Abstract
Using nanopores to sequence biopolymers was proposed more than a decade ago1. Recent advances in enzyme-based control of DNA translocation2 and in DNA nucleotide resolution using modified biological pores3 have satisfied two technical requirements of a functional nanopore DNA sequencing device. Nanopore sequencing of proteins was also envisioned1. Although proteins have been shown to move through nanopores4, 5, 6, a technique to unfold proteins for processive translocation has yet to be demonstrated. Here we describe controlled unfolding and translocation of proteins through the α-hemolysin (α-HL) pore using the AAA+ unfoldase ClpX. Sequence-dependent features of individual engineered proteins were detected during translocation. These results demonstrate that molecular motors can reproducibly drive proteins through a model nanopore—a feature required for protein sequence analysis using this single-molecule technology.
Protein sequencing using nanopores is technically challenging for two reasons: (i) both tertiary and secondary structures must be unfolded to allow the denatured protein to thread through the nanopore sensor with amino acid residues in single-file order; and (ii) processive unidirectional translocation of the denatured polypeptide through the nanopore electric field must be achieved despite a nonuniform charge along the polypeptide chain. To address these challenges, we have devised a general method for enzyme-controlled unfolding and translocation of native proteins through a nanopore sensor using the protein unfoldase ClpX. ClpX is a component of the ClpXP proteasome-like complex that is responsible for the targeted degradation of numerous protein substrates in Escherichia coli and other organisms7. Within this complex, ClpX forms a homohexameric ring that uses ATP hydrolysis to unfold and translocate proteins through its central pore and into a proteolytic chamber (ClpP) for degradation. We chose E. coli ClpX for this nanopore application because it generates sufficient mechanical force (~20 pN) to denature stable protein folds, and because it translocates along proteins at a rate suitable for primary sequence analysis by nanopore sensors (up to 80 amino acids per second)8, 9.
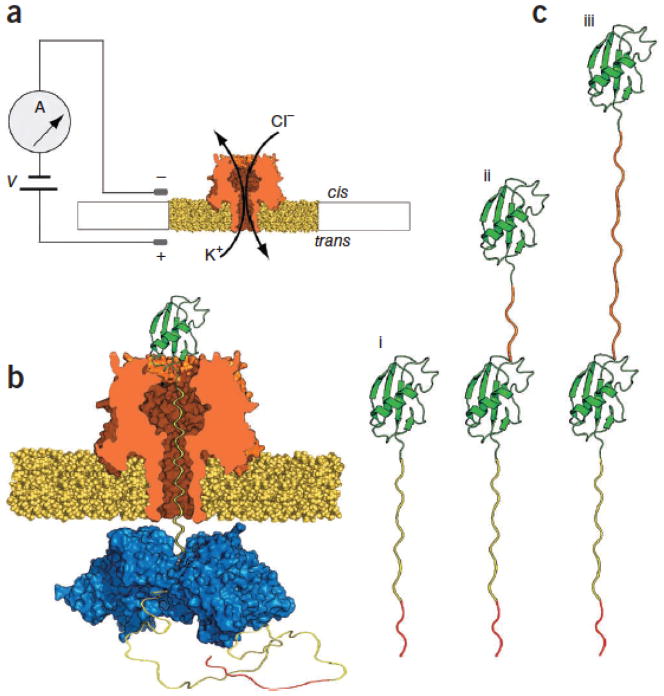
For each experiment a single α-HL nanopore was inserted into a 30-μm-diameter lipid bilayer between two compartments (termed cis and trans), each filled with 100 μl of a 200 mM KCl buffer optimized for ClpX function and supplemented with 5 mM ATP (Fig. 1a). A patch-clamp amplifier applied a constant 180 mV potential between two Ag/AgCl electrodes (trans side +) and recorded ionic current through the nanopore as a function of time. This electrophysiology setup is fundamentally the same as one used in earlier experiments to identify a protein-conducting channel in the endoplasmic reticulum10. Substrate proteins were added to the cis solution at ~1 μM final concentration, and ~100 nM of a linked ClpX variant (Online Methods) was present in the trans solution.
Figure 1.
