Summary
Ornithine lipids (OLs) are phosphorus-free membrane lipids that are widespread among Gram-negative bacteria. Their basic structure consists of a 3-hydroxy fatty acyl group attached in amide linkage to the α-amino group of ornithine and a second fatty acyl group ester-linked to the 3-hydroxy position of the first fatty acid. It has been shown that OLs can be hydroxylated within the amide-linked fatty acyl moiety, the secondary fatty acyl moiety or within the ornithine moiety. These modifications have been related to increased stress tolerance and symbiotic proficiency in different organisms such as Rhizobium tropici or Burkholderia cenocepacia. Analysing the membrane lipid composition of the plant pathogen Agrobacterium tumefaciens we noticed that it forms two different OLs. In the present work we studied if OLs play a role in stress tolerance and pathogenicity in A. tumefaciens. Mutants deficient in the OLs biosynthesis genes olsB or olsE were constructed and characterized. They either completely lack OLs (ΔolsB) or only form the unmodified OL (ΔolsE). Here we present a characterization of both OL mutants under stress conditions and in a plant transformation assay using potato tuber discs. Surprisingly, the lack of agrobacterial OLs promotes earlier tumour formation on the plant host.
Introduction
Ornithine lipids (OLs) are phosphorus-free membrane lipids that are widespread among Gram-negative bacteria and also present in some Gram-positive bacteria, especially in actinomycetes species, but seem to be absent from Archaea and Eukarya (López-Lara et al., 2003; Geiger et al., 2010; Vences-Guzmán et al., 2012). They contain a 3-hydroxy fatty acyl group that is attached in amide linkage to the α-amino group of ornithine. A second fatty acyl group is ester-linked to the 3-hydroxy position of the first fatty acid. The genes olsB and olsA encoding the two enzymes essential for OL biosynthesis from ornithine and acyl-ACPs have been first described in Sinorhizobium meliloti 1021 (Fig. S1) (Weissenmayer et al., 2002; Gao et al., 2004; Geiger et al., 2010). For a long time it has been known that the ester-linked fatty acid of OLs can be hydroxylated at the C-2 position in some organisms (Asselineau, 1991). Recently, two more OL modifications have been described. In Burkholderia cenocepacia the amide-linked fatty acid can be hydroxylated in an unknown position by the hydroxylase OlsD (González-Silva et al., 2011) and in Rhizobium tropici the ornithine headgroup can be hydroxylated in a still unknown position (Vences-Guzmán et al., 2011). All three modifications have been related to increased stress tolerance or symbiotic proficiency (Taylor et al., 1998; Rojas-Jiménez et al., 2005; González-Silva et al., 2011; Vences-Guzmán et al., 2011).
Although OLs are probably found in both membranes of Gram-negative bacteria, they seem to be enriched in the outer membrane (OM) as was shown in the acid-resistant species Thiobacillus thiooxidans and R. tropici (Dees and Shively, 1982; Vences-Guzmán et al., 2011). It has been speculated that OL hydroxylation may increase hydrogen bonding between neighbouring OL molecules similarly as has been suggested for LpxO-hydroxylated lipid A in Salmonella and hydroxylated sphingolipids (Gibbons et al., 2000; Nikaido, 2003; Murata et al., 2007). These additional hydrogen bonds should result in bilayer stabilization and a decrease in membrane permeability which could help to explain the observed stress resistance phenotype related to hydroxylated OLs.
Ornithine lipids together with other phosphorus-free membrane lipids such as diacylglycerol-N,N,N-trimethylhomoserine (DGTS) or sulfolipids (SLs) accumulate under phosphate-limiting conditions in several bacterial species while at the same time phospholipids are actively degraded (Benning et al., 1995; López-Lara et al., 2003; Geiger et al., 2010; Zavaleta-Pastor et al., 2010). In S. meliloti it has been shown that this remodelling is under the control of the response regulator PhoB (Geiger et al., 1999; Zavaleta-Pastor et al., 2010). As OLs and phosphatidylethanolamine (PE) both are zwitterionic lipids, it has been speculated that OLs might replace PE under these conditions (Benning, 1998), although clear experimental evidence is lacking so far. Some other bacteria such as Brucella abortus (Thiele and Schwinn, 1973; Comerci et al., 2006; Bukata et al., 2008), Mesorhizobium loti (Devers et al., 2011), B. cenocepacia (González-Silva et al., 2011) and R. tropici (Rojas-Jiménez et al., 2005; Sohlenkamp et al., 2007; Vences-Guzmán et al., 2011) also form significant amounts of OLs during growth in standard laboratory media such as Luria–Bertani (LB) which contain phosphate in concentrations that are not growth-limiting. It is not clear if OLs biosynthesis is regulated by PhoB in these organisms. We had observed that OLs apparently play an important role in R. tropici both under abiotic stress conditions such as heat stress and acid stress but also under symbiotic conditions (Vences-Guzmán et al., 2011).
Agrobacterium tumefaciens is α-proteobacterium able to form tumours on many dicotyledonous plant species. Analysing the membrane lipid composition of the plant pathogen A. tumefaciens we noticed that it forms two different OLs, corresponding to the unmodified OL S1 and the hydroxylated OL S2 (Fig. S1). In the present work we wanted to study if OLs play a role in stress tolerance and pathogenicity in A. tumefaciens.
Agrobacterium tumefaciens mutants deficient in the OLs biosynthesis genes olsB or olsE were constructed and characterized. They either completely lack OLs (ΔolsB) or only form the unmodified OL (ΔolsE). Here we present a characterization of both OL mutants under different abiotic stress conditions and in a plant transformation assay using potato tuber discs.
Results
Agrobacterium tumefaciens forms two different ornithine lipids
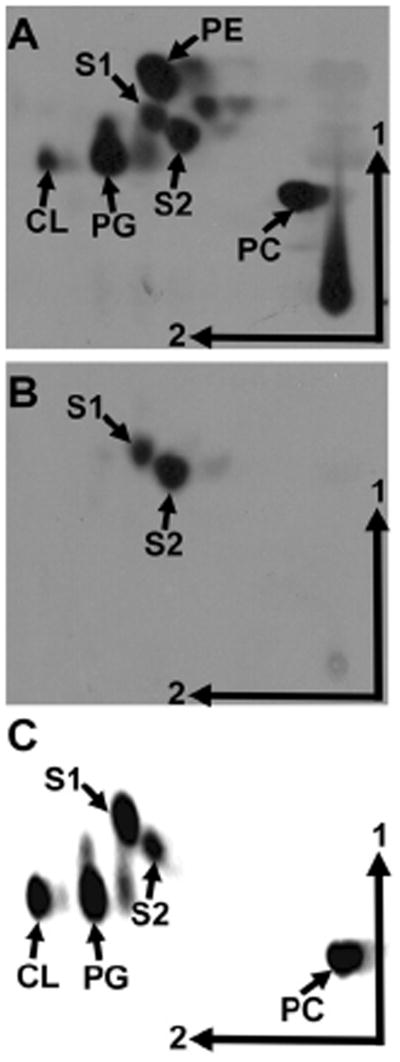
When analysing the lipid composition of [14C]acetate-labelled A. tumefaciens C58 by two-dimensional thin layer chromatography (TLC), in addition to the phospholipids phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), dimethyl PE, cardiolipin (CL), and the unmodified OL S1, a lipid migrating like the OL S2 from R. tropici CIAT899 (Fig. S1) (Rojas-Jiménez et al., 2005; Vences-Guzmán et al., 2011) canbe observed (Fig. 1A). In order to confirm that this unknown lipid is biosynthetically derived from the amino acid ornithine, A. tumefaciens C58 cells were labelled with [14C]ornithine and the lipids were extracted and analysed by TLC. Apparently two ornithine-derived lipids are present in A. tumefaciens C58 (Fig. 1B). An identical result was obtained with A. tumefaciens strain A208 (data not shown).
