Summary
Class-1 PI3-kinases are major regulators of the actin cytoskeleton, whose precise contributions to chemotaxis, phagocytosis and macropinocytosis remain unresolved. We used systematic genetic ablation to examine this question in growing Dictyostelium cells. Mass spectroscopy shows that a quintuple mutant lacking the entire genomic complement of class-1 PI3-kinases retains only 10% of wild-type PtdIns(3,4,5)P3 levels. Chemotaxis to folate and phagocytosis of bacteria proceed normally in the quintuple mutant but macropinocytosis is abolished. In this context PI3-kinases show specialized functions, only one of which is directly linked to gross PtdIns(3,4,5)P3 levels: macropinosomes originate in patches of PtdIns(3,4,5)P3, with associated F-actin-rich ruffles, both of which depend on PI3-kinase 1/2 (PI3K1/2) but not PI3K4, whereas conversion of ruffles into vesicles requires PI3K4. A biosensor derived from the Ras-binding domain of PI3K1 suggests that Ras is activated throughout vesicle formation. Binding assays show that RasG and RasS interact most strongly with PI3K1/2 and PI3K4, and single mutants of either Ras have severe macropinocytosis defects. Thus, the fundamental function of PI3-kinases in growing Dictyostelium cells is in macropinocytosis where they have two distinct functions, supported by at least two separate Ras proteins.
Key words: Dictyostelium; Macropinocytosis; PI3-kinase signalling; PtdIns(3,4,5)P3; Ras
Introduction
Class-1 phosphoinositide 3-kinases (PI3-kinases; PI3K in the context of single genes or proteins) play a pivotal role in many cellular responses to external stimuli (Vanhaesebroeck et al., 2001). A key event of activation is binding of Ras–GTP to a characteristic Ras-binding domain (RBD) in the protein, which in turn stimulates enzyme activity and leads to the accumulation of phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3] in the plasma membrane (Rodriguez-Viciana et al., 1994; Rodriguez-Viciana et al., 1996; Pacold et al., 2000). The activity of mammalian PI3-kinases can be modulated further, when the enzyme binds with its regulatory subunit phosphorylated tyrosines or Gβγ subunits as provided by activated G-protein-coupled receptors (Pacold et al., 2000; Vanhaesebroeck et al., 2010). PtdIns(3,4,5)P3 acts as a docking site for effector proteins, such as those with specific PH-domains, which are recruited to the membrane and orchestrate an appropriate cellular response. As one of the main outlets for activated Ras, class-1 PI3-kinases have a major and well-understood function in growth factor signalling in vertebrate cells, but they are also involved in a set of F-actin-driven processes – chemotaxis, phagocytosis and macropinocytosis, where their role and specific mode of regulation are less clear.
The role of PI3-kinase in chemotaxis – the movement of cells along chemical gradients – has been the subject of major debate and revision over the last decade. Initial, very striking results demonstrated that chemotactic gradients could induce the formation of matching, but sharper PtdIns(3,4,5)P3 gradients in the plasma membrane, leading to the idea that PtdIns(3,4,5)P3 acts as a ‘chemotactic compass’, guiding cell movement (Parent et al., 1998; Janetopoulos et al., 2004; Weiner, 2002). However, genetic and inhibitor experiments in both Dictyostelium amoebae and neutrophils showed that PtdIns(3,4,5)P3 gradients are dispensable for chemotaxis in strong gradients, though defects can become apparent in more stringent conditions (Loovers et al., 2006; Hoeller and Kay, 2007; Ferguson et al., 2007; Takeda et al., 2007; Bosgraaf et al., 2008). In Dictyostelium cells, PtdIns(3,4,5)P3 is currently viewed as participating in one of a number of partially redundant signalling pathways controlling chemotaxis to cyclic-AMP (Kay et al., 2008; Chen et al., 2007; Veltman et al., 2008; Swaney et al., 2010), but its role in folate chemotaxis is unclear, although current evidence suggests it may not be required (Srinivasan et al., 2013; Kortholt et al., 2011).
Phagocytosis and macropinocytosis are related ways for cells to take in large volumes of extracellular material (Swanson, 2008). Both processes are utilized by immune cells to destroy invading microorganisms, clear apoptotic corpses and gather antigens for presentation (Lim and Gleeson, 2011). In many other cell types, macropinocytosis can be stimulated by the addition of growth factors (Willingham et al., 1983; Dowrick et al., 1993; Maniak, 2001) or induced by oncogenic mutations such as in H-Ras (Bar-Sagi and Feramisco, 1986).
In phagocytosis, a solid particle, such as a bacterium or yeast, is engulfed. After the particles engage with the cell surface, actin-driven processes are extended around it and eventually fuse, thus taking the particle into an internal vesicle, which is acidified and the contents processed further by delivery of lysosomal enzymes. During the engulfment phase, and for a short period afterwards, the phagocytic vesicle is highly enriched in PtdIns(3,4,5)P3 as part of a series of phosphoinositide metabolic events, which include increases and then depletion of PtdIns(4,5)P2 and appearance of PtdIns(3,4)P2 (Clarke et al., 2010; Botelho et al., 2000; Yeung et al., 2006). PtdIns(3,4,5)P3 production seems to be important for engulfing large particles (Cox et al., 1999; Marshall et al., 2001; Chen et al., 2012) but its role in taking up smaller particles, such as bacteria in the case of Dictyostelium amoebae, is unresolved (Dormann et al., 2004; Peracino et al., 2010; Cardelli, 2001).
Macropinosomes derive from F-actin driven ruffles on the cell surface, which can form into a cup, then fuse to engulf a volume of medium. During this process the membrane is highly enriched in PtdIns(3,4,5)P3, and it appears that macropinocytosis requires PtdIns(3,4,5)P3 signalling (Dormann et al., 2004; Rupper et al., 2001). The precise function this signal mediates, however, remains unclear. While in some cell types PI3-kinase signalling is required for ruffle formation (Wennström et al., 1994; Araki et al., 2007), in others a role later, during cup closure, has been proposed (Araki et al., 1996).
Both phagocytosis and macropinocytosis, as well as chemotaxis, are central to the life-style of Dictyostelium amoebae, and all three processes can occur in growing cells. Wild-type amoebae track bacteria by chemotaxis to folic acid, and consume them by phagocytosis. Additionally, a single recessive mutation at the axeB locus, whose protein product is currently unknown, makes macropinocytosis constitutive and allows cells to grow in liquid medium. These axenic cells can therefore grow either by phagocytosis or by macropinocytosis making them a convenient genetic vehicle to investigate the role of PI3-kinases in all three actin-driven processes.
Mammalian genomes encode four class-1 PI3-kinases and each protein exists as a heterodimer with an adapter subunit. In contrast, Dictyostelium has five class-1 PI3-kinases, and although homology searches detect no adaptor subunit in the genome (Eichinger et al., 2005) they have a clear RBD as do their mammalian counterparts. Much of the knowledge of the roles of the PI3-kinases in chemotaxis, phagocytosis and macropinocytosis comes from the use of enzyme inhibitors, such as LY294002, which might give incomplete inhibition (Loovers et al., 2006; Kortholt et al., 2011), do not discriminate between isoforms and have off-target effects (Gharbi et al., 2007). Initial genetic studies in Dictyostelium showed that a double mutant of PI3K1 and PI3K2 had little if any defect in chemotaxis or phagocytosis, but was unable to grow in liquid medium, with a strong defect in fluid uptake, thus genetically linking PI3-kinases to macropinocytosis for the first time (Zhou et al., 1995; Buczynski et al., 1997; Zhou et al., 1998). However, later work that concentrated on the involvement of PI3-kinases in chemotaxis cast doubt on the phenotype of this early mutant (Funamoto et al., 2001; Funamoto et al., 2002). In addition, the involvement of further PI3-kinases remains to be explored.
To allow a comprehensive dissection of the role of PI3-kinases in cell movement, phagocytosis and macropinocytosis we produced and analysed a complete set of single PI3-kinase knockout mutants in Dictyostelium, supplemented by multiple knockouts, including a quintuple mutant of all five PI3-kinase genes (Hoeller and Kay, 2007). Our results clearly establish a crucial role for class-1 PI3-kinases in macropinocytosis, and allow us to resolve two separate steps: one involves PI3K1/2, which produce patches of PtdIns(3,4,5)P3 that go along with ruffle formation; the other involves PI3K4 and is required at a later stage of vesicle formation, for the conversion of PtdIns(3,4,5)P3 patches into macropinosomes. Mechanistically, we identify two specific Ras proteins, RasG and RasS that are likely to regulate PI3-kinases in this context.
Results
PtdIns(3,4,5)P3 labelled structures in growing cells
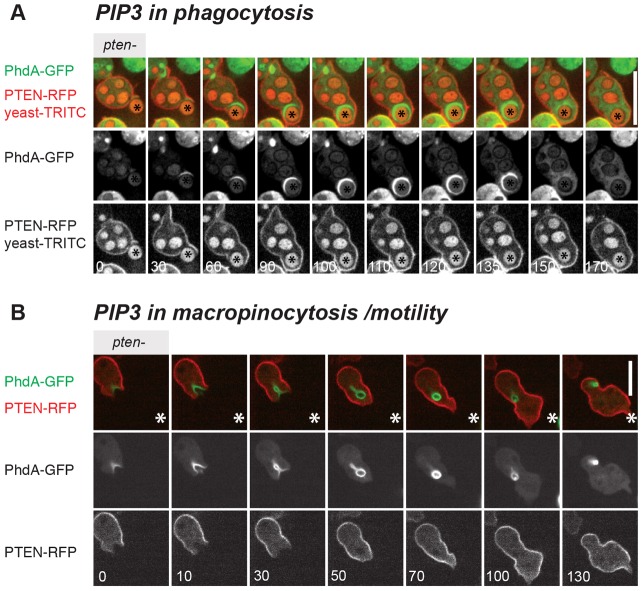
As a first step towards identifying the roles of PI3-kinase signalling in growing Dictyostelium cells, we surveyed the structures labelled with PtdIns(3,4,5)P3 using PhdA–GFP as a reporter for PtdIns(3,4,5)P3 (Funamoto et al., 2001; Funamoto et al., 2002) and PTEN–RFP, a 3′ phosphatase that degrades PtdIns(3,4,5)P3, which generally displays a reciprocal pattern to PtdIns(3,4,5)P3 reporters (Funamoto et al., 2002; Iijima and Devreotes, 2002).
