Abstract
Atoh1 function is required for the earliest stages of inner ear hair cell development, which begins during the second week of gestation. Atoh1 expression in developing hair cells continues until early postnatal ages, but the function of this late expression is unknown. To test the role of continued Atoh1 expression in hair cell maturation we conditionally deleted the gene in the inner ear at various embryonic and postnatal ages. In the organ of Corti, deletion of Atoh1 at E15.5 led to the death of all hair cells. In contrast, deletion at E16.5 caused death only in apical regions, but abnormalities of stereocilia formation were present throughout the cochlea. In the utricle, deletion at E14.5 or E16.5 did not cause cell death but led to decreased expression of myosin VIIa and failure of stereocilia formation. Furthermore, we show that maintained expression of Barhl1 and Gfi1, two transcription factors implicated in cochlear hair cell survival, depends upon continued Atoh1 expression. However, maintained expression of Pou4f3 and several hair cell-specific markers is independent of Atoh1 expression. These data reveal novel late roles for Atoh1 that are separable from its initial role in hair cell development.
Keywords: Hearing, Cochlea, Development, Vestibular system, Differentiation
Introduction
Hair cell production begins in the ventromedial otocyst, where prosensory cells are specified and subsequently give rise to hair and supporting cells (Barald and Kelley, 2004; Fritzsch et al., 2011; Groves and Fekete, 2012). In the mouse organ of Corti these cells become post-mitotic from E12.5 to E15.5 in an apical-to-basal gradient (Ruben, 1967). Paradoxically, cochlear hair cell differentiation begins in the mid-basal turn and progresses in the opposite direction (Jahan et al., 2010, 2013; Rubel, 1978). Hair cells of the vestibular system are born over a much longer time period (E12.5–P2) and differentiate in a roughly central-to-peripheral direction from E13.5 through the first postnatal week (Burns et al., 2012; Ruben, 1967; Sans and Chat, 1982; Takumida and Harada, 1984). Sox2, Eya1, Six1 and Atoh1 (Math1) have been implicated in initial hair cell specification, while Barhl1, Gfi1, Hes6, NeuroD and Pou4f3 (Brn3c, Brn3.1) are expressed by and/or required for hair cell maturation and survival (Ahmed et al., 2012; Kim et al., 2001; Qian et al., 2006; Wallis et al., 2003; Xiang et al., 1998).
Atoh1 encodes a basic helix-loop-helix (bHLH) transcription factor necessary for hair cell production but not required for establishment of the sensory epithelium (Bermingham et al., 1999; Chen et al., 2002; Kawamoto et al., 2003; Woods et al., 2004; Zheng and Gao, 2000). Atoh1 expression in the cochlea begins at E13.5, progressing in basal-to-apical and medial-to-lateral directions (Dabdoub et al., 2008; Jahan et al., 2010; Matei et al., 2005; Pan et al., 2012; Woods et al., 2004). Interestingly, mouse cochlear hair cells express Atoh1 until P3-P7 and possibly into adulthood (Lanford et al., 2000; Lumpkin et al., 2003; Matei et al., 2005; Scheffer et al., 2007). Atoh1 expression in the vestibular system starts at E12.5, spreads throughout the sensory epithelium by E14.5, and is down-regulated during the first postnatal week (Bermingham et al., 1999; Shailam et al., 1999). In Atoh1-null mice, sensory precursor cells of the cochlea remain undifferentiated and undergo apoptosis from E15.5 to E17.5 in a basal-to-apical gradient that follows that of normal maturation (Basch et al., 2011; Chen et al., 2002; Fritzsch et al., 2005; Pan et al., 2011). Recent use of conditional deletion techniques in “self-terminating” Atoh1Cre/flox mice showed that limited cochlear hair cell differentiation occurs even with transient Atoh1 expression (Pan et al., 2012). However, this approach was limited by inability to control the duration of Atoh1 expression and by stochastic effects secondary to the Cre driver that was used.
To explore additional functions of Atoh1 during inner ear development we used a tamoxifen-inducible Cre allele to conditionally delete Atoh1 in developing hair cells at different embryonic and postnatal ages, allowing us to determine how varying durations of Atoh1 expression affected hair cell survival/differentiation. We demonstrate that Atoh1 deletion following specification of hair cells leads to cell death or disrupts differentiation depending upon the timing of Atoh1 gene deletion. These data demonstrate that Atoh1 functions in multiple phases of hair cell development and suggest that late expression is crucial for normal hair cell maturation and viability.
Material and methods
Mice
All mice were housed at the Case Western Reserve University or Children’s Hospital of Pittsburgh of UPMC Animal Care Facilities in accordance with IACUC guidelines. Generation of Atoh1CreERT2, Atoh1flox and ROSALacZ (JAX #007914) mice was described previously (Fujiyama et al., 2009; Shroyer et al., 2007; Soriano, 1999). ROSALacZ mice were maintained on a congenic C57BL/6J strain background, while Atoh1CreERT2 and Atoh1flox mice were maintained on mixed genetic backgrounds. Timed matings were used to obtain embryonic ages, with noon of the plug date defined as E0.5. Conditional deletion of Atoh1 occurred in Atoh1CreERT2/flox mice following tamoxifen administration; Atoh1+/flox littermates served as controls.
Tamoxifen administration
Pregnant dams received a single 250 mg/kg oral gavage dose of tamoxifen (Sigma) at E14.5, E15.5, E16.5, E17.5, P1 or P3. Embryos were subsequently harvested 1–4 days later at E16.5, E18.5, P3, or P5.
Tissue preparation
Pregnant dams were sacrificed and embryos harvested into cold 1X phosphate-buffered saline (PBS). Temporal bones were either dissected directly into cold 4% paraformaldehyde (PFA) (embryonic, P0–P5) or dissected after transcardial perfusion with 4% PFA (embryonic, P0–P5 and adult), fixed overnight in 4% PFA, washed in 1X PBS and stored at 4 °C in PBS (for immunohistochemistry) or 0.4% PFA (for in situ hybridization). Cochleae and utricles were dissected and processed for frozen sectioning or were analyzed in wholemount. Three–seven cochleae and 3–5 utricles from at least 3 animals of each genotype were examined for each experiment.
Immunohistochemistry
Whole cochleae or whole utricles were blocked for 30 min at room temperature (RT) in 1X PBS/ 0.3% Triton X-100/3% non-fat dry milk (PBST-NFDM) (cochleae) or 2% bovine serum albumin/0.8% normal goat serum/0.5% Triton X-100 (utricles) and incubated at 4 °C overnight in the same solution containing primary antibodies at the following dilutions: rabbit anti-myosin VI (25–6790, Proteus Bioscience) 1:500; rabbit anti-myosin VIIa (Proteus Biosciences, Ramona, CA) 1:500; chicken anti-β-galactosidase (ab9361, Abcam) 1:500; Alexa Fluor 488-conjugated phalloidin (A12379, Invitrogen) 1:100; Alexa Fluor 647-conjugated phalloidin (A22287, Invitrogen) 10 μg/ml; rabbit anti-cleaved caspase 3 (9661S, Cell Signaling) 1:100. Donkey Cy3-, Alexa 488- and Alexa 594-conjugated secondary antibodies (Jackson ImmunoResearch, 1:500 dilution) in blocking solution were applied for 30 min at RT. Some tissue was counterstained with 1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole; Sigma-Aldrich) for 10 min. Slides were mounted in Prolong Gold (Invitrogen) and imaged using Zeiss LSM 510 confocal, Olympus FV-1000 confocal, Nikon Eclipse 800 epifluorescence or Leica DM5500B epifluorescence microscopes. For confocal imaging, Z-series were constructed from images obtained with a 20 ×, 40 × or 63 × lens. Images were converted to RGB stacks using ImageJ (NIH).
