutant spermatocytes also arrest at a pachytene-like stage. In these spermatocytes, there is a high degree of homologous synapsis and most bivalents show SCs that are approximately half as long as those found in wild-type pachytene spermatocytes (Revenkova et al., 2004; Novak et al., 2008). However, in these mutant spermatocytes homologous synapsis is rarely complete, and some partially synapsed bivalents and asynapsed univalent AEs are also found. Our results showed that SMC6 colocalized with SCs, but not on asynapsed AEs (Fig. 8H). Then we tested the distribution of SMC5 on Smc1βrepro11 pachytene-like spermatocytes, which have the same phenotype as Smc1β−/− mutants. In Smc1βrepro11 spermatocytes, SMC5 colocalized with TEX12 along the shortened SCs. However, SMC5 and TEX12 were not present at asynapsed AEs (Fig. 9A,B). Given that SMC6 Ab detected the heterochromatic chromocenters during prophase I, we tested whether its localization was dependent on SMC1β-containing cohesin axes. Our results showed that SMC6 Ab recognized the chromocenters at the end of SCs and asynapsed AEs in Smc1βrepro11 pachytene-like spermatocytes (Fig. 9C,D). Thus, the presence of SMC6 at the heterochromatic chromocenters is independent of SMC1β-containing cohesin axes. Altogether, our results indicate that the loading of SMC5/6 complexes onto the synaptonemal complex central region is independent of meiosis-specific cohesin axes containing the REC8 and/or SMC1β subunits.
Fig. 9.
The distribution of SMC5 at the synaptonemal complex central region and SMC6 Ab at chromocenters is independent of SMC1β-containing cohesin axes. Triple immunolabeling of SMC5 Ge and SMC6 Ab (green), SYCP3 (red) and TEX12 (light blue) or γH2AX (light blue) on spread Smc1β+/+ wild-type pachytene (A,C,E,G) and Smc1βrepro11 pachytene-like (B,D,F,H) mutant spermatocytes. The sex body (XY) and sex univalents (X,Y) are indicated. Boxed areas in the merged images are enlarged in the insets at the right column. Arrowheads in B,D indicate asynapsed AEs. Scale bar: 10 µm.
In wild-type late pachytene spermatocytes SMC5 Ge and SMC6 Ab colocalize with γH2AX at the sex body (Fig. 9E,G). Most Smc1βrepro11 pachytene-like spermatocytes contained 19 bivalents, and the only asynapsed chromosomes were within a γH2AX-rich region (Fig. 9F,H), suggesting that they are the sex chromosomes. SMC5 and SMC6 fail to localize to the γH2AX-rich chromatin in the Smc1βrepro11 spermatocytes (Fig. 9F,H). This is an expected result since in the absence of SMC1β the spermatocytes arrest at an early pachytene-like stage (Revenkova et al., 2004) when SMC5 and SMC6 are still not detected at the sex body.
The redistribution of SMC6 at the centromeres during the diplotene to metaphase I transition is independent of SGOL2
Our results showed that SMC6 Ab appeared at the heterochromatic chromocenters during most prophase I stages (Fig. 3; Fig. 4A,B), and that two signals appeared at each metaphase I centromere (Fig. 4C). Thus, SMC6 redistributes at the centromeres during the diplotene to metaphase I transition.
During mammalian meiosis I, the cohesin complexes present at centromeres are protected from separase cleavage by the shugoshin SGOL2 (Llano et al., 2008). In this way, the recombined chromatids of each homolog are connected at the centromere, and when separase cleaves the deprotected cohesin complexes at metaphase II centromeres it allows their accurate segregation to opposite poles during meiosis II (Gómez et al., 2007). It has been reported that during mammalian mitosis and meiosis SGOL2 facilitates the loading of MCAK at the inner centromere domain (Huang et al., 2007; Llano et al., 2008; Parra et al., 2009; Tanno et al., 2010). Because SGOL2 is recruited to the centromeres by late diplotene during mouse meiosis (Gómez et al., 2007), we tested whether the redistribution of SMC6 Ab at centromeres was dependent on SGOL2. Our results showed that in Sgol2−/− metaphase I spermatocytes SMC6 appeared as two signals at each centromere as in wild-type spermatocytes (Fig. 10A,B). Moreover, a positive SMC6 signal was observed at the centromeres of the individualized chromatids present in metaphase II-like spermatocytes (Fig. 10C,D). These results indicate that the centromere redistribution of SMC6 during the diplotene to metaphase I transition is not dependent on SGOL2.
Fig. 10.