Dictyostelium cells can feed by engulfing solid nutrient particles and readily phagocytose yeast cells. When they do so, the PtdIns(3,4,5)P3 reporter first localizes to where the yeast cell makes initial contact with Dictyostelium, initially faintly, but increasing in intensity and stretching around the entire yeast as it becomes fully engulfed (Fig. 1A); PTEN–RFP localizes reciprocally – excluded from the PtdIns(3,4,5)P3 areas but staining the rest of the plasma membrane (Dormann et al., 2004). Once the yeast is engulfed, PtdIns(3,4,5)P3 is lost from the vesicle, but PTEN does not relocalise to it.
Fig. 1.
PtdIns(3,4,5)P3 (PIP3)-labelled structures in growing cells. (A) Phagocytosis: a Dictyostelium cell takes up TRITC-labelled yeast. Some yeast cells (red particles) have already been taken up. At 0 seconds contact with a yeast cell (marked with an asterisk) is established and as it is subsequently taken up, PhdA–GFP extends from the base of the cup all around the particle. (B) Macropinocytosis/motility: PhdA–GFP forms patches, and even those aligned with the direction of general movement, develop into macropinocytic vesicles. PhdA–GFP stays associated with the forming vesicle, but is lost from the leading edge. An asterisk serves as a spatial landmark. The green structures in the lower right are macropinosomes formed in a cell not expressing PTEN–RFP. Pten− cells were rescued with PTEN–RFP and co-express PhdA–GFP as a marker for PtdIns(3,4,5)P3 and possibly other lipid products of PI3-kinases. Cells were plated on coverslips in buffer and behaviour recorded. Composite images are shown in the top rows. GFP and RFP/TRITC channels are shown in the rows beneath. Numbers indicate time in seconds. Scale bars: 10 µm.
Alternatively, Dictyostelium cells can grow by taking in fluid nutrient by macropinocytosis. In this case, a patch of PtdIns(3,4,5)P3 forms in the plasma membrane and F-actin-rich projections extend from it, which in some cases fuse to form a macropinosome. The sealed macropinosome is initially still decorated with PtdIns(3,4,5)P3, which is lost as the vesicle moves towards the centre of the cell (Dormann et al., 2004).
Cells moving randomly under buffer project pseudopods in the direction of travel, but these are not usually enriched with PtdIns(3,4,5)P3. When PtdIns(3,4,5)P3 does form patches in these cells, they can decorate the leading edge of pseudopods but are usually destined to develop into macropinosomes. Motility and endocytosis present conflicting demands for the machinery that builds cellular protrusions (Maniak et al., 1995; Chubb et al., 2000). Previous studies focused on spatial competition between pseudopods and endocytic cups (Maniak et al., 1995), but our observations suggests a temporal aspect to this as well: PtdIns(3,4,5)P3 patches that label translocating pseudopods mostly end up forming macropinosomes. PtdIns(3,4,5)P3 then stays associated with the incipient vesicle, but is lost from the leading edge as the cell continues to move along its path (Fig. 1B).
We did not consistently observe PtdIns(3,4,5)P3 localized on other intracellular structures, but did not examine localizations during cell division (Janetopoulos et al., 2005). Thus in what follows, we focus on the involvement of the PI3-kinases in phagocytosis, macropinocytosis and cell movement.
PI3K mutants and PtdIns(3,4,5)P3 measurements
We created an isogenic set of PI3-kinase knockout mutants by homologous recombination using our strain of Ax2 as parent (wild type for this work), disrupting singly each of the five class-1 PI3-kinase genes present in the Dictyostelium genome (supplementary material Fig. S1) as well as using our previously described PI3-kinase double mutant [PI3K(1-2)−] and a quintuple mutant [PI3K(1-5)−] in which all five PI3-kinase genes are knocked out (Hoeller and Kay, 2007). Several independent clones of each single knockout were examined for consistency of phenotype, but generally the results for only one are presented. A list of genes, the corresponding proteins and knockout strains is given in supplementary material Table S1.
As a first step to understand the contribution of individual PI3-kinase isoforms, we took advantage of a new mass spectroscopy method to directly measure the relative levels of PtdIns(3,4,5)P3 (Clark et al., 2011) in wild-type and mutant cells after growth on bacteria. Growth on bacteria was chosen because some of the mutants grow very poorly in liquid medium (see later). None of the single gene mutants have a greater than 30% reduction in PtdIns(3,4,5)P3 levels compared to wild type, but PtdIns(3,4,5)P3 levels are substantially reduced in the PI3K(1-2)− mutant and further in the PI3K quintuple mutant, which has only about 10% of wild-type PtdIns(3,4,5)P3 remaining (Fig. 2; also see Discussion).
Fig. 2.
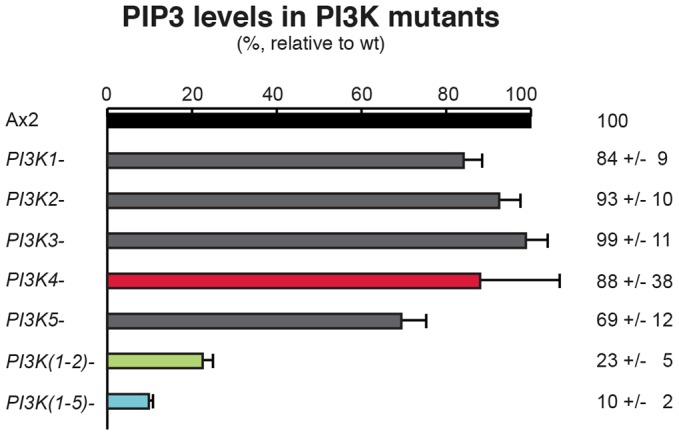
PtdIns(3,4,5)P3 (PIP3) levels in PI3-kinase mutants. PtdIns(3,4,5)P3 levels (%; mean ± s.e.m.) in PI3-kinase mutants relative to wild-type (Ax2) cells were measured by mass spectroscopy. Data are the means of three to five biological experiments, with triplicate samples taken in each and analysed in multiple spectra.
Chemotaxis to folic acid
All mutants showed comparable speed when moving randomly under buffer and chemotaxed efficiently to a needle releasing folic acid, with no clear differences in either speed or accuracy from wild type (supplementary material Table S2). Although only strong chemotactic gradients were used, this result eliminates a major, non-redundant role for PI3-kinases in folate chemotaxis, consistent with previous inhibitor studies (Kortholt et al., 2011).
Growth on bacteria and phagocytosis
We examined the growth of cells on shaken suspensions of heat-killed bacteria, which, like yeast, are taken up by phagocytosis. Wild-type cells grow with a doubling time of 3.4 hours, and there were no significant differences between wild type and any of the mutants. Even the PI3K quintuple mutant grew at nearly the same rate on bacteria as the wild type. Consistent with this, the uptake of 1.0 µm latex beads was very similar in all mutants compared to wild type, although there was some day to day variability in the results (supplementary material Table S3).
Growth on liquid medium and macropinocytosis
Wild-type cells grow much more slowly on liquid medium than on bacteria with a doubling time of 8.6 hours. The growth rates of the single mutants are only modestly impaired in shaken suspension compared to wild type, except for the PI3K4-null mutant, which grew at less than half the speed of wild type, with a mean generation time (MGT) of 21.3 hours. To confirm this result, we examined the growth of three other independent PI3K4 mutants, two made by insertion into the PI3K4 gene and one carrying a substantial deletion, and found that all grew with a doubling time of around 20 hours (mean for all four strains of 22 hours). Conversely, transformation of PI3K4– cells with a multi-copy plasmid carrying the PI3K4 coding sequence driven by its own promoter largely rescued the growth defect (MGT: 11.1 hours). The PI3K(1-2)– double mutant grew very slowly as well, indicating that this pair of genes is also crucial for growth, whereas the quintuple mutant barely grew at all, with a doubling time of more than 4 days (Fig. 3).
Fig. 3.
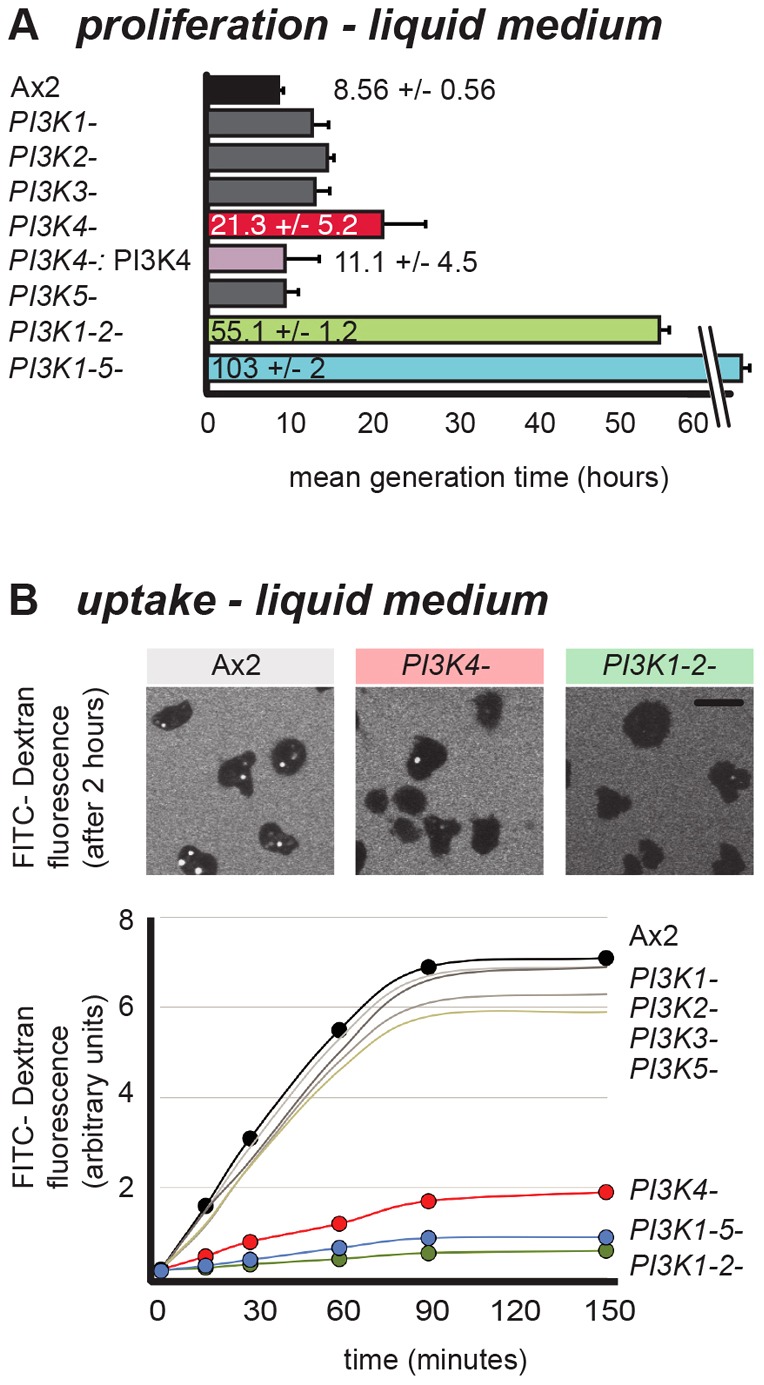
PI3K1/2 and PI3K4 control fluid-phase uptake. (A) Doubling times in liquid medium of PI3-kinase mutants and PI3K4− cells rescued with overexpressed PI3K4 (hours; mean ± s.d.; 4–17 independent experiments). (B) Uptake of FITC–dextran. Images of cells incubated for 2 hours with FITC–dextran show intracellular accumulation of this fluid-phase tracer. A representative time course of uptake for PI3-kinase mutants is shown. Rates of uptake can be found in the supplementary material Table S4. Scale bar: 10 µm.
