SUMMARY
Increasing evidence links the gut microbiota with colorectal cancer. Metagenomic analyses indicate that commensal Fusobacterium spp. are associated with human colorectal carcinoma but whether this is an indirect or causal link remains unclear. We find that Fusobacterium spp. are enriched in human colonic adenomas relative to surrounding tissues and in stool samples from colorectal adenoma and carcinoma patients compared to healthy subjects. Additionally, in the ApcMin/+ mouse model of intestinal tumorigenesis, Fusobacterium nucleatum increases tumor multiplicity and selectively recruits tumor-infiltrating myeloid cells, which can promote tumor progression. Tumors from ApcMin/+ mice exposed to F. nucleatum exhibit a pro-inflammatory expression signature that is shared with human fusobacteria-positive colorectal carcinomas. However, unlike other bacteria linked to colorectal carcinoma, F. nucleatum does not exacerbate colitis, enteritis or inflammation-associated intestinal carcinogenesis. Collectively, these data suggest that, through recruitment of tumor-infiltrating immune cells, fusobacteria, generate a pro-inflammatory microenvironment that is conducive for colorectal neoplasia progression.
INTRODUCTION
There is accumulating evidence that members of the gut microbiota contribute to colorectal cancer, the second most incident cancer worldwide (Jemal et al., 2011). The majority of studies have focused on a small subset of colorectal cancers, colitis-associated colorectal cancers, and employed rodent pre-clinical models. Antibiotic-treatment or absence of the gut microbiota reduced tumor incidence in several mouse colitis-associated colorectal cancer models (Garrett et al., 2009; Uronis et al., 2009). Recently, two bacterial pathogens have been identified that promote colitis-associated colorectal cancer. Enterotoxigenic Bacteroides fragilis induces colitis and colonic tumors in ApcMin/+ mice by triggering a T helper type 17 (Th17) inflammatory response (Wu et al., 2009), while adherent-invasive Escherichia coli strain NC101 promotes colitis-associated colorectal cancer in monocolonized, azoxymethane-injected Il10−/− mice (Arthur et al., 2012). However, the majority of human colorectal cancers do not arise in the setting of inflammatory bowel disease.
The gut microbiota may be a driver in colorectal cancers that are not associated with colitis. Metagenomic analyses using whole-genome sequencing (Kostic et al., 2012), transcriptome sequencing (Castellarin et al., 2012), or bacterial 16S ribosomal RNA gene DNA sequencing (Kostic et al., 2012; Marchesi et al., 2011) have shown enrichment of Fusobacterium species in colorectal cancers relative to adjacent normal tissue. However, it has been unclear whether these findings represent an indirect association, or whether Fusobacterium spp. functionally contributes to colorectal cancer (CRC) tumorigenesis (Tjalsma et al., 2012).
Here, we assess the association of F. nucleatum in stool and colonic tissue from patients with colorectal adenomas and adenocarcinomas and analyze the affect of F. nucleatum on intestinal cancer progression and on tumor inflammation in mouse models. Our data are consistent with the possibility that F. nucleatum potentiates non-colitis associated intestinal tumorigenesis.
RESULTS
Fusobacteria are enriched in colonic adenomas and in stools samples from patients with adenomas and colorectal carcinomas
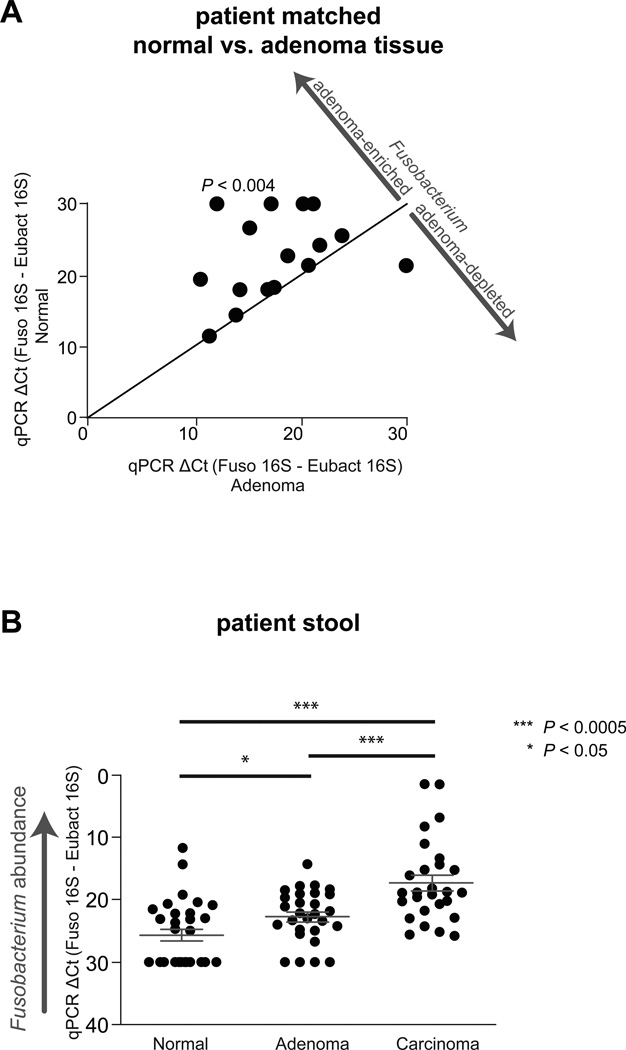
Colonic adenomas are neoplastic epithelial lesions that can become malignant and are precursors of the majority of sporadic colorectal cancers. Given the enrichment of Fusobacterium spp. in colorectal tumor versus adjacent normal tissue, we examined if there was a similar enrichment in adenomas, suggesting the involvement of Fusobacterium in neoplastic initiation or progression, prior to the establishment of carcinoma. A few recent studies have performed case control studies of colorectal adenomas using 16S rRNA gene surveys with one noting fusobacterial enrichments in their patient cohort (McCoy et al., 2013; Sanapareddy et al., 2012; Scanlan et al., 2008; Shen et al., 2010). We measured Fusobacterium spp. abundance in paired adenoma tissue versus adjacent normal tissue from the same patient drawing samples from several geographic locations and registries: the Cooperative Human Tissue Network (Eastern, Southern, and Western divisions of the United States), Massachusetts General Hospital, and University of Aberdeen School of Medicine. None of these patients had a history of inflammatory bowel disease (IBD). We found that Fusobacterium was detectable by qPCR in 48% of adenomas (n=29), and in those cases that were positive, Fusobacterium was enriched in adenomas relative to surrounding tissue (P < 0.004) (Figure 1A and Table S1). These data suggest that Fusobacterium begin to accumulate at early stages of colonic tumorigenesis in some groups of patients.