(a) Nanopore sensor. A single α-HL pore is embedded in a lipid bilayer separating two polytetrafluoroethylene wells each containing 100 μl of 0.2 M KCl solution at 30 °C. Voltage is applied between the wells (trans side +180 mV), causing ionic current flow through the channel. Current diminishes in the presence of a captured protein molecule. (b) Protein capture in the nanopore. A model protein bearing an Smt3 domain (green) at its N terminus is coupled to a charged flexible linker (yellow) with an ssrA tag (red) at its C terminus. As a result of the applied voltage, the charged, flexible tag is threaded through the pore into the trans-side solution until the folded Smt3 domain prevents complete translocation of the captured protein. ClpX present in the trans solution binds the C-terminal ssrA sequence. Fueled by ATP hydrolysis, ClpX translocates along the protein tail toward the channel, and subsequently catalyzes unfolding and translocation of the Smt3 domain through the pore. (c) Engineered proteins used in this study. S1, a protein bearing a single N-terminal Smt3-domain coupled to a 65-amino-acid-long charged flexible segment capped at its carboxy terminus with the 11 amino acid ClpX-targeting domain (ssrA tag) (i); S2-35, similar to S1 but appended at its N terminus by a 35-amino-acid linker and a second Smt3 domain (ii); S2-148, identical to S2-35 except for an extended 148-amino-acid linker between the Smt3 domains (iii). The linker lengths in this panel are not to scale.
For our initial experiments, we used a modified version of the ubiquitin-like protein Smt3 (ref. 11) as the substrate. Smt3 comprises 98 amino acids arranged into four β-strands and a single α-helix. To facilitate nanopore analysis, we modified the engineered Smt3 protein, designated ‘S1’, in two ways. First, it was appended with a 65-amino-acid-long glycine/serine tail including 13 interspersed negatively charged aspartate residues (Supplementary Fig. 1). This unstructured polyanion was designed to promote capture and retention of S1 in the electric field across the nanopore. Based on its crystal structure12, the Smt3 folded domain is predicted to sit on top of the α-HL vestibule. Second, the appended polyanion was capped at its C terminus with the ssrA tag, an 11-amino-acid ClpX-targeting motif13. This ssrA peptide tag allowed ClpX to specifically bind to the C terminus of the protein when it threaded through the pore into the trans compartment (Fig. 1b and c, i). Experiments conducted in bulk phase confirmed that ClpX unfolds and translocates proteins appended with this unique polyanion tag in an ATP-dependent manner (Supplementary Fig. 2).
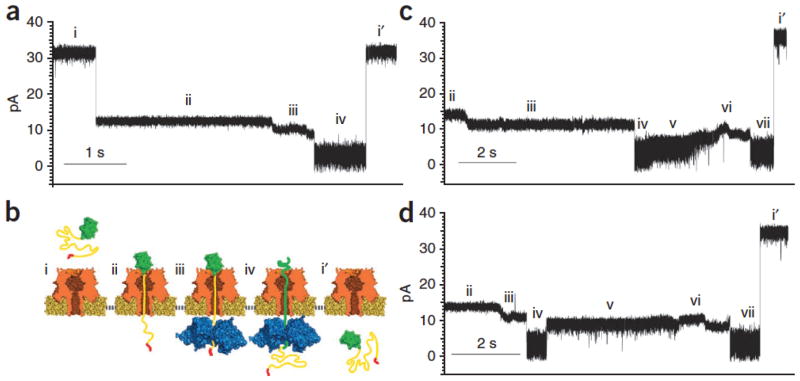
Representative ionic current traces for capture and translocation of protein S1 in the presence of ClpX and ATP are shown in Figure 2a and Supplementary Figure 3a. From the open channel current of ~34 ± 2 pA (Fig. 2a, i), S1 capture resulted in a current drop to ~14 pA (Fig. 2a, ii). This stable current lasted for tens of seconds and was observed in the presence or absence of ClpX and ATP added to the trans compartment. This is consistent with the Smt3 structure held stationary atop the pore vestibule by the electrical force acting on the charged polypeptide tail in the pore electric field. In the presence of ClpX and ATP, this initial ionic current state was often followed by a progressive downward current ramp reaching an average of ~11 pA with a median duration of 4.2 s (Fig. 2a, iii and Supplementary Fig. 4). This current ramp was observed with protein S1 a total of 45 times over ~5.5 h of experimentation when ClpX and ATP were present; in contrast, the ramp was never observed after ionic current state ii (Fig. 2a, ii) when ClpX or ATP was absent from the trans solution over ~2.3 and ~1.7 h of experimentation, respectively. In a majority of events, the ClpX-dependent ramping state terminated with an abrupt ionic current decrease to ~3.9 pA (Fig. 2a, iv). The median duration for state iv was ~700 ms before it ended in a rapid increase to open the channel current (Fig. 2a, i′). As an additional control, we constructed a variant of S1 appended with three additional amino acid residues at the C terminus (protein S1-RQA, Supplementary Fig. 1). Because ClpX recognition of the ssrA tag is dependent upon the C terminus α-carboxyl group, the additional residues placed between the tag sequence and the C terminus thereby inhibit ClpX binding14. In the presence of ATP and ClpX, we never observed the ramping state with protein S1-RQA over ~1.7 h of experimentation (data not shown).