Certain mutants, such as those lacking myosin II, are unable to grow in suspension because they cannot complete cytokinesis and form giant multi-nucleate cells, which eventually lyse (De Lozanne and Spudich, 1987). Although there was a modest increase in the number of nuclei per cell in the mutants with severely impaired growth, we did not observe giant cells. This and the absence of proliferation defects when grown on a bacterial diet indicates that impaired cytokinesis is not the cause of the poor growth of PI3K4– cells and the double and quintuple mutants in liquid medium (Fig. 3A).
Growth on liquid medium depends on nutrient uptake by macropinocytosis, in which fluid is internalized through large vesicles, processed through an intracellular pathway involving acidification and digestion, before the undigested remnants are expelled (Maniak, 2001). We therefore examined fluid-phase uptake, which is largely a measure of macropinocytosis, since only small volumes are internalized by clathrin-mediated endocytosis (Aguado-Velasco and Bretscher, 1999; Traynor and Kay, 2007). Cells were incubated with FITC–dextran, which is taken up at a constant rate for about 90 minutes, before a steady state is reached where exocytosis balances endocytosis. Of the single mutants, all were virtually indistinguishable from wild type, except for PI3K4– cells, which were severely impaired in fluid uptake (Fig. 3B). Defects in uptake can be rescued by expressing PI3K4 from a plasmid. Ras can allosterically activate PI3-kinases (Rodriguez-Viciana et al., 1996; Pacold et al., 2000; Funamoto et al., 2002), and abolishing this interaction leaves only limited basal enzymatic activity (Funamoto et al., 2002). A PI3K4 gene carrying a point mutation expected to block Ras binding, rescued much less efficiently, pointing to the importance of Ras in regulating PI3K4 activity in this context (see later; supplementary material Fig. S2).
Consistent with its poor growth, the double PI3K(1-2)– mutant also took up very little FITC–dextran, suggesting that PI3K1 and PI3K2 may have a partially redundant function (Fig. 3). Finally, and as expected, the PI3-kinase quintuple mutant was also very poor at fluid uptake.
These results show that PI3-kinases are necessary for proliferation of cells in rich medium in suspension and specifically for fluid uptake by macropinocytosis. It appears that PI3K4 and the pair PI3K1/2 play crucial, yet separate, roles in both processes.
PI3Ks have distinct roles in macropinocytosis
In order to understand the cellular basis for the distinct genetic requirement for PI3K1/2 and PIK4 in macropinocytosis, we first measured overall PtdIns(3,4,5)P3 levels in PI3K4– mutant cells growing in liquid medium. We found that also under these conditions, where cells attempt macropinocytosis, PtdIns(3,4,5)P3 levels were, if anything, greater in the mutant than wild type (151.9±36.5% of Ax2 cell levels; mean ± s.d., n = 3). Clearly, the PI3K4– mutant phenotype cannot be explained by reduced overall PtdIns(3,4,5)P3 levels.
We reasoned that differences in spatial patterns of PtdIns(3,4,5)P3 accumulation might be responsible for the phenotype of PI3K4– mutants. In PI3K(1-2)– cells, the PtdIns(3,4,5)P3 patches that mark incipient macropinosomes of the wild type are entirely missing. Only rarely are PhdA–GFP-labelled vesicles observed and these apparently do not derive from the plasma membrane. In contrast and surprisingly, PI3K4– cells produce PtdIns(3,4,5)P3 patches that are virtually indistinguishable from those of the wild type (Fig. 4A). Patches in Ax2 cells are typically 1.8±0.6 µm long with a PtdIns(3,4,5)P3-reporter intensity of 261±112% above cytoplasmic background (n = 32) and in PI3K4– cells they are 2±1.1 µm long with an intensity of 328±79% above cytoplasmic background (n = 33; Fig. 4B).
Fig. 4.
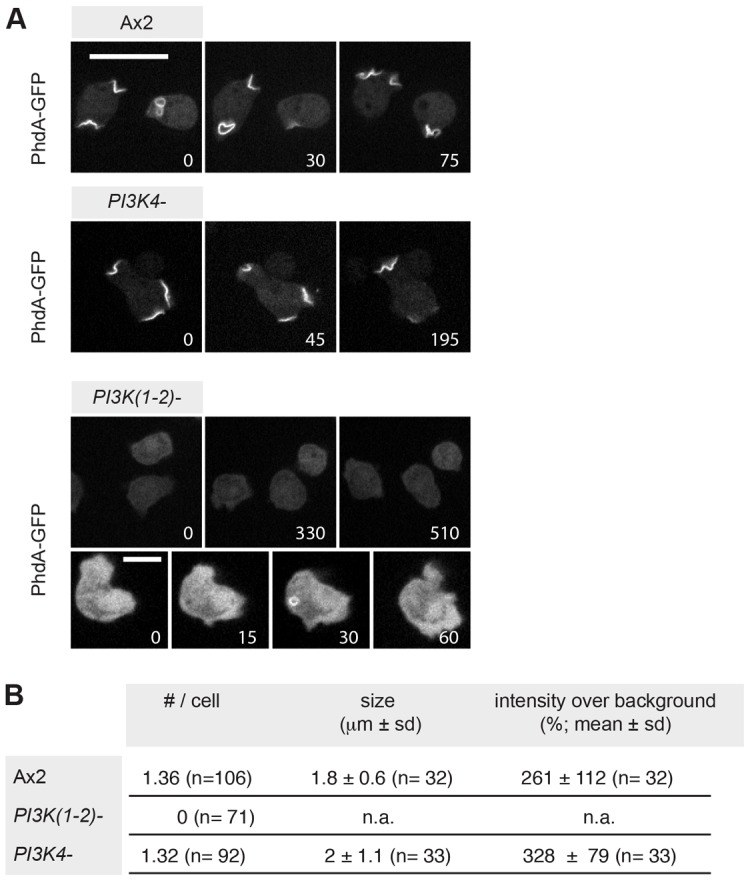
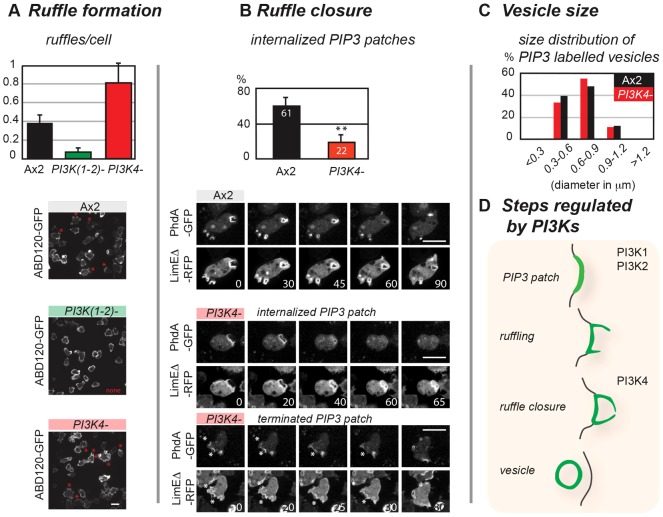
PI3K1 and PI3K2, but not PI3K4, are required to form PtdIns(3,4,5)P3 patches in the plasma membrane. (A) PtdIns(3,4,5)P3 patches are formed by Ax2 and PI3K4– cells, but PI3K(1-2)– cells do not form any such structures. Very rarely, labelled vesicles can be observed in these cells, but they do not seem to derive from the plasma membrane. PtdIns(3,4,5)P3 patches are identifiable using the PhdA–GFP reporter; numbers indicate time in seconds. Scale bars: 10 µm. (B) Properties of PtdIns(3,4,5)P3 patches formed by Ax2 and PI3K4– cells. Data extracted from images taken on several different days.
Wild-type cells constantly form thin sheet-like, actin-rich protrusions that can mature into closed cups and become internalized. These crowns and ruffles are labelled with the F-actin reporters ABD120–GFP (Pang et al., 1998; Lee and Knecht, 2002) or LimEΔ–RFP, which binds newly formed F-actin (Schneider et al., 2003), and colocalise with PtdIns(3,4,5)P3. We found on average 0.4 ruffles per cell at any one instant (n = 120) in wild-type but very few (7 in 120 cells) in PI3K(1-2)– cells. In contrast, PI3K4– cells had at least as many ruffles as wild type (average of 0.8 per cell; n = 120), showing that there is no defect in the earliest stage of macropinocytosis (Fig. 5A,D).
Fig. 5.