In situ hybridization
Plasmids containing cDNAs for Atoh1 and Pou4f3 were used to generate antisense RNA probes by in vitro transcription. Following fixation cochleae were dehydrated in graded methanol and stored at −20 °C. Rehydrated cochleae were digested with 20 μg/ml proteinase K (Ambion, Austin, TX, USA) for 20 min and stored overnight at 60 °C with riboprobe in hybridization solution containing 50% (v/v) formamide, 50% (v/v) 2X saline sodium citrate (Roche) and 6% (w/v) dextran sulfate. Ears of mutant mice and control littermates were processed in the same reaction tubes to maintain consistency. Cochleae were washed and incubated overnight with anti-digoxigenin antibody (Roche Diagnostics GmbH, Mannheim, Germany) conjugated with alkaline phosphatase. Signal was developed in Nitroblue phosphate/5-bromo, 4-chloro, 3-indolyl phosphate (BM purple substrate, Roche Diagnostics, Germany) and cochleae were mounted flat in glycerol and viewed on a Nikon Eclipse 800 microscope using differential interference contrast microscopy; images were captured with Metamorph software.
Cell counts, length and density measurements
Total numbers of cells were counted on photographs of immunostained, wholemount tissues; no correction factors were applied. Cochlear length was determined by using ImageJ to draw a line along the row of inner hair cells from the base of the cochlea past the apical-most hair cells to the cochlear apex (“cochlear length line”). Organ of Corti length was calculated using the same method by tracing from the basal-most to the apical-most myosin VI+ hair cell. For counts by cochlear region the “cochlear length line” was divided into equal basal, middle and apical portions and the total number of hair cells was counted in each region.
Whole utricles were triple-labeled for myosin VIIa, phalloidin and DAPI or β-galactosidase, phalloidin and DAPI. Cell density was determined by counting the number of DAPI-stained nuclei in 3 centrally- and 3 peripherally-located 1120 μm2 boxes drawn on z-stack photographs within the myosin VIIa+ territory (examples of box locations are shown in Fig. 5). The number of myosin VIIa+ cells was counted in these boxes and divided by the total DAPI+ nucleus number to calculate the percentage of cells that expressed myosin VIIa. Phalloidin-labeled hair bundles on myosin VIIa+ cells were qualitatively categorized as long, short or absent and the percentage of myosin VIIa+ cells in each category was calculated.
Fig. 5.
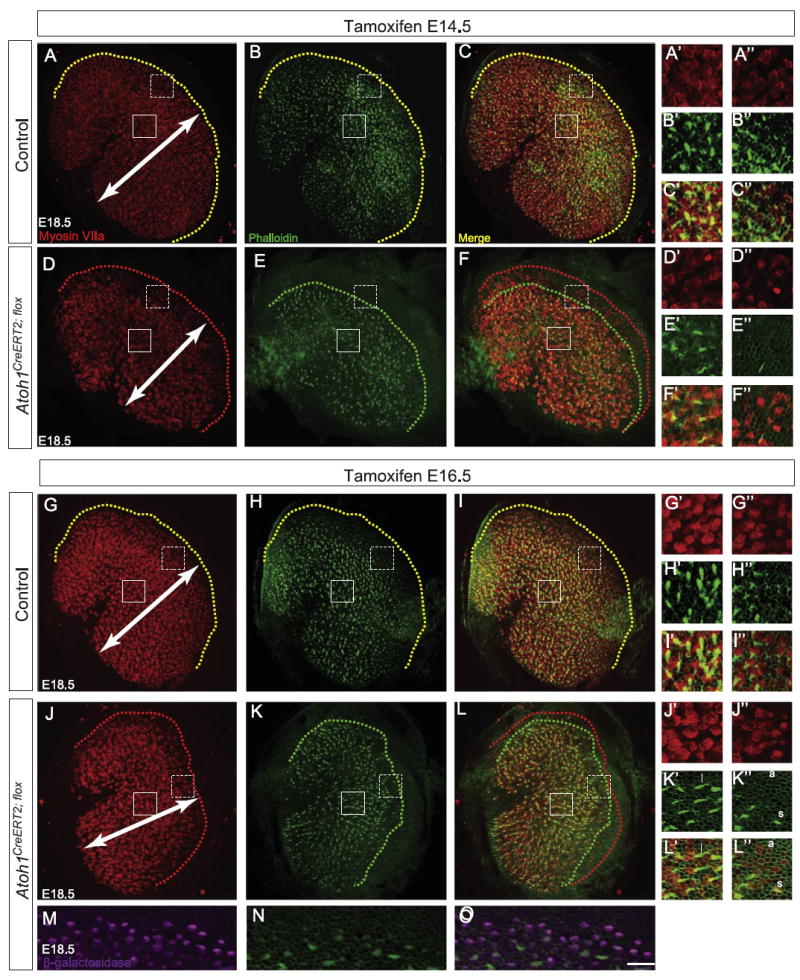
Atoh1CreERT2/flox embryos have fewer myosin VIIa+ cells and fewer stereociliary bundles in the utricle. Pregnant dams were gavaged with tamoxifen at either E14.5 (A–F″) or E16.5 (G–L″) and utricles harvested at E18.5. Immunostaining for myosin VIIa (A, D, G, and J) and labeling with phalloidin (B, E, H, and K) revealed decreased mediolateral extent (arrows) of the hair cell field in Atoh1CreERT2/flox embryos (D–F, J–L) compared to control littermates (A–C, G–I). Yellow dotted lines in utricles from control littermates (A–C, G–I) show that hair cells with stereocilia are found right up to the border of the hair cell field. In contrast, red (D, F, J, and L) and green (E, F, K, and L) dotted lines in utricles from Atoh1CreERT2/flox embryos show that many peripherally-located hair cells lack stereocilia. Solid and dotted boxes indicate regions shown in A′–L′ and A″ –L″, respectively, and are representative of the locations of “central” and “peripheral” regions of interest for cell counts (see text). Fewer myosin VIIa+ hair cells are seen in these regions (A′, A″, D′, D″, G′, G″, J′, and J″), and more of these hair cells lack stereocilia in Atoh1CreERT2/flox embryos than in controls (B′–C″, E′–F″, H′–I″, and K′–L″). Examples of long (l), short (s) and absent (a) stereociliary bundles are shown in K′–L″. Immunostaining for β-galactosidase (M) shows that Atoh1CreERT2/flox; ROSALacZ embryos have many cells that have undergone recombination, but only a small number of these have phalloidin+ hair bundles (N, O). Scale bar: 30 μm (A–L); 12 μm (M–O); 10 μm (A′–L″).
Statistical comparisons were made using unpaired, two-tailed Student’s t-tests and single factor ANOVA (Microsoft Excel).
RNA extraction, cDNA synthesis, and RT-PCR
Fresh organs of Corti were dissected away from the spiral ganglion and greater epithelial ridge, pooled by genotype (14–22/age) and homogenized using a mortar and pestle. RNA was extracted (74104, Qiagen) and reverse-transcribed (205311, Qiagen). PCR primers (Table 1) were designed to target exon sequences and cDNA was amplified using the following protocols:
| Atoh1: 94 °C (30 s), 60 °C (30 s), 72 °C (60 s) for 10 cycles |
| 94 °C (30 s), 50 °C (30 s), 72 °C (60 s) for 30 cycles |
| All other genes: 95 °C (45 s), 58 °C (45 s), 72 °C (90 s) for 40 cycles. |
Table 1.