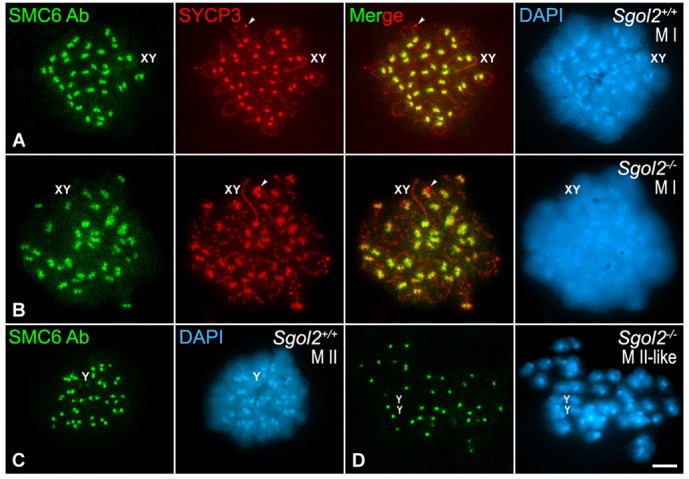
The distribution of SMC6 Ab at metaphase I and metaphase II centromeres is independent of SGOL2. Double-immunolabeling of SMC6 Ab (green) and SYCP3 (red) and counterstaining of the chromatin with DAPI (blue) on spread wild-type (Sgol2+/+) metaphase I (A) and metaphase II (C), and Sgol2−/− metaphase I (B) and metaphase II-like (D) spermatocytes. The sex bivalent (XY) and Y chromosome (Y) or chromatids (YY) are indicated. Arrowheads in A,B indicate SYCP3 agglomerates in the cytoplasm. Scale bar: 10 µm.
Discussion
The SMC5/6 complex has been implicated in DNA repair, chromosome replication and chromosome segregation in somatic cells (De Piccoli et al., 2009; Kegel and Sjögren, 2010; Wu and Yu, 2012), but its possible functions during mammalian meiosis remain unexplored. Here, we have determined the pattern of expression of the different subunits of the SMC5/6 complex during the first wave of mouse spermatogenesis. We have also analyzed the dynamics of SMC5 and SMC6 during male mouse meiosis in wild-type individuals and knockout mice lacking components of the SC, cohesin subunits and the cohesin regulator shugoshin SGOL2, in order to infer their function during both meiotic divisions. Our results are consistent with the participation of SMC5/6 complexes in homologous chromosome pairing and/or recombination during prophase I, chromosome and centromere structure during meiosis I and mitosis, and centromere cohesion, together with Topo IIα, during meiosis II.
Involvement of SMC5/6 complexes in SC assembly
We observed that the different subunits of the SMC5/6 complex are expressed increasingly from zygotene on. This result is consistent with the finding that SMC5 and SMC6 are first detected during zygotene, together with SYCP1 and TEX12, at the synaptonemal complex central region. The similar central region distribution and dynamics of both SMC5 and SMC6 from zygotene up to diplotene suggest that these proteins could dimerize and function in the synaptonemal complex central region. Because the synaptonemal complex central region is composed of the transverse filament protein SYCP1, which connects the homologous LEs, and by a central element, which is formed by the proteins SYCE1, SYCE2, SYCE3 and TEX12 (de Boer and Heyting, 2006; Bolcun-Filas et al., 2009; Fraune et al., 2012), we tested whether the distribution of SMC6 at the synaptonemal complex central region was dependent on the presence of transverse filaments or central element proteins. Our results on Syce3−/− and Sycp1−/− pachytene-like spermatocytes support that the loading of SMC5/6 at the synaptonemal complex central region depends on the presence of the transverse filament protein SYCP1. The absence of SMC5 and SMC6 along the unsynapsed AEs of the sex chromosomes, and their absence along the unsynapsed autosomal AEs in Smc1β−/− and Smc1βrepro11 mutant spermatocytes, further support the idea that the localization of SMC5 and SMC6 at the synaptonemal complex central region depends on SYCP1. Moreover, a role for SYCP1 in loading of SMC5 and SMC6 at the central region is also supported by localization of SMC6 in Rec8−/− mutant spermatocytes along the chromosome cores of univalents exhibiting inappropriate sister chromatid synapsis and presence of SYCP1 (Bannister et al., 2004; Xu et al., 2005). Because SMC6 is observed along the chromosome cores in the absence of the SMC1β or REC8, we conclude that SMC6 loading is independent of cohesin complexes containing these meiosis-specific subunits.
Analyses of fission yeast and C. elegans mutants for subunits of the SMC5/6 complex have suggested that these complexes are involved in meiotic homologous recombination (Pebernard et al., 2004; Bickel et al., 2010; Wehrkamp-Richter et al., 2012). Our results on localization of SMC5 and SMC6 at mammalian SCs are consistent with this idea. Nevertheless, other possibilities are not excluded; for example, SMC5/6 complexes at the central region might recruit central element proteins that facilitate the formation and/or stabilization of the SC, or they might function in SUMOylating or ubiquitylating target proteins acting at the SC. In fact, the non-SMC subunit Nse2–Mms21 has a SUMO-E3 ligase activity mediating repair of double-strand breaks (De Piccoli et al., 2009), and it is known that SUMOylation plays an important role in the assembly of the SC (de Carvalho and Colaiácovo, 2006).