Figure 2.
(a) S1 translocation. Open channel current through the α-HL nanopore under standard conditions (~34 ± 2 pA, RMS noise 1.2 ± 0.1 pA) (i). Capture of the S1 substrate. Upon protein capture, the ionic current drops to ~14 pA (~0.7 pA RMS noise) (ii). ClpX-mediated ramping state. The ionic current decreases to ~10 pA and is characterized by one or more gradual amplitude transitions. This pattern is only observed in the presence of ClpX and ATP (trans compartment) (iii). Smt3 domain unfolding and translocation through the nanopore (~3.8 pA, 1.7 pA RMS noise) (iv). Return to open channel current upon completion of substrate translocation to the trans compartment (i′). (b) Working model of ClpX-mediated translocation of S1. Roman numerals used to label panels correspond to ionic current states in a. (c) S2-35 translocation. Open channel current (i) is not shown. States ii–iv are identical to states ii–iv in a. Gradual increase in ionic current to ~10 pA. In our working model this corresponds to a transition from Smt3 domain translocation to linker region translocation (v). A second putative ramping state that closes resembles ramping state iii (vi). A second putative Smt3 translocation state with ionic current properties that closely resemble state iv (vii). Return to open channel current (i′). (d) S2-148 translocation. Ionic current states i–iv and vi–i′ were nearly identical to those states for S2-35 translocation in c. (v) In our working model, this ionic current state corresponds to translocation of the 148-amino-acid linker. Its amplitude is ~3 pA higher than the S2-35 linker amplitude (~9 pA), and it has a median duration ~2.5 fold longer than the comparable S2-35 state v. Translocation events that included ramping state iii were observed 62 times for protein S2-35 (7.3 h of experimentation), and 66 times for protein S2-148 (4.3 h of experimentation), when ClpX and ATP were present. In the absence of ClpX, these ramping states were never observed for S2-35 (1.7 h of experimentation) and S2-148 (1.2 h of experimentation).
Based on these data we hypothesized that ClpX served as a molecular machine that used chemical energy derived from ATP hydrolysis to pull the S1 protein through the nanopore. In the proposed process, an open channel (Fig. 2b, i) captures protein S1 with the Smt3 segment perched above the pore vestibule with the slender, charged polypeptide tail segment extended into the pore lumen, and the ssrA tag in the trans compartment (Fig. 2b, ii). In this ionic current state, ClpX is not bound to S1 or, alternatively, is bound but is still distant from the pore; ClpX advances along the S1 strand toward the trans-side orifice of the α-HL pore until it makes contact (Fig. 2b, iii). At this time, ionic current decreases owing to proximity of ClpX to the pore; under the combined force exerted by ClpX and the pore electric field, the Smt3 structure atop the pore is sequentially denatured, thus allowing the polypeptide to advance through the nanopore (Fig. 2b, iv). In this state, the ionic current has decreased because larger amino acids (or Smt3 secondary structures) have entered the pore lumen. This ionic current state persists until the S1 protein is completely pulled into the trans compartment resulting in a return to the open channel current (Fig. 2b, i′).