RT-PCR primers.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Atoh1 | TCTGCTGCATTCTCCCGAGC | CTCTGGGGGTTACTCGGTGC |
| Barhl 1 | GAACCGCAGGACTAAATGGAA | GAGCGCCGAGTAATTGCCT |
| Cadherin 23 | ACTCCCTGGTCACTTGCTATG | TGGTGAAGAAAGGTAGTCGGT |
| Eya1 | TAACAGCTCGCCGTATCCAG | GTCCCAGATGAACACTCTCTCA |
| GAPDH | GTCATCATCTCCGCCCCTTCTGC | GATGCCTGCTTCACCACCTTCTTG |
| Gfi1 | AGAAGGCGCACAGCTATCAC | GGCTCCATTTTCGACTCGC |
| Hes1 | TCAACACGACACCGGACAAAC | ATGCCGGGAGCTATCTTTCTT |
| Jag1 | ATGCAGAACGTGAATGGAGAG | GCGGGACTGATACTCCTTGAG |
| Myo6 | CCACAATGTCAAAGTTCGGTACA | GGCATCGTCCCAAGAGATTTTC |
| Myo7a | GGGGACTATGTATGGATGGACC | TGATGTGCGTGGCATTCTGAG |
| Pou4f3 | ATGCGCCGAGTTTGTCTCC | GGGCTTGAACGGATGATTCTTG |
| Prox1 | TCGAACGTACTCCGCAAGC | TGCCACCGTTTTTGTTCATGT |
| Six1 | GAAAGGGAGAACACCGAAAACA | GTGGCCCATATTGCTCTGGA |
| Sox2 | CCTACTCACTGCTCGACCTG | TTGCACGGGATCATGTAGCC |
Results
To study Atoh1 function in the late embryonic and early postnatal inner ear we conditionally deleted the gene using an Atoh1CreERT2 knock-in mouse line that faithfully recapitulates endogenous Atoh1 expression (Fujiyama et al., 2009). A single tamoxifen dose administered to pregnant dams at E15.5 resulted in β-galactosidase expression in 97 ± 3% of cochlear hair cells (n=3 cochleae) in E18.5 Atoh1CreERT2; ROSALacZ embryos (Fig. 1A). Furthermore, tamoxifen administration at E16.5 efficiently excised an Atoh1flox allele (Shroyer et al., 2007): Atoh1 mRNA transcripts were undetectable by in situ hybridization on whole cochleae (Fig. 1C and D′) and vestibular organs (Fig. 1E and F) and RT-PCR performed on organ of Corti total RNA (Fig. 1B) obtained from E18.5 Atoh1CreERT2/flox embryos. These data indicate that the Atoh1CreERT2 allele we used for these experiments produces complete or near complete excision of floxed sequences at the ROSA and Atoh1 loci at the ages we tested.
Fig. 1.
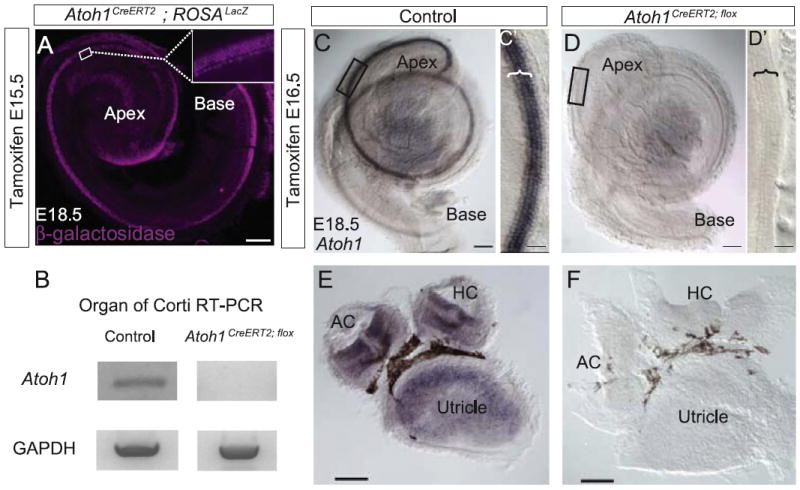
Tamoxifen administration activates Atoh1CreERT2 in nearly all cochlear hair cells and efficiently deletes an Atoh1flox allele. (A) Cochlea from E18.5 Atoh1CreERT2/+; ROSALacZ embryo treated with tamoxifen at E15.5 and immunostained for β-galactosidase. Box shows location of inset. Nearly every hair cell is labeled. (B) RT-PCR for Atoh1 on organ of Corti total RNA isolated from Atoh1CreERT2/flox verifies Atoh1 deletion. (C–F) In situ hybridization for Atoh1 on cochleae (C, D) and vestibular organs (E, F) from E18.5 control (C, C′, and E) and Atoh1CreERT2/flox (D, D′, and F) embryos following tamoxifen administration at E16.5. No labeling is present in the Atoh1CreERT2/flox embryonic cochlea or vestibular system. Boxes show locations of C′ and D′; brackets identify organ of Corti. AC – anterior crista, HC – horizontal crista. Scale bar: 500 μm (A); 100 μm (C–F); 25 μm (C′, D′).
Continued Atoh1 expression is necessary for survival and differentiation of cochlear hair cells
Cochlear hair cells become post-mitotic between E12.5 and E15.5, and the earliest-born hair cells (those in the mid-basal turn) begin to differentiate at E13.5 (Ruben, 1967; Woods et al., 2004). To determine the role of Atoh1 just after the last cochlear hair cells became post-mitotic we administered tamoxifen to pregnant dams at E15.5 and harvested embryos at E16.5 (Fig. 2). E16.5 Atoh1CreERT2/flox embryos had 25% fewer myosin VI+ hair cells compared to control littermates (1163 ± 714, n=3 cochleae vs. 1544 ± 80, n=4 cochleae, p=0.003), and the length of the organ of Corti was decreased by 9% (2630 ± 50 μm vs. 2876 ± 60 μm; n=3 cochleae/genotype, p=0.035). This cell loss was not secondary to changes in overall cochlear length (Atoh1CreERT2/flox 3436 ± 62 μm vs. control 3497 ± 59 μm; n=3 cochleae/genotype, p=0.51). To determine whether hair cell loss was regionally variable we divided cochleae longitudinally into three equal parts and counted the number of myosin VI+ hair cells in each region. We found 19% (491 ± 31 vs. 616 ± 13; p=0.009), 23% (391 ± 18 vs. 507 ± 21; p=0.007) and 33% (281 ± 24 vs. 422 ± 39; p=0.025) decrease in hair cell number in the base, middle and apex of Atoh1CreERT2/flox (n=3 cochleae) vs. control littermate (n=4 cochleae) embryos (Fig. 2D). Third row outer hair cells and scattered inner hair cells were most obviously affected (Fig. 2B–B″). In addition, the regular spacing between outer hair cells was disrupted in Atoh1CreERT2/flox embryos (Fig. 2B–B″). Strikingly, cochleae (n=7) from E18.5 Atoh1CreERT2/flox embryos that received tamoxifen at E15.5 had no myosin VI+ cells and lacked DAPI +nuclei in the typical hair cell position (Fig. 2C–C″ and data not shown). These data suggested that Atoh1-null hair cells died between E16.5 and E18.5. To verify this, cochleae from E16.5 Atoh1CreERT2/flox embryos and littermate controls dosed with tamoxifen at E15.5 were immunostained for activated caspase 3. Multiple activated caspase 3+ cells were seen throughout the organ of Corti of Atoh1CreERT2/flox embryos, but no hair cells were labeled in control littermates (Fig. 2E–F″″). Thus, deletion of Atoh1 at E15.5 led to rapid hair cell demise throughout the cochlea, suggesting that Atoh1 is necessary for hair cell survival shortly (1–3 days) after hair cells become post-mitotic.
