To the Editor:
Evidence from genomic research, in vitro cell signaling, and animal studies strongly implicates the interleukin 4/13 (IL4/13) pathways in the pathogenesis of allergic asthma.1-7 However, there is limited data on pharmacological modulation of these pathways. We performed a descriptive pharmacogenetic analysis using data from an early phase II mechanistic trial of the drug pitrakinra, a novel IL4/13 dual antagonist, in patients with allergic asthma (Clinicaltrials.gov identifier NCT00535431).8 Pitrakinra is similar to the native IL4 molecule, though specifically designed with two amino acid changes (R121D/Y124D), rendering it an effective competitive antagonist of interleukin 4 receptor α (IL4Rα).8 Pitrakinra reduces late phase responses to inhaled antigen based on Phase II clinical studies to date8 and genomic DNA samples were obtained from 29 participants in this clinical trial. In addition, we obtained DNA from 23 Cynomolgus monkeys from pre-clinical antigen-challenge studies, in which monkeys were treated with an IL4/IL13 antagonist similar to pitrakinra and with the same mechanism of action.9 We hypothesized that single nucleotide polymorphisms (SNPs) in the IL4Rα gene (IL4RA) would predict treatment response in the human studies and there would be a similar effect of IL4RA gene variation in non-human primates.
The protocol and results of the early phase II trials have been reported previously and are described in brief below.8 The primary clinical outcome was change in late phase antigen response to inhaled allergen challenge in patients with asthma. Acute and late phase responses were measured at screening and after four weeks of twice-daily therapy with nebulized pitrakinra or placebo. After allergen challenge, Forced Expiratory Volume in 1 second (FEV1) was measured over a 10 hour period to assess late antigen response (LAR; 4-10 hours). The outcome for this genetic analysis was the ratio of LAR after four weeks of treatment with either placebo or pitrakinra compared to baseline levels. Therefore, an increased LAR ratio corresponds to a reduction in late phase FEV1 response to antigen following treatment, or an improvement in lung function relative to baseline measurements.
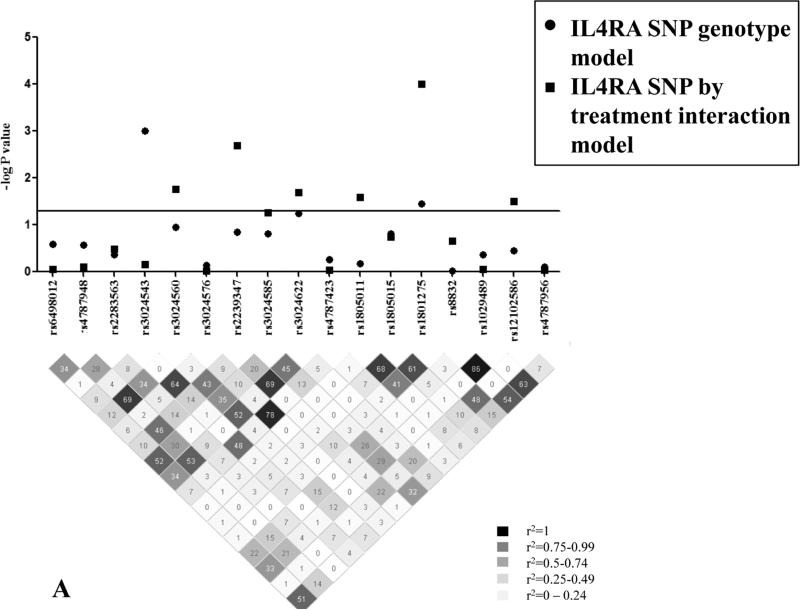
Pharmacogenetic analysis was performed on genomic DNA from predominantly non-Hispanic white (83%) male (63%) subjects with asthma randomized to placebo (n=14) or pitrakinra (n=15) with complete data on the primary outcome. We selected 14 IL4RA tagging single nucleotide polymorphisms (SNPs) and 3 non-synonymous amino acid coding changes from the HapMap CEU reference population for genotyping using the Sequenom MassARRAY iPLEX platform. We evaluated 17 IL4RA SNPs in Hardy-Weinberg equilibrium (HWE; P > 0.01) with minor allele frequency (MAF) >5% (Figure 1A). General linear models with treatment group as a covariate were developed to assess the genetic contribution of each SNP to LAR ratio (genotype model) or with a multiplicative SNP by treatment term to evaluate pharmacogenetic interaction. All statistical analyses were performed in SAS v9.1 (SAS Institute Inc., Cary, NC) or Plink v1.06. We did not correct for multiple comparisons in this exploratory analysis of human and Cynomolgus IL4RA variation.
Figure 1.