SMC5/6 complexes at the sex body
We have found that SMC5, SMC6 and NSMCE1, as well as NSMCE2 and NSMCE4A (data not shown), are present at the sex body during pachytene and diplotene colocalizing with γH2AX. Thus, SMC5/6 complexes might have an unexpected role at the sex body. It is widely accepted that the sex body forms following a process known as meiotic sex chromosome inactivation (MSCI), which involves the transcriptional silencing of the sex chromosomes during the pachytene and diplotene stages of prophase I. MSCI is accompanied by a series of structural and epigenetic changes, which include histone modifications, and the recruitment of specific histone variants, non-histone proteins and RNAs to the sex body (Page et al., 2012). However, an alternative possibility recently proposed assumes that MSCI represents the maintenance of a prior repressed state (Page et al., 2012). Given that the SMC5/6 complexes may have a role in chromosome topology (Carter and Sjögren, 2012), it is possible that these complexes could mediate the changes in chromatin conformation that occur at the sex body.
Involvement of SMC5/6 complexes in centromere cohesion
We have found that the SMC6 Ab antibody, recognizing the C-terminal region of the protein, as well as the NSMCE1 antibody, recognized the chromocenters during all prophase I stages. By contrast, the SMC6 Le and Sc antibodies against the more N-terminal and central domains, respectively, labeled primarily the synaptonemal complex central region. Since the spreading technique and the conditions for fixing the spermatocytes were always the same, the differences in localization pattern most likely reflect differing accessibility of the relevant epitopes in different chromatin structures (i.e. SC, chromocenters and centromeres). Alternatively, the antibodies may recognize different protein isoforms generated by alternative splicing, or different post-translational modified forms, although this seems unlikely at least for SMC6, because the Le and Ab antibodies detected a single SMC6 band with the same electrophoretic mobility (140 kDa).
Our results, using SMC6 Le, SMC6 Sc, SMC5 Le and SMC5 Ge antibodies, showed SMC6 and SMC5 at heterochromatic chromocenters during late diplotene, after their disappearance from the synaptonemal complex central region during desynapsis. Thus, SMC6 and SMC5 might have a dual role at the synaptonemal complex central region and at centromeres. Interestingly, this is the first description of localization of subunits of the SMC5/6 complex at the centromeres of mammalian cells. Our results support previous reports describing the presence of Smc5 and Smc6 at the centromeres of both budding yeast (Lindroos et al., 2006; Yong-Gonzales et al., 2012) and fission yeast (Pebernard et al., 2008) during mitosis, and taken together, these data indicate that SMC5/6 complexes play a conserved role at centromeres.
We found that although SMC6 and SMC5 localized at the chromocenters during prophase I stages, they were observed distal to the closely associated sister kinetochores at metaphase I centromeres. Moreover, the partial colocalization of SMC6 with SYCP3 at metaphase I centromeres indicates that SMC6 is present at the inner centromeric domain. Thus, SMC6 and SMC5 redistribute from heterochromatic chromocenters to the inner centromeric domain during the late diplotene to metaphase I transition. We demonstrated that this redistribution within the centromere is not dependent on the shugoshin SGOL2, which by contrast, is involved in the loading of other centromere proteins (Parra et al., 2009; Tanno et al., 2010).
It is striking that SMC6 and Topo IIα colocalize, not only during prophase I stages, but also during meiotic and mitotic divisions, a finding that is relevant regarding the function of SMC5/6 complexes at centromeres. In this regard, the persistence of SMC6 at a strand joining sister kinetochores at metaphase II centromeres, after the redistribution of Aurora-B (Parra et al., 2006), and the ‘stretching’ of that strand, which still associates the kinetochores of segregating anaphase II chromatids, where Topo IIα is also present, suggests that the SMC5/6 complexes regulate sister-chromatid centromere cohesion. Notably, it has been recently reported that the SMC5/6 complexes might be involved in sister-chromatid cohesion and dissolution of aberrant DNA-mediated sister-chromatid linkages after DNA replication in budding and fission yeasts (Outwin et al., 2009; Bermúdez-López et al., 2010), chicken cells (Stephan et al., 2011) and HeLa cells (Behlke-Steinert et al., 2009). Moreover, evidence suggests a close relationship between SMC5/6 and cohesin complexes in unperturbed cells, where both complexes require the participation of Scc2 to load on the chromosomes during interphase (Lindroos et al., 2006), and, in addition, the SMC5/6 subunit NSE2 sumoylates the cohesin subunits RAD21/Scc1, Smc1 and Smc3 in human and budding yeast cells (Takahashi et al., 2008; McAleenan et al., 2012; Wu et al., 2012). SMC5/6 complexes seem to be functionally related to Topo IIα in the resolution of topological stress during replication (Tapia-Alveal et al., 2010; Kegel and Sjögren, 2011). In fact, the fission yeast smc6-74 mutant is synthetically lethal with temperature-sensitive top2-191 at semipermissive temperatures (Verkade et al., 1999), and budding yeast nse2 and smc6-56 mutants show synthetic growth defects when combined with mutations in Top2 (Takahashi et al., 2008). Here we augment these observations by showing that SMC6 and Topo IIα colocalize at ‘stretched’ strands that connect lagging chromatids at anaphase II following Etoposide-mediated Topo IIα inhibition, together suggesting that the SMC5/6 complexes could be involved in sister-chromatid centromere cohesion. Our immunoprecipitation data indicate that SMC6 and Topo IIα apparently do not form a stable proteinaceous complex since Topo IIα does not co-immunoprecipitate with SMC6. However, the SMC5/6 complex might be involved in centromere cohesion by regulating the decatenating activity of Topo IIα through the SUMO-E3 ligase activity of the subunit NSE2. In this regard, it has been proposed that the SUMOylation of Topo IIα is required to allow its decatenating activity at centromeres during anaphase (Dawlaty et al., 2008).