Figure 1. Fusobacterium is enriched in adenoma versus adjacent normal tissue and detected at a higher abundance in stool from CRC and adenoma cases than from healthy controls.
(A)Fusobacterium abundance for normal tissue (x-axis) vs adenoma (y-axis) is plotted. 29 matched adenoma normal tissues pairs were tested. Each symbol represents data from one patient (adenoma and normal tissue). (B) Fecal Fusobacterium abundance from healthy subjects (n=30), subjects with colorectal adenomas (n=29), and colorectal cancer (n = 27). See also Table S1.
Next, we determined whether fusobacteria have a higher overall abundance in the fecal microbiota of CRC patients relative to healthy controls in a case-control experiment. We compared the levels of Fusobacterium spp. to universal Eubacteria 16S by quantitative qPCR of stool. Subjects provided their stool samples to the Early Detection Research Network (EDRN) prior to bowel preparation and screening colonoscopy and had no prior history of colorectal cancer or gastrointestinal disease (Figure 1B and Table S1). Fusobacterium spp. were enriched in CRC patients (P < 1×10−5) and were also enriched in stool from subjects with adenomas as compared to healthy controls (P < 5×10−3). These results indicate that increased abundance of Fusobacterium species in the gut microbiota may be a general feature of colonic tumorigenesis.
Fusobacterium nucleatum promotes intestinal tumorigenesis
The enrichment of Fusobacterium spp. in colorectal adenomas and the previously reported enrichment in colorectal carcinomas (Castellarin et al., 2012; Kostic et al., 2012; Marchesi et al., 2011) prompted us to examine whether Fusobacterium could accelerate tumorigenesis in mice. We utilized mice which develop intestinal tumors because of a mutation in one copy of the tumor suppressor gene Apc (C57BL/6 ApcMin/+) or genetic defects resulting in chronic intestinal inflammation (BALB/c Il10−/− and BALB/c T-bet−/− X Rag2−/−). We introduced human isolates of F. nucleatum, Streptococcus species (S. anginosus, S. parasanguinis and S. sanguinis, n=4 mice per strain), or tryptic soy broth (TSB) to ApcMin/+ mice; and F. nucleatum or TSB to IL10−/− and T-bet−/− X RAG2−/− mice. Streptococci were used as control strains because of the longstanding association of streptococcal species with occult colonic malignancies.
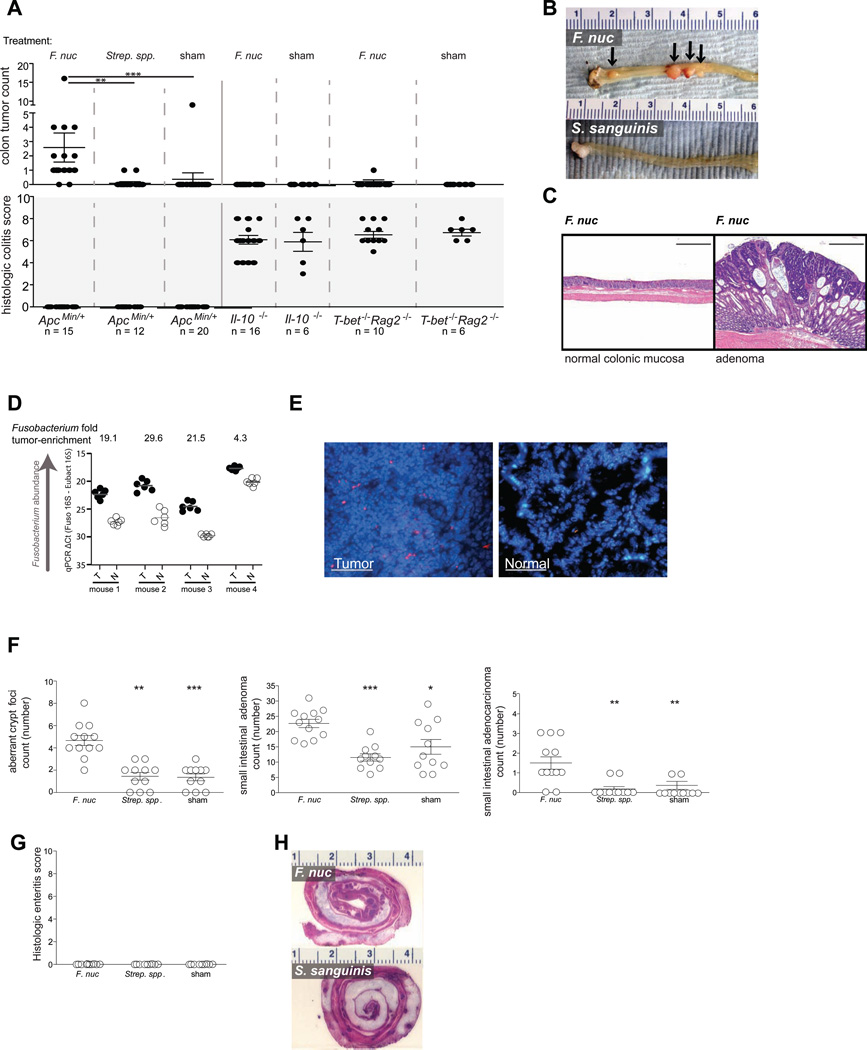
Introduction of F. nucleatum into ApcMin/+ mice accelerated onset of colonic tumors. ApcMin/+ mice fed F. nucleatum developed a significantly higher number of colonic tumors as compared to ApcMin/+ mice fed Streptococcus (Strep.) spp., or TSB (P < 0.001, P < 0.0001) (Figure 2A – C). However, F. nucleatum did not induce colitis (Figure 2A – C) in contrast with enterotoxigenic Bacteroides fragilis, which causes colitis and accelerates tumorigenesis in ApcMin/+ mice (Wu et al., 2009). Also, F. nucleatum neither exacerbated intestinal inflammation nor accelerated tumorigenesis in the two mouse models of colitis-associated colorectal cancer examined (Figure 2A). Consistent with prior findings in human colorectal carcinoma (Castellarin et al., 2012; Kostic et al., 2012), F. nucleatum was culturable from ApcMin/+ tumors and enriched in tumor tissue relative to adjacent normal tissue in F. nucleatum fed ApcMin/+ mice as assayed by qPCR (Figure 2D) and fluorescence in situ hybridization using a Fusobacterium 16S rRNA-directed probe (Figure 2E). However, we did not observe an overall increase in bacterial tumor load in F. nucleatum fed ApcMin/+ mice (Figure S1).