PI3-kinases regulate distinct steps in vesicle formation. (A) Ruffles depend on PI3K1 and PI3K2; ruffles (thin F-actin-rich projections) and open F-actin-rich cups were identified using ABD120–GFP as an F-actin reporter and counted (number/cell at any one time; mean ± s.d.) in Ax2 (0.4±0.1, n = 120), PI3K(1-2)– (0.01±0.05, n = 120) and PI3K4– (0.8±0.2, n = 120) cells on three different days. In the images, red asterisks label representative structures identified as ruffles and cups. (B) Transformation of PtdIns(3,4,5)P3 (PIP3) patches into closed macropinosomes depends on PI3K4: PtdIns(3,4,5)P3 patches, reported by PhdA–GFP, and used as a marker for nascent macropinosomes, were identified and their fate analysed in three independent time-lapse movies (1 frame/5 seconds) on at least two separate days. In Ax2 cells 61±9.9% (mean ± s.d.; n = 90) of those patches were clearly internalized but only 22±8.8% (n = 70) in PI3K4– cells. **P<0.01 using Student's two-tailed t-test. Still images of Ax2 show a PtdIns(3,4,5)P3 patch during internalization. PhdA–GFP colocalises with LimEΔ–RFP (a reporter for new F-actin) at sites where vesicles form. Once a vesicle has closed PhdA–GFP is lost before LimEΔ–RFP. Two cases are shown for PI3K4– cells: internalized PtdIns(3,4,5)P3 patches show the same sequence of events as in Ax2 cells, however, often PtdIns(3,4,5)P3 patches are terminated instead. In this case PtdIns(3,4,5)P3 patches do not continue on to form a vesicle, but disintegrate at the plasma membrane. Asterisks show two sites where a PtdIns(3,4,5)P3 patch does not develop further. (C) Vesicle size: this was determined by measuring the diameter of internalized PtdIns(3,4,5)P3-labelled vesicles. The histogram shows the frequency distribution of combined data from three different days (Ax2, n = 33; PI3K4–, n = 27). (D) Schematic diagram showing the sequence of events in macropinocytosis and the stages regulated by distinct PI3Ks. Numbers in time-lapse images indicate time in seconds. Scale bars: 10 µm.
To narrow down the macropinocytic defect of PI3K4– mutants, we examined the fate of PtdIns(3,4,5)P3. In wild-type cells at least 60% of all PtdIns(3,4,5)P3 patches were unambiguously converted to macropinosomes, but only a little over 20% in PI3K4– cells. Both PtdIns(3,4,5)P3 and F-actin also stayed associated with the vesicle as it is taken up, but after internalization, PtdIns(3,4,5)P3 disappeared from the vesicle first (Fig. 5B). The same sequence of events holds true for ‘productive’ PtdIns(3,4,5)P3 patches (those being internalized) in PI3K4– cells. In the case of ‘unproductive’ PtdIns(3,4,5)P3 patches, F-actin-rich ruffles also formed and colocalised, but no vesicle was pinched off. These data indicate that PI3K4 has a distinct role in macropinosome formation. It is required at some step in the conversion of F-actin-rich ruffles into sealed macropinosomes, after PI3K1/2 have formed patches of PtdIns(3,4,5)P3 (Fig. 5D). The diameter of PtdIns(3,4,5)P3-labelled vesicles after uptake was similar in wild-type and PI3K4– cells, suggesting that PI3K4 is not involved in determining vesicle size, but is probably involved in forming vesicles of any size (Fig. 5C).
PI3-kinase–Ras interaction map
We next set out to gain a better understanding of how these PI3K functions are regulated in the context of macropinocytosis. Each of the class-1 PI3Ks carries a RBD through which it may be activated by Ras–GTP. We have shown earlier that interactions with the RBD are required for the activity of PI3K4, and a similar result had been obtained independently for PI3K1 (Funamoto et al., 2002; Sasaki et al., 2007). Therefore, we sought to determine which Ras proteins regulate particular PI3-kinases, using pull-down assays between activated Ras and the PI3-kinase RBDs. The Dictyostelium genome encodes 14 Ras proteins, not all of which have been characterized, and so we focused on the six best-studied ones.
Each Ras isoform was expressed in the active, GTP-bound state, tagged with poly-histidine, pulled down with the appropriate recombinant RBD, and detected with a poly-histidine specific antibody. Results are standardised to Ras levels pulled-down by the promiscuous RBD of Byr2, with constitutively GDP-bound Ras serving as a control. The results of a typical blot are shown in Fig. 6A and the averaged values for several experiments in Fig. 6B. Of the Ras proteins, RasC bound very little to any of the PI3K RBDs, suggesting that it may couple to different effector proteins such as the Tor complex (Cai et al., 2010). PI3K1 and PI3K2 had similar interaction patterns, both strongly preferring (activated) RasG and RasS over the other Ras proteins. PI3K3 had a distinctly different pattern, binding to RasB, D and G and Rap1 similarly, but little to RasS. PI3K4 also had a distinct binding pattern, with a strong preference for RasG, but also some binding to the other Ras proteins (apart from RasC). PI3K5-RBD only bound at low levels to any of the Ras proteins.
Fig. 6.
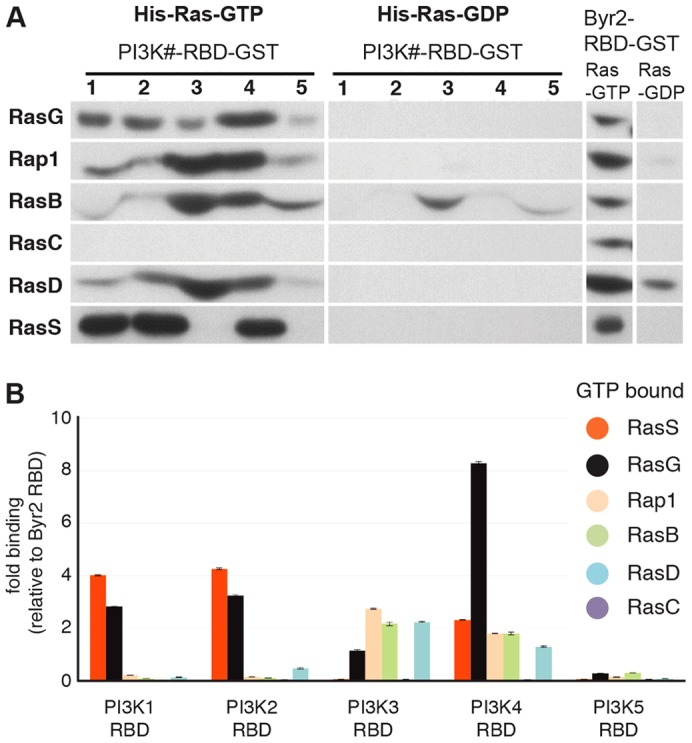
Regulators of PI3-kinases: a Ras–PI3K interaction map. (A) Representative blots showing the pull-down of His-tagged Ras proteins by GST-tagged Ras-binding domains (RBDs) of the individual PI3-kinases. (B) Analysis of the relative binding strengths of constitutively GTP-bound Ras–RBD pairs. Data are means ± s.d. of four independent experiments. Recombinant His-tagged Ras proteins (constitutively GTP- or GDP-bound versions; see Materials and Methods) were allowed to bind in vitro to GST-tagged, bacterially expressed RBDs of the individual PI3-kinases. The amount of bound Ras was detected by western blotting with an anti-His antibody and binding normalized to that of the unrelated, promiscuous RBD of Byr2 run as a standard in the same experiment.
These results point to RasG and RasS as the Ras proteins most likely to regulate the PI3-kinases genetically implicated in macropinocytosis (PI3K1, PI3K2 and PI3K4). We therefore tested macropinocytosis in null mutants of these two Ras genes. RasS-null cells have previously been reported to be defective in macropinocytosis (Chubb et al., 2000) yet strain-background-dependent inconsistencies (Pollitt et al., 2006) encouraged us to confirm this result with a different isolate (Ax2 background against DH1 background of the original isolate). The effect was smaller than in the original report yet still clear. We also found that rasG-null cells take up FITC at a reduced rate compared to wild type (38–51% of wild type, depending on genetic background). In contrast, a rasC mutant, took up fluid at wild-type rates (supplementary material Table S4).
Ras activity at sites of macropinocytosis
We used RBD-Raf1–GFP, a pan-Ras reporter (Sasaki et al., 2004), to determine whether Ras becomes activated at sites of macropinocytosis (Srinivasan et al., 2013). Indeed active Ras mirrors PtdIns(3,4,5)P3 and forms patches at sites where FITC–dextran is taken up. Similar to PtdIns(3,4,5)P3, it also only disappeared after the vesicle had pinched off from the plasma membrane (Fig. 7A).
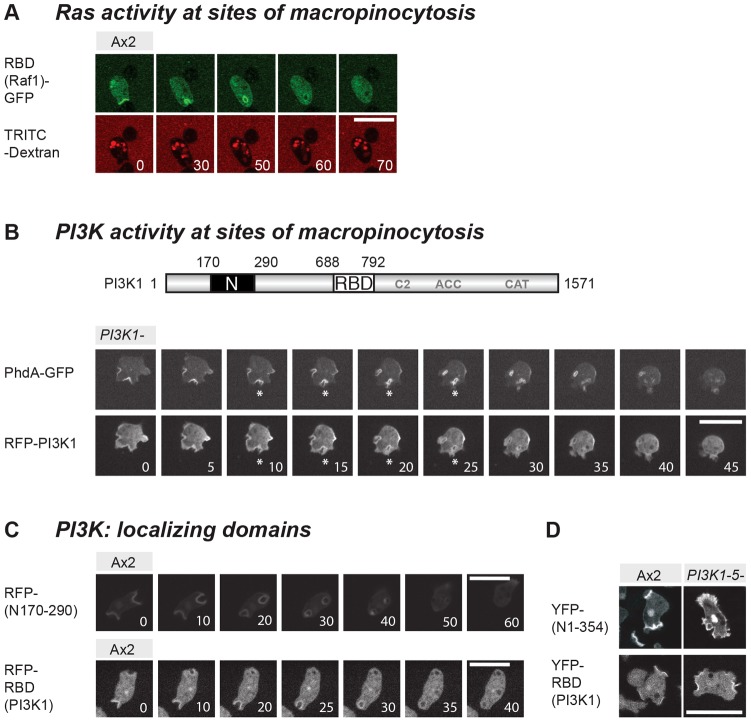
Fig. 7.