Involvement of SMC5/6 complexes in chromosome structure
Taken together, the results reported here implicate the SMC5/6 complexes in many chromosome processes during meiosis. Indeed, we have also observed SMC6 along the chromatid axes in metaphase I and anaphase I chromosomes, as well as in mitotic chromosomes. This distribution of SMC6 at chromatid axes has not previously been reported, but might well be relevant, because SMC6 colocalizes with Topo IIα on the axes. This suggests that the SMC5/6 complexes may have another chromosomal role in addition to their function in SCs and at centromeres. It has been recently proposed that condensin I complexes together with the kinesin motor protein KIF4 are involved in the lateral compaction of mitotic chromosomes, whereas condensin II complexes and Topo IIα orchestrate their axial shortening (Hirano, 2012; Samejima et al., 2012). Our findings suggest that the SMC5/6 complexes could additionally be implicated in chromosome structure and condensation.
In summary, our results show that some subunits of the SMC5/6 complex are present at the synaptonemal complex central region during prophase I, and at the centromeres and chromatid axes in condensed meiotic and mitotic chromosomes. These intriguing findings indicate that SMC5/6 complexes could be involved in sister-chromatid cohesion, in concert with SMC1/3-based cohesin complexes, as well as in chromosome condensation, with SMC2/4-based condensin complexes. Given that these three SMC complexes are all also involved in DNA repair, their interdependence seems a likely possibility. One important question is whether there are different SMC5/6 complexes acting at different stages of the cell cycle and with distinct functions, as previously suggested for human cells (Behlke-Steinert et al., 2009). Likewise, the existence of mitotic and meiotic paralogs for each of the subunits remains to be determined. Future genetic tests for the requirement of SMC5/6 complexes, particularly during meiosis, will shed further light on the biological function of these interesting proteins.
Materials and Methods
Animals
Mice used in this study were as follows: Wild-type (C57BL/6 and C57BL/6×SJL F1 hybrids), Syce3−/− knockout (Schramm et al., 2011), Sycp1−/− knockout (Hamer et al., 2008), Rec8−/− knockout (Bannister et al., 2004), Smc1β−/− knockout (Revenkova et al., 2004), mutant Smc1βrepro11 mice produced by the Reproductive Genomics Program at The Jackson Laboratory by ENU mutagenesis of C57BL6/J germ cells, and maintained on an C3Heb/FeJ background, and Sgol2−/− knockout (Llano et al., 2008). Animals were handled according with relevant regulatory standards and experiments were approved by The Jackson Laboratory (JAX) Institutional Animal Care and Use Committee (IACUC) and UAM Ethics Committee.
Cell culture
Mouse 3T3 cells were grown in Dulbecco's Modified Eagle's Medium with 4.5 g/l glucose (DMEM, Cambrex) supplemented with 10% fetal bovine serum (Gibco) and 2 mM L-glutamine (Cambrex), at 37°C in a humidified atmosphere containing 5% CO2. Cells were fixed in 2% formaldehyde in PBS (137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.7 mM KH2PO4, pH 7.4) containing 0.05% Triton X-100 (Sigma-Aldrich) for 10 minutes, and then processed for immunofluorescence.
Inhibition of Topo IIα activity by Etoposide
Etoposide (Sigma-Aldrich, E1383) was prepared in a solution of 50 mg/kg (12.5 mg/ml) diluted in DMSO (Sigma-Aldrich, D5879). After tempering the drug to body temperature, it was administered to anesthetised adult C57BL/6 mice by intraperitoneal injections of 0.5 ml. Specimens were monitored and sacrificed after 48 hours following all the recommendations of the Animal Experimentation UAM Ethics Committee.
Squashing of seminiferous tubules and spreading of spermatocytes
Squashing of seminiferous tubules was conducted as previously described (Page et al., 1998; Parra et al., 2002). Spreading of spermatocyte chromatin was prepared as previously reported (Peters et al., 1997).