Figure 2. Fusobacterium nucleatum promotes small and large intestinal tumorigenesis and is enriched in tumor tissues of ApcMin/+ mice.
(A) Colon tumor counts and histologic colitis scores from ApcMin/+, Il10–/– and T-bet–/– X Rag2–/– mice fed F. nucleatum (F. nuc), Streptococcus spp., or TSB control. Mice were started on the 8-week feeding regimen at 6 weeks of age. (B) Representative images of colons (ruler numbers in cm) and (C) colonic histological analysis of ApcMin/+ mice (100 icrometer scale bar). (D) Fusobacterium abundance in matched tumor (T) versus normal (N) tissues from colons of ApcMin/+ mice fed F. nucleatum measured by quantitative PCR. (E) Representative FISH images of colonic tumor and matched normal colonic tissue from an ApcMin/+ mouse fed F. nucleatum using a Fusobacterium 16S rDNA-directed probe (50 icrometer scale bar). (F) Small intestinal aberrant crypt foci, adenoma, and adenocarcinoma counts and (G) histologic enteritis scores in ApcMin/+ mice fed F. nucleatum, Streptococcus species, or TSB control. (H) Representative sections of small intestines from ApcMin/+ mice (ruler numbers in cm). *** P < 0.0001, ** P < 0.001, * P < 0.01. See also Fig. S1.
ApcMin/+ mice fed F. nucleatum also had a significantly higher count of small intestinal aberrant crypt foci, adenomas, and adenocarcinonas, versus Strep. spp, or TSB–fed controls. In addition, we did not observe any small intestinal enteritis in these mice. (Figure 2F – H) Together, these results indicate that F. nucleatum may accelerate tumorigenesis in the absence of enteritis, or macroscopic inflammation, in ApcMin/+ mice.
F. nucleatum selectively expands myeloid-derived immune cells
Immune cells and their effectors are key components of tumors and promote neoplastic progression. To address whether F. nucleatum contributes to tumorigenesis by affecting intratumoral immune cells, we characterized tumor infiltrating immune cells from the intestinal tumors of ApcMin/+ mice that were fed F. nucleatum or S. sanguinis for 8 weeks, or were not fed bacteria over the same time frame. Small intestinal rather than colonic tumors were used, because controls did not develop sufficient numbers of colon tumors within the experimental timeframe.
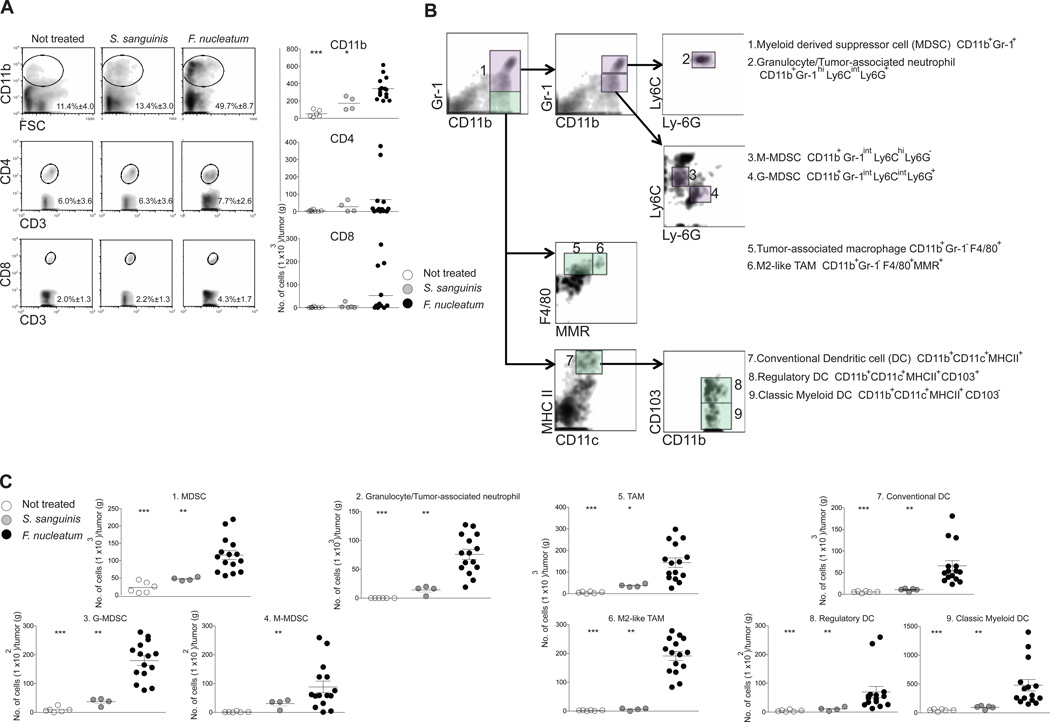
We observed an increase in infiltrating cells of the myeloid lineage in the tumors from Fusobacterium-treated mice. CD11b+ myeloid cells (mean 3.2X higher cell number, mean 4.0X higher % population) increased in the tumors of ApcMin/+ mice fed F. nucleatum as compared to controls, while numbers of CD3+CD4+ and CD3+CD8+ T lymphocytes were not significantly different (Figure 3A).