Fig. 1.
Agrobacterium tumefaciens forms an OL migrating similarly as S2 from R. tropici. [14C]acetate- (A) and [14C]ornithine- (B) labelled lipids from A. tumefaciens wild-type C58 grown at 30°C in 20E medium and [14C]acetate-labelled lipids from S. meliloti CS111.pNG25.pERMAV14 grown in TY medium (C) were separated by two-dimensional TLC. The phospholipids PE, monomethyl PE (MMPE), PC, phosphatidylglycerol (PG), cardiolipin (CL), and the OLs S1 and S2 are indicated.
The plant pathogen Agrobacterium tumefaciens also forms the ornithine lipid S2
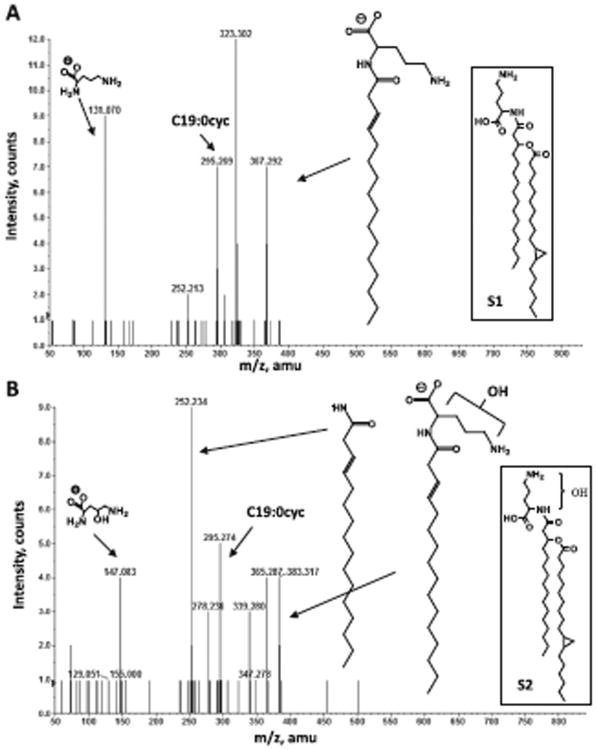
Three different hydroxyl group modifications of OLs have been described in bacteria. To identify in which part of the second agrobacterial OL the modification is encountered, lipids were extracted according to Bligh and Dyer from a 1 l culture of A. tumefaciens wild-type A208. Both putative OLs were purified from the total lipid extract and analysed by normal phase LC/ESI-MS/MS in the negative ion mode. The [M-H]− molecular ions of OL S1 and S2 were detected at m/z 663.6 and 679.6 respectively (Fig. S2). The mass difference of 16 amu suggests the presence of an additional oxygen atom indicating the presence of an additional hydroxyl group in S2. Comparing the fragmentation patterns of both lipids it was determined that a hydroxyl group was present within the ornithine moiety in the case of the unknown OL but not in the fatty acyl chains (Fig. 2). Agrobacterium tumefaciens therefore forms the OLs S1 and S2 (Fig. S1). The fragment ion spectra of the [M-H]− ions of S1 and S2 at m/z 663.6 and 679.6 respectively, revealed that the amide-linked fatty acid is almost exclusively a C16 (3-OH) fatty acid indicating a substrate specificity of the agrobacterial OlsB for this fatty acid.
Fig. 2.
Collision-induced dissociation mass spectra of OL S1 and S2 detected in lipid extract of A. tumefaciens. Negative ion collision induced dissociation mass spectra of [M-H]− ions at m/z 663.6 (A) obtained from OL S1 and m/z 679.6 (B) obtained from OL S2. The structures of major fragment ions are indicated. The structures of the parental lipids are shown in the boxes.
Ninhydrin staining of the A. tumefaciens membrane lipids separated by two-dimensional TLC confirmed our earlier observation of the OLs from R. tropici. S2 reacts with a delay in comparison to S1 and the developed colour by S2 is different. While S1 upon reaction with ninhydrin develops a red-to-purple colour, the reaction of S2 causes the formation of an orange colour (Fig. S3).
Atu0318 is responsible for the synthesis of the second ornithine lipid
The presence of the OL S2 indicates that the A. tumefaciens genome should encode an OlsE homologue. One of the closest homologues of OlsE from R. tropici CIAT899 is indeed Atu0318 which presents 61% identity and 71% similarity on amino acid level. BLAST searches show (Altschul et al., 1997) that other close OlsE homologues are present in all sequenced Agrobacterium strains, Rhodospirillum centenum SE, Azospirillum amazonense, Verrucomicrobium spinosum and Parvibaculum lavamentivorans DS-1. Further homologues are present in several actinobacteria, a few γ-proteobacteria and a few other α-proteobacteria (Vences-Guzmán et al., 2012). To confirm that Atu0318 serves the same function as OlsE from R. tropici it was cloned into the broad host range plasmid pRK404 and subsequently conjugated into S. meliloti CS111.pNG25. In this strain a lipid migrating similarly as S2 from R. tropici was present that was absent in the negative control strain CS111.pNG25 (Fig. 1C and data not shown).
Lipid composition analysis of olsE and olsB mutants
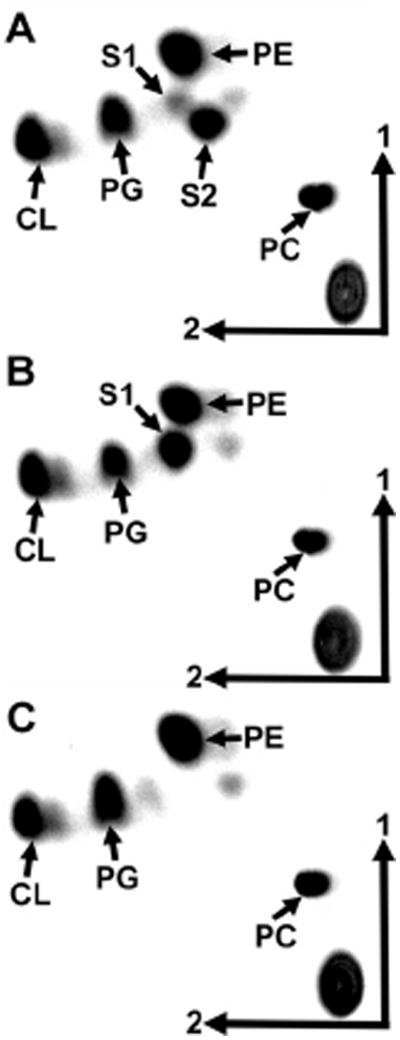
We had observed a role for OLs in R. tropici stress resistance and symbiosis (Vences-Guzmán et al., 2011) and we therefore considered it a possibility that OLs might play a role in stress resistance and pathogenesis in A. tumefaciens. To study the role of OLs in A. tumefaciens in more detail, mutants deficient in olsE and mutants deficient in olsB were constructed. Their lipid compositions were compared with the wild-type strain A. tumefaciens A208 (Fig. 3 and Table 1). As expected the olsE-deficient mutant MAV07 lacked the OL S2, and the olsB-deficient mutant MAV08 lacked S1 and S2. The total amount of OLs, being the sum of S1 and S2, is around 15% in both, the A. tumefaciens wild-type and the olsE-deficient mutant MAV07 when grown in complex LB medium at 30°C (Table 1). In the absence of OLs in the mutant MAV08, more PE is formed, whereas no major differences are observed in the relative amounts of the phospholipids, PC, PG and CL between the different strains (Table 1). This suggests that the amounts of PE and OLs are inversely proportional.