RasS and RasG regulate PI3K1 at sites of macropinocytosis. (A) The pan-Ras activity reporter RBD(Raf1)–GFP localizes to sites of macropinocytosome formation and remains associated with the vesicle for a period after it has closed. TRITC–dextran was used as a tracer for the fluid phase. (B) PI3K1 is localized to sites of macropinocytosis. Full-length PI3K1 colocalizes with its 3-phosphorylated lipid products (labelled with PhdA–GFP) at sites of macropinocytosis and remains associated for a period with the internalized vesicle. One example is labelled with an asterisk. RFP-tagged PI3K1 was expressed in a strain lacking endogenous PI3K1. On the domain map of PI3K1: N, N-terminal domain; RBD, Ras-binding domain; C2, C2 domain; ACC, accessory domain; CAT, catalytic domain. (C) Two distinct domains of PI3K1 localize to macropinosomes. RFP fusions of a minimal N-terminal stretch of 120 aa (aa 170–290) and of the Ras-binding domain (RBD: aa 688–792) both target macropinosomes. (D) Targeting of PI3K1 domains localizing to membrane patches does not depend on class-1 PI3-kinase activity. The N-terminal domain and the RBD were expressed in PI3K(1-5)– cells. Numbers indicate time in seconds. Scale bars: 10 µm.
In order to tie this activated Ras localization more closely to the PI3-kinases required for macropinocytosis, we created new reporters from PI3K1. Full-length PI3K1 fused to RFP colocalises with its product PtdIns(3,4,5)P3 both at the plasma membrane, and throughout the early life of a macropinosome (Fig. 7B). An N-terminal stretch of 350 amino acids (aa) is reported to target PI3K1 to the plasma membrane as without it, the truncated enzyme remains cytoplasmic. The N-terminus alone localizes to the cell periphery (Funamoto et al., 2002) and we narrowed down the targeting domain to an 120 aa stretch that still decorates a forming vesicle. Further dissection revealed the RBD as a second domain that is capable of localizing independently. RFP–RBD (PI3K1) labels macropinosomes, in a similar way to the pan-Ras reporter (Fig. 7C). We found that both the N-terminal domain as well as the RBD localize to the cell periphery independently of PI3-kinase activity (Fig. 7D), yet whereas recruitment of the N-terminus requires F-actin (lost in the presence of latrunculin A), the RBD accumulates independently of F-actin (supplementary material Fig. S3).
The association of the RFP–RBD (PI3K1) reporter with macropinosomes thus provides more direct evidence that PI3K1 is activated there by Ras, and (from the binding studies) most likely by RasG and/or RasS. Similar experiments involving PI3K4 were not successful because its isolated RBD failed to express in Dictyostelium cells.
Redundancy in Ras regulation of PI3-kinases in macropinocytosis
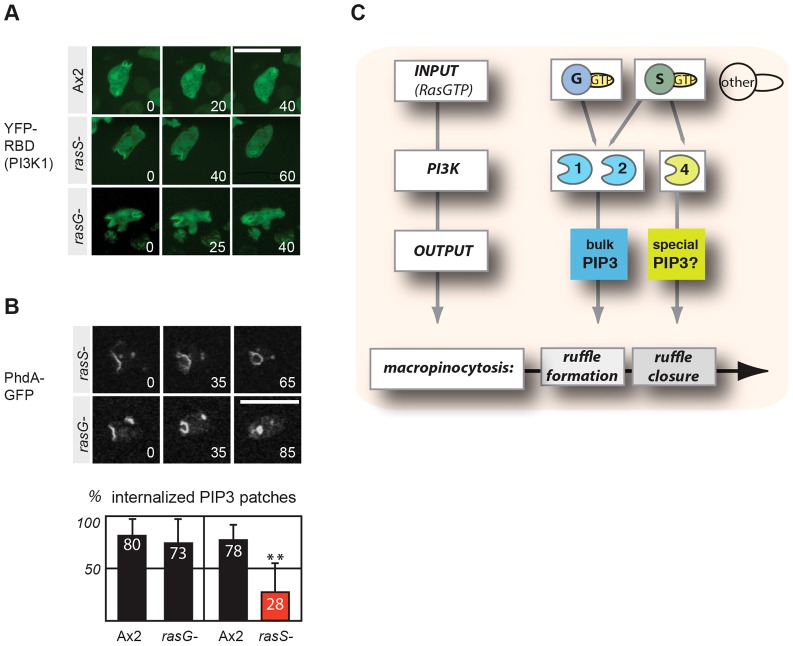
Finally, we attempted to separate the roles of RasG and RasS in macropinocytosis. First, we tested whether both Ras proteins are active at the site of macropinocytosis by examining the localization of YFP–RBD (PI3K1) in rasS- and rasG-null mutants, and found that it still localized to macropinosomes in both cases (Fig. 8A). Similarly, PtdIns(3,4,5)P3 patches still formed at sites of fluid-phase uptake in both rasG- and rasS-null cells. However rasS-null cells, like PI3K4– cells, converted fewer PtdIns(3,4,5)P3 patches into endosomes compared to Ax2, whereas rasG-null cells converted just as efficiently as wild-type cells (Fig. 8B). Hence, RasG and RasS show at least partial redundancy in supporting the formation of PtdIns(3,4,5)P3 patches, or perhaps compensate when one of them is missing, but the evidence suggests that RasS is the main regulator of PI3K4 in converting PtdIns(3,4,5)P3 patches into vesicles. We conclude by summarizing our findings in Fig. 8C, and also include the possibility that additional Ras proteins participate.
Fig. 8.
Redundancy in the regulation of PI3Ks in macropinocytosis. (A) YFP–RBD(PI3K1) reports sites of Ras input into PI3K1 and is still localized in rasS-null (rasS–) and rasG-null (rasG–) cells. (B) PtdIns(3,4,5)P3 (PIP3) patches form in both rasS– and rasG– cells and label sites of macropinocytosis. The fate of PtdIns(3,4,5)P3 patches (marked by phdA–GFP) was followed over time to calculate the percentage internalized (mean ± s.d.) in corresponding pairs of mutant and parent (Ax2: 80±17%, n = 66; rasG–: 73±23%, n = 26; Ax2: 78±14%, n = 50; rasS–: 28±28%, n = 75). **P<0.01(Student's two-tailed t-test) between means of Ax2 and rasS– cells. Data are from at least five independent movies recorded on three different days. Representative images are shown. Numbers indicate time in seconds. Scale bars: 10 µm. (C) A diagram of the proposed interactions of Ras and PI3K in the context of macropinocytosis.
Discussion
Our work shows that the fundamental role of class-1 PI3-kinases in growing Dictyostelium cells is in macropinocytosis, rather than in chemotaxis or phagocytosis of bacteria-sized particles. Macropinosomes are strongly decorated with PtdIns(3,4,5)P3 as they form, and macropinocytosis and proliferation on liquid medium are blocked in a mutant lacking all five class-1 PI3-kinases, in agreement with earlier work examining just a subset of these enzymes (Zhou et al., 1995; Zhou et al., 1998; Buczynski et al., 1997). Phagosomes forming around a yeast cell are also strongly decorated with PtdIns(3,4,5)P3, and it is therefore surprising to find that cells lacking all class-1 PI3-kinases can still phagocytose bacteria and 1 µm diameter particles as efficiently as wild-type cells, but in agreement with earlier work (Buczynski et al., 1997). There is evidence from inhibitor studies that uptake of larger particles (Cox et al., 1999) and yeast (Dormann et al., 2004) depends on PI3-kinase activity. The discrepancy between the uptake of small (bacteria) and larger (yeast) particles and the role that particular PI3Ks play in this context is currently under investigation although the situation appears to be complex and a resolution is beyond the scope of the present paper.
In contrast, we find that PtdIns(3,4,5)P3 patches are only sporadically associated with pseudopods in moving cells and that random movement, or chemotaxis to folic acid, is little affected in a mutant lacking all five class-1 PI3-kinases, consistent with inhibitor studies (Kortholt et al., 2011).
The multiplicity of PI3-kinases encoded in Dictyostelium and mammalian cells begs the question of whether these proteins have specialized functions or are generally redundant with each other (Vanhaesebroeck et al., 2010). Our work reveals that PI3-kinases participate at two genetically separable steps in macropinocytosis. Macropinosomes originate in patches of PtdIns(3,4,5)P3 on the plasma membrane, which throw off F-actin-rich crowns and ruffles, most likely from their margins rather than their body (Gerisch, 2010). We find that formation of these patches depends on PI3K1 and PI3K2, which appear to act redundantly, and are jointly responsible for producing the bulk of cellular PtdIns(3,4,5)P3 under the conditions used for measurement. Without these two enzymes, PtdIns(3,4,5)P3 patches are not formed, the number of F-actin ruffles is greatly reduced and few macropinosomes form, leaving the cells almost unable to grow on liquid medium (Zhou et al., 1995; Zhou et al., 1998; Buczynski et al., 1997; Rupper et al., 2001; Dormann et al., 2004).
PtdIns(3,4,5)P3-containing F-actin ruffles and crowns can go on to produce sealed macropinosomes. This process requires the appropriate extension of the ruffles, their bringing together, closure and eventual fusion of the membranes by means that are currently unknown. We postulate that PI3K4 is required at some point in this process, but do not know which. Without this enzyme, cells produce at least wild-type levels of PtdIns(3,4,5)P3 in liquid medium and this is organized into PtdIns(3,4,5)P3 patches as in the wild type. However, although membrane ruffles and crowns form efficiently in the mutant, they rarely go on to produce macropinosomes, resulting in greatly impaired fluid uptake and slow growth of cells on liquid medium.
These genetic results echo earlier inhibitor studies, which suggested that PI3-kinases might be involved in two distinct processes in macropinocytosis: in some cell types PI3-kinase inhibitors blocked the early events of actin polymerization and ruffling (Wennström et al., 1994), whereas in others, including Dictyostelium, it was suggested that macropinocytic cups still form, but recede without closing (Araki et al., 1996; Li et al., 1997; Rupper et al., 2001).
The role of PI3K4 in macropinocytosis is a continuing puzzle. The presence of at least wild-type levels of PtdIns(3,4,5)P3 in PI3K4– cells, both globally as well as locally, suggests a specialized function for PI3K4. Discrepancies between phenotypes of knockout and kinase dead gene replacements of mammalian PI3Kgamma revealed the potential for kinase independent functions of PI3Ks (Patrucco et al., 2004). However, preliminary experiments suggest that catalytic activity is necessary for PI3K4 to rescue the fluid-phase uptake in PI3K4– cells (unpublished observation).
Assuming that PI3K4 makes PtdIns(3,4,5)P3, this can only be a very small proportion of the total cellular PtdIns(3,4,5)P3 production (unless there is compensatory upregulation of other PI3-kinases in the PI3K4 mutant) and its disproportionate importance for macropinocytosis must depend on action at a specific time or place that is different from PI3K1 and PI3K2. PI3K1 colocalises with PtdIns(3,4,5)P3 throughout the formation of a macropinosome. Similarly, a specific localization of PI3K4 could give insight into its function: however, we found that GFP-tagged versions of the protein, although able to rescue growth and macropinocytosis of PI3K4– cells, had an uninformative distribution, being uniformly dispersed throughout the cytoplasm (unpublished observation).