Figure 3. F. nucleatum selectively expands myeloid-derived immune cells, but not lymphoid immune cells in the intestinal tumor microenvironment.
Multicolor flow cytometric analyses: (A) Percentage (left panel) with representative density plots and the number (right panel) of intratumoral myeloid cells and lymphoid cells. Data for CD11b+ myeloid cells, CD3+CD4+ T cells, or CD3+CD8+ T cells are shown. Mean percentages ± s.e.m are shown within each plot. n= 6, 4, or 15 for not treated, S. sanguinis, or F. nucleatum. *** P < 0.001, ** P < 0.01, * P < 0.05 (B) A schematic of the gating strategy for subset identification of intratumoral myeloid cells. Representative density plots of myeloid cells are shown. The initial analysis region used CD11b vs. Gr-1 to identify CD11b+Gr-1+ MDSC population (violet boxes) and CD11b+Gr-1macrophage population (light green boxes). Arrows show the gating progression used to discriminate the 9 subsets. Each number on the density plot represents the corresponding subset of myeloid cells. (C) Cell number/gram tumor for myeloid cells from the treatment groups. Each symbol represents data from an individual mouse. See also Fig. S2.
Differentiated CD11b+ myeloid cells such as macrophages, dendritic cells (DCs) and granulocytes play an important role in promoting tumor progression and angiogenesis (Coussens and Pollard, 2011). Thus, we defined the different subsets of tumor infiltrating myeloid cells by multicolor flow cytometry using a gating strategy that employed 9 cell surface molecules (Figure 3B) as well as functional properties including arginase-1 and inducible nitric oxide synthase (iNOS) expression and T cell suppressive activity (Figure S2). Myeloid-derived suppressor cells (MDSCs) are tumor-permissive myeloid cells with potent immune suppressive activity (Gabrilovich et al., 2012). MDSCs were enriched (mean 3.7X increased cell number) in F. nucleatum fed mice vs controls (Figure 3C). There are two principal MDSC subsets, monocytic (M-MDSC) and granulocytic (G-MDSC), both of which increased in the tumors of F. nucleatum fed mice (M-MDSCs, mean 6.7X and G-MDSCs, mean 8.9X increased cell number) compared to controls (Figure 3C). These MDSC subsets suppress CD4 T cells predominantly via expression of arginase-1 and iNOS (Gabrilovich et al., 2009). Consistent with published phenotypes (Gabrilovich et al., 2009), M-MDSC expressed higher levels of arginase-1 and iNOS than G-MDSC (Figure S2A). Additionally, MDSC showed significant T cell suppressive activity (Figure S2C). Recent studies support that tumor-associated neutrophils (TANs) play a role in tumor progression and angiogenesis and in the modulation of the antitumor immunity (Mantovani et al., 2011). We found that numbers of granulocytes/TANs increased by a mean 13.4 X in tumors from F. nucleatum fed mice compared to controls (Figure 3C).
Substantial experimental data from clinical and pre-clinical studies indicate that tumor-associated macrophages (TAMs) promote tumor progression and metastasis (Mantovani and Sica, 2010; Qian and Pollard, 2010). Both TAMs and M2-like TAMs were enriched in F. nucleatum fed mice as compared to controls (mean 7.8X and 50.8X increased cell number, respectively) (Figure 3C). TAMs inhibit T cell responsiveness via expression of arginase-1 (Ruffell et al., 2012). M2-like TAMs had two-fold higher arginase-1 levels compared to TAMs and exhibited significant suppressive activity on CD4 T cells (Figure S2B–C).
Within tumors, DCs can either dampen or promote anti-tumor immunity (Mellman et al., 2011). The intestine possesses a specific subset of DCs expressing CD103 integrin. These cells play a role in regulation of immune responses by promoting the expansion of Foxp3+ regulatory T cells, a CD4+ T cell subset that suppresses cytotoxic and effector T cells and thus dampen anti-tumor immunity (Coombes et al., 2007; Josefowicz et al., 2012). We observed that numbers of conventional DCs were increased in tumors from F. nucleatum fed mice (mean 8.4X higher cell number) and found increases in classical myeloid DCs and CD103+ regulatory DCs (mean 4.5X and mean 6.9X increased cell number, respectively) compared to tumors from control mice (Figure 3C). To determine whether the changes in TAMs, MDSCs, and DCs were affecting CD4+ T cell subsets implicated in colon tumorigenesis (Grivennikov et al., 2012), we examined both intratumoral T-helper 17 (Th17) cells and CD4+ Foxp3+ regulatory T cells. While there was a trend towards an increase in Foxp3+ regulatory T cells (Treg) and Th17 cells in F. nucleatum fed mice, there was significant heterogeneity within this group, that did not correlate with intratumoral F. nucleatum abundance, and differences were not statistically significant (Figure S2D). Additionally, there was no positive or negative correlation between Th17 and Treg cell numbers in the same mice. Collectively, these data support that F. nucleatum modulates the tumor immune microenvironment and results in expansion of myeloid-derived immune cell types, that have been reported to promote tumor progression (Qian and Pollard, 2010).