Fig. 2.
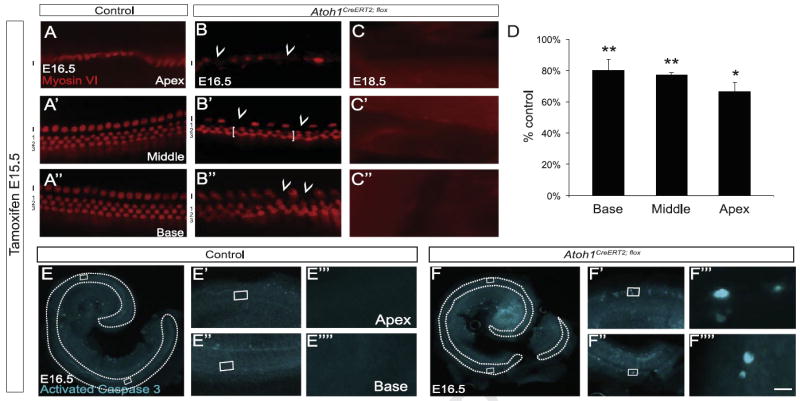
Cochlear hair cells die following Atoh1 deletion at E15.5. Pregnant dams received tamoxifen at E15.5. (A–C″) Cochleae from control littermate (A) and Atoh1CreERT2/flox (B–C″) embryos immunostained for myosin VI. (B–B″) E16.5 Atoh1CreERT2/flox mice lacked outer hair cells (1–3, brackets in B′) predominantly in the third row and scattered inner hair cells (I, arrowheads) throughout the cochlea. No hair cells were present at E18.5 (C–C″). (D) Myosin VI+ hair cell counts by region in Atoh1CreERT2/flox embryos as percent of control littermate numbers (n=3–4 cochleae/genotype). *p<0.05, **p<0.01 compared to control. (E–F″″) Activated caspase 3 immunostaining of cochleae from E16.5 embryos demonstrated apoptotic cell death in the organ of Corti (outlined) of Atoh1CreERT2/flox (F–F″″) but not littermate controls (E–E″″). Scale bar: 15 μm (A–C″), 500 μm (E, F); 50 μm (E′, E″, F′, and F″); 5 μm (E′″, E″″, F′″, and F″″).
To determine whether Atoh1 was necessary for hair cell survival at later embryonic ages we administered tamoxifen to pregnant dams at E16.5 and harvested embryos at E18.5 (Fig. 3). The number of myosin VI+ hair cells was decreased by 50% (1099 ± 81 vs. 2160 ± 56; n=3 cochleae/genotype, p=0.0004) in Atoh1CreERT2/flox vs. control littermates; we verified that these hair cells underwent Cre-mediated recombination in Atoh1CreERT2/flox; ROSALacZ embryos (Fig. 3E–E″). The length of the organ of Corti was decreased by 15% (3557 ± 55 μm vs. 4212 ± 51 μm, n=3 cochleae/genotype, p=9.7 × 10−4), but overall cochlear length was unchanged (4504 ± 31 μm vs. 4458 ± 24 μm, n=3 cochleae/genotype, p=0.30). Looking by region, we found that hair cell numbers were reduced by 56% (323 ± 73 vs. 728 ± 19; p=0.0059) in the middle turn and 88% (83 ± 4 vs. 716 ± 10; p=5.4 × 10−7) in the apex of cochleae from Atoh1CreERT2/flox embryos compared to control littermates (n=3 cochleae/genotype) (Fig. 3F). Activated caspase 3 immunostaining was also seen in these regions in Atoh1CreERT2/flox embryos but not in control littermates (Fig. 3G, G′, G′″, H, H′, and H′″), suggesting that these cells were undergoing apoptosis. In contrast, myosin VI immunostaining was present in four rows of hair cells in the lower middle and basal turns (Fig. 3A′, A″, C′, C″), and we found no decrease in the number of myosin VI+ hair cells in the basal turn of Atoh1CreERT2/flox embryos (693 ± 13 vs. 716 ± 37, n=3 cochleae/genotype; p=0.60; Fig. 2F). However, phalloidin labeling revealed that essentially all of the remaining hair cells in the middle and basal turns had grossly disorganized and fragmented hair bundles while those in the apical turn lacked hair bundles completely (compare Fig. 3B–B″ to D–D″). This phenotype could not be explained by activation of cell death programs in the lower middle and basal turns since there were no activated caspase 3+ cells in these regions (Fig. 3G″, G″″, H″, and H″″). Cell loss and stereocilia phenotypes were also seen in E18.5 embryos that received tamoxifen at E17.5 except that four continuous rows of hair cells extended farther apically than in those that received tamoxifen at E16.5 (data not shown). Taken together, these data suggest that apical hair cells still require Atoh1 for survival at E16.5 and E17.5, while basal hair cells require Atoh1 for stereocilia formation at those ages.
Fig. 3.
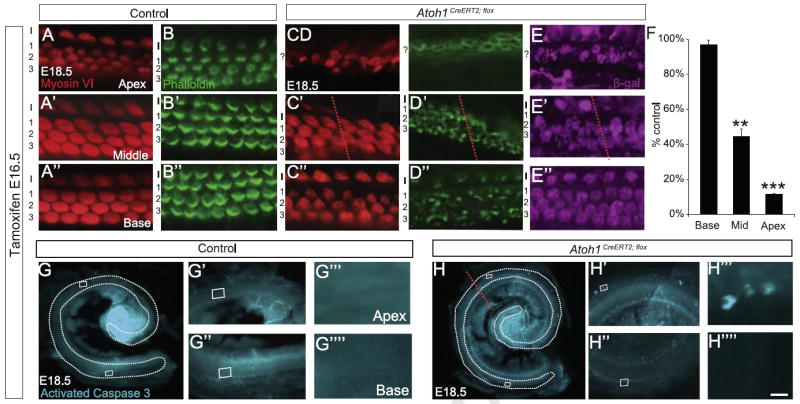
Atoh1 deletion at E16.5 causes hair cell loss and disrupts cochlear hair cell stereocilia formation. (A–D″) Cochleae from E18.5 littermate control (A–B″) and Atoh1CreERT2/flox (C–D″) embryos treated with tamoxifen at E16.5 and immunostained with myosin VI, β-galactosidase or labeled with phalloidin (A–A″, B–B″, C–C″, D–D″ and E–E″ are from different animals). Atoh1CreERT2/flox embryos have fewer hair cells from the mid-middle turn (red lines in C′, D′, E′ and H) to the apex while these cells are present in normal numbers from the mid-middle turn to the base. However, remaining hair cells have disorganized or no stereocilia (D–D″). Remaining hair cells also show evidence of recombination (E–E″). (F) Hair cell counts by cochlear region graphed as percent of control littermate numbers show significant loss of myosin VI+ hair cells in the apex and middle turn but not the base of Atoh1CreERT2/flox embryos (n=3 cochleae/genotype). **p<0.001, ***p<1 × 10−6. G–H″″) Immunostaining for activated caspase 3 shows cell death in the apex, but not the base, of Atoh1CreERT2/flox embryos. Scale bar: 15 μm (A–E″); 500 μm (G, H); 50 μm (G′, G″, H′, and H″); 5 μm (G′″, G″″, H′″, and H″″).