Finally, in addition to its lipid kinase activity, PI3-kinases can also have protein kinase activity, which could be the relevant function of PI3K4 in macropinocytosis (Dhand et al., 1994; Bondeva et al., 1998). Although we can currently only speculate on the nature of the special function of PI3K4, it evidently depends on the regulation of the enzyme by Ras proteins.
Interaction with a specific Ras regulator may be important to determine the time, place and extent of PI3-kinase activity. A systematic investigation of this idea is only just beginning. In mammalian cells a subset of constitutively active Ras proteins stimulates both PI3Kgamma and PI3Kalpha activity, but not other PI3-kinase isoforms (Rodriguez-Viciana et al., 2004). Moreover, in vitro, GTP-bound Ras isoforms have distinct propensities to stimulate PtdIns(3,4,5)P3 formation by PI3Kgamma (Suire et al., 2002).
Our evidence suggests that Ras proteins regulate PI3-kinase activity during macropinosome formation. A general reporter for activated Ras locates to macropinosomes from their inception to just after vesicle closure, mirroring the localization of PtdIns(3,4,5)P3 itself, and a more specific reporter derived from the PI3K1 Ras-binding domain behaves in the same way. Previous work has implicated RasG and RasS in growth on liquid medium (Tuxworth et al., 1997; Chubb et al., 2000) and we find that mutants in either gene are impaired in macropinocytosis. Although pull-down experiments are semi-quantitative at best, they suggest that the activated forms of RasG and RasS bind preferentially to the Ras-binding domains of PI3K1, PI3K2 and PI3K4. It is difficult to further disentangle the precise roles of these two Ras proteins in macropinocytosis, but the mutant phenotype of RasS suggests that it may be a main regulator of PI3K4 in converting PtdIns(3,4,5)P3 patches to endocytic vesicles.
At a technical level, we have applied a new, sensitive mass-spectroscopic method (Clark et al., 2011) to measure PtdIns(3,4,5)P3 levels in Dictyostelium cells. Surprisingly, using this method we find that about 10% of wild-type levels of PtdIns(3,4,5)P3 remain in cells lacking all class-1 PI3-kinases. The source of the remaining PtdIns(3,4,5)P3 is currently unknown, but PikH, an unusual PI3-kinase with a PH-domain, is unlikely to be responsible, as a sextuple mutant with this gene also knockedout, has a similar residual level of PtdIns(3,4,5)P3 (unpublished observation). We hypothesize that, as in fission yeast, a metabolic pathway starting from PtdIns(3)P, generated by PikE, the Dictyostelium homologue of Vps34, may be responsible (Mitra et al., 2004).
In the course of this work we revisited domains that may localize Dictyostelium PI3-kinases. We found that the Ras-binding domain of PI3K1 can localize independently, a characteristic that has been overlooked previously (Funamoto et al., 2002). Unlike the previously reported N-terminal domain, which could bind to F-actin and whose targeting is inhibited by latrunculin A (Funamoto et al., 2002), recruitment of RFP–RBD is independent of F-actin. It is possible that the existence of these two binding domains in PI3K1 gives an additional layer of regulation. The RBD may target the enzyme to membranes, where its substrate PtdIns(4,5)P2 can be found, thus initiating PtdIns(3,4,5)P3 production, while the other domain may modulate this response once F-actin forms. Targeting by the RBD would provide a satisfying explanation for why, in latrunculin-A-treated cells, cyclic-AMP triggers normal PtdIns(3,4,5)P3 accumulation (Loovers et al., 2006).
In summary, our results show that macropinocytosis depends on two genetically separable steps mediated by PI3-kinases, which are in turn regulated by a subset of Ras proteins. This provides a firm genetic basis for future work in Dictyostelium and mammalian cells.
Materials and Methods
Cell growth and transformation
Dictyostelium strains were grown at 22°C in HL5 medium (Watts and Ashworth, 1970) either in tissue culture dishes or flasks shaken at 180 r.p.m. Growth curves were obtained in suspension using pre-formulated HL5 (Formedium, Hunstanton, UK) with cells counted using a Coulter Counter. Cells were grown on bacteria, either on SM plates with Klebsiella aerogenes or, for growth curves, in suspensions of heat-killed Escherichia coli B/r, which were grown overnight at 37°C in LB medium, harvested (3000 g, 15 minutes), washed three times in KK2 (20 mM KH2PO4/K2HPO4, 2 mM MgSO4, 0.1 mM CaCl2, pH 6.2), re-suspended at 1010 cells/ml in KK2 + 200 µg/ml di-hydrostreptomycin and heated to 60°C for 15 minutes. Dictyostelium cells, already growing on bacteria, were inoculated at 4×105 cells/ml, shaken at 180 rpm and growth monitored using a haemocytometer.
All PI3-kinase knockout strains were created in Ax2 cells (Kay lab) using methods described previously (Hoeller and Kay, 2007). Homologous recombinants were identified by screening for the loss of a smaller sized wild-type band in favour of a larger fragment, using the same primers as previously (Hoeller and Kay, 2007). The resulting strains are pikA (HM1135), pikB (HM1128), pikC (HM1161), pikF (HM1148) are pikG (HM1151). Double and quintuple PI3K mutants, as well as the Ras mutants have been described before (Hoeller and Kay, 2007; Chubb et al., 2000; Pollitt et al., 2006; Khosla et al., 2005; Bolourani et al., 2006).
Lipid measurements
PtdIns(3,4,5)P3 was measured by mass spectroscopy with each biological replicate consisting of triplicate samples of ∼1.7×106 cells in 170 µl of either KK2 or HL5 growth medium. Incubations were terminated by adding 750 µl of CHCl3/methanol/I M HCl (242/484/23.55 v/v) to create a single primary extraction phase. Lipids were then extracted, derivatised with trimethylsilyl diazomethane, and analysed by LC-ESI mass spectrometry, as described previously (Clark et al., 2011). The MRM transition for the PtdIns(3,4,5)P3 measured [1-O-hexadecyl-2-(11Z-octadecenoyl)-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′,5′-trisphosphate)] was m/z 1161.5>563.5 and the MRM transition for the PtdIns(4,5)P2 measured [1-O-hexadecyl-2-(11Z-octadecenoyl)-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate)] was m/z 1053.55>563.5. Data for the integrated ion currents for each species were corrected for protein concentration determined in parallel samples.
DNA constructs
PhdA-GFP, RBD-Raf1-GFP, RFP-LimEΔcoil, ABD120-GFP (Loovers et al., 2006; Sasaki et al., 2004; Fischer et al., 2004; Pang et al., 1998) and PI3K knockout constructs (Hoeller and Kay, 2007) were described previously. All other constructs were made by standard methods. The PI3K4 open reading frame was cloned into pDXA-3C containing an extended polylinker (pOH120); and PI3K4 (K515E; pOH141) was made from it. The PTEN ORF cloned into pDM451 gave PTEN-RFP (pHO313). The ORF of PI3K1 cloned into a pDXA-YFP variant, (YFP replaced with RFP) gave RFP-PI3K1 (pOH271). Fragments of PI3K1 were cloned into pDXA-YFP: RBD (aa 688–792) gave YFP-RBD(PI3K1) (pOH250) and similarly YFP-(N1-354) (pOH241) and YFP-(N170-290) (pHO57). An RFP-tagged variant of the minimal N-terminal localization domain was cloned into pDXA-RFP giving RFP-(N170-290) (pHO58). The RBD of PI3K1 was cloned into the same backbone giving RFP-RBD(PI3K1) (pHO62). The RBDs of Byr2 (aa 1–237), PI3K1 (aa 613–866), PI3K2 (aa 753–976), PI3K3 (aa 737–823), PI3K4 (aa 467–578), and PI3K5 (aa 692–840) were cloned into pGEX4T1. To make the 6xHis-tagged Rases: RasG (G12T) and RasG (aa 1–188); RasC (G13T) and RasC (aa 1–188); RasB (G15T) and RasB (aa 1–196); RasS (G12T) RasS (aa 1–193); Rap1 (Q65E) and Rap1 (aa 1–185); RasD (G12T) and RasD (aa 1–186) were cloned into pET-21a (Novagen) yielding His-Ras-GTP and His-Ras-GDP, respectively.
His–Ras–RBD(PI3K)–GST interaction analysis
All fusion proteins were expressed in E. coli BL21 DE3 (Invitrogen). His-tagged Ras fusion proteins were prepared from frozen bacterial pellets by resuspension at 2 ml per gram wet weight in 1× PB lysis buffer [50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0, 1% Triton X-100, 10% glycerol, 10 mM MgCl2, with two protease inhibitor tablets (Roche complete) per 50 ml], followed by lysozyme treatment (1 mg/ml, 30 minutes on ice) and sonication. Lysates were cleared by centrifugation and 1.5 ml of 50% Ni-NTA slurry (Qiagen) added per 4 ml suspension, then mixed for 2 hours at 4°C, packed into a column, washed twice with ice-cold wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and eluted four times with 500 µl elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). Eluted protein was dialyzed against PBS (2.7 mM KCl, 1.4 mM KH2PO4, 4.3 mm Na2HPO4, 137 mM NaCl, pH 7.5) with two exchanges overnight at 4°C and protein concentrations determined. GST–RBD fusion proteins were prepared essentially as described for other RBDs (Kae et al., 2004).
400 µg of purified His-tagged Ras protein was then incubated with 100 µg of GST–RBD on glutathione–Sepharose beads (Amersham Biosciences) and tumbled in binding buffer [10 mM sodium phosphate pH 7.2, 1% Triton X-100, 10% glycerol, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM Na3VO4, 5 mM NaF, with one tablet of protease inhibitor (Roche complete) per 50 ml buffer] at 4°C overnight. Beads were washed three times in ice-cold binding buffer and analysed by SDS-PAGE and western blots probed with anti-His monoclonal antibody (Santa Cruz Biotechnology, sc-8036).