A Fusobacterium-associated human colorectal cancer gene signature shared and validated in mice
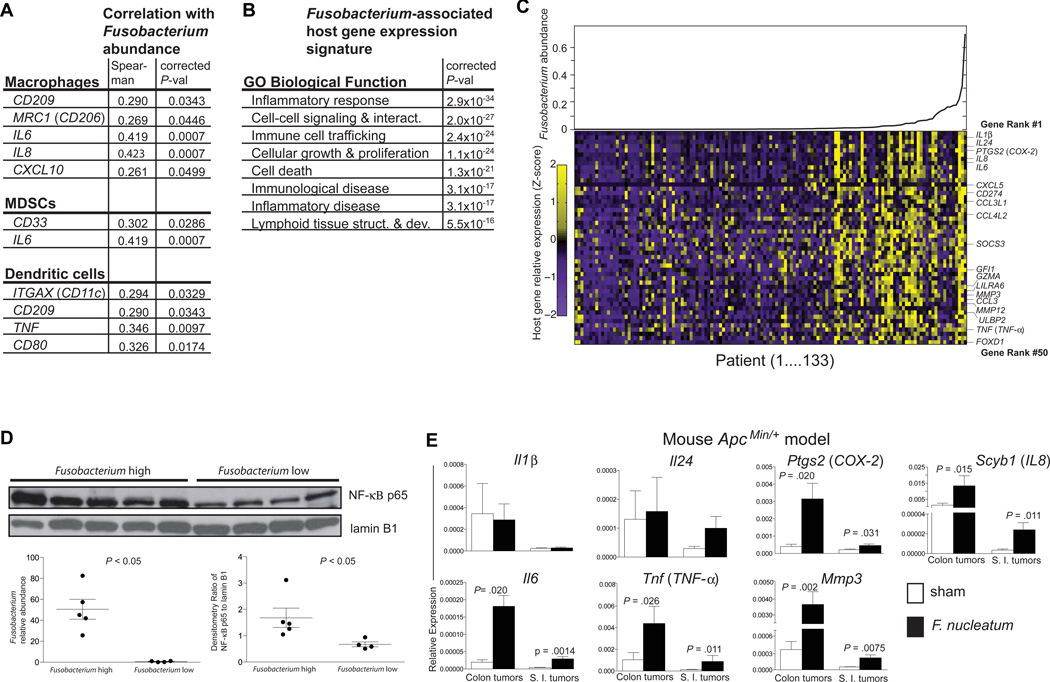
Given our findings of F. nucleatum-induced myeloid-derived cell expansion in mouse intestinal tumors, we asked if there would also be a similar immunological profile in human Fusobacterium-associated colon tumor transcriptomic data. We utilized a data set of deep transcriptome sequencing (i.e. RNA-Seq) of 133 colon tumors generated by The Cancer Genome Atlas (Cancer Genome Atlas Network, 2012). These patients all had colorectal cancer, including stage I–IV, and none of these patients had IBD-associated colorectal cancer or a history of IBD. All non-human sequencing reads were applied to PathSeq, a computational tool that identifies and quantifies the abundance of all bacteria present in each tumor http://www.broadinstitute.org/software/pathseq/ (Kostic et al., 2011). By calculating the Spearman’s rank correlation coefficient of the relative abundance of Fusobacterium spp. transcripts with host gene expression in this dataset, we identified a Fusobacterium-associated human CRC gene expression signature. We found a correlation of immune cell marker genes associated with TAM (CD209, CD206/MRC1, IL-6, IL-8, and CXCL10), MDSC (CD33 and IL-6), and DC (CD11c/ITGAX, CD209, TNF, CD80) with Fusobacterium abundance in human Fusobacterium-associated tumors by RNA-Seq, similar to our findings with Fusobacterium in mouse flow cytometry experiments (Figure 4A). Analysis using the Ingenuity IPA gene ontology module revealed that the Fusobacterium-associated human CRC gene expression signature was highly enriched for the inflammatory response gene ontology category (corrected P < 1×10−33; Ingenuity analysis) as well as other categories also related to immune and inflammatory disease (Figure 4B). To test the specificity of this correlation, we performed the same analysis with the other top-four highly abundant genera besides Fusobacterium (Bacteroides, Escherichia, Streptococcus, and Propionobacterium), but none of these other genera were associated with enrichment for inflammation-related gene functions, nor were they associated with comparably high gene ontology enrichments in any other functional categories (Figure S3). The relative abundance of Fusobacterium spp. transcripts are shown for each of the 133 TCGA colon tumors with scaled expression values for the top 50 ranked genes denoted as the row Z-score in a heat map (Figure 4C). Many of the the top ranked Fusobacterium-associated genes, PTGS2 (COX-2), IL1β, IL6, IL8, and TNF (TNF- α), have not only been investigated in colorectal carcinogenesis but also are induced by Fusobacterium in co-culture with human and mouse cell lines in vitro (Dharmani et al., 2011; Trinchieri, 2012).
Figure 4. A Fusobacterium-associated human colorectal cancer gene signature shared and validated in mice.
(A) Immune cell types enriched in Fusobacterium-associated mouse tumors are shown with the human marker gene utilized to determine abundance in the TCGA CRC RNA-seq data set. (B) Ingenuity Pathway Analysis Biological Function Gene Ontology categories enriched for Fusobacterium abundance-correlating gene sets are shown. (C) Fusobacterium spp. transcript relative abundance are plotted for the 133 TCGA colon tumors (upper panel and lower panel x-axis) and scaled expression values for the top 50 ranked genes denoted as the row Z-score (y-axis) are shown in a heat map (lower panel) with a purple (low expression)–yellow (high expression) color scale. (D) Western blot of nuclear extracts from human colon cancer with a high or low Fusobacterium relative abundance. Fusobacterium relative abundance and western blot densitometry are shown in the lower panel. (E) qPCR analysis of a selection of the top 50 ranked genes in (B) in colon and small intestinal tumors from F. nucleatum vs TSB fed ApcMin/+ mice. Data are represented as mean +/− SEM. Tumors from 6–9 mice per group were used. See also Fig. S3.