This model makes a testable prediction. If the observed states are due to processive movement of polypeptide segments into the pore lumen driven in part by ClpX, then changing the protein primary structure should result in sequential ClpX- and ATP-dependent changes in the ionic current pattern. In particular, addition of a second Smt3 domain should result in a second ramping state (Fig. 2a, iii) followed by a second Smt3 translocation state centered at ~4 pA (Fig. 2a, iv). As a test, we fused a flexible glycine/serine-rich 35 amino acid linker to the N terminus of the S1 protein and capped this with a second Smt3 domain (protein S2-35, Fig. 1c, ii and Supplementary Fig. 1). Thus, the single folded-component sequence of S1 (that is, Smt3) is repeated twice in S2-35.
When protein S2-35 was captured in the nanopore with ClpX and ATP present in the trans compartment, an ionic current pattern with eight reproducible states was observed (Fig. 2c and Supplementary Fig. 3b). The first four states (Fig. 2c, i–iv) were identical to states i iv caused by S1 translocation (compare Fig. 2a and c). This similarity included ramping state iii that is diagnostic for ClpX engagement, and the Smt3-dependent state iv. However, beginning at state v, the S2-35 pattern diverged from the S1 pattern (compare Fig. 2a and c). That is, following Smt3 translocation state iv, a typical S2-35 ionic current trace did not proceed to the open channel current but instead transitioned to a ~6.3-pA state with a median duration of 1.5 s (Fig. 2c, v). This was followed by a ~8.5-pA state (Fig. 2c, vi) that closely resembled ramping state iii, and a subsequent ionic current state that closely resembled the putative Smt3 translocation state iv (Fig. 2c, vii). In other words, consistent with our model, the putative ClpX-bound and Smt3-dependent states that were observed once during S1 events (Fig. 2a) were observed twice during S2-35 events (Fig. 2c). These analogous states for the two constructs shared nearly identical amplitudes, root mean square (RMS) noise values and durations. This dependence of ionic current on protein structure is consistent with ClpX-driven protein translocation through the nanopore. As an additional test, we re-examined ionic current state v observed during S2-35 translocation. This state is consistent with movement of the 35-amino-acid linker through the nanopore based on two observations: (i) its average ionic current is measurably higher than that of surrounding states (Fig. 2c) as expected for an amino acid sequence with few bulky side chains; and (ii) in the time domain, state v occurs between Smt3-dependent states iv and vi as expected, given its position along the S2-35 primary sequence (Fig. 1c, ii and Supplementary Fig. 1).
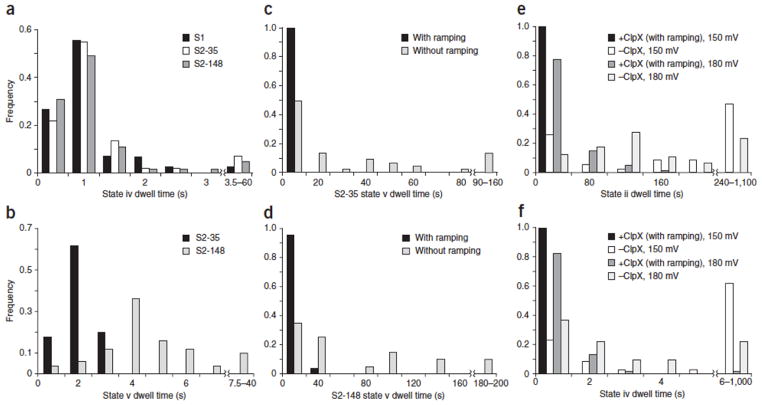
If state v corresponds to translocation of the polypeptide linker under ClpX control, then changes in the length and composition of this linker should result in duration and current amplitude changes. To test this, we designed a third protein in which the S2-35 linker region was appended with an additional 113 amino acids, yielding a final construct consisting of two Smt3 domains separated by an extended 148-amino-acid flexible linker (protein S2-148, Fig. 1c, iii and Supplementary Fig. 1). As predicted, when this protein was captured in the nanopore under standard conditions in the presence of ClpX and ATP, eight reproducible states similar to S2-35 events were observed (Figs. 2d,3a and Supplementary Figs. 3c,4–6). Importantly, however, the S2-35 and S2-148 events differed substantially at state v (compare Fig. 2c and d). That is, the S2-148 state v had a higher mean residual current than did S2-35 (~9 versus ~6 pA, respectively), and a median duration ~2.5 fold longer than that of S2-35 state v (Fig. 3b). The increased duration of S2-148 state v relative to S2-35 state v was anticipated and consistent with the model described in Figure 2. The increased current level was likely due to differences in linker amino acid composition between the two proteins (S2-35 linker: 51% Gly, 34% Ser, 15% other; S2-148 linker: 34% Gly, 32% Ser, 19% Ala, 15% other). However, confirmation of this hypothesis will require systematic testing of the relationship between amino acid identity and resistance to ionic current through the pore lumen.