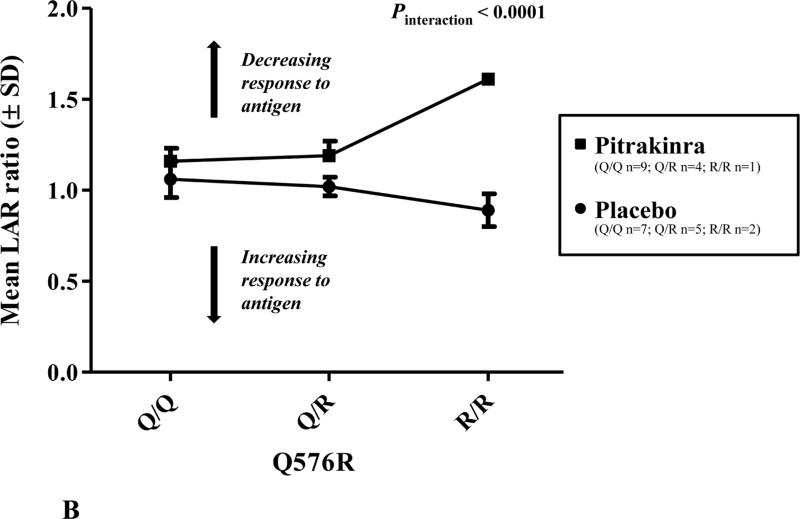
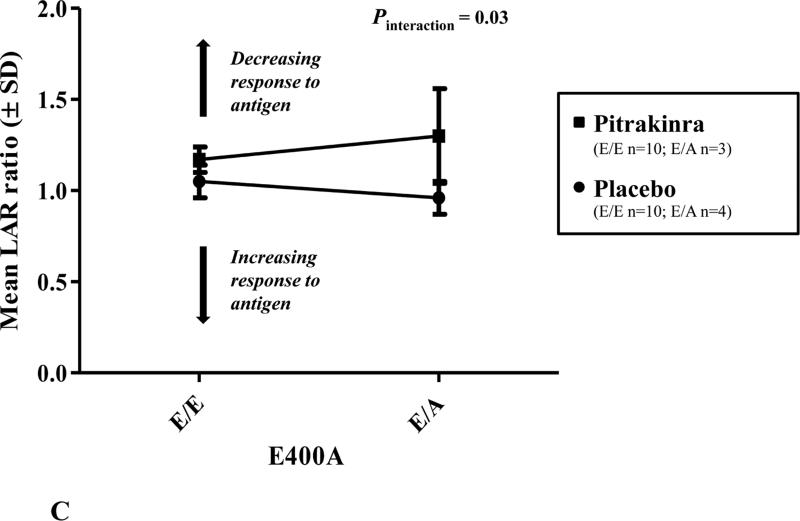
A, Human IL4RA SNP LD structure (r2) and associations with LAR ratio. Highly correlated SNPs (r2 > 0.7) have similar P values for association. The line indicates significance at P < 0.05; LD plot generated in Haploview v4.1. B, Mean (± standard deviation; SD) LAR ratio by IL4RA rs1801275 (Q576R) genotype and treatment group. C, Mean (±SD) LAR ratio by IL4RA rs1805011 (E400A) genotype and treatment group.
Figure 1 shows the linkage disequilibrium (LD) structure for the human IL4RA SNPs analyzed in this study and SNP associations with LAR ratio. Notably, the non-synonymous polymorphism rs1801275 (Q576R) was a significant predictor of mean difference in LAR ratio by genotype (P = 0.04) and interaction with treatment (P < 0.0001). As shown in Figure 1B, one individual homozygous for the R/R variant had an increased LAR ratio, indicating improved lung function relative to baseline, in the pitrakinra-treated group only. However, there was only one individual homozygous for the R/R variant, which may be primarily responsible for the significant association with LAR. If Q/R and R/R individuals were analyzed together, there was a borderline significant interaction with treatment (P = 0.06). Similarly, rs1805011 (E400A) E/A heterozygotes treated with pitrakinra had a higher mean LAR ratio than E/E homozygotes (Figure 1C).
The complementary outcomes in the primate pre-clinical trials were bronchial hyperresponsiveness (BHR) to methacholine (provocative challenge or PC100) and bronchoalveolar lavage eosinophilia in response to Ascaris allergen exposure. An IL4/13 antagonist similar to pitrakinra or vehicle control was administered in a cross-over design; details are presented in Tomkinson et al.9 Briefly, aerosolized Ascaris suum solution was delivered via respirator and BHR to methacholine was measured in anesthetized, ventilated animals. Bronchoalveolar lavage was performed with saline using standard methods. We used general linear models with an additive genetic term to evaluate Cynomolgus IL4RA polymorphisms associated with the log-transformed post-allergen IL4/13 antagonist treatment to control ratio (Table I) for both BHR (median ratio 2.5, range 0.4-8.7) and eosinophil counts (median ratio 0.95, range 0.15-9.9).