Fluid-phase uptake and phagocytosis assays
Cells at 6×106/ml were shaken at 160 r.p.m. for 1 hour in HL5, then either FITC–dextran (molecular mass, 70 kDa; Sigma) was added to 1 mg/ml or 1.0 µm amine-modified polystyrene, fluorescent yellow-green latex beads (Sigma) were added to 300 beads/cell. At each time point, 2.0 ml aliquots were added to 10 ml ice-cold KK2, the cells pelleted (2000 r.p.m. for 5 minutes), washed once with 1 ml ice-cold KK2 containing 40 µg/ml Trypan Blue to quench external fluorescence and then twice with KK2. The final pellet was lysed in 0.2% Triton X-100 in 50 mM Na2HPO4 and fluorescence measured using a microplate fluorometer (Molecular Device, Sunnyvale) set at λEx = 485 nm and λEm = 535 nm. A variant method without quenching of external fluorescence was used for supplementary material Fig. S2.
Live-cell confocal microscopy and analysis of PtdIns(3,4,5)P3 patches
Cells were typically seeded in 200 µl HL-5 at 2–8×104 cells/chamber in a Lab-Tek II 8 well chamber (Nunc) and imaged at room temperature using either a Zeiss LSM510 confocal microscope with a Plan-Apochromat 63× 1.4 NA or a Nikon Eclipse Ti spinning disc, using excitation at the 488 nm and 543 nm laser lines for GFP and TRITC, respectively. TRITC–dextran (molecular mass 65–85 kDa; T1162, Sigma) was used at 1.7 µg/ml. Where necessary, contrast was adjusted uniformly using ImageJ or Photoshop and a uniform Gaussian Blur applied.
To follow the fate of PtdIns(3,4,5)P3 patches, cells were recorded for 100 frames at 1 frame/5 seconds. A patch is defined as a continuous stretch of pixels where each pixel >1.5× the intensity of the cytoplasm. Patches were measured in ImageJ with the curved line tool and metrics such as size and mean intensity of pixels over background extracted (Kortholt et al., 2011).
Random motility and folate chemotaxis
Random motility was measured with cells plated in HL5 medium in eight-well chambers (Nunc) and DIC movies recorded for 15 minutes (1 frame/15 seconds). Custom software was used to track cells (Zhu et al., 2011) with only cells that did not touch and could be tracked for >9 frames included in the analysis. For folate chemotaxis, cells were plated in 20% HL5 and at time 0 seconds, an Eppendorf Femtotip micropipette (Hamburg, Germany) filled with 25 mM folate was positioned in the field of view and cell movements monitored by time-lapse microscopy using an Olympus IX-70 inverted microscope fitted with a 20× objective at 2 frames/minute for 100 minutes and analysed using Volocity Software (Improvision, Coventry, England). Motility parameters were computed from the centroid of each cell: the instantaneous velocity was the distance travelled to the folate source divided by the tracking time and the chemotactic index as the net distance travelled towards the source of folate divided by the total distance travelled in that period.
Supplementary Material
Acknowledgments
We thank Anna Kielkowska for help with PtdIns(3,4,5)P3 measurements, Dictybase for strains and the Japanese cDNA consortium for cDNA clone SLF434.
Footnotes
Author contributions
O.H, G.W. and R.R.K. conceived the project. O.H., L.R.S., P.T.H., J.C., P.B., G.W. and R.R.K. designed the experiments. O.H., P.B., J.C., P.T.H. and R.R.K. executed experiments. All authors interpreted the data. O.H., G.W. and R.R.K. wrote the paper with help from P.B.
Funding
This work was supported by an EMBO Long Term fellowship [grant number ALTF 169-2008 to O.H.]; the National Institutes of Health [grant number GM084040 to O.D.W.]; the Canadian Institute for Health Research (G.W.); core support from the BBSRC in the form of a strategic programme grant to the Signalling ISP at Babraham [grant number BB J004456/1 to L.R.S. and P.T.H.]; and core funding from the Medical Research Councils [Reference U105115237 to R.R.K.]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.134015/-/DC1
References
- Aguado-Velasco C., Bretscher M. S. (1999). Circulation of the plasma membrane in Dictyostelium. Mol. Biol. Cell 10, 4419–4427 10.1091/mbc.10.12.4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki N., Johnson M. T., Swanson J. A. (1996). A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol. 135, 1249–1260 10.1083/jcb.135.5.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki N., Egami Y., Watanabe Y., Hatae T. (2007). Phosphoinositide metabolism during membrane ruffling and macropinosome formation in EGF-stimulated A431 cells. Exp. Cell Res. 313, 1496–1507 10.1016/j.yexcr.2007.02.012 [DOI] [PubMed] [Google Scholar]
- Bar-Sagi D., Feramisco J. R. (1986). Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 233, 1061–1068 10.1126/science.3090687 [DOI] [PubMed] [Google Scholar]
- Bolourani P., Spiegelman G. B., Weeks G. (2006). Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum. Mol. Biol. Cell 17, 4543–4550 10.1091/mbc.E05-11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeva T., Pirola L., Bulgarelli-Leva G., Rubio I., Wetzker R., Wymann M. P. (1998). Bifurcation of lipid and protein kinase signals of PI3Kgamma to the protein kinases PKB and MAPK. Science 282, 293–296 10.1126/science.282.5387.293 [DOI] [PubMed] [Google Scholar]
- Bosgraaf L., Keizer-Gunnink I., Van Haastert P. J. M. (2008). PI3-kinase signaling contributes to orientation in shallow gradients and enhances speed in steep chemoattractant gradients. J. Cell Sci. 121, 3589–3597 10.1242/jcs.031781 [DOI] [PubMed] [Google Scholar]
- Botelho R. J., Teruel M., Dierckman R., Anderson R., Wells A., York J. D., Meyer T., Grinstein S. (2000). Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 151, 1353–1368 10.1083/jcb.151.7.1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczynski G., Grove B., Nomura A., Kleve M., Bush J., Firtel R. A., Cardelli J. (1997). Inactivation of two Dictyostelium discoideum genes, DdPIK1 and DdPIK2, encoding proteins related to mammalian phosphatidylinositide 3-kinases, results in defects in endocytosis, lysosome to postlysosome transport, and actin cytoskeleton organization. J. Cell Biol. 136, 1271–1286 10.1083/jcb.136.6.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Das S., Kamimura Y., Long Y., Parent C. A., Devreotes P. N. (2010). Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J. Cell Biol. 190, 233–245 10.1083/jcb.201001129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardelli J. (2001). Phagocytosis and macropinocytosis in Dictyostelium: phosphoinositide-based processes, biochemically distinct. Traffic 2, 311–320 10.1034/j.1600-0854.2001.002005311.x [DOI] [PubMed] [Google Scholar]
- Chen L., Iijima M., Tang M., Landree M. A., Huang Y. E., Xiong Y., Iglesias P. A., Devreotes P. N. (2007). PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev. Cell 12, 603–614 10.1016/j.devcel.2007.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-L., Wang Y., Sesaki H., Iijima M. (2012). Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci. Signal. 5, ra10 10.1126/scisignal.2002446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J. R., Wilkins A., Thomas G. M., Insall R. H. (2000). The Dictyostelium RasS protein is required for macropinocytosis, phagocytosis and the control of cell movement. J. Cell Sci. 113, 709–719 [DOI] [PubMed] [Google Scholar]
- Clark J., Anderson K. E., Juvin V., Smith T. S., Karpe F., Wakelam M. J. O., Stephens L. R., Hawkins P. T. (2011). Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 8, 267–272 10.1038/nmeth.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M., Engel U., Giorgione J., Müller-Taubenberger A., Prassler J., Veltman D., Gerisch G. (2010). Curvature recognition and force generation in phagocytosis. BMC Biol. 8, 154 10.1186/1741-7007-8-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D., Tseng C. C., Bjekic G., Greenberg S. (1999). A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J. Biol. Chem. 274, 1240–1247 10.1074/jbc.274.3.1240 [DOI] [PubMed] [Google Scholar]
- De Lozanne A., Spudich J. A. (1987). Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236, 1086–1091 10.1126/science.3576222 [DOI] [PubMed] [Google Scholar]
- Dhand R., Hiles I., Panayotou G., Roche S., Fry M. J., Gout I., Totty N. F., Truong O., Vicendo P., Yonezawa K. et al. (1994). PI 3-kinase is a dual specificity enzyme: autoregulation by an intrinsic protein-serine kinase activity. EMBO J. 13, 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Weijer G., Dowler S., Weijer C. J. (2004). In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium phagocytosis and chemotaxis. J. Cell Sci. 117, 6497–6509 10.1242/jcs.01579 [DOI] [PubMed] [Google Scholar]
- Dowrick P., Kenworthy P., McCann B., Warn R. (1993). Circular ruffle formation and closure lead to macropinocytosis in hepatocyte growth factor/scatter factor-treated cells. Eur. J. Cell Biol. 61, 44–53 [PubMed] [Google Scholar]
- Eichinger L., Pachebat J. A., Glöckner G., Rajandream M-A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q. et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 10.1038/nature03481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson G. J., Milne L., Kulkarni S., Sasaki T., Walker S., Andrews S., Crabbe T., Finan P., Jones G., Jackson S. et al. (2007). PI(3)Kgamma has an important context-dependent role in neutrophil chemokinesis. Nat. Cell Biol. 9, 86–91 10.1038/ncb1517 [DOI] [PubMed] [Google Scholar]
- Fischer M., Haase I., Simmeth E., Gerisch G., Müller-Taubenberger A. (2004). A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium. FEBS Lett. 577, 227–232 10.1016/j.febslet.2004.09.084 [DOI] [PubMed] [Google Scholar]
- Funamoto S., Milan K., Meili R., Firtel R. A. (2001). Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in dictyostelium. J. Cell Biol. 153, 795–810 10.1083/jcb.153.4.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S., Meili R., Lee S., Parry L., Firtel R. A. (2002). Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109, 611–623 10.1016/S0092-8674(02)00755-9 [DOI] [PubMed] [Google Scholar]
- Gerisch G. (2010). Self-organizing actin waves that simulate phagocytic cup structures. PMC Biophys 3, 7 10.1186/1757-5036-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi S. I., Zvelebil M. J., Shuttleworth S. J., Hancox T., Saghir N., Timms J. F., Waterfield M. D. (2007). Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 404, 15–21 10.1042/BJ20061489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller O., Kay R. R. (2007). Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 17, 813–817 10.1016/j.cub.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Iijima M., Devreotes P. (2002). Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109, 599–610 10.1016/S0092-8674(02)00745-6 [DOI] [PubMed] [Google Scholar]