Cochlear hair cells stop expressing detectable amounts of Atoh1 mRNA between P3 and P7 (Lanford et al., 2000; Lumpkin et al., 2003; Scheffer et al., 2007). To determine whether Atoh1 was required for auditory hair cell survival or maturation following birth, Atoh1CreERT2/flox pups and control littermates were given tamoxifen at P1 or P3 and cochleae were harvested two days later (P3 or P5, respectively). No hair cell loss or hair bundle abnormalities were seen in Atoh1CreERT2/flox pups at either age (Fig. 4 and data not shown). These data indicate that the function of postnatal Atoh1 expression is different from that of late embryonic expression in that postnatal expression is not necessary for short-term hair cell survival or stereocilia formation/maintenance.
Fig. 4.
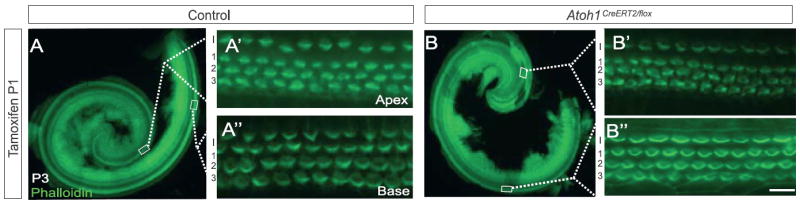
Atoh1 deletion at P1 does not affect hair cell number or stereocilia formation. Pups were injected with tamoxifen at P1 and cochleae harvested at P3. Phalloidin labeling revealed no loss of hair cells or stereocilia abnormalities. Scale bar: 500 μm (A, B); 15 μm (A′, A″, B′, and B″).
Continued Atoh1 expression is necessary for differentiation of vestibular hair cells
We next examined the utricle to determine whether Atoh1 deletion produced similar effects in vestibular hair cells. In contrast to the cochlea, utricular hair cells become post-mitotic over a long period of time (E12.5–P2) in a predominantly central-to-peripheral gradient (Burns et al., 2012; Ruben, 1967; Sans and Chat, 1982). We first verified that tamoxifen administration to Atoh1CreERT2; ROSALacZ embryos at E14.5 labeled essentially all hair cells by E18.5 (1538 ± 60 β-gal+, n=2 utricles vs. 1559 ± 17 myosin VIIa+ cells, n=4 utricles; p=0.67). To determine potential effects of Atoh1 deletion at times when some hair cells were being born while others were differentiating, we administered tamoxifen at E14.5 or E16.5 and harvested utricles at E18.5. We found that Atoh1CreERT2/flox embryos exposed to tamoxifen at either age had statistically significant decreases in the number of myosin VIIa+ cells and the number of cells with a phalloidin+stereociliary bundle compared to littermate controls (Table 2; Fig. 5). These effects were more prominent with tamoxifen administration at E14.5 (Table 2; Fig. 5D–F, J–L). In addition, the percentage of myosin VIIa+ cells with a stereociliary bundle was decreased to about the same extent compared to controls following tamoxifen administration at either age (Table 2). Furthermore, hair bundles of surviving hair cells visualized by phalloidin labeling appeared thinner in Atoh1CreERT2/flox embryos than in littermate controls (Fig. 5B′, B″, E′, E″, H′, H″, K′, and K″). These data suggest that conditional deletion of Atoh1 disrupts utricular hair cell differentiation.
Table 2.
Utricular hair cells require Atoh1 for survival and stereocilia formation. Total numbers of immunostained cells were counted in the indicated number of utricles harvested from E18.5 embryos and reported as average number ± SEM. Reported decreases are relative to littermate controls. Values for E14.5 and E16.5 tamoxifen administration are compared within genotype at the bottom of the Table.
| Control littermate | Atoh1CreERT2/flox | Decrease | p-value (t-test) | |
|---|---|---|---|---|
| Tamoxifen at E14.5 | n=4 utricles | n=4 utricles | ||
| Myosin VIIa+ | 1559 ± 17 | 881 ± 56 | 43% | 2.5 × 10−5 |
| Phalloidin+hair bundle | 965 ± 25 | 394 ± 17 | 59% | 1.4 × 10−6 |
| % Phalloidin+/Myosin VIIa+ | 62 ± 1% | 45 ± 1% | 17% | 8.4 × 10−5 |
| Tamoxifen at E16.5 | n=4 utricles | n=4 utricles | ||
| Myosin VIIa+ | 1525 ± 20 | 1076 ± 36 | 29% | 3.6 × 10−5 |
| Phalloidin+hair bundle | 946 ± 24 | 544 ± 23 | 42% | 2.1 × 10−5 |
| % Phalloidin+/Myosin VIIa+ | 62 ± 1% | 51 ± 3% | 11% | 0.014 |
| Tamoxifen E14.5 vs. E16.5 | p-value (t-test) | p-value (t-test) | ||
| Myosin VIIa+ | 0.25 | 0.03 | ||
| Phalloidin+hair bundle | 0.60 | 0.0020 | ||
| % Phalloidin+/Myosin VIIa+ | 0.97 | 0.13 |
We next sought to determine whether hair cell survival was affected and/or whether there was regional variability akin to that seen in the cochlea. To this end, we chose to expand our analysis of E18.5 Atoh1CreERT2/flox embryos exposed to tamoxifen at E14.5 because more pronounced effects were seen at this age (Table 2). We analyzed 6 regions of equal size (3 central, 3 peripheral; see Fig. 5 for examples of locations) in utricles from Atoh1CreERT2/flox embryos and control littermates to determine whether regional variability was present. We found no differences in the numbers of DAPI+cell nuclei/100 μm2 located in the central or peripheral regions of Atoh1CreERT2/flox embryos and control littermates (Table 3). In contrast, the number of myosin VIIa+ cells was decreased both centrally and peripherally (13% vs. 27%) in Atoh1CreERT2/flox embryos. We also found that the number of β-galactosidase+cells (Fig. 5M; 1495 ± 39, n=2 utricles) was much higher than the number of myosin VIIa+ cells (1076 ± 36, n=4 utricles; p=0.0021) in E18.5 Atoh1CreERT2/flox; ROSALacZ embryos that received tamoxifen at E16.5 but similar to the number of myosin VIIa+ cells in control littermates (1525 ± 20, n=4 utricles; p=0.48). These data suggest that cell death was not responsible for the decreased numbers of myosin VIIa+ cells seen in Atoh1CreERT2/flox embryos. We next qualitatively classified myosin VII+ hair cells into three groups based on the length (long, short or absent; examples are shown in Fig. 5K′-L″) of their stereociliary bundles. We found no difference in the percentage of myosin VIIa+ cells with long stereocilia in central or peripheral regions (Table 3). However, decreases of 13% (central) and 28% (peripheral) in the number of hair cells with short stereocilia were found in Atoh1CreERT2/flox embryos. In addition, the percentage of myosin VIIa+ cells without stereocilia increased by 15% and 24%, respectively. Thus, the effects of Atoh1 deletion on myosin VIIa expression and hair bundle production/development were more severe in peripheral than in central regions.
Table 3.