Table I.
Cynomolgus IL4RA SNPs significantly associated (P < 0.05) with BHR and eosinophil counts (post-treatment with IL4/13 antagonist compared to control).
| Cynomolgus IL4RA polymorphism | Rhesus chromosome 20 location | IL4RA SNP location | Major/minor allele; minor allele frequency | BHR P value | Eosinophil P value |
|---|---|---|---|---|---|
| PR2 | 25763581 | 5’ proximal | C/T; 0.26 | 0.03 | 0.61 |
| PR11 | Gap | 5’ proximal | C/T; 0.02 | 0.14 | 0.05 |
| PR20 | 25763996 | 5’ proximal | T/A; 0.02 | 0.14 | 0.05 |
| In1_5 | Gap | intron | G/C; 0.27 | 0.04 | 0.65 |
| In3_2 | 25794379 | Coding Syn Ser | C/T; 0.07 | 0.04 | 0.60 |
| In3_3 | 25794391 | intron | T/C; 0.08 | 0.02 | 0.44 |
| Ex5_2 | 25799301 | Coding Syn Asn | C/T; 0.26 | 0.03 | 0.61 |
| In5_7 | 25799454 | intron | C/T; 0.02 | 0.03 | 0.01 |
| In7_2 | 25803175 | intron | G/C; 0.08 | 0.64 | 0.05 |
| In8_3 | 25806353 | intron | G/A; 0.02 | 0.14 | 0.05 |
| Ex9_3 | 25809423 | Coding Syn Ile | C/T; 0.34 | 0.04 | 0.74 |
| In10_4 | 25811349 | intron | G/A; 0.33 | 0.05 | 0.91 |
| In10_5 | 25811415 | intron | C/T; 0.36 | 0.05 | 0.82 |
BHR, bronchial hyperresponsiveness
Since Cynomolgus genomic IL4RA DNA sequence was not available, we directly sequenced 100 base pairs surrounding IL4RA exons, the proximal promoter and 3’ untranslated (UTR) regions using DNA from 23 monkeys. Cynomolgus IL4RA genomic and predicted mRNA sequence was compared to Rhesus Macaca mulatta using Sequencher DNA analysis software and UCSC genome browser (genome.ucsc.edu) BLAT alignment tool; information is available on request. We identified 110 novel IL4RA polymorphisms, predominantly in intronic or promoter regions as well as seven synonymous, nine non-synonymous coding changes and six insertion/deletion polymorphisms. All SNPs tested for association (n=83) had ≥70% genotyping data, a MAF of ≥ 2% and were concordant with HWE. The genomic position and MAF for 13 polymorphisms significantly associated with BHR or eosinophilia are presented in Table I and the LD structure is shown in Figure 2. For the majority of significant SNPs, P values for association were generally nominal, and the minor allele was a predictor of increased BHR ratio, which corresponds to decreased hyperresponsiveness in pitrakinra-treated animals compared to controls. Only one relatively low frequency SNP (In5_7; MAF = 0.02), was associated with both endpoints (Table I), though this result was mainly due to one (C/T) heterozygote with a higher eosinophil ratio and a lower BHR ratio than (C/C) homozygotes. Though this result is somewhat contradictory, the function of this novel intronic SNP is currently unknown and the mechanism of SNP modulation of these allergic outcomes may form the basis of future study.
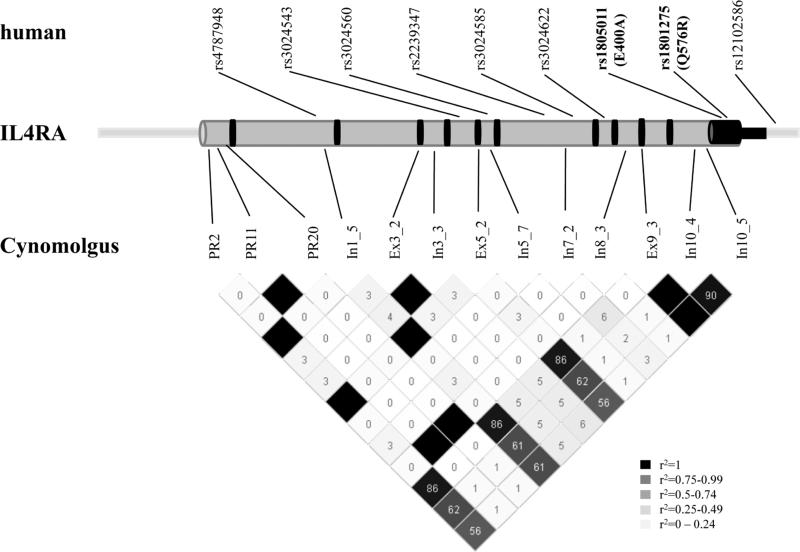
Figure 2.