The expression signature, notable for PTGS2 (COX-2), IL1β, IL6, IL8, TNF (TNF- α) and MMP3, was suggestive of an NF-κB-driven pro-inflammatory response (Hayden et al., 2012 and http://www.bu.edu/nf-kb/gene-resources/target-genes/). As NF-κB has been identified as a central link between inflammation and cancer (DiDonato et al., 2012), we assessed whether there was a correlation between increased NF-κB activation and Fusobacterium abundance in human colorectal cancers. We obtained freshly resected human colorectal cancer samples and generated nuclear extracts from these samples. After stratifying the samples by their abundance of Fusobacterium spp., we performed western blots on the nuclear extracts from Fusobacterium high and low tumors to examine NF-κB activation. NF-κB was indeed more activated (increased nuclear translocation of the p65 NF-κB subunit) in tumors with a high vs low Fusobacterium abundance (Figure 4D). Both Fusobacterium relative abundance) (left lower panel) and the densitometry ratio of NF-κB to laminB1 (right lower panel) for each individual’s tumor are shown.
While our RNA sequencing and immunoblotting analyses suggested a strong association between intratumoral Fusobacterium abundance and specific immune pathways and genes, we wanted to assess whether this association is direct, by examining gene expression patterns of small intestinal and colonic tumors from ApcMin/+ mice exposed to F. nucleatum, compared to tumors from mice without Fusobacterium exposure. We found that most of the mouse homologs of Fusobacterium-associated pro-inflammatory genes identified in human tumors, including Ptgs2 (COX-2 mouse homolog), Scyb1 (IL8 mouse homolog), Il6, Tnf (TNF, and Mmp3, were more highly expressed in both small intestinal and colonic tumors from mice that were treated with F. nucleatum compared to tumors from mice treated with TSB (Figure 4E). In general, expression levels of these human Fusobacterium-associated pro-inflammatory genes were higher in the colonic tumors than small intestinal tumors; this may be intrinsic to the anatomic site and be related to the fact that the gene list was derived from human colorectal cancer.
DISCUSSION
Entry of microbes and microbial products into the evolving tumor microenvironment potentiates tumor growth by eliciting tumor-promoting immune cell responses (Jobin, 2012). Our results demonstrate that Fusobacterium spp., rare gut microbiome constituents in the healthy human population (Segata et al., 2012), are found at increased abundance in the stool of patients with adenomas and colorectal cancer, and are enriched in adenomas and adenocarcinomas relative to non-involved colonic tissues. Introduction of F. nucleatum to ApcMin/+ mice resulted in accelerated small intestinal and colonic tumorigenesis, infiltration of specific myeloid cell subsets into tumors, and an NF-κB proinflammatory signature. This pro-inflammatory signature was shared with human colorectal cancer tissue with a high Fusobacterium abundance.
In contrast to other bacterial-driven models of intestinal tumorigenesis (Arthur et al., 2012; Wu et al., 2009), we have found a specific bacterial strain that accelerates intestinal tumorigenesis in the absence of colitis. While ApcMin/+ mice fed F. nucleatum exhibited enhanced intestinal tumorigenesis, neither Il10−/− nor T-bet−/− X Rag2−/− mouse models of colitis showed accelerated tumorigenesis or exacerbated colitis upon introduction of F. nucleatum. This may suggest that the tumorigenic effects of fusobacteria operate downstream of the loss of the tumor suppressor APC and the resulting intestinal dysplasia that occurs in ApcMin/+ mice. This is relevant to most cases of human CRC, as only 2% of CRC cases are linked to colitis, but greater than 80% of non-hypermutated CRC tumors bear APC mutations (Cancer Genome Atlas Network, 2012). Our Fusobacterium findings are also relevant to adenomas because mutations in APC are among the earliest molecular alterations that occur in an epithelium as it transitions to become an adenoma (Cho and Vogelstein, 1992). Therefore, early tumor-initiating somatic mutations likely precede the tissue enrichment of Fusobacterium spp. These mutations may contribute to the development of epithelial barrier defects, featuring the loss of tight junctions, cell-to-cell contacts, epithelial polarity and the mucus layer (Grivennikov et al., 2012). Intestinal barrier defects at local sites of dysplasia may promote the infiltration of Fusobacterium spp., among other bacteria and microbial products, allowing fusobacteria to take up residence in the tumor environment. This may represent a crucial stage in colorectal neoplasia wherein myeloid cell-mediated immune responses provide the driving force for inflammatory genotoxic and epigenetic changes that lead to cancer.
Although barrier defects expose the intestinal mucosa to the entire luminal microbial milieu, Fusobacterium spp. become the most highly enriched bacterium in colorectal tumors relative to adjacent tissue (Castellarin et al., 2012; Kostic et al., 2012; Tjalsma et al., 2012). This enrichment may be attributable to the strong adhesive and invasive abilities of fusobacteria for epithelial cells (Han et al., 2000; Strauss et al., 2011). The tumor enrichment may also result from the growth advantage that fusobacteria provide to the tumor by eliciting pro-tumorigenic responses from myeloid immune cells. Alternatively, metabolic specializations may endow fusobacteria with a competitive advantage in the evolving tumor milieu. Fusobacterium nucleatum is an asaccharolytic bacterium so, unlike the Enterobacteriaceae, it will not compete for glucose, a preferred substrate for tumor metabolism (Vander Heiden et al., 2009). Instead fusobacteria can utilize amino acids and peptides as nutrient sources in the tumor microenvironment. Products of amino acid metabolism generated by fusobacteria, including formyl-methionyl-leucyl-phenylalanine and short chain fatty acids, are myeloid cell chemoattractants, which may explain the intratumoral myeloid cell expansion we observed and interconnect tumor metabolism, bacterial metabolism, and immune cell function within the tumor microenvironment.