- Janetopoulos C., Ma L., Devreotes P. N., Iglesias P. A. (2004). Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 101, 8951–8956 10.1073/pnas.0402152101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetopoulos C., Borleis J., Vazquez F., Iijima M., Devreotes P. (2005). Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell 8, 467–477 10.1016/j.devcel.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Kae H., Lim C. J., Spiegelman G. B., Weeks G. (2004). Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 5, 602–606 10.1038/sj.embor.7400151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay R. R., Langridge P., Traynor D., Hoeller O. (2008). Changing directions in the study of chemotaxis. Nat. Rev. Mol. Cell Biol. 9, 455–463 [DOI] [PubMed] [Google Scholar]
- Khosla M., Spiegelman G. B., Weeks G. (2005). The effect of the disruption of a gene encoding a PI4 kinase on the developmental defect exhibited by Dictyostelium rasC(-) cells. Dev. Biol. 284, 412–420 10.1016/j.ydbio.2005.05.037 [DOI] [PubMed] [Google Scholar]
- Kortholt A., Kataria R., Keizer-Gunnink I., Van Egmond W. N., Khanna A., Van Haastert P. J. M. (2011). Dictyostelium chemotaxis: essential Ras activation and accessory signalling pathways for amplification. EMBO Rep. 12, 1273–1279 10.1038/embor.2011.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Knecht D. A. (2002). Visualization of actin dynamics during macropinocytosis and exocytosis. Traffic 3, 186–192 10.1034/j.1600-0854.2002.030304.x [DOI] [PubMed] [Google Scholar]
- Li G., D'Souza-Schorey C., Barbieri M. A., Cooper J. A., Stahl P. D. (1997). Uncoupling of membrane ruffling and pinocytosis during Ras signal transduction. J. Biol. Chem. 272, 10337–10340 10.1074/jbc.272.16.10337 [DOI] [PubMed] [Google Scholar]
- Lim J. P., Gleeson P. A. (2011). Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol. Cell Biol. 89, 836–843 10.1038/icb.2011.20 [DOI] [PubMed] [Google Scholar]
- Loovers H. M., Postma M., Keizer-Gunnink I., Huang Y. E., Devreotes P. N., van Haastert P. J. M. (2006). Distinct roles of PI(3,4,5)P3 during chemoattractant signaling in Dictyostelium: a quantitative in vivo analysis by inhibition of PI3-kinase. Mol. Biol. Cell 17, 1503–1513 10.1091/mbc.E05-09-0825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniak M. (2001). Fluid-phase uptake and transit in axenic Dictyostelium cells. Biochim. Biophys. Acta 1525, 197–204 10.1016/S0304-4165(01)00105-2 [DOI] [PubMed] [Google Scholar]
- Maniak M., Rauchenberger R., Albrecht R., Murphy J., Gerisch G. (1995). Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell 83, 915–924 10.1016/0092-8674(95)90207-4 [DOI] [PubMed] [Google Scholar]
- Marshall J. G., Booth J. W., Stambolic V., Mak T., Balla T., Schreiber A. D., Meyer T., Grinstein S. (2001). Restricted accumulation of phosphatidylinositol 3-kinase products in a plasmalemmal subdomain during Fc γ receptor-mediated phagocytosis. J. Cell Biol. 153, 1369–1380 10.1083/jcb.153.7.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P., Zhang Y., Rameh L. E., Ivshina M. P., McCollum D., Nunnari J. J., Hendricks G. M., Kerr M. L., Field S. J., Cantley L. C. et al. (2004). A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J. Cell Biol. 166, 205–211 10.1083/jcb.200404150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold M. E., Suire S., Perisic O., Lara-Gonzalez S., Davis C. T., Walker E. H., Hawkins P. T., Stephens L., Eccleston J. F., Williams R. L. (2000). Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–944 10.1016/S0092-8674(00)00196-3 [DOI] [PubMed] [Google Scholar]
- Pang K. M., Lee E., Knecht D. A. (1998). Use of a fusion protein between GFP and an actin-binding domain to visualize transient filamentous-actin structures. Curr. Biol. 8, 405–408 10.1016/S0960-9822(98)70159-9 [DOI] [PubMed] [Google Scholar]
- Parent C. A., Blacklock B. J., Froehlich W. M., Murphy D. B., Devreotes P. N. (1998). G protein signaling events are activated at the leading edge of chemotactic cells. Cell 95, 81–91 10.1016/S0092-8674(00)81784-5 [DOI] [PubMed] [Google Scholar]
- Patrucco E., Notte A., Barberis L., Selvetella G., Maffei A., Brancaccio M., Marengo S., Russo G., Azzolino O., Rybalkin S. D. et al. (2004). PI3Kgamma modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell 118, 375–387 10.1016/j.cell.2004.07.017 [DOI] [PubMed] [Google Scholar]
- Peracino B., Balest A., Bozzaro S. (2010). Phosphoinositides differentially regulate bacterial uptake and Nramp1-induced resistance to Legionella infection in Dictyostelium. J. Cell Sci. 123, 4039–4051 10.1242/jcs.072124 [DOI] [PubMed] [Google Scholar]
- Pollitt A. Y., Blagg S. L., Ibarra N., Insall R. H. (2006). Cell motility and SCAR localisation in axenically growing Dictyostelium cells. Eur. J. Cell Biol. 85, 1091–1098 10.1016/j.ejcb.2006.05.014 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Warne P. H., Dhand R., Vanhaesebroeck B., Gout I., Fry M. J., Waterfield M. D., Downward J. (1994). Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370, 527–532 10.1038/370527a0 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Warne P. H., Vanhaesebroeck B., Waterfield M. D., Downward J. (1996). Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 15, 2442–2451 [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Sabatier C., McCormick F. (2004). Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 24, 4943–4954 10.1128/MCB.24.11.4943-4954.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupper A., Lee K., Knecht D., Cardelli J. (2001). Sequential activities of phosphoinositide 3-kinase, PKB/Aakt, and Rab7 during macropinosome formation in Dictyostelium. Mol. Biol. Cell 12, 2813–2824 10.1091/mbc.12.9.2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. T., Chun C., Takeda K., Firtel R. A. (2004). Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167, 505–518 10.1083/jcb.200406177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. T., Janetopoulos C., Lee S., Charest P. G., Takeda K., Sundheimer L. W., Meili R., Devreotes P. N., Firtel R. A. (2007). G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 178, 185–191 10.1083/jcb.200611138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider N., Weber I., Faix J., Prassler J., Müller-Taubenberger A., Köhler J., Burghardt E., Gerisch G., Marriott G. (2003). A Lim protein involved in the progression of cytokinesis and regulation of the mitotic spindle. Cell Motil. Cytoskeleton 56, 130–139 10.1002/cm.10139 [DOI] [PubMed] [Google Scholar]
- Srinivasan K., Wright G. A., Hames N., Housman M., Roberts A., Aufderheide K. J., Janetopoulos C. (2013). Delineating the core regulatory elements crucial for directed cell migration by examining folic-acid-mediated responses. J. Cell Sci. 126, 221–233 10.1242/jcs.113415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suire S., Hawkins P., Stephens L. (2002). Activation of phosphoinositide 3-kinase gamma by Ras. Curr. Biol. 12, 1068–1075 10.1016/S0960-9822(02)00933-8 [DOI] [PubMed] [Google Scholar]
- Swaney K. F., Huang C-H., Devreotes P. N. (2010). Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys 39, 265–289 10.1146/annurev.biophys.093008.131228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. A. (2008). Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9, 639–649 10.1038/nrm2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Sasaki A. T., Ha H., Seung H-A., Firtel R. A. (2007). Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium. J. Biol. Chem. 282, 11874–11884 10.1074/jbc.M610984200 [DOI] [PubMed] [Google Scholar]
- Traynor D., Kay R. R. (2007). Possible roles of the endocytic cycle in cell motility. J. Cell Sci. 120, 2318–2327 10.1242/jcs.007732 [DOI] [PubMed] [Google Scholar]
- Tuxworth R. I., Cheetham J. L., Machesky L. M., Spiegelmann G. B., Weeks G., Insall R. H. (1997). Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 138, 605–614 10.1083/jcb.138.3.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. (2001). Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70, 535–602 10.1146/annurev.biochem.70.1.535 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. (2010). The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329–341 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- Veltman D. M., Keizer-Gunnik I., Van Haastert P. J. M. (2008). Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J. Cell Biol. 180, 747–753 10.1083/jcb.200709180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D. J., Ashworth J. M. (1970). Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 119, 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner O. D. (2002). Regulation of cell polarity during eukaryotic chemotaxis: the chemotactic compass. Curr. Opin. Cell Biol. 14, 196–202 10.1016/S0955-0674(02)00310-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennström S., Hawkins P., Cooke F., Hara K., Yonezawa K., Kasuga M., Jackson T., Claesson-Welsh L., Stephens L. (1994). Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr. Biol. 4, 385–393 10.1016/S0960-9822(00)00087-7 [DOI] [PubMed] [Google Scholar]
- Willingham M. C., Haigler H. T., Fitzgerald D. J., Gallo M. G., Rutherford A. V., Pastan I. H. (1983). The morphologic pathway of binding and internalization of epidermal growth factor in cultured cells. Studies on A431, KB, and 3T3 cells, using multiple methods of labelling. Exp. Cell Res. 146, 163–175 10.1016/0014-4827(83)90334-8 [DOI] [PubMed] [Google Scholar]
- Yeung T., Terebiznik M., Yu L., Silvius J., Abidi W. M., Philips M., Levine T., Kapus A., Grinstein S. (2006). Receptor activation alters inner surface potential during phagocytosis. Science 313, 347–351 10.1126/science.1129551 [DOI] [PubMed] [Google Scholar]
- Zhou K., Takegawa K., Emr S. D., Firtel R. A. (1995). A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vps34p PI 3-kinase homologs during growth and development. Mol. Cell. Biol. 15, 5645–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Pandol S., Bokoch G., Traynor-Kaplan A. E. (1998). Disruption of Dictyostelium PI3K genes reduces [32P]phosphatidylinositol 3,4 bisphosphate and [32P]phosphatidylinositol trisphosphate levels, alters F-actin distribution and impairs pinocytosis. J. Cell Sci. 111, 283–294 [DOI] [PubMed] [Google Scholar]
- Zhu J. W., Doan K., Park J., Chau A. H., Zhang H., Lowell C. A., Weiss A. (2011). Receptor-like tyrosine phosphatases CD45 and CD148 have distinct functions in chemoattractant-mediated neutrophil migration and response to S. aureus. Immunity 35, 757–769 10.1016/j.immuni.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.