Figure 3.
(a) Comparison of putative Smt3 translocation (state iv) dwell times for three model proteins. Values are from events that included the ClpX-dependent ramping state (Fig. 2,iii). Black bars: median = 0.71 s, interquartile range (IQR) = 0.41, n = 45. Gray bars: median = 0.64 s, IQR = 0.47, n = 60. White bars: median = 0.63 s, IQR = 0.40, n = 65. (b) Comparison of putative linker region (state v) dwell times for S2-35 and S2-148 proteins. Values are from events that included the ClpX-dependent ramping state (Fig. 2,iii). Black bars: median = 1.52 s, IQR = 0.68, n = 50. Gray bars: median = 3.62 s, IQR = 2.03, n = 50. (c) State v translocation dwell times for S2-35 events. Black bars: median = 1.52 s, IQR = 0.68, n = 50. Gray bars: median = 11.45 s, IQR = 39.53, n = 45. (d) State v translocation dwell times for S2-148 translocation events. Black bars: median = 3.62 s, IQR = 2.03, n = 50. Gray bars: median = 37.07 s, IQR = 89.80, n = 20. (e) State ii dwell times for protein substrates at two voltages. Black bars: median = 4.89 s, IQR = 6.72, n = 104. White bars: median = 165.50 s, IQR = 351.75, n = 34. Gray bars: median = 17.26 s, IQR = 31.08, n = 173. Light gray bars: median = 90.0 s, IQR = 112.0, n = 72. (f) State iv dwell times for the S1 protein substrate at two voltages. Black bars: median = 0.33 s, IQR = 0.40, n = 104. White bars: median = 8.25 s, IQR = 26.82, n = 34. Gray bars: median = 0.65 s, IQR = 0.45, n = 52. Light gray bars: median = 1.74 s, IQR = 3.12, n = 41.
Prior studies have shown that proteins can translocate through nanopores under an applied voltage without the assistance of processive enzymes4, 5, 6, 15, 16, 17. This was also the case in our experiments, that is, translocation of the three model proteins was observed in the absence of ClpX- or ATP-dependent mechanical work performed on captured strands (Supplementary Fig. 7). However, these ClpX- and ATP-independent translocation events lacked the diagnostic ramping states (Fig. 2), and they were measurably longer and more variable in duration than were ClpX-mediated translocation events (Fig. 3c–f). This is consistent with an unregulated translocation process that depends upon random structural fluctuations of the captured protein molecule and intermittent electrical force acting on amino acid segments with variable charge density in the pore electric field. This model predicts that ClpX-dependent translocation will be relatively unaffected by changes in applied voltage. This proved to be true. State ii and iv dwell times for ClpX- and ATP-dependent S1 translocation events acquired at 150 mV were comparable to those acquired at 180 mV (Fig. 3e, f). At both voltages, these events were consistently faster and more narrowly distributed than ClpX- or ATP-independent events. Thus, ClpX activity (not voltage) is dominating the unfolding and translocation process. In summary, we have demonstrated enzymatic control of protein unfolding and translocation through the α-HL nanopore. Segments of each protein could be discerned based on sequence-dependent features as the protein passed through the ~50-Angstrom-long trans-membrane pore lumen. This proof-of-concept study is analogous to early work on nanopore DNA sequencing in which T7 DNA polymerase moved model DNA templates through the α-HL pore over short distances18. As was true for DNA translocation control in that early work, protein translocation control for bench-top sequencing applications will require technical improvements. This is worthwhile because an optimized nanopore device could provide long reads of individual native protein strands.