In addition, F. nucleatum strains, unlike many strict anaerobes of the intestinal lumen, possess a rudimentary electron transport chain, endowing them with a limited ability to respire oxygen (Kapatral et al., 2002). Thus, F. nucleatum may be able to persist and slowly replicate in the hypoxic tumor microenvironment. Adhesive molecules that contribute to invasivity in F. nucleatum can promote bacterial aggregation and biofilm formation that also enhance oxygen tolerance (Gursoy et al., 2010). Products of fusobacterial metabolism may make the tumor microenvironment more tumor-permissive over time by directly promoting tumor cell proliferation, blood vessel growth, or immune cell infiltration.
We have shown that, in both human and mouse intestinal tumors, the pro-inflammatory gene expression signature associated with Fusobacterium features the upregulation of PTGS2. Epidemiological and clinical data suggest that non-steroidal anti-inflammatory drugs (NSAIDs) may be effective as a primary and secondary preventative measure in colorectal neoplasia (Chan et al., 2012). Our findings on Fusobacterium spp. and the intrinsic inflammation that they elicit may further explain why anti-inflammatory strategies such as NSAIDs are an effective colorectal cancer prevention strategy.
If our results demonstrating that fusobacteria potentiate tumorigenesis can be extended to human CRC, then targeted reduction of Fusobacterium populations in the oral cavity where they are most abundant (Consortium, 2012), or in the gastrointestinal tract may work to delay or prevent tumor progression for patients at increased risk for CRC. These findings suggest that it will be important to consider the contribution of members of the tumor microbiota, such as Fusobacterium nucleatum, and the intersection between microbial gene function and the host response for understanding colorectal cancer risk, development and progression.
EXPERIMENTAL PROCEDURES
Bacterial strains and culturing
F. nucleatum (EAVG_002; 7/1) (Strauss et al., 2011) was a gift from the laboratory of E. Allen-Vercoe (U. Guelph). Streptococcus species were isolated from freshly resected human colon tissues. Please see supplemental methods for additional information on bacterial identification methods and culturing techniques.
Mice
All mice (C57BL/6J–ApcMin/J, BALB/c Il-10−/− and BALB/c T-bet−/− Rag2−/−) were maintained in the barrier facility at the Harvard School of Public Health (HSPH) and all experimentation was carried out in accordance with institutional guidelines. ApcMin/+ mice were initially ordered from Jackson Laboratory and then bred at HSPH.
Bacterial feeding experiments were performed for a period of 8 weeks, beginning at 6 wks of age. Bacteria were fed at 108 CFU per day. Sham treatment consisted of TSB.
Histopathology
Colons and small intestines were prepared for histologic analysis and assessed for colitis or enteritis as previously described (Garrett et al., 2007). Adenomas and adenocarcinomas were counted in colons and small intestines submitted in their entirety. ACF were identified in the same paraffin sections as foci of 1–3 crypts in flat mucosa showing increased size, and nuclear enlargement and hyperchromasia.
Human specimen collection
Please see supplemental methods for details on colonic adenocarcinoma, adenomas and stool samples.
DNA preparation and bacterial quantification by qPCR
Human and mouse tissues or stools DNA preparation and qPCR are described in detail in supplemental methods.
RNA preparation and gene expression by qPCR
Total RNA was extracted using the RNeasy® kit (QIAGEN), DNase treated with the DNA-free™ kit (Ambion), and cDNA was generated using the iScript™ cDNA Synthesis kit (Bio-Rad). Relative gene expression was calculated using the ∆∆CT method. Primers were designed using MGH primerbank: pga.mgh.harvard.edu/primerbank/.
Microbial FISH analysis
Microbial FISH was performed as described (Kostic et al., 2012).
RNA-Seq processing and analysis
All primary sequence data on colonic adenoma (COAD) samples (https://tcgadata.nci.nih.gov/docs/publications/coadread_2012/) were downloaded from The Cancer Genome Atlas via dbGAP and the Data Coordinating Center (http://cancergenome.nih.gov/). For additional details, please see supplemental methods.
Detection of NF-κB activation
Fresh or fresh-frozen colon tumor tissue (20− 80mg per sample) was processed for nuclear protein isolation with the NE-PER® Kit (Thermo Scientific). Protein lysates were resolved, transferred to PVDF membrane, and blots were probed with antibodies directed against p65 and lamin B1 (Cell Signaling Technology). Samples were stratified as Fusobacterium-high (qPCR Fusobacterium abundance greater than 40.0 [calculated as 1.8^(-CT<Fusobacterium> - CT<Eubacteria>)*1000], or 16S sequencing Fusobacterium abundance greater than 0.25 [calculated as described (Kostic et al., 2012)]) or Fusobacterium-low (qPCR Fusobacterium abundance less than 1.0, or 16S sequencing Fusobacterium abundance less than 0.015). Wester blot densitometry analysis was performed using Image-J software (http://rsbweb.nih.gov/ij/).
Isolation of intestinal tumor infiltrating cells
Small intestine was removed, opened and washed with Ca+- and Mg+-free DPBS. Tumors were dissected, weighed, minced and incubated in HBSS with 0.1 mg/ml collagenase D (Roche), 50 U/ml DNase I (Roche) and 50 µg/mg Dispase (StemCell Technologies) for 30 min at 37°C. Single cell suspensions were resuspend ed in cell staining solution (PBS with 2% FCS) for flow cytometry.
Multicolor Flow cytometry
Cells were incubated with Fc blocking antibody (BioLegend) and stained with fluorochrome-conjugated monoclonal antibodies of cell surface markers. Antibodies and clone information are provided in supplemental methods.
Expression of B220 (clone RA3–6B2) and PDCA-1 (clone 927) was also examined. These two antigens on CD11c low expressing cells are used to identify plasmacytoid DCs. However, cell numbers were very low in tumors (average 2 × 103 cells/tumor gram with a range from 0–16 ×103 cells/tumor gram in F. nucleatum treated mice). Thus these data are not shown.
Methods describing intracellular staining for IL-17A, Foxp3, Arginase-1, and iNOS and cell sorting are provided in supplemental methods.