Immunocytology
Cells and squashed or spread spermatocyte preparations were rinsed three times for 5–10 minutes in PBS or 10% antibody dilution buffer (ADB: 10% horse serum, 3% BSA and 0.05% Triton-X in PBS) and incubated in a humid slide chamber for 2 hours at 37°C or overnight at 4°C with the corresponding primary antibodies (supplementary material Table S1) diluted in PBS or ADB. Following three washes in PBS or ADB for 5–10 minutes, the slides were incubated for 30 minutes at room temperature with the corresponding secondary antibodies (shown in supplementary material Table S1). The slides were subsequently rinsed in PBS and counterstained for 3 minutes with 10 µg/ml DAPI (4′,6-diamidino-2-phenylindole). After a final rinse in PBS, the slides were mounted with Vectashield (Vector Laboratories) and sealed with nail polish.
Immunofluorescence image stacks or single images were collected on an Olympus BX61 microscope equipped with epifluorescence optics, a motorized z-drive, and an Olympus DP71 digital camera controlled by analySIS software (Soft Imaging System) or on a Zeiss Axio Observer Z1 linked to an AxioCam MR Rev3 camera controlled by the AxioVision V4.8 image software. Stacks were analyzed and processed using the public domain ImageJ software (National Institutes of Health, USA; http://rsb.info.nih.gov/ij). Final images were processed with Adobe Photoshop 7.0 or CS5 softwares.
Mouse germ cell isolation and short-term culture
Isolation of mixed germ cells from testes was performed using techniques previously described (Bellvé, 1993; La Salle et al., 2009). Pachytene spermatocytes were enriched using a 2–4% BSA gradient generated in a STAPUT sedimentation chamber (ProScience) as previously described (Bellvé, 1993; La Salle et al., 2009; Sun and Handel, 2008). Highly enriched pachytene spermatocytes (2.5×106 cells/ml) were cultured for 10 hours at 32°C in 5% CO2 in HEPES (25 mM)-buffered MEMα culture medium (Sigma-Aldrich) supplemented with 25 mM NaHCO3, 5% fetal bovine serum (Atlanta Biologicals), 10 mM sodium lactate, 59 µg/ml penicillin, and 100 µg/ml streptomycin. To initiate the G2–MI transition, cultured pachytene spermatocytes were treated with 5 µM okadaic acid (OA) (CalBiochem). For protein analyses spermatocytes were harvested at 0, 2.5 and 5 hours after OA treatment for protein extraction as described below.
Transcript expression analysis
mRNA was prepared from enriched germ cells using the Qiagen RNA purification kit. Following purification, RNA was treated with DNase and the RNA purity and concentration was assessed using a ND1000 Nanodrop spectrophotometer. cDNA was synthesized via a reverse transcriptase reaction (Qiagen). 10 ng of cDNA was used as a template for standard PCR (58°C annealing, 70°C, 30 second extension for 25 cycles) and reactions were assessed using standard agarose gel electrophoresis. Primers used are presented in supplementary material Table S2.
Protein analyses
Protein was extracted from germ cells using RIPA buffer (Santa Cruz Biotechnology) or NP40 buffer (150 mM NaCl, 50 mM Tris-HCl, 1% NP40, pH 8.0) containing 1× protease inhibitor cocktail (Roche). Protein concentration was calculated using a BCA protein assay kit (Pierce). For enriched germ cells 20 µl of 1 mg/ml protein extract were loaded per lane on SDS polyacrylamide gels. For STAPUT purified pachytene cells 20 µl of 0.1 mg/ml were loaded per lane on SDS polyacrylamide gels. Following protein separation via standard SDS-PAGE, proteins were transferred to PVDF membranes using semi-dry western blotting apparatus (Bio-Rad). Primary antibodies and dilutions used are presented in supplementary material Table S1. The presence of antibodies on the PVDF membranes was detected via treatment with Pierce ECL western blotting substrate (Thermo Scientific) and exposure to Kodak Blue XB film.
Testis extracts and immunoprecipitation
To prepare testis extracts, testes were removed from male C57BL/6 mice, detunicated and washed in PBS. Nuclear and cytoplasmic extracts were obtained using the NE-PER Nuclear and Cytoplasmic Extraction Reagent Kit (Pierce) according to the manufacturer's instructions. The testis nuclear extract was stored at −80°C until used for immunoprecipitation (IP).
For IP of SMC6, 10 µl of anti-SMC6 Ab antibody was coupled to the nuclear extract and incubated for 2 hours at 4°C, followed by incubation with the nProtein A Sepharose for 1 hour at 4°C with rotation. Then, co-immunoprecipitation SMC6 complexes were washed and eluted from beads following instructions for Immunoprecipitation Started Pack (GE Healthcare, 71-5017-54 AD). SMC6 co-immunoprecipitated proteins were run at the same time as the nuclear extract, and first supernatant of IP experiment in 8% SDS-PAGE. Western blotting was probed with anti-SMC6 Ab for positive control, with anti-SMC5 (Abcam, ab-18038) and anti-Topo IIα (dilutions are indicated in supplementary material Table S1). Visualization was performed using ECL detection system (GE Healthcare).