Atoh1CreERT2/fox embryonic utricles have regionally-variable differentiation defects but no change in cell density. Pregnant dams received tamoxifen at E14.5 and embryos were harvested at E18.5. Total numbers of DAPI+ nuclei in the regions of interest (450−500 cells; see Methods) were counted for cell density calculations (DAPI+nuclei/100 μm2). Hair bundles were qualitatively scored as long, short or absent. All numbers are reported as average ± SEM; percent change is relative to littermate controls.
| Control littermate | Atoh1CreERT2/flox | % Change | p-value (t-test) | |
|---|---|---|---|---|
| DAPI+ nuclei/100 μm2 | n=2 utricles | n=2 utricles | ||
| Central | 7.1 ± 0.2 | 7.1 ± 0.3 | 0% | 0.97 |
| Peripheral | 7.1 ± 0.3 | 6.8 ± 0.2 | −4% | 0.43 |
| Myosin VIIa+/DAPI+ | n=4 utricles | n=4 utricles | ||
| Central | 74 ± 4% | 61 ± 3.8% | −13% | 0.04 |
| Peripheral | 70 ± 3.8% | 43 ± 3.2% | −27% | 3.4 × 10−4 |
| Hair bundles (phalloidin+/myosin VIIa+) | n=4 utricles | n=4 utricles | ||
| Long | ||||
| Central | 29 ± 1.6% | 27 ± 5.9% | −2% | 0.70 |
| Peripheral | 4.9 ± 2.1% | 8.2 ± 2.1% | +3.3% | 0.27 |
| Short | ||||
| Central | 36 ± 1.6% | 23 ± 3.3% | −13% | 0.003 |
| Peripheral | 50 ± 3.9% | 22 ± 2.5% | −28% | 6.6 ×10−6 |
| Absent | ||||
| Central | 35 ± 1.6% | 50 ± 3.4% | +15% | 6.7 × 10−4 |
| Peripheral | 45 ± 4.0% | 69 ± 2.6% | +24% | 4.5 × 10−5 |
Conditional Atoh1 deletion affects cochlear hair cell gene expression
Given the stereocilia abnormalities seen in surviving hair cells following Atoh1 deletion at E14.5 (utricle), E16.5 and E17.5 (cochlea) we next sought to determine whether there were qualitative changes in the expression of candidate genes involved in hair and supporting cell specification, differentiation and maturation. We administered tamoxifen to pregnant dams at E16.5, isolated RNA from pooled E18.5 Atoh1CreERT2/flox or control embryo organs of Corti, and performed RT-PCR (Fig. 6A). Genes upstream of Atoh1 (Eya1, Six1, Sox2), those expressed by supporting cells (Jag1, Hes1, Prox1), hair cell markers (Myo6, Myo7A, CDH23) and the hair cell differentiation factor Pou4f3 continued to be expressed in cochleae from treated Atoh1CreERT2/flox embryos. In contrast, expression of Barhl1 and Gfi1 was absent, suggesting that Atoh1 is required for continued expression of these genes. We verified these results for Barhl1 and Pou4f3 using in situ hybridization (n=4 cochleae/genotype). No expression of Barhl1 was detected in cochleae from E18.5 Atoh1CreERT2/flox embryos that received tamoxifen at E16.5 (Fig. 6B and C), while Pou4f3 was expressed by remaining hair cells in the cochlea and vestibular system (Fig. 6D–E′″).
Fig. 6.
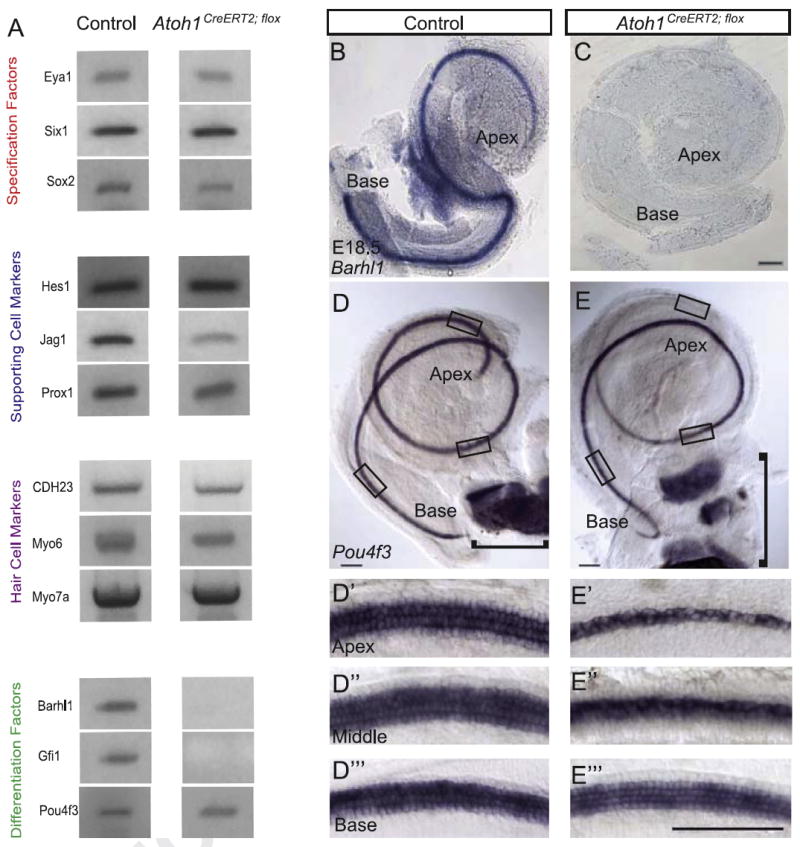
Atoh1 deletion at E16.5 abolishes cochlear expression of Barhl1 and Gfi1 but not several other genes. Pregnant dams received tamoxifen at E16.5 and embryos were harvested at E18.5. (A) Qualitative RT-PCR of organ of Corti mRNA from control and Atoh1CreERT2/flox embryos demonstrates that expression of Barhl1 and Gfi1 is lost in Atoh1CreERT2/flox mice. Upstream specification factors, hair cell markers, supporting cell markers and Pou4f3 continue to be expressed. (B, C) In situ hybridization for Barhl1 confirms absence of expression in Atoh1CreERT2/flox embryonic cochleae. (D–E′″) Pou4f3 continues to be expressed in all regions of Atoh1CreERT2/flox embryonic cochleae where hair cells are present. Pou4f3 expression is also maintained in the vestibular system (brackets). Scale bar: 100 μm.
Discussion
Atoh1 has been viewed to function as a proneural gene important for specification and/or as a differentiation initiation factor in various cell types (Bermingham et al., 1999; Matei et al., 2005; Pan et al., 2012). Our data suggest that Atoh1 serves at least two additional functions during cochlear hair cell development: an intermediate stage function in hair cell survival and a late stage function in hair cell differentiation/maturation. These latter two functions need not be mutually exclusive and are likely to overlap temporally because of the maturation gradients (basal-to-apical, medial-to-lateral) inherent to cochlear development. These gradients also likely explain the increasing basal-to-apical phenotypic severity seen in the cochleae of Atoh1CreERT2/flox mice. Alternatively, Atoh1 might function primarily as a survival factor after specification is complete and differentiation has started, and the disorganized stereocilia we observe in the cochlea of late knockout embryos may represent a non-specific marker of hair cell degeneration. This interpretation is consistent with data from other models of hair cell loss (Jahan et al., 2012; Soukup et al., 2009) but seems unlikely here because basal hair cells of Atoh1CreERT2/flox mice are negative for activated caspase 3, unlike their apical counterparts. However, given the short survival times in our studies we cannot rule out the possibility that these defective hair cells degenerate postnatally as they do in another model of delayed Atoh1 deletion (Pan et al., 2012). Unfortunately, treated Atoh1CreERT2/flox mice survived only 2–3 days postnatally secondary to near complete Atoh1 deletion in the gut, which ultimately resulted in their death (data not shown).