Fig. 3.
Analysis of membrane lipid composition of A. tumefaciens wild-type A208 (A), olsE-deficient mutant MAV07 (B) and olsB-deficient mutant MAV08 (C). Lipids were labelled with [14C]acetate during growth in 20E medium at 30°C and separated using two-dimensional TLC. The phospholipids phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), cardiolipin (CL) and the OLs S1 and S2 are indicated.
Table 1.
Membrane lipid composition of A. tumefaciens wild-type A208, olsE-deficient mutant MAV07 and olsB-deficient mutant MAV08 after growth on complex LB medium at 30°C.
| Lipid | Composition (% of total 14C) | ||
|---|---|---|---|
|
| |||
| A208 | MAV07 (ΔolsE) | MAV08 (ΔolsB) | |
| PC | 21.3 ± 0.5 | 26.0 ± 1.2 | 24.2 ± 0.5 |
| S1 | 5.8 ± 0.2 | 15.9 ± 0.3 | n.d. |
| S2 | 9.5 ± 0.2 | n.d. | n.d. |
| PE | 41.5 ± 0.4 | 35.7 ± 1.3 | 53.2 ± 0.2 |
| PG | 18.8 ± 0.4 | 20.5 ± 0.2 | 20.2 ± 0.9 |
| CL | 3.1 ± 0.5 | 1.9 ± 0.1 | 2.4 ± 0.2 |
The values shown are mean values ± standard deviation derived from at least three independent experiments.
S1: unmodified ornithine lipid; S2: hydroxylated ornithine lipid; n.d.: not detected.
Both mutants, MAV07 (ΔolsE) and MAV08 (ΔolsB), were also complemented. When introducing a functional copy of olsB in trans into the mutant MAV08 again formation of S1 and S2 was observed, while when introducing a functional copy of olsE in trans into the mutant MAV07 again formation of S2 was observed (data not shown).
Hydroxylated OL S2 accumulates when A. tumefaciens is grown at 15°C
We had reported earlier that in R. tropici no OlsE-dependent hydroxylation could be observed at 42°C (Vences-Guzmán et al., 2011). We therefore wondered if OlsE might be more active at lower temperatures than at higher temperatures. When comparing the membrane lipid composition of cells grown at 15°C to cells grown at 30°C several differences can be observed (Table 2). The relative amounts of the phospholipids PE and PC seem to increase, whereas CL is drastically reduced and the amount of PG does not change when going from 30°C to 15°C. Interestingly, virtually all OL seems to be in the hydroxylated form at 15°C indicating that OlsE is sufficiently active to convert all OL S1 into hydroxylated S2. When studying the growth of both mutants and the wild-type at 15°C, all strains grow with similar generation times indicating that the presence or absence of the OLs does not affect growth of A. tumefaciens under these conditions (data not shown).
Table 2.
Membrane lipid composition of A. tumefaciens wild-type, olsE-deficient mutant MAV07 and olsB-deficient mutant MAV08 after labelling for 48 h with [14C]acetate in Sherwood minimal medium at 30°C or 15°C.
| Composition (% of total 14C) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| 30°C | 15°C | |||||
|
|
|
|||||
| Lipid | A208 | MAV07 | MAV08 | A208 | MAV07 | MAV08 |
| PC | 13.6 | 14.7 | 19.3 | 28.4 | 34.1 | 33.3 |
| S1 | 6.8 | 20.2 | n.d. | n.d. | 16.0 | n.d. |
| S2 | 14.3 | n.d. | n.d. | 16.8 | n.d. | n.d. |
| PE | 29.7 | 30.8 | 43.0 | 39.0 | 31.3 | 52.6 |
| PG | 12.2 | 11.5 | 18.9 | 11.6 | 15.0 | 11.7 |
| CL | 23.4 | 22.8 | 18.8 | 4.2 | 3.6 | 2.4 |
The values shown are mean values derived from at least two independent experiments.
S1: unmodified ornithine lipid; S2: hydroxylated ornithine lipid; n.d.: not detected.
Absence of OLs does not affect growth of A. tumefaciens under conditions of high osmolarity
Agrobacterium tumefaciens wild-type A208 and both mutants deficient in OL biosynthesis were cultivated in complex 20E medium supplemented with up to 1 M NaCl. Up to 0.8 M NaCl no significant differences in generation time could be observed between the three strains. At 0.9 M NaCl only residual growth was observed for all strains and in the presence of 1.0 M NaCl none of the strains was able to grow. Under all conditions studied both mutants behaved very similar to the wild-type indicating that OLs do not play an important role in protecting the bacteria against osmotic stress (data not shown).
Growth of A. tumefaciens under phosphate limitation
Agrobacterium tumefaciens wild-type A208 and mutants deficient in OL biosynthesis were grown in Sherwood minimal medium with high (1.3 mM) or low phosphate (20 μM) concentrations. Under high phosphate conditions the phospholipids PC, PG, PE, dimethyl PE and CL were detected in the wild-type (Table 3). In addition, as observed earlier in cells grown on complex medium the two OLs S1 and S2 were detected. Under low phosphate conditions all phospholipids were reduced and both OLs increased to a total of about 45–50% in the wild-type. In addition, several new lipids were observed in the phosphate-deplete grown cells. In addition to DGTS several unknown lipids were detected. As Benning and colleagues (1995) and Devers and colleagues (2011) had observed the formation of glycolipids in R. sphaeroides and M. loti under low phosphate conditions we considered it a possibility that the unknown lipids were glycolipids. Naphthol staining of membrane lipids separated by TLC confirmed the presence of at least four different glycolipids. In contrast to S. meliloti and R. sphaeroides no sulfolipid was detected under phosphate starvation conditions which is consistent with the absence of genes coding for homologues involved in their biosynthesis in the A. tumefaciens genome. In the olsB-deficient mutant unable to form OLs increased amounts of DGTS can be detected in addition to increased amounts of the unknown lipids (Table 3).
Table 3.
Analysis of A. tumefaciens strains grown in Sherwood medium containing 20 μM or 1.3 mM phosphate.
| Composition (% of total 14C) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| High phosphate | Low phosphate | |||||
|
|
|
|||||
| A208 | MAV07 | MAV08 | A208 | MAV07 | MAV08 | |
| PC | 13.6 ± 0.5 | 14.7 ± 0.5 | 19.3 ± 0.7 | 7.3 ± 0.6 | 9.5 ± 0.5 | 10.0 ± 1.0 |
| PG | 12.2 ± 0.2 | 11.5 ± 0.5 | 18.9 ± 0.7 | 3.6 ± 0.6 | 4.5 ± 0.1 | 5.3 ± 0.6 |
| PE | 29.7 ± 0.6 | 30.8 ± 1.4 | 43.0 ± 1.2 | 2.7 ± 0.4 | 2.0 ± 0.6 | 6.6 ± 0.5 |
| CL | 23.5 ± 1.0 | 22.8 ± 1.1 | 18.8 ± 0.9 | 3.9 ± 0.9 | 5.9 ± 0.1 | 7.0 ± 0.1 |
| S1 + Gl4 | 6.8 ± 1.0 | 20.2 ± 0.3 | n.d. | 21.8 ± 0.8 | 42.9 ± 0.9 | 6.4 ± 0.5 |
| S2 | 14.2 ± 0.3 | n.d. | n.d. | 30.0 ± 0.1 | n.d. | n.d. |
| GL1 | n.d | n.d. | n.d. | 12.9 ± 0.9 | 20.9 ± 0.5 | 17.1 ± 0.3 |
| GL2 | n.d | n.d. | n.d. | 2.6 ± 0.5 | 2.9 ± 0.3 | 11.3 ± 0.8 |
| GL3 | n.d. | n.d. | n.d. | 1.5 ± 0.4 | 2.5 ± 0.1 | 2.6 ± 0.5 |
| DGTS | n.d. | n.d. | n.d. | 13.7 ± 0.5 | 8.9 ± 0.5 | 33.7 ± 1.4 |
A208 – wild-type, MAV07 – ΔolsE mutant, MAV08 – ΔolsB mutant. The values shown are mean values ± standard deviation derived from at least three independent experiments. The bacterial strains were labelled for 48 h with [14C]acetate.