Supplementary Material
Acknowledgments
We express our sincere thanks to Dr Alan R. Lehmann for providing the SMC5 and SMC6 antibodies, Dr José Luis Barbero for the SYCP1 antibody, Dr Akihiko Kikuchi for the Topo IIα antibody, Dr John C. Schimenti and Dr Kerry Schimenti for providing spreads from knockout REC8 mice, Maria Fernanda Ruíz Lorenzo for advice and Lorena Barreras for technical assistance.
Footnotes
Author contributions
R.G., P.W.J., M.A.H. and J.A.S. conceived and designed the experiments. R.G., P.W.J., M.A.H. and J.A.S. performed the experiments. R.G., P.W.J., A.V., M.A., T.F., R.J., E.L., A.M.P., M.A.H. and J.A.S. analysed data. R.G., P.W.J., M.A.H. and J.A.S. wrote the manuscript.
Funding
This work was supported by Ministerio de Economía y Competitividad (Spain) [grant number SAF2011-28842-C02-01 to J.A.S. and SAF2011-25252 to A.M.P.]; a UK-US Fulbright Distinguished Scholar Award; the US National Institutes of Health [grant number HD069458 to P.W.J.; HD33816 to M.A.H.; HD42137 to John Eppig, M.A.H. and J.C.S.]; the Priority Program SPP 1384 ‘Mechanisms of genome haploidization’ (to M.A. and R.J.) from the German Science Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the NIH. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.130195/-/DC1
References
- Bannister L. A., Reinholdt L. G., Munroe R. J., Schimenti J. C. (2004). Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis 40, 184–194 10.1002/gene.20085 [DOI] [PubMed] [Google Scholar]
- Behlke-Steinert S., Touat-Todeschini L., Skoufias D. A., Margolis R. L. (2009). SMC5 and MMS21 are required for chromosome cohesion and mitotic progression. Cell Cycle 8, 2211–2218 10.4161/cc.8.14.8979 [DOI] [PubMed] [Google Scholar]
- Bellvé A. R. (1993). Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 225, 84–113 10.1016/0076-6879(93)25009-Q [DOI] [PubMed] [Google Scholar]
- Bermúdez-López M., Ceschia A., de Piccoli G., Colomina N., Pasero P., Aragón L., Torres-Rosell J. (2010). The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res. 38, 6502–6512 10.1093/nar/gkq546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel J. S., Chen L., Hayward J., Yeap S. L., Alkers A. E., Chan R. C. (2010). Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. 6, e1001028 10.1371/journal.pgen.1001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E., Hall E., Speed R., Taggart M., Grey C., de Massy B., Benavente R., Cooke H. J. (2009). Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 5, e1000393 10.1371/journal.pgen.1000393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S. D., Sjögren C. (2012). The SMC complexes, DNA and chromosome topology: right or knot? Crit. Rev. Biochem. Mol. Biol. 47, 1–16 10.3109/10409238.2011.614593 [DOI] [PubMed] [Google Scholar]
- Cobb J., Miyaike M., Kikuchi A., Handel M. A. (1999). Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase II α localization and chromosome condensation. Chromosoma 108, 412–425 10.1007/s004120050393 [DOI] [PubMed] [Google Scholar]
- Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J. M. (2008). Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell 133, 103–115 10.1016/j.cell.2008.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E., Heyting C. (2006). The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115, 220–234 10.1007/s00412-006-0057-5 [DOI] [PubMed] [Google Scholar]
- de Carvalho C. E., Colaiácovo M. P. (2006). SUMO-mediated regulation of synaptonemal complex formation during meiosis. Genes Dev. 20, 1986–1992 10.1101/gad.1457806 [DOI] [PubMed] [Google Scholar]
- De Piccoli G., Torres-Rosell J., Aragón L. (2009). The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 17, 251–263 10.1007/s10577-008-9016-8 [DOI] [PubMed] [Google Scholar]
- de Vries F. A., de Boer E., van den Bosch M., Baarends W. M., Ooms M., Yuan L., Liu J. G., van Zeeland A. A., Heyting C., Pastink A. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389 10.1101/gad.329705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Yang Y., Chen Y. H., Arenz J., Rangi G. K., Zhao X., Ye H. (2009). Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5-6 subcomplex and the hinge regions of Smc5 and Smc6. J. Biol. Chem. 284, 8507–8515 10.1074/jbc.M809139200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S., San-Segundo P. A., Aragón L. (2011). The Smc5-Smc6 complex is required to remove chromosome junctions in meiosis. PLoS ONE 6, e20948 10.1371/journal.pone.0020948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firooznia A., Revenkova E., Jessberger R. (2005). From the XXVII North American Testis Workshop: the function of SMC and other cohesin proteins in meiosis. J. Androl. 26, 1–10 [PubMed] [Google Scholar]
- Fraune J., Schramm S., Alsheimer M., Benavente R. (2012). The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp. Cell Res. 318, 1340–1346 10.1016/j.yexcr.2012.02.018 [DOI] [PubMed] [Google Scholar]