Our data are consistent with and extend those obtained in an Atoh1 “self-terminating” mutant, where hair cell death and stereo-cilia disorganization were observed when a constitutively-expressed transgenic Atoh1Cre allele was used to delete Atoh1 expression (Pan et al., 2012). An advantage of our system was that it allowed us to exert precise temporal control of Atoh1 deletion by using a strong inducible knock-in Atoh1CreERT2 allele, and to therefore separate early from late effects of Atoh1 deletion in a way that was impossible to achieve in that and other previously-described models (Pan et al., 2011).
Interestingly, our data suggest that continued expression of Atoh1 plays slightly different roles in the utricle and cochlea. In both areas Atoh1CreERT2/flox mice have stereocilia abnormalities ranging from abnormal appearance to failure of formation. On the other hand utricular hair cells, unlike their cochlear counterparts, do not require continued Atoh1 expression for survival. This disparity could arise from the different rate at which hair cells differentiate in the two structures. Some apical cochlear hair cells begin to differentiate 4–5 days after their terminal mitoses (Jahan et al., 2010, 2013; Rubel, 1978), and our data suggest that continued Atoh1 expression during this “limbo” period is crucial for their survival. In contrast, stereocilia development in the utricle begins at E13.5, just one day after the first utricular hair cells become post-mitotic (Takumida and Harada, 1984). Thus, it could be that Atoh1 is a survival factor for only as long as it takes to initiate differentiation, an interpretation consistent with our observation that basal cochlear hair cells die after E15.5 but not E16.5 or E17.5 tamoxifen administration. Another possibility is that deletion of Atoh1 changes cellular interactions in the utricle, leading to changes in precursor allocation. For example, lateral inhibition plays an important role in supporting cell specification (Kelley, 2006), so disrupted differentiation of hair cells could inhibit supporting cell differentiation, leading to persistence of precursors that subsequently become “reassigned” to a hair cell identity. This explanation presupposes utricular hair cell death followed by transdifferentiation of supporting cells and/or increased mitotic activity of precursors. The former explanation is plausible given that utricular hair cells continue to be generated for two weeks after mitotic activity ceases, likely through trans-differentiation of supporting cells (Burns et al., 2012). However, in our system this seems unlikely given that we did not see decreased overall cell numbers (Table 3) and/or evidence of more cell divisions (data not shown).
Our data also provide further evidence for central-to-peripheral progression of utricular hair cell development. We saw larger numbers of hair cells with mature stereocilia in the central regions of E18.5 control and Atoh1CreERT2/flox embryos, consistent with this interpretation. Furthermore, peripheral areas of Atoh1CreERT2/flox embryonic utricles had fewer myosin VIIa+ hair cells than central areas, and fewer of the peripherally located hair cells had stereociliary bundles (Table 3). Central regions of Atoh1CreERT2/flox embryonic utricles also had decreased numbers of myosin VIIa+ hair cells, and fewer of these cells had stereociliary bundles, than in central regions of control littermates. So, while hair cell differentiation in general progresses from center to periphery, new cells must continue to be generated and differentiate centrally even after hair cell generation has begun in the periphery. This interpretation agrees well with previous birth-dating studies (Sans and Chat, 1982).
Another important aspect of our study is the demonstration that Barhl1 and Gfi1, two transcription factors important for cochlear hair cell survival and differentiation, rely on Atoh1 for sustained expression. Previous evidence suggested that both genes lie downstream of Atoh1 (Chellappa et al., 2008; Shroyer et al., 2005). Barhl1 is first expressed in hair cells at E14.5 (Li et al., 2002; Pauley et al., 2008) and Gfi1 at E15.5 (Hertzano et al., 2004; Wallis et al., 2003); thus, expression of both genes initiates prior to Atoh1 deletion in Atoh1CreERT2/flox mice that received tamoxifen at E16.5. Because most middle turn and all basal turn hair cells survive until E18.5 in this paradigm yet no Barhl1 or Gfi1 expression is seen at E18.5, we conclude that Atoh1 is necessary for maintained expression of these genes. This conclusion is in line with a recent knock-in mouse model where Atoh1 was replaced by Neurog1 and no expression of Barhl1 was observed in surviving hair cells (Jahan et al., 2012). Gfi1 has been described as a downstream target of Pou4f3 because it is not expressed in hair cells of Pou4f3ddl/ddl mice (Hertzano et al., 2007; Hertzano et al., 2004). Our data show that Gfi1 expression is lost in Atoh1CreERT2 mice treated with tamoxifen at E16.5 despite continued expression of Pou4f3. This shows that Pou4f3 expression alone is not sufficient to maintain Gfi1 expression, suggesting either that Pou4f3 is necessary only for initiation of Gfi1 expression or that both Atoh1 and Pou4f3 are required for maintenance of expression. This second possibility could be tested by conditionally deleting Pou4f3 during late embryonic development. Interestingly, our data also suggest that Gfi1’s role in hair cell development might, like that of Atoh1, change over time. Previous evidence suggested that Gfi1 acts as a hair cell survival factor because Gfi1-null mice lose virtually all hair cells in a basal-to-apical gradient of increasing severity between E17.5 and P3 (Pauley et al., 2008; Wallis et al., 2003). Our analysis was limited to embryonic ages, but we saw no evidence of hair cell death in the middle and basal turns of Atoh1CreERT2/flox mice that received tamoxifen at E16.5 or later. These observations suggest that a short period of Gfi1 expression might be sufficient for hair cell survival, an interpretation supported by experiments conducted in cultured organ of Corti where downregulation of Gfi1 by Prox1 overexpression at E15.5, but not P1, caused hair cell death (Kirjavainen et al., 2008). As with Atoh1 itself, we interpret this to mean that initial expression of Gfi1 is required for survival, but the requirement for Gfi1 changes over developmental time. Hair cell death in Barhl1-null mice begins after P6 and is slowly progressive over several months (Li et al., 2002); further experiments will be necessary to determine if this occurs in Atoh1CreERT2/flox mice.
Our data also provide in vivo evidence that several cochlear hair cell-specific genes including Pou4f3 do not rely on Atoh1 for maintained expression. Previous work demonstrated that sequences upstream of the Pou4f3 locus can drive luciferase expression in the presence of Atoh1, suggesting that Atoh1 might directly regulate Pou4f3 (Masuda et al., 2011; Masuda et al., 2012). However, another in vitro study demonstrated that ectopic expression of Eya1 and Six1 in cochlear explants directed expression of Pou4f3 either alone or together with Atoh1, suggesting that regulation of the two genes might be independent of one another (Ahmed et al., 2012). Our results agree with and extend this second study by showing that Atoh1 is not necessary for maintained Pou4f3 expression in vivo, demonstrating that Pou4f3 operates in a parallel pathway. Furthermore, we demonstrate that Atoh1 expression is not necessary for maintained expression of CDH23, Myo6, or Myo7a, all of which are first expressed in hair cells between E14.5 and E16.5 (Lagziel et al., 2005; Xiang et al., 1998). This leaves three possibilities. First, Atoh1 might be directly required for initial expression of these markers, but maintenance is controlled by other transcription factors. Certainly this is possible in our system, as Atoh1 is expressed throughout the organ of Corti by E15.5 (Woods et al., 2004) and we looked at marker gene expression following Atoh1 deletion at E16.5. Second, Atoh1 might upregulate a set of transcription factors that then, in turn, directly control expression of CDH23, Myo6, and Myo7a. Finally, it is possible that Atoh1 does not regulate these genes at all. This possibility is supported by the finding that myosin VI expression was induced in Atoh1-positive and -negative “hair cell-like” cells induced by ectopic Eya1 and Six1 expression (Ahmed et al., 2012). Furthermore, in that study essentially all of the myosin VI-positive cells also expressed Pou4f3, suggesting that this gene is capable of regulating at least some aspects of hair cell differentiation in the absence of Atoh1. This interpretation is also consistent with the finding that small numbers of “hair cell-like” cells can form even in the complete absence of Atoh1 expression (Du et al., 2007; Pan et al., 2011). Future work will need to address the genetic pathways that directly control marker expression in hair cells.