S1: unmodified ornithine lipid; S2: hydroxylated ornithine lipid; GL1, GL2, GL3, GL4: unknown glycolipids; n.d.: not detected.
Bioinformatic prediction of the PhoB regulon
A bioinformatic search of the A. tumefaciens C58 genome was performed for the presence of Pho boxes to find out if the OL biosynthesis genes are under the control of PhoB and at the same time understand the phosphate starvation response in A. tumefaciens. A position-specific scoring matrix (PSSM) was constructed using Pho box sequences from S.meliloti 1021 identified in an earlier study (Yuan et al., 2006). The presence of a Pho box was predicted for the olsB, but not for the olsE promoter from A. tumefaciens (data not shown). Additionally, our bioinformatic analysis predicted PhoB-dependent regulation for DGTS biosynthesis as had been shown for S. meliloti. Putative Pho boxes were also predicted to precede the genes encoding the putative phosphate transport regulator Atu4634, the putative phospholipase C PlpC (Atu1649), and the putative phosphohydrolase Atu0877. Interestingly, four genes (Atu0842, Atu1808, Atu2222, Atu2297) are preceded by putative Pho boxes that might encode glycosyltransferases involved in the biosynthesis of the several unknown and putative glycolipids that accumulate under conditions of phosphate starvation in A. tumefaciens.
Phenocopying the OL composition of R. tropici in A. tumefaciens
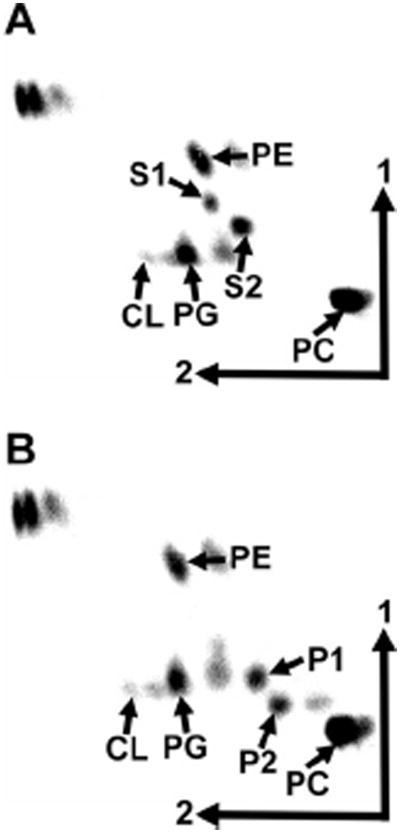
We had observed earlier that the presence of the hydroxylase OlsC responsible for the synthesis of P1 was important to confer acid resistance to R. tropici (Vences-Guzmán et al., 2011). Agrobacterium tumefaciens forms the OLs S1 and S2 and by introducing the gene olsC we would phenocopy or mimic the OL composition of R. tropici. We speculated that A. tumefaciens harbouring olsC might show an increase in acid resistance. Wild-type strain A208, mutants deficient in olsB or olsE and the wild-type expressing olsC from R. tropici were grown at pH 5.0, pH 5.5 and pH 7.0. Although OLs P1 and P2 could be detected in the strain expressing olsC, the presence of OlsC did not improve the growth of A. tumefaciens at low pH (Fig. 4, data not shown).
Fig. 4.
Heterologous expression of olsC from Rhizobium tropici in Agrobacterium tumefaciens A208. Lipids of the wild-type A. tumefaciens A208 harbouring the empty plasmid pERMAV06 (A) or the olsC-containing plasmid pERMAV15 (B) were labelled with [14C]acetate for 48 h during growth in LB medium at 30°C and separated using two-dimensional TLC. The phospholipids phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), cardiolipin (CL) and the OLs S1, and S2 are indicated. OLs P1 and P2 derived from OlsC-dependent hydroxylation of S1 and S2 are also indicated.
Plant assays of tumours in A. tumefaciens
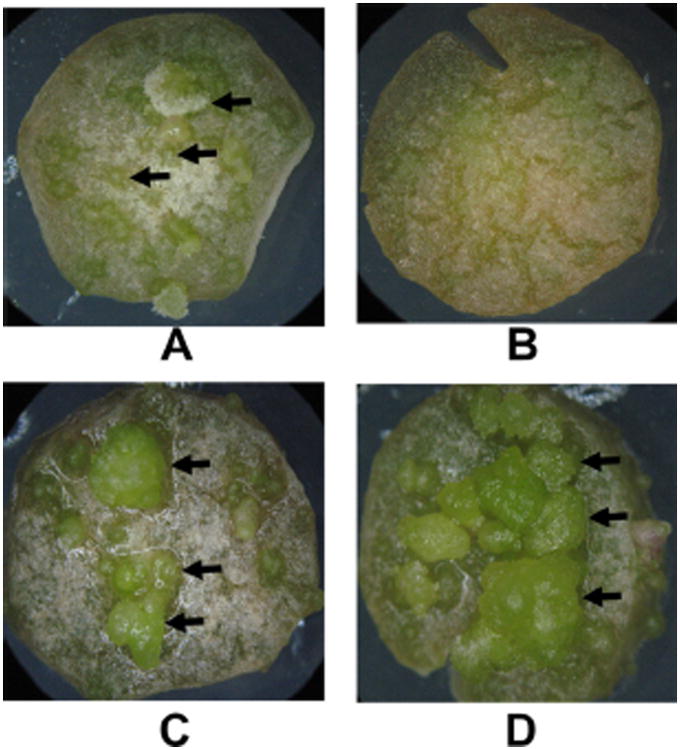
The virulence of A. tumefaciens mutants deficient in the formation of OLs was assayed using a potato tuber disc system (Shurvinton and Ream, 1991; Tsai et al., 2009). All strains caused the formation of tumours on potato tuber discs. Interestingly, the mutants deficient in the formation of OLs behaved differently from the wild-type. Tumours appeared within 18 days on discs inoculated with MAV08 or MAV07 while tumours appeared 6 days later on discs inoculated with the wild-type A. tumefaciens A208 (Fig. 5 and Table 4). Complementation of both mutants with the respective gene in trans restored the wild-type phenotype. The tumours on potato tuber discs inoculated with the mutant strains are also larger than the tumours on tuber discs inoculated with the wild-type. This size difference is probably due to the longer development of the tumours on tuber discs inoculated with the OL-deficient mutant strains.
Fig. 5.
Assay of tumour formation by Agrobacterium tumefaciens using potato tuber discs. Potato discs were inoculated with (A) A. tumefaciens A208, (B) no bacteria, (C) OlsE-deficient mutant MAV07, (D) OlsB-deficient mutant MAV08. Potato tuber discs were scored every 3 days for tumour formation. The tuber discs have a diameter of 1 cm. Images were taken 48 days after inoculation. Tuber discs were incubated at 28°C. The arrows highlight some tumours on the potato discs.