- Gómez R., Valdeolmillos A., Parra M. T., Viera A., Carreiro C., Roncal F., Rufas J. S., Barbero J. L., Suja J. A. (2007). Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep. 8, 173–180 10.1038/sj.embor.7400877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez R., Viera A., Berenguer I., Llano E., Pendás A. M., Barbero J. L., Kikuchi A., Suja J. A. (2013). Cohesin removal precedes Topoisomerase IIα-dependent decatenation at centromeres in male mammalian meiosis II. Chromosoma. In press. [DOI] [PubMed] [Google Scholar]
- Hamer G., Gell K., Kouznetsova A., Novak I., Benavente R., Höög C. (2006). Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J. Cell Sci. 119, 4025–4032 10.1242/jcs.03182 [DOI] [PubMed] [Google Scholar]
- Hamer G., Novak I., Kouznetsova A., Höög C. (2008). Disruption of pairing and synapsis of chromosomes causes stage-specific apoptosis of male meiotic cells. Theriogenology 69, 333–339 10.1016/j.theriogenology.2007.09.029 [DOI] [PubMed] [Google Scholar]
- Handel M. A., Lessard C., Reinholdt L., Schimenti J., Eppig J. J. (2006). Mutagenesis as an unbiased approach to identify novel contraceptive targets. Mol. Cell Endocrinol. 250, 201–205 10.1016/j.mce.2005.12.046 [DOI] [PubMed] [Google Scholar]
- Hirano T. (2012). Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 26, 1659–1678 10.1101/gad.194746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Feng J., Famulski J., Rattner J. B., Liu S. T., Kao G. D., Muschel R., Chan G. K., Yen T. J. (2007). Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J. Cell Biol. 177, 413–424 10.1083/jcb.200701122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A., Sjögren C. (2010). The Smc5/6 complex: more than repair? Cold Spring Harb. Symp. Quant. Biol. 75, 179–187 10.1101/sqb.2010.75.047 [DOI] [PubMed] [Google Scholar]
- La Salle S., Sun F., Handel M. A. (2009). Isolation and short-term culture of mouse spermatocytes for analysis of meiosis. Methods Mol. Biol. 558, 279–297 [DOI] [PubMed] [Google Scholar]
- Lindroos H. B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. (2006). Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22, 755–767 10.1016/j.molcel.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Llano E., Gómez R., Gutiérrez-Caballero C., Herrán Y., Sánchez-Martín M., Vázquez-Quiñones L., Hernández T., de Alava E., Cuadrado A., Barbero J. L. et al. (2008). Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 22, 2400–2413 10.1101/gad.475308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleenan A., Cordon-Preciado V., Clemente-Blanco A., Liu I. C., Sen N., Leonard J., Jarmuz A., Aragón L. (2012). SUMOylation of the α-kleisin subunit of cohesin is required for DNA damage-induced cohesion. Curr. Biol. 22, 1564–1575 10.1016/j.cub.2012.06.045 [DOI] [PubMed] [Google Scholar]
- Murray J. M., Carr A. M. (2008). Smc5/6: a link between DNA repair and unidirectional replication? Nat. Rev. Mol. Cell Biol. 9, 177–182 10.1038/nrm2309 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H. (2009). Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Nitiss J. L. (2009). Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 10.1038/nrc2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I., Wang H., Revenkova E., Jessberger R., Scherthan H., Höög C. (2008). Cohesin Smc1β determines meiotic chromatin axis loop organization. J. Cell Biol. 180, 83–90 10.1083/jcb.200706136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outwin E. A., Irmisch A., Murray J. M., O'Connell M. J. (2009). Smc5-Smc6-dependent removal of cohesin from mitotic chromosomes. Mol. Cell. Biol. 29, 4363–4375 10.1128/MCB.00377-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page J., Suja J. A., Santos J. L., Rufas J. S. (1998). Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res. 6, 639–642 10.1023/A:1009209628300 [DOI] [PubMed] [Google Scholar]
- Page J., de la Fuente R., Manterola M., Parra M. T., Viera A., Berríos S., Fernández-Donoso R., Rufas J. S. (2012). Inactivation or non-reactivation: what accounts better for the silence of sex chromosomes during mammalian male meiosis? Chromosoma 121, 307–326 10.1007/s00412-012-0364-y [DOI] [PubMed] [Google Scholar]
- Parra M. T., Page J., Yen T. J., He D., Valdeolmillos A., Rufas J. S., Suja J. A. (2002). Expression and behaviour of CENP-E at kinetochores during mouse spermatogenesis. Chromosoma 111, 53–61 10.1007/s00412-002-0185-5 [DOI] [PubMed] [Google Scholar]
- Parra M. T., Viera A., Gómez R., Page J., Benavente R., Santos J. L., Rufas J. S., Suja J. A. (2004). Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J. Cell Sci. 117, 1221–1234 10.1242/jcs.00947 [DOI] [PubMed] [Google Scholar]