Acknowledgments
The authors thank Matt Kelley and members of the Maricich lab for thoughtful discussions of the data and critical reading of the manuscript, and Andy Groves for discussing unpublished data. This work was supported by the Child Neurology Society (SMM), American Hearing Research Foundation (SMM), Health Hearing Foundation (IJ), NIDCD R01 DC003696 (JS), NIDCD P30 Core Grant DC004661 (JS), NIH T32 GM008056 (MCW), NIDCD R01 DC005590 (IJ and BF) and P30 Core Grant DC010362 (IJ and BF). Some of the confocal imaging was performed at the Case Western Reserve University Neurosciences Imaging Center.
Footnotes
Publisher's Disclaimer: This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barald KF, Kelley MW. From placode to polarization: new tunes in inner ear development. Development. 2004;131:4119–4130. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- Basch ML, Ohyama T, Segil N, Groves AK. Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J Neurosci. 2011;31:8046–8058. doi: 10.1523/JNEUROSCI.6671-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Burns JC, On D, Baker W, Collado MS, Corwin JT. Over half the hair cells in the mouse utricle first appear after birth, with significant numbers originating from early postnatal mitotic production in peripheral and striolar growth zones. J Assoc Res Otolaryngol. 2012;13:609–627. doi: 10.1007/s10162-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappa R, Li S, Pauley S, Jahan I, Jin K, Xiang M. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol Cell Biol. 2008;28:1905–1914. doi: 10.1128/MCB.01454-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, Kelley MW. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Jensen P, Goldowitz D, Hamre KM. Wild-type cells rescue genotypically Math1-null hair cells in the inner ears of chimeric mice. Dev Biol. 2007;305:430–438. doi: 10.1016/j.ydbio.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, Beisel KW, Wang VY. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Jahan I, Pan N, Kersigo J, Duncan J, Kopecky B. Dissecting the molecular basis of organ of Corti development: where are we now? Hear Res. 2011 doi: 10.1016/j.heares.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y, Kawaguchi Y, Nabeshima Y, Hoshino M. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136:2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–257. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yucel R, Frankel WN, Rechavi G, Moroy T, Friedman TB, Kelley MW, Avraham KB. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- Hertzano R, Dror AA, Montcouquiol M, Ahmed ZM, Ellsworth B, Camper S, Friedman TB, Kelley MW, Avraham KB. Lhx3, a LIM domain transcription factor, is regulated by Pou4f3 in the auditory but not in the vestibular system. Eur J Neurosci. 2007;25:999–1005. doi: 10.1111/j.1460-9568.2007.05332.x. [DOI] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One. 2010;5:e11661. doi: 10.1371/journal.pone.0011661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Calisto LE, Morris KA, Kopecky B, Duncan JS, Beisel KW, Fritzsch B. Expression of neurog1 instead of atoh1 can partially rescue organ of corti cell survival. PLoS One. 2012;7:e30853. doi: 10.1371/journal.pone.0030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Beyond generalized hair cells: molecular cues for hair cell types. Hear Res. 2013;297:30–41. doi: 10.1016/j.heares.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto K, Ishimoto S, Minoda R, Brough DE, Raphael Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J Neurosci. 2003;23:4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nature reviews. Neuroscience. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirjavainen A, Sulg M, Heyd F, Alitalo K, Yla-Herttuala S, Moroy T, Petrova TV, Pirvola U. Prox1 interacts with Atoh1 and Gfi1, and regulates cellular differentiation in the inner ear sensory epithelia. Dev Biol. 2008;322:33–45. doi: 10.1016/j.ydbio.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Lagziel A, Ahmed ZM, Schultz JM, Morell RJ, Belyantseva IA, Friedman TB. Spatiotemporal pattern and isoforms of cadherin 23 in wild type and waltzer mice during inner ear hair cell development. Dev Biol. 2005;280:295–306. doi: 10.1016/j.ydbio.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Lanford PJ, Shailam R, Norton CR, Gridley T, Kelley MW. Expression of Math1 and HES5 in the cochleae of wildtype and Jag2 mutant mice. J Assoc Res Otolaryngol. 2000;1:161–171. doi: 10.1007/s101620010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Price SM, Cahill H, Ryugo DK, Shen MM, Xiang M. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development. 2002;129:3523–3532. doi: 10.1242/dev.129.14.3523. [DOI] [PubMed] [Google Scholar]
- Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expression Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- Masuda M, Dulon D, Pak K, Mullen LM, Li Y, Erkman L, Ryan AF. Regulation of POU4F3 gene expression in hair cells by 5′ DNA in mice. Neuroscience. 2011;197:48–64. doi: 10.1016/j.neuroscience.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Pak K, Chavez E, Ryan AF. TFE2 and GATA3 enhance induction of POU4F3 and myosin VIIa positive cells in nonsensory cochlear epithelium by ATOH1. Dev Biol. 2012;372:68–80. doi: 10.1016/j.ydbio.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear Res. 2011;275:66–80. doi: 10.1016/j.heares.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Duncan JS, Kopecky B, Fritzsch B. A novel atoh1 “self-terminating” mouse model reveals the necessity of proper atoh1 level and duration for hair cell differentiation and viability. PLoS One. 2012;7:e30358. doi: 10.1371/journal.pone.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Kopecky B, Beisel K, Soukup G, Fritzsch B. Stem cells and molecular strategies to restore hearing. Panminerva Med. 2008;50:41–53. [PMC free article] [PubMed] [Google Scholar]
- Qian D, Radde-Gallwitz K, Kelly M, Tyrberg B, Kim J, Gao WQ, Chen P. Basic helix-loop-helix gene Hes6 delineates the sensory hair cell lineage in the inner ear. Dev Dyn. 2006;235:1689–1700. doi: 10.1002/dvdy.20736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubel EW. Ontogeny of Structure and Function in the Vertebrate Auditory System. Springer; New York: 1978. [Google Scholar]
- Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. 1967;(220):221–244. [PubMed] [Google Scholar]
- Sans A, Chat M. Analysis of temporal and spatial patterns of rat vestibular hair cell differentiation by tritiated thymidine radioautography. J Comp Neurol. 1982;206:1–8. doi: 10.1002/cne.902060102. [DOI] [PubMed] [Google Scholar]
- Scheffer D, Sage C, Corey DP, Pingault V. Gene expression profiling identifies Hes6 as a transcriptional target of ATOH1 in cochlear hair cells. FEBS Lett. 2007;581:4651–4656. doi: 10.1016/j.febslet.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Shailam R, Lanford PJ, Dolinsky CM, Norton CR, Gridley T, Kelley MW. Expression of proneural and neurogenic genes in the embryonic mammalian vestibular system. J Neurocytol. 1999;28:809–819. doi: 10.1023/a:1007009803095. [DOI] [PubMed] [Google Scholar]
- Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumida M, Harada Y. Development of the utricular macula in the mouse. Arch Oto-Rhino-Laryngology. 1984;241:9–15. doi: 10.1007/BF00457911. [DOI] [PubMed] [Google Scholar]
- Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, Zoghbi HY, Orkin SH, Bellen HJ. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- Xiang M, Gao WQ, Hasson T, Shin JJ. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Development. 1998;125:3935–3946. doi: 10.1242/dev.125.20.3935. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]