Map of human and Cynomolgus monkey IL4RA polymorphisms significantly associated with allergen-induced clinical outcomes. IL4RA introns and promoter are depicted in grey; exons and 3’ untranslated region in black. Genomic information primarily adapted from the human and Rhesus UCSC genome Browser. Rhesus sequence (the closest publically available sequence to Cynomolgus) contains gaps, therefore map is not to scale. Cyno IL4RA SNP LD plot and SNP correlation (r2) generated in Haploview v4.1.
Genetic association with response to allergen-induced clinical outcomes was found across the IL4RA gene (Figure 2). There was a great deal of variation in the Cynomolgus IL4RA gene and SNPs associated with allergic outcomes were not the same as in the human. However, replication for pharmacogenetic studies is challenging and this analysis provides separate lines of evidence that IL4RA variation may modulate pharmacogenetic response to allergen challenge. Admittedly, the sample sizes for this unique analysis are small. In humans, the lowest P values were found with amino acid changes at the 3’ end of IL4RA, which have potential functional implications. The IL4RA 576R variant has been shown to synergize with the signal transducer and activator of transcription 6 molecule to enhance expression of IL4 and IL13 regulated genes and mediate allergic inflammation in a mouse model of asthma.7 We speculate that the variant at position 400 as well as 576 may also affect signaling through other intracellular mediators such as janus kinase 1.
This analysis suggests that IL4RA amino acid variations may predict a therapeutic treatment response to anti-IL4/IL13 pathway inhibitors, though larger studies are required to confirm these findings. Pharmacogenetic studies such as these may become increasingly important to predict individual response to invasive and possibly expensive novel asthma therapies.
Capsule summary.
The IL4RA Q576R polymorphism may predict treatment response to an IL4/IL13 antagonist in patients with asthma. Pharmacogenetic analysis of IL4RA in humans and non-human primates was concordant and supports further studies in larger populations.
Acknowledgments
This work was funded by Aerovance Inc., Bayer Healthcare, and the National Institute of Heart, Lung, and Blood (NHLBI) grant HL65899. RES is supported by the NHLBI K12 grant HL089992, PI Deborah A. Meyers, PhD.
Abbreviations
- BHR
Bronchial hyperresponsiveness
- HWE
Hardy-Weinberg equilibrium
- FEV1
Forced Expiratory Volume in 1 second
- IL4
Interleukin 4
- IL4RA
Interleukin 4 receptor α gene
- IL13
Interleukin 13
- LAR
Late antigen response
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- PC100
Provocative challenge
- SD
Standard deviation
- SNP
Single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–5. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 2.Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, et al. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. 2006;117:269–74. doi: 10.1016/j.jaci.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 3.Vercelli D. Genetics of IL-13 and functional relevance of IL-13 variants. Curr Opin Allergy Clin Immunol. 2002;2:389–93. doi: 10.1097/00130832-200210000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Wenzel SE, Balzar S, Ampleford E, Hawkins GA, Busse WW, Calhoun WJ, et al. IL4R alpha mutations are associated with asthma exacerbations and mast cell/IgE expression. Am J Respir Crit Care Med. 2007;175:570–6. doi: 10.1164/rccm.200607-909OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, et al. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet. 2002;70:230–6. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard TD, Whittaker PA, Zaiman AL, Koppelman GH, Xu J, Hanley MT, et al. Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol. 2001;25:377–84. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 7.Tachdjian R, Mathias C, Al Khatib S, Bryce PJ, Kim HS, Blaeser F, et al. Pathogenicity of a disease-associated human IL-4 receptor allele in experimental asthma. J Exp Med. 2009;206:2191–204. doi: 10.1084/jem.20091480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenzel S, Wilbraham D, Fuller R, Getz EB, Longphre M. Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: results of two phase 2a studies. Lancet. 2007;370:1422–31. doi: 10.1016/S0140-6736(07)61600-6. [DOI] [PubMed] [Google Scholar]
- 9.Tomkinson A, Tepper J, Morton M, Bowden A, Stevens L, Harris P, et al. Inhaled vs subcutaneous effects of a dual IL-4/IL-13 antagonist in a monkey model of asthma. Allergy. 2010;65:69–77. doi: 10.1111/j.1398-9995.2009.02156.x. [DOI] [PubMed] [Google Scholar]