Table 4.
Kinetics of tumour formation by A. tumefaciens on potato tuber discs.
| Strain | Genotype | Days after inoculation | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| 3 | 6 | 9 | 12 | 15 | 18 | 21 | 24 | 27 | 30 | 33 | 36 | 39 | 42 | 45 | 48 | ||
| No bacteria | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| MAV07 | olsE− | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| MAV07.pERMAV05 | olsE− | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| MAV07.pERMAV28 | olsE+ | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + |
| MAV08 | olsB− | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| MAV08.pERMAV06 | olsB− | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| MAV08.pERMAV30 | olsB+ | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + |
| A. tumefaciens A208 | Wild-type | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + |
Discs were inculated with 108 bacteria on day 0 and bacteria were killed by timentin treatment after 48 h. Discs were scored for tumour formation every 3–4 days. The experiment was repeated independently three times. Each experiment contained 30 replicates of each strain.
Discussion
Ornithine lipids are widespread in eubacteria. In addition to the unmodified OL consisting of a 3-hydroxy fatty acid linked by an amide bond to the α-amino group of ornithine and a second fatty acid bound in ester linkage to the first, several hydroxylated forms of OL have been described. The exact function of these modifications is not known yet, but several studies now hint at functions related to abiotic stress resistance and during interactions with eukaryotic hosts (Dees and Shively, 1982; Asselineau, 1991; Taylor et al., 1998; Rojas-Jiménez et al., 2005; González-Silva et al., 2011; Vences-Guzmán et al., 2011).
Here, we wanted to study the role of OLs in the plant pathogen A. tumefaciens. We characterized A. tumefaciens mutants deficient in OL biosynthesis under a set of abiotic stress conditions: low growth temperatures, high salt concentrations, low phosphate concentration in the growth medium, and acid stress.
It has been speculated that the 2-hydroxylation of the ester-linked fatty acid in OL may increase hydrogen bonding between neighbouring OL molecules similarly as has been suggested for LpxO-hydroxylated lipid A in Salmonella and hydroxylated sphingolipids (Gibbons et al., 2000; Nikaido, 2003; Murata et al., 2007). These additional hydrogen bonds should result in bilayer stabilization and a decrease in membrane permeability. The fact that the OL S2 with OlsE-dependent modification accumulates at low growth temperatures in A. tumefaciens indicates that introducing a hydroxyl group into the OL headgroup might have the opposite effect than introducing it into the ester-linked fatty acid.
Agrobacterium tumefaciens forms two OLs even under high phosphate conditions. Together both OLs make up about 15% of the membrane lipids. Under low phosphate conditions we observed an accumulation of OLs to up to 45–50% of total lipids. This is consistent with our bioinformatic analysis indicating that agrobacterial olsB is preceded by a Pho box. Apparently although olsB is constitutively expressed in A. tumefaciens, the response regulator PhoB can further induce the levels of olsB transcription under conditions of phosphate limitation. No Pho box was predicted for the olsE promoter.
The presence of the OL hydroxylase OlsC has been shown to be important in R. tropici to resist conditions of acid stress and high temperature. Agrobacterium tumefaciens wild-type grows in complex media at pH 5.2 whereas R. tropici is distinctively more acid-resistant and grows at pH 4.0. Expressing olsC from R. tropici in A. tumefaciens would enable A. tumefaciens to synthesize the OL P1 which has an important role under these conditions in R. tropici (Vences-Guzmán et al., 2011). Although OlsC is clearly active in the Agrobacterium strain harbouring the olsC-containing plasmid, no effect on acid stress resistance can be observed. This indicates that the stress resistance observed in R. tropici and that depended on the presence of OlsC might not be a direct effect of the presence of hydroxylated OL, but rather an indirect effect. The hydroxylated OL might somehow influence the enzyme activity of a protein important in stress resistance present in R. tropici but absent in A. tumefaciens. An alternative explanation is that OlsC might be able to hydroxylate a second substrate in addition to OLs and that this hydroxylation is responsible for the observed stress resistance.
Potato tuber discs inoculated with the mutant MAV08 (ΔolsB) deficient in OL formation or with the mutant MAV07 (ΔolsE) deficient in formation of the OL S2, show tumour formation about 1 week earlier than plants inoculated with wild-type bacteria. Complementation of both mutants (MAV07 and MAV08) with the respective genes in trans restores the slow tumour growth phenotype. A consequence of the earlier onset of tumour formation is the increased size of the tumours that is observed. Crown gall tumour formation occurs because A. tumefaciens can transfer an oncogenic T-DNA from its tumour-inducing (Ti) plasmid into plant cells. After integration of the T-DNA into the plant host genome, T-DNA-encoded proteins cause cytokine and auxin synthesis eventually leading to crown gall formation. T-DNA transfer from A. tumefaciens to the plant cell requires activation of the vir regulon present on the Ti plasmid (Gelvin, 2006). Salicylic acid and ethylene, two plant signal molecules involved in the regulation of defence-regulated gene expression during plant–microbe interaction (Ecker and Davis, 1987; Prithiviraj et al., 2005) have been shown to inhibit expression of the vir regulon (Yuan et al., 2007; Nonaka et al., 2008). One possible hypothesis for the earlier appearance of tumours is that (hydroxylated) OLs can be detected by the plant and that the presence of (hydroxylated) OLs causes some kind of plant defence leading to the production of ethylene or salicylic acid. In the absence of OLs this response is not as strong or absent and the infection process is accelerated. Interestingly, OLs share a 3-acyl-oxyacylamide structure with lipid A from Gram-negative bacteria, which has been shown to work as an elicitor in plant–microbe interactions (Scheidle et al., 2005; Silipo et al., 2008; 2010; Madala et al., 2011). Both, OLs and lipid A are OM lipids. Several studies have shown that plants can sense lipid A but owing to the limited number of lipid A structures determined from plant-associated bacteria, no clear structure–activity relationship exists as for lipid A effects in animals (Madala et al., 2011). Our observation that the absence of an agrobacterial membrane lipid affects tumour formation is not unprecedented. Wessel and colleagues (2006) had observed that A. tumefaciens mutants not able to form PC are deficient in tumour formation because they completely lack the type IV secretion machinery (Wessel et al., 2006).
The exact functions of OLs are still unknown but the present study adds more evidence to an important role of these lipids in bacteria–host interactions.
Experimental procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in the present work and their relevant characteristics are shown in Table S1. Agrobacterium tumefaciens strains were grown in complex LB medium at 30°C, in complex 20E medium at 30°C (Werner et al., 1975), or in Sherwood minimal medium (Sherwood, 1970) at 30°C or 15°C. Escherichia coli strains were grown in LB medium at 37°C. Sinorhizobium meliloti CS111.pNG25 was grown in complex tryptone-yeast extract medium (TY) (Beringer, 1974) supplemented with 10 mM CaCl2. When needed, antibiotics were added at the following final concentrations (mg ml−1): kanamycin (Km) 50; carbenicillin (Cb) 100; tetracycline (Tc) 8; timentin (Tim) 100, and rifampicin (Rif) 10.
DNA manipulations
Recombinant DNA techniques were performed according to standard protocols (Russell and Sambrook, 2001). Oligonucleotide primer sequences are listed in Table S2.