In vitro T cell suppression assay
Splenic CD3ε+CD4+ T cells were sorted from ApcMin/+ mice and labeled with Cell Trace Violet (Invitrogen) as per manufacturers instructions. Labeled CD3ε+CD4+ T cells were plated at 2 × 105 cells/wells with sorted MDSCs, TAMs, and M2-like TAMs (2×105) in the presence of 1µg/ml anti-CD3 antibody and 1µg/ml anti-CD28 antibody. After 72h, cells were collected and analyzed by flow cytometry.
Statistical analysis
Generally, data is displayed in dot-plot format, with the center-line indicating the mean and SEM represented by the error bars. All two-group comparisons were performed using the non-parametric Mann-Whitney U test, except for Figure 1A for which the non-parametric Wilcoxon matched-pairs signed rank test was performed. For RNA-Seq data, genus relative abundance was correlated to host gene expression using the Spearman’s rank correlation coefficient, and corrected P-values were obtained by correcting for multiple hypothesis testing using the False Discovery Rate method. Statistical analysis for Table S1 was performed using the Fisher’s exact test.
Supplementary Material
HIGHLIGHTS.
Fusobacterium is enriched in human adenomas, suggesting an early role in tumorigenesis
Fusobacterium nucleatum accelerates tumorigenesis in ApcMin/+ mice
F. nucleatum drives myeloid cell infiltration in intestinal tumors
Fusobacterium is associated with a proinflammatory signature in mouse and human tumors
ACKNOWLEDGEMENTS
We thank members of the Garrett and Meyerson labs for discussion and E. Allen-Vercoe for Fusobacterium strains and culturing advice. Adenomas and matched normal tissue samples were provided by the Cooperative Human Tissue Network, a NCI supported resource. These studies were supported by R01CA154426 (NCI), K08AI078942 (NIAID), a Burroughs Wellcome Career in Medical Sciences Award, a Searle Scholars Award and a Cancer Research Institute Investigator Award to WSG, U54HG003067 (National Genome Research Institute) P50CA127003 (NCI), RC2CA148317 (NCI), R01CA154426 (NCI) and a Starr Cancer Consortium Award to MM, and P50CA127003 (SPORE) to CSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutaga K, van Winkelhoff AJ, Vandenbroucke-Grauls CMJE, Savelkoul PHM. Periodontal pathogens: A quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol. Med. Microbiol. 2005;45:191–199. doi: 10.1016/j.femsim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AT, Arber N, Burn J, Chia WK, Elwood P, Hull MA, Logan RF, Rothwell PM, Schrör K, Baron JA. Aspirin in the Chemoprevention of Colorectal Neoplasia: An Overview. Cancer Prevention Research. 2012;5:164–178. doi: 10.1158/1940-6207.CAPR-11-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KR, Vogelstein B. Genetic alterations in the adenoma--carcinoma sequence. Cancer. 1992;70:1727–1731. doi: 10.1002/1097-0142(19920915)70:4+<1727::aid-cncr2820701613>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Consortium THMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Garrett W, Siddiqui KRR, Arancibia-Cárcamo CV, Hall J, Sun C-M, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Pollard JW. Leukocytes in mammary development and cancer. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmani P, Strauss J, Ambrose C, AllenVercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor-{alpha} Infection and Immunity. 2011 doi: 10.1128/IAI.05118-11. IAI.05118–11v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato JA, Ruffell B, Saleh M, Mantovani A, Segata N, Mercurio F, Affara NI, Trinchieri G, Allavena P, Haake SK, et al. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Arthur JC, Ostrand-Rosenberg S, Perez-Chanona E, Bronte V, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Campbell BJ, et al. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Castellarin M, Nagaraj S, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, et al. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Punit S, Gallini CA, Michaud M, Zhang D, Sigrist KS, Lord GM, Glickman JN, Glimcher LH. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16:208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu G-Y, Österreicher CH, Hung KE, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012 doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursoy UK, Hanahan D, Pöllänen M, Weinberg RA, Könönen E, Uitto V-J. Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. J. Periodontol. 2010;81:1084–1091. doi: 10.1902/jop.2010.090664. [DOI] [PubMed] [Google Scholar]
- Han YW, Shi W, Huang GTJ, Kinder Haake S, Park N-H, Kuramitsu H, Genco RJ. Interactions between Periodontal Bacteria and Human Oral Epithelial Cells: Fusobacterium nucleatum Adheres to and Invades Epithelial Cells. Infection and Immunity. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Hayden MS, Ghosh S, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes & Development. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jobin C. Colorectal cancer: CRC-all about microbial products and barrier function? Nat Rev Gastroenterol Hepatol. 2012;9:694–696. doi: 10.1038/nrgastro.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapatral V, Kapatral V, Anderson I, Anderson I, Ivanova N, Ivanova N, Reznik G, Reznik G, Los T, Los T, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 2002;184:2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RGW, Getz G, Meyerson M. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat. Biotechnol. 2011;29:393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Current Opinion in Immunology. 2010;22:231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- Marchesi JR, Dutilh BE, Hall N, Peters WHM, Roelofs R, Boleij A, Tjalsma H. Towards the Human Colorectal Cancer Microbiome. PLoS ONE. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AN, Araujo-Perez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium Is Associated with Colorectal Adenomas. PLoS ONE. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B-Z, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, Randall TA, Galanko J, Benson A, Sandler RS, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. Isme J. 2012;6:1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, Clune Y, Collins JK, O’Sullivan GC, O’Riordan M, Holmes E, Wang Y, Marchesi JR. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ. Microbiol. 2008;10:789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, Jovov B, Abdo Z, Sandler RS, Keku TO. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver–passenger model for colorectal cancer: beyond the usual suspects. Nature Reviews Microbiology. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu. Rev. Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- Uronis JM, Mühlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Grivennikov SI, Han YW, Cantley LC, Wang K, Shi W, Thompson CB, Mucida D, Huang GT, Stewart CA, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.