- Parra M. T., Gómez R., Viera A., Page J., Calvente A., Wordeman L., Rufas J. S., Suja J. A. (2006). A perikinetochoric ring defined by MCAK and Aurora-B as a novel centromere domain. PLoS Genet. 2, e84 10.1371/journal.pgen.0020084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra M. T., Gómez R., Viera A., Llano E., Pendás A. M., Rufas J. S., Suja J. A. (2009). Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet. 5, e1000417 10.1371/journal.pgen.1000417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S., McDonald W. H., Pavlova Y., Yates J. R., 3rd, Boddy M. N. (2004). Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol. Biol. Cell 15, 4866–4876 10.1091/mbc.E04-05-0436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S., Schaffer L., Campbell D., Head S. R., Boddy M. N. (2008). Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 27, 3011–3023 10.1038/emboj.2008.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelttari J., Hoja M. R., Yuan L., Liu J. G., Brundell E., Moens P., Santucci-Darmanin S., Jessberger R., Barbero J. L., Heyting C. et al. (2001). A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell. Biol. 21, 5667–5677 10.1128/MCB.21.16.5667-5677.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H., Plug A. W., van Vugt M. J., de Boer P. (1997). A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 10.1023/A:1018445520117 [DOI] [PubMed] [Google Scholar]
- Potts P. R., Yu H. (2005). Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25, 7021–7032 10.1128/MCB.25.16.7021-7032.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E., Eijpe M., Heyting C., Hodges C. A., Hunt P. A., Liebe B., Scherthan H., Jessberger R. (2004). Cohesin SMC1 β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6, 555–562 10.1038/ncb1135 [DOI] [PubMed] [Google Scholar]
- Samejima K., Samejima I., Vagnarelli P., Ogawa H., Vargiu G., Kelly D. A., de Lima Alves F., Kerr A., Green L. C., Hudson D. F. et al. (2012). Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J. Cell Biol. 199, 755–770 10.1083/jcb.201202155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm S., Fraune J., Naumann R., Hernandez-Hernandez A., Höög C., Cooke H. J., Alsheimer M., Benavente R. (2011). A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 7, e1002088 10.1371/journal.pgen.1002088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A. K., Kliszczak M., Dodson H., Cooley C., Morrison C. G. (2011). Roles of vertebrate Smc5 in sister chromatid cohesion and homologous recombinational repair. Mol. Cell. Biol. 31, 1369–1381 10.1128/MCB.00786-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Handel M. A. (2008). Regulation of the meiotic prophase I to metaphase I transition in mouse spermatocytes. Chromosoma 117, 471–485 10.1007/s00412-008-0167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Dulev S., Liu X., Hiller N. J., Zhao X., Strunnikov A. (2008). Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet. 4, e1000215 10.1371/journal.pgen.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno Y., Kitajima T. S., Honda T., Ando Y., Ishiguro K., Watanabe Y. (2010). Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 24, 2169–2179 10.1101/gad.1945310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Alveal C., Outwin E. A., Trempolec N., Dziadkowiec D., Murray J. M., O'Connell M. J. (2010). SMC complexes and topoisomerase II work together so that sister chromatids can work apart. Cell Cycle 9, 2065–2070 10.4161/cc.9.11.11734 [DOI] [PubMed] [Google Scholar]
- Taylor E. M., Moghraby J. S., Lees J. H., Smit B., Moens P. B., Lehmann A. R. (2001). Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell 12, 1583–1594 10.1091/mbc.12.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. M., Copsey A. C., Hudson J. J., Vidot S., Lehmann A. R. (2008). Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol. Cell. Biol. 28, 1197–1206 10.1128/MCB.00767-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade H. M., Bugg S. J., Lindsay H. D., Carr A. M., O'Connell M. J. (1999). Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10, 2905–2918 10.1091/mbc.10.9.2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrkamp-Richter S., Hyppa R. W., Prudden J., Smith G. R., Boddy M. N. (2012). Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex. Nucleic Acids Res. 40, 9633–9646 10.1093/nar/gks713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Yu H. (2012). The Smc complexes in DNA damage response. Cell Biosci. 2, 5 10.1186/2045-3701-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Kong X., Ji Z., Zeng W., Potts P. R., Yokomori K., Yu H. (2012). Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev. 26, 1473–1485 10.1101/gad.193615.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Beasley M. D., Warren W. D., van der Horst G. T., McKay M. J. (2005). Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell 8, 949–961 10.1016/j.devcel.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Yong-Gonzales V., Hang L. E., Castellucci F., Branzei D., Zhao X. (2012). The Smc5-Smc6 complex regulates recombination at centromeric regions and affects kinetochore protein sumoylation during normal growth. PLoS ONE 7, e51540 10.1371/journal.pone.0051540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.