Expression of the candidate ORF Atu0318 from Agrobacterium tumefaciens
The candidate ORF for OlsE (Atu0318) was amplified using genomic DNA from A. tumefaciens A208 as a template and XL-PCR polymerase (Applied Biosystems). Specific oligonucleotide primers oC58OlsEN and oC58OlsEH incorporating NdeI and HindIII sites into the PCR product were used. After digestion with the respective enzymes, the PCR product was cloned as NdeI/HindIII fragment into pET17b yielding pEMAV27. Subsequently, the plasmid was linearized with HindIII and cloned into the HindIII site of pRK404 yielding pERMAV28. As a negative control pET17b was linearized with HindIII and cloned into the HindIII site of pRK404 yielding pERMAV05.
In vivo labelling of S. meliloti and A. tumefaciens with [14C]acetate or [14C]ornithine and quantitative analysis of lipid extracts
The lipid compositions of bacterial strains were determined following labelling with [1-14C]acetate (Amersham Biosciences). In the case of A. tumefaciens, cells were also labelled with [14C]ornithine to observe specific incorporation into OLs. Cultures (1 ml) of wild-type and mutant strains were inoculated from precultures grown in the same medium. After addition of 0.5 mCi [14C]acetate (60 mCi mmol−1) or 1 mCi [14C]ornithine (56 mCi mmol−1) to each culture, the cultures were incubated for 4 h if not indicated otherwise. Under phosphate starvation conditions cells were labelled for 48 h in Sherwood minimal medium (Sherwood, 1970) containing 20 μM phosphate. Lipid extraction and analysis are described in detail in Vences-Guzmán and colleagues (2011).
Construction of the olsB and olsE deletion mutants
Oligonucleotide primers oC58OlsEar1 and oC58OlsEar2 were used in a PCR (XL-PCR kit; Applied Biosystems) to amplify about 1.0 kb of genomic DNA upstream of the putative olsE (Atu0318) gene from A. tumefaciens, introducing EcoRI and BamHI sites into the PCR product. Similarly, primers oC58OlsEab1 and oC58OlsEab2 were used to amplify about 1.0 kb of genomic DNA downstream of the putative olsE gene from A. tumefaciens, introducing BamHI and XbaI sites into the PCR product.
After digestion with the respective enzymes, PCR products were cloned as EcoRI/BamHI or BamHI/XbaI fragments into pUC18 to yield the plasmids pUMAV18 and pUMAV19 respectively. Then, the BamHI/XbaI fragment from pUMAV19 was subcloned into pUMAV18 to yield pUMAV20. Plasmid pUMAV20 was digested with EcoRI and XbaI to subclone the regions usually flanking the olsE gene into the suicide vector pK18mobsacB (Schäfer et al., 1994) to yield pPMAV21. Via diparental mating using E. coli S17-1 as a mobilizing strain, pPMAV21 was introduced into the wild-type strain A. tumefaciens A208. Transconjugants were selected on LB medium containing neomycin and rifampicin to select for single recombinants in a first step. The plasmid pK18mobsacB contains the sacB gene (Selbitschka et al., 1993), which confers sucrose sensitivity to many bacteria. Growth of the single recombinants on high sucrose will therefore select for double recombinants and the loss of the vector backbone of pK18mobsacB from the bacterial genome. Single recombinants were grown under non-selective conditions in complex medium for 1 day before being plated on LB medium containing 12% (w/v) sucrose. Several large and small colonies grew after 4 days, and the membrane lipids of eight candidates were analysed by in vivo labelling during growth on complex medium with [14C]acetate and subsequent TLC (data not shown). Southern blot analysis confirmed that the S2-deficient strains were indeed double recombinants in which the gene olsE was deleted (data not shown).
Similarly, an olsB deletion mutant was constructed. Oligonucleotide primers oC58OlsBar1 and oC58OlsBar2 were used in a PCR (XL-PCR kit; Applied Biosystems) to amplify about 1.0 kb of genomic DNA upstream of the putative olsB gene from A. tumefaciens, introducing SmaI and BamHI sites into the PCR product. Similarly, primers oC58OlsBab1 and oC58OlsBab2 were used to amplify about 1.0 kb of genomic DNA downstream of the putative olsB gene from A. tumefaciens, introducing BamHI and HindIII sites into the PCR product. After digestion with the respective enzymes, PCR products were cloned as SmaI/BamHI or BamHI/HindIII fragments into pUC18 to yield the plasmids pUMAV23 and pUMAV24 respectively. The A. tumefaciens BamHI/HindIII fragment from pUMAV24 was subcloned into pUMAV23 to yield pUMAV25. Plasmid pUMAV25 was digested with SmaI and HindIII to subclone the regions usually flanking the olsB gene into the suicide vector pK18mobsacB (Schäfer et al., 1994) to yield pPMAV26. Via diparental mating using E. coli S17-1 (Simon et al., 1983) as a mobilizing strain, pPMAV26 was introduced into the wild-type strain A. tumefaciens. Subsequent steps were performed as described above for the construction of the olsE mutant. Four clones lacking OLs S1 and S2 were identified. Southern blot analysis confirmed that the S1- and S2-deficient strains were indeed double recombinants in which the gene olsB was deleted (data not shown).
Complementation of the A. tumefaciens mutants MAV07 and MAV08
The olsE (Atu0318) gene was amplified using genomic DNA from A. tumefaciens A208 as template and XL-PCR polymerase (Applied Biosystems). Specific oligonucleotide primers incorporating NdeI and HindIII sites into the final PCR product were used (oC58OlsXN and oC58OlsXH). The digested PCR product was cloned into pET17b to yield the plasmid pEMAV27. Plasmid pEMAV27 was linearized with HindIII and cloned into HindIII-digested pRK404 to yield pERMAV28. As a negative control pET17b was linearized with HindIII and cloned into the HindIII site of pRK404 yielding pERMAV05. In this construct olsE is expressed under control of the T7 promoter. In earlier work we had observed that this promoter causes the constitutive expression of the genes under control in different Rhizobiaceae (Gao et al., 2004; Sohlenkamp et al., 2007; Vences-Guzmán et al., 2011).
Similarly, the olsB-deficient mutant MAV08 was complemented with an intact copy of olsB in trans. The gene olsB (Atu0344) was amplified using genomic DNA from A. tumefaciens A208 as template and XL-PCR polymerase (Applied Biosystems). Specific oligonucleotide primers incorporating NdeI and BamHI sites into the final PCR product were used (oC58OlsBN and oC58OlsBB). The digested PCR product was cloned into pET9a to yield the plasmid pEMAV29. Plasmid pEMAV29 was linearized with BamHI and cloned into BamHI-digested pRK404 to yield pERMAV30. As a negative control pET9a was linearized with BamHI and cloned into the BamHI site of pRK404 yielding pERMAV06. In this construct olsB is expressed under control of the T7 promoter.
ESI-MS/MS analysis of ornithine lipids
In order to identify in which part of the second OL the modification is encountered, a 1 l culture A. tumefaciens was grown to an optical density of 1.2 at 620 nm in LB medium, and lipids were extracted according to Bligh and Dyer (1959). The lipids were separated by one-dimensional TLC and the area of the TLC plate corresponding to the OLs was identified by iodine and ninhydrin staining as described above. Silica containing OLs was scraped from the TLC plates and the OLs were extracted from the silica. OL-containing fractions were dried under N2 stream and redissolved in methanol/chloroform (1:1, v/v). Normal phase LC-MS/MS of lipids was made as described in detail in Vences-Guzmán and colleagues (2011).