Materials
A covalently linked trimer of an N-terminal truncated ClpX variant (ClpX-ΔN3) was used for all ClpX nanopore experiments19. This variant maintains near-WT ClpX activity levels on ssrA-tagged substrates while staying associated at lower working concentrations. The ClpX-ΔN3 BLR expression strain was obtained from A. Martin (UC Berkeley), as was a his-tagged ClpP expression strain. ClpX protein expression was induced at an A600 of ~1 by addition of 0.5 mM IPTG, and incubated at 23 °C with shaking for 3–4 h. Cultures were pelleted, resuspended in lysis buffer (50 mM NaH2PO4 pH 8, 300 mM NaCl, 100 mM KCl, 20 mM imidazole, 10% glycerol, 10 mM BME) and lysed by vortexing with glass beads. After centrifugation and filtration of the lysate, the protein was purified on a Ni2+-NTA affinity column (Thermo) and an Uno-Q anion exchange column (Bio-Rad). His-tagged ClpP protein expression was induced at an A600 of ~0.6 by addition of 0.5 mM IPTG, and incubated at 30 °C with shaking for 3–4 h; purification was done on a Ni2+-NTA affinity column. To construct substrate protein S1, DNA encoding the 76 amino acid tail(GGSSGGSGGSGSSGDGGSSGGSGGSGSSGDGGSSGGSGG-DGSSGDGGSDGDSDGSDGDGDSDGDDAANDENYALAA)was constructed by PCR and cloned into pET-SUMO vector (Invitrogen) at the T/A-cloning site, fusing the tail sequence onto the Smt3 sequence 3′ end. S2-35 was constructed by PCR-based addition of DNA encoding the 35 amino acid linker (GGSGSGGSGSGGSGSQNEYRSGGSGSGGSGSGGSG) to the 5′ end of the S1 Smt-3 sequence. This linker-modified S1 gene was then cloned into pE-SUMO vector (LifeSensors) at the BsaI site, fusing the added linker and S1 sequence to the 3′ end of the pE-SUMO Smt3 sequence. DNA for the S2-148 linker addition(GGSGSAGSGASGSSGSEGSGASGSAGS-GSAGSRGSGASGSAGSGSAGSGGAEAAKEAAKEAAKEAAKEAAKAGGSGSAGSAGSASSGSDGSGASG SAGSGSAGSKGSGASGSAGSGSSGS) was constructed by PCR, and cloned into the S2-35 vector within the 35 amino acid linker region by the Gibson assembly method. DNA for the GFP and titin-I27 domains of fluorescent protein S2-GT were extracted by PCR from a GFP-titin-I27V15P-ssrA20 expression vector obtained from A. Martin, and cloned into the S2-35 vector by Gibson assembly. These engineered proteins were expressed in BL21 (DE3)*. Expression was induced at ~0.6 A600 by addition of 0.5 mM IPTG, and incubated at 37 °C with shaking for 4–6 h. Cultures were pelleted, resuspended in lysis buffer and lysed via vortexing with glass beads. After centrifugation and filtration of the lysate, the protein was purified on a Ni2+-NTA affinity column (Thermo). The protein substrates and enzymes were flash frozen in small aliquots following purification and stored at −80 °C.
Nanopore experiments
All experiments were performed in PD buffer containing 200 mM KCl, 5 mM MgCl2, 10% glycerol and 25 mM HEPES-KOH pH 7.6. Setup of the nanopore device and insertion of an α-HL nanopore into a lipid bilayer have been described2. Briefly, a single α-HL nanopore was inserted into a lipid bilayer that separates two wells that each contained 100 μl of PD buffer. A constant 180 mV potential was applied across the bilayer and ionic current was measured through the nanopore between Ag/AgCl electrodes in series with an integrating patch clamp amplifier (Axopatch 200B, Molecular Devices) in voltage clamp mode. Data were recorded using an analog-to-digital converter (Digidata 1440A, Molecular Devices) at 100 kHz bandwidth in whole-cell configuration then filtered at 2 kHz using an analog low-pass Bessel filter. Experimental conditions were prepared by the daily preparation of PD/ATP 5 mM and PD/ATP 4 mM. ClpX was diluted 1:10 in PD/ATP 5 mM for a final concentration of 30–100 nM ClpX6 in 4.5 mM ATP final. ClpX solution was used to fill the entire system before isolation of a single α-HL nanopore. Upon insertion, the cis well was perfused with ~6 mL PD/ATP 4 mM. Experiments were conducted at 30 °C with 1–2 μM substrate added to the cis well. In the presence of ClpX, protein substrate capture events were ejected with reverse polarity due to pore clogs or after a predetermined duration. Those capture events that remained in state ii for a duration of ~90 s or longer were considered to be inaccessible to ClpX and were ejected. Voltage-induced translocations were frequently ejected to prevent clogging and to increase the efficiency of data collection. A single nanopore experiment is defined as the time during which ionic current data were acquired from one α-HL nanopore in an intact bilayer before termination by bilayer rupture, loss of channel conductance or completion of a preset number of translocation events. Experiments were conducted over multiple days for each substrate and condition tested. Prior to each experiment, protein samples that were to be used on that day were freshly thawed before the start of the experiment.