Identification of potential Pho boxes in the Agrobacterium tumefaciens genome
To study if OL biosynthesis and/or modification are under control of PhoB the genome sequence of A. tumefaciens C58 was searched for the presence of putative Pho boxes. A PSSM was constructed using Pho box sequences reported previously for S. meliloti 1021 (Yuan et al., 2006). Escherichia coli sequences were excluded for matrix construction. Sequences used for matrix construction are listed in Table S3. The program Info-Gibbs (Defrance and van Helden, 2009) was used for matrix construction. One motif per sequence was expected and the desired matrix size was fixed to 18 bp. As background model the regions upstream of A. tumefaciens C58 genes were employed. The resulting matrix has a maximum score for putative Pho boxes of 12.4, and the threshold used for the search was set to 7. The program Matrix-scan (Turatsinze et al., 2008) was used to search the A. tumefaciens C58 genome for the presence of Pho boxes. The Markov order used was 0. Again the upstream regions of genes from A. tumefaciens C58 were employed as the background model. The search was performed scanning the upstream regions of predicted genes in 18 bp windows and comparing them with the PSSM. This search was performed for both strands. The list of genes presenting putative upstream Pho boxes was analysed for the presence of OL biosynthesis genes olsB, olsE and genes coding for glycosyltransferases (Cantarel et al., 2009).
Tumour assays using potato tuber discs
The tumour assays were performed according to Tsai and colleagues (2009) (Shurvinton and Ream, 1991; Tsai et al., 2009). Potato tubers were peeled and then immersed for 5 min in 1.05% sodium hypochlorite (17.5% bleach) solution. Tubers were rinsed three times with sterile water. A sterile cork borer was used to cut 1.0 cm diameter cylinders of potato tubers. About 1 cm pieces were removed from each end of a cylinder and the remaining cores were cut into 2–3 mm thick discs and placed on water agar plates. Each disc was inoculated on one side with 106 cells suspended in 100 μl PBS. Discs were placed on 1% water agar plates and incubated for 2 days in a controlled growth chamber at 28°C with a 15 h day/9 h night cycle. Potato discs were treated with timentin to kill all bacteria and then transferred to 1% (w/v) water agar plates containing timentin. Potato discs were incubated for 48 days under the conditions described above. Discs were scored every 3 days for the presence of tumours. After 48 days images were taken of the potato discs using a stereomicroscope equipped with a digital camera.
Supplementary Material
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Biosynthesis of OLs in Agrobacterium tumefaciens. The genes coding for OlsB and OlsA have been first identified in Sinorhizobium meliloti. Here we describe the identification of the gene encoding the OL hydroxylase OlsE introducing a hydroxyl group in the ornithine moiety of OL S1 leading to the formation of OL S2. LOL, lyso-ornithine lipid.
Fig. S2. Negative ion mode mass spectra of OL S1 (A) and OL S2 (B) purified from A. tumefaciens A208. The major OL [M-H]− ion species are labelled with arrows.
Fig. S3. Ornithine lipid S2 shows a different ninhydrin staining compared with OL S1. Unlabelled lipids were extracted according to Bligh and Dyer from 50 ml cultures of A. tumefaciens A208 (A) and R. tropici CIAT899 (B) grown in LB medium and separated in duplicate by two-dimensional TLC. Lipids were subjected to ninhydrin staining. The phospholipids PE, monomethyl PE (MMPE), and the OLs S1, S2, P1 and P2 are indicated.
Table S1. Bacterial strains and plasmids used in this study.
Table S2. Oligonucleotides used in this study. Introduced restriction sites are underlined.
Table S3. Sequences used for PSSM construction using the program Info-Gibbs. Sequences were from taken from Yuan and colleagues (2006).
Acknowledgments
This research was supported by grants from CONACyT-Mexico (46020-N and 153200) and DGAPA/UNAM (IN217907 and IN201310). Z.G. and the mass spectrometry facility in the Department of Biochemistry, Duke University Medical Center, were supported by a LIPID MAPS glue grant (GM-069338) from the National Institutes of Health. M.Á.V.-G. is a PhD student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México and is a recipient of a scholarship from the Consejo Nacional de Ciencia y Tecnología, Mexico.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselineau J. Bacterial lipids containing amino acids or peptides linked by amide bonds. Fortschr Chem Org Naturst. 1991;56:1–85. doi: 10.1007/978-3-7091-9084-5_1. [DOI] [PubMed] [Google Scholar]
- Benning C. Membrane lipids in anoxygenic photosynthetic bacteria. In: Siegenthaler PA, Murata N, editors. Lipids in Photosynthesis: Structure, Function and Genetics. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1998. pp. 83–101. [Google Scholar]
- Benning C, Huang ZH, Gage DA. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bukata L, Altabe S, de Mendoza D, Ugalde RA, Comerci DJ. Phosphatidylethanolamine synthesis is required for optimal virulence of Brucella abortus. J Bacteriol. 2008;190:8197–8203. doi: 10.1128/JB.01069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerci DJ, Altabe S, de Mendoza D, Ugalde RA. Brucella abortus synthesizes phosphatidylcholine from choline provided by the host. J Bacteriol. 2006;188:1929–1934. doi: 10.1128/JB.188.5.1929-1934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees C, Shively JM. Localization of quantitation of the ornithine lipid of Thiobacillus thiooxidans. J Bacteriol. 1982;149:798–799. doi: 10.1128/jb.149.2.798-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defrance M, van Helden J. Info-Gibbs: a motif discovery algorithm that directly optimizes information content during sampling. Bioinformatics. 2009;25:2715–2722. doi: 10.1093/bioinformatics/btp490. [DOI] [PubMed] [Google Scholar]
- Devers EA, Wewer V, Dombrink I, Dörmann P, Hölzl G. A processive glycosyltransferase involved in glycolipid synthesis during phosphate deprivation in Mesorhizobium loti. J Bacteriol. 2011;193:1377–1384. doi: 10.1128/JB.00768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Davis RW. Plant defense genes are regulated by ethylene. Proc Natl Acad Sci USA. 1987;84:5202–5206. doi: 10.1073/pnas.84.15.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JL, Weissenmayer B, Taylor AM, Thomas-Oates J, López-Lara IM, Geiger O. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol Microbiol. 2004;53:1757–1770. doi: 10.1111/j.1365-2958.2004.04240.x. [DOI] [PubMed] [Google Scholar]
- Geiger O, Röhrs V, Weissenmayer B, Finan TM, Thomas-Oates JE. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol Microbiol. 1999;32:63–73. doi: 10.1046/j.1365-2958.1999.01325.x. [DOI] [PubMed] [Google Scholar]
- Geiger O, González-Silva N, López-Lara IM, Sohlenkamp C. Amino acid-containing membrane lipids in bacteria. Prog Lipid Res. 2010;49:46–60. doi: 10.1016/j.plipres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Gelvin SB. Agrobacterium virulence gene induction. Methods Mol Biol. 2006;343:77–84. doi: 10.1385/1-59745-130-4:77. [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Lin S, Cotter RJ, Raetz CR. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J Biol Chem. 2000;275:32940–32949. doi: 10.1074/jbc.M005779200. [DOI] [PubMed] [Google Scholar]
- González-Silva N, López-Lara IM, Reyes-Lamothe R, Taylor AM, Sumpton D, Thomas-Oates J, Geiger O. The dioxygenase-encoding olsD gene from Burkholderia cenocepacia causes the hydroxylation of the amide-linked fatty acyl moiety of ornithine-containing membrane lipids. Biochemistry. 2011;50:6396–6408. doi: 10.1021/bi200706v. [DOI] [PubMed] [Google Scholar]
- López-Lara IM, Sohlenkamp C, Geiger O. Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions. Mol Plant Microbe Interact. 2003;16:567–579. doi: 10.1094/MPMI.2003.16.7.567. [DOI] [PubMed] [Google Scholar]
- Madala NE, Leone MR, Molinaro A, Dubery IA. Deciphering the structural and biological properties of the lipid A moiety of lipopolysaccharides from Burkholderia cepacia strain ASP B 2D, in Arabidopsis thaliana. Glycobiology. 2011;21:184–194. doi: 10.1093/glycob/cwq146. [DOI] [PubMed] [Google Scholar]
- Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Yuhashi K, Takada K, Sugaware M, Minamisawa K, Ezura H. Ethylene production in plants during transformation suppresses vir gene expression in Agrobacterium tumefaciens. New Phytol. 2008;178:647–656. doi: 10.1111/j.1469-8137.2008.02400.x. [DOI] [PubMed] [Google Scholar]
- Prithiviraj B, Bais HP, Weir T, Suresh B, Najarro EH, Dayakar BV, et al. Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect Immun. 2005;73:5319–5328. doi: 10.1128/IAI.73.9.5319-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Jiménez K, Sohlenkamp C, Geiger O, Martínez-Romero E, Werner D, Vinuesa P. A ClC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol Plant Microbe Interact. 2005;18:1175–1185. doi: 10.1094/MPMI-18-1175. [DOI] [PubMed] [Google Scholar]
- Russell DW, Sambrook JG. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schäfer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Scheidle H, Gross A, Niehaus K. The Lipid A substructure of the Sinorhizobium meliloti lipopolysaccharides is sufficient to suppress the oxidative burst in host plants. New Phytol. 2005;165:559–565. doi: 10.1111/j.1469-8137.2004.01214.x. [DOI] [PubMed] [Google Scholar]
- Selbitschka W, Niemann S, Pühler A. Construction of gene replacement vectors for Gram- bacteria using a genetically modified sacRN gene as a positive selection marker. Appl Microbiol Biotechnol. 1993;38:615–618. [Google Scholar]
- Sherwood MT. Improved synthetic medium for the growth of Rhizobium. J Appl Bacteriol. 1970;33:708–713. doi: 10.1111/j.1365-2672.1970.tb02253.x. [DOI] [PubMed] [Google Scholar]
- Shurvinton CE, Ream W. Stimulation of Agrobacterium tumefaciens T-DNA transfer by overdrive depends on a flanking sequence but not on helical position with respect to the border repeat. J Bacteriol. 1991;173:5558–5563. doi: 10.1128/jb.173.17.5558-5563.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A, Sturiale L, Garozzo D, Erbs G, Jensen TT, Lanzetta R, et al. The acylation and phosphorylation pattern of lipid a from Xanthomonas campestris strongly influence its ability to trigger the innate immune response in Arabidopsis. Chembiochem. 2008;9:896–904. doi: 10.1002/cbic.200700693. [DOI] [PubMed] [Google Scholar]
- Silipo A, Erbs G, Shinya T, Dow JM, Parrilli M, Lanzetta R, et al. Glyco-conjugates as elicitors or suppressors of plant innate immunity. Glycobiology. 2010;20:406–419. doi: 10.1093/glycob/cwp201. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- Sohlenkamp C, Galindo-Lagunas KA, Guan Z, Vinuesa P, Robinson S, Thomas-Oates J, et al. The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to polymyxin B under acidic growth conditions. Mol Plant Microbe Interact. 2007;20:1421–1430. doi: 10.1094/MPMI-20-11-1421. [DOI] [PubMed] [Google Scholar]
- Taylor CJ, Anderson AJ, Wilkinson SG. Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology. 1998;144:1737–1745. doi: 10.1099/00221287-144-7-1737. [DOI] [PubMed] [Google Scholar]
- Thiele OW, Schwinn G. The free lipids of Brucella melitensis and Bordetella pertussis. Eur J Biochem. 1973;34:333–344. doi: 10.1111/j.1432-1033.1973.tb02764.x. [DOI] [PubMed] [Google Scholar]
- Tsai YL, Wang MH, Gao C, Klusener S, Baron C, Narberhaus F, Lai EM. Small heat-shock protein HspL is induced by VirB protein(s) and promotes VirB/D4-mediated DNA transfer in Agrobacterium tumefaciens. Microbiology. 2009;155:3270–3280. doi: 10.1099/mic.0.030676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- Vences-Guzmán MA, Guan Z, Ormeño-Orrillo E, González-Silva N, López-Lara IM, Martínez-Romero E, et al. Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol Microbiol. 2011;79:1496–1514. doi: 10.1111/j.1365-2958.2011.07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vences-Guzmán MA, Geiger O, Sohlenkamp C. Ornithine lipids and their structural modifications: from A to E and beyond. FEMS Microbiol Lett. 2012 doi: 10.1111/j.1574-6968.2012.02623.x. [DOI] [PubMed] [Google Scholar]
- Weissenmayer B, Gao JL, López-Lara IM, Geiger O. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol Microbiol. 2002;45:721–733. doi: 10.1046/j.1365-2958.2002.03043.x. [DOI] [PubMed] [Google Scholar]
- Werner D, Wilcockson J, Zimmermann E. Adsorption and selection of rhizobia with ion-exchange papers. Arch Microbiol. 1975;105:27–32. doi: 10.1007/BF00447108. [DOI] [PubMed] [Google Scholar]
- Wessel M, Klüsener S, Gödeke J, Fritz C, Hacker S, Narberhaus F. Virulence of Agrobacterium tumefaciens requires phosphatidylcholine in the bacterial membrane. Mol Microbiol. 2006;62:906–915. doi: 10.1111/j.1365-2958.2006.05425.x. [DOI] [PubMed] [Google Scholar]
- Yuan ZC, Zaheer R, Morton R, Finan TM. Genome prediction of PhoB regulated promoters in Sinorhizobium meliloti and twelve proteobacteria. Nucleic Acids Res. 2006;34:2686–2697. doi: 10.1093/nar/gkl365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZC, Edlind MP, Liu P, Saenkham P, Banta LM, Wise AA, et al. The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone-quenching genes in Agrobacterium. Proc Natl Acad Sci USA. 2007;104:11790–11795. doi: 10.1073/pnas.0704866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaleta-Pastor M, Sohlenkamp C, Gao JL, Guan Z, Zaheer R, Finan TM, et al. Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc Natl Acad Sci USA. 2010;107:302–307. doi: 10.1073/pnas.0912930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Biosynthesis of OLs in Agrobacterium tumefaciens. The genes coding for OlsB and OlsA have been first identified in Sinorhizobium meliloti. Here we describe the identification of the gene encoding the OL hydroxylase OlsE introducing a hydroxyl group in the ornithine moiety of OL S1 leading to the formation of OL S2. LOL, lyso-ornithine lipid.
Fig. S2. Negative ion mode mass spectra of OL S1 (A) and OL S2 (B) purified from A. tumefaciens A208. The major OL [M-H]− ion species are labelled with arrows.
Fig. S3. Ornithine lipid S2 shows a different ninhydrin staining compared with OL S1. Unlabelled lipids were extracted according to Bligh and Dyer from 50 ml cultures of A. tumefaciens A208 (A) and R. tropici CIAT899 (B) grown in LB medium and separated in duplicate by two-dimensional TLC. Lipids were subjected to ninhydrin staining. The phospholipids PE, monomethyl PE (MMPE), and the OLs S1, S2, P1 and P2 are indicated.
Table S1. Bacterial strains and plasmids used in this study.
Table S2. Oligonucleotides used in this study. Introduced restriction sites are underlined.
Table S3. Sequences used for PSSM construction using the program Info-Gibbs. Sequences were from taken from Yuan and colleagues (2006).