ClpX bulk phase assays
The protein S1 degradation time course was performed in PD buffer with 6 μM S1, 150 nM (ClpX-ΔN3)2, 300 nM ClpP14 and an ATP-regeneration mix (4 mM ATP, 16 mM creatine phosphate, and 0.16 mg/mL creatine phosphokinase) at 37 °C. Samples were visualized by SDS-PAGE gel (Any kD Mini-PROTEAN TGX, Bio-Rad) and stained with Coomassie blue. Fluorescence assays were performed in PD buffer with 1 μM S2-GT, 150 nM (ClpX-ΔN3)2, 300 nM ClpP14 and an ATP-regeneration mix. Reactions were incubated at 37 °C in a Stratagene Mx3005P measuring emission at 516 nm (excitation at 492 nm).
Supplementary Material
Acknowledgments
The authors thank Oxford Nanopore Technologies (Oxford, UK) for supplying α-HL heptamers, and A. Martin (UC Berkeley) for supplying ClpX-related expression plasmids and for helpful discussions on their use. R. Abu-Shumays, D. Bernick, K. Lieberman and H. Olsen commented on drafts of the manuscript. This work was supported by a UC startup grant to M.A., and by equipment purchased previously using National Human Genome Research Institute grant R01HG006321. The ClpX-ΔN3 BLR expression strain was obtained from A. Martin (UC Berkeley), as was a his-tagged ClpP expression strain.
Footnotes
Contributions
J.N. conceived and designed the project, performed the protein engineering and production, conceived and performed experiments and co-wrote the manuscript. D.B.M. performed and conceived experiments, analyzed data and co-wrote the manuscript. M.A. co-wrote the manuscript, designed the nanopore platform and directed the project.
Competing financial interests
M.A. is a consultant to Oxford Nanopore Technologies, Oxford, UK.
References
- 1.Church GM, Deamer DW, Branton D, Baldarelli R, Kasianowicz J. Characterization of individual polymer molecules based on monomer-interface inter- action. 5,795,782. US patent. 1998
- 2.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammad MM, Prakash S, Matouschek A, Movileanu L. Controlling a single protein in a nanopore through electrostatic traps. J Am Chem Soc. 2008;130:4081–4088. doi: 10.1021/ja710787a. [DOI] [PubMed] [Google Scholar]
- 5.Talaga DS, Li J. Single-molecule protein unfolding in solid state nanopores. J Am Chem Soc. 2009;131:9287–9297. doi: 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merstorf C, et al. Wild type, mutant protein unfolding and phase transition detected by single-nanopore recording. Acs Chem Biol. 2012;7:652–658. doi: 10.1021/cb2004737. [DOI] [PubMed] [Google Scholar]
- 7.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aubin-Tam ME, et al. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- 11.Johnson ES, Schweinhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. Embo J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng W, Liao X. Solution structure of a yeast ubiquitin-like protein Smt3: the role of structurally less defined sequences in protein-protein recognitions. Protein Sci. 2002;11:1482–1491. doi: 10.1110/ps.0201602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, et al. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 15.Christensen C, et al. Effect of charge, topology and orientation of the electric field on the interaction of peptides with the α-hemolysin pore. J Pept Sci. 2011;17:726–734. doi: 10.1002/psc.1393. [DOI] [PubMed] [Google Scholar]
- 16.Movileanu L. Interrogating single proteins through nanopores: challenges and opportunities. Trends Biotechnol. 2009;27:333–341. doi: 10.1016/j.tibtech.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Oukhaled G, et al. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys Rev Lett. 2007;98:158101. doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 18.Olasagasti F, et al. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnol. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 20.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.