Abstract
One way to enhance therapeutic development is through the identification and development of evaluative tools such as biomarkers. This review focuses on putative diagnostic, pharmacodynamic, and predictive biomarkers for smoking cessation. These types of biomarkers may be used to more accurately diagnose a disease, personalize treatment, identify novel targets for drug discovery, and enhance the efficiency of drug development. Promising biomarkers are presented across a range of approaches including metabolism, genetics, and neuroimaging. A preclinical viewpoint is also offered, as are analytical considerations and a regulatory perspective summarizing a pathway toward biomarker qualification.
Tobacco remains the leading preventable cause of death worldwide. Up to one-half of regular tobacco users will eventually die of a tobacco-related disease. Globally, smoking kills more than 5 million people each year and is a risk factor for six of the eight leading causes of death.1 Domestically, smoking takes the lives of more than 440,000 individuals each year and is the leading cause of premature death.2 It is estimated to cost the United States more than $200 billion each year in health-care and other related costs, such as diminished worker productivity. Whereas preventive and regulatory strategies, including harm warnings and product taxation, have significantly decreased smoking incidence and prevalence over the past 50 years, smoking prevalence has remained close to 20% over the past decade.3 In 2009 alone, there were more than 45 million active smokers, or approximately one of every five US adults, 70% of whom said they wanted to quit.4 Several approved drug products are available to aid in smoking cessation, including bupropion (Zyban), varenicline (Chantix), and various types of nicotine-replacement therapy (NRT); however, these treatments confer only a two- to threefold increase in quit-success rates over placebo, and only ~15–20% of treatment-seeking smokers are able to remain abstinent after 1 year.5,6 is suggests that more effective antismoking treatments are needed to reduce the lingering prevalence of smoking.
In general, the development of safe and effective medications for indications such as nicotine dependence poses significant challenges. Less than 7% of central nervous system (CNS) drug candidates have reached the market in the past decade.7 As such, there is a need to enhance the decision-making efficiency by which potential therapeutic options can be more easily translated to the marketplace. One of the most important ways to enhance drug development processes is via the identification and qualification of novel evaluative tools such as biomarkers. A biomarker has been defined as “a characteristic that is objectively measured…as an indicator of normal biological processes, pathogenic processes, or biological responses to a therapeutic intervention.”8 Biomarkers in particular are well suited for “high-risk” translational areas involving mechanistically heterogeneous diseases7—nicotine dependence being a prime example.
Biomarkers are often identified from a mechanistic understanding of the disease pathophysiology and observed correlations with a particular clinical outcome. the Institute of Medicine proposes a three-part iterative framework for the development of biomarkers, including the following (Figure 1):
Figure 1.
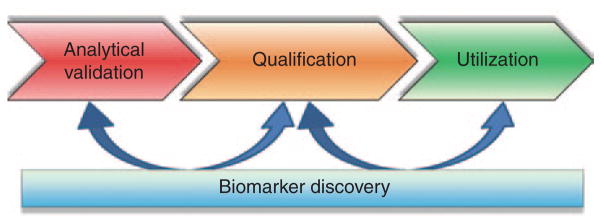
Biomarker development is interdependent. Initially, a measure is “analytically validated” for its precision and accuracy. An optimal measure would be practical (e.g., robust and cost effective) and easy to collect (e.g., noninvasive, measured within a surrogate tissue (e.g., saliva)). A validated test is required before “qualification,” which is the evidentiary linkage of the measure with its clinical outcome. Finally, independently corroborated qualification of biomarkers can be “utilized” for more general purposes. Data from concurrent biomarker discovery efforts (e.g., “omic”-type projects) can be used to hone further efforts along each step of development (Table 2). Adapted from ref. 73.
Analytical validation (i.e., is the biomarker able to be accurately, precisely, and reliably measured?)
Qualification (i.e., assessment of available evidence on associations between the biomarker and disease states)
Utilization (i.e., is there adequate evidence to support applying the biomarker for a specific use?)
The US Food and Drug Administration (FDA) has established a process for biomarker development and qualification in the Center for Drug Evaluation and Research that differs from that recommended by the Institute of Medicine and is applicable to any use of biomarkers in the drug development process. In the FDA framework, the biomarker is qualified for a specific “context of use”. A context of use is defined as a “comprehensive and clear statement that describes the manner of use, interpretation, and the purpose of a biomarker in drug development” and includes a detailed description of its measurement, purpose, and application. Importantly, the context of use determines the evidence needed for qualifying a biomarker and also delineates its application for future studies and drug development programs.
As defined in the Diagnostic and Statistical Manual of Mental Disorders, fourth edition, a substance use disorder such as nicotine dependence (§305.1) is “a maladaptive pattern of substance use, leading to clinically significant impairment or distress” characterized by key features such as tolerance, withdrawal, craving, and persistent use—despite its known harmful consequences. is review focuses specifically on three types of biomarkers thought to potentially have the most significant short-term translational impact for nicotine dependence, including (i) diagnostic biomarkers, which can be used to identify the presence or absence of a specific disease state and aid in patient selection; (ii) pharmacodynamic biomarkers, which can be used to report on the downstream effects of a pharmacotherapy; and (iii) predictive biomarkers, which can be used to select patients who are likely to respond to therapy (Box 1; ref. 7). is is not a comprehensive review of biomarkers for nicotine dependence, nor is it a description of biomarkers for tobacco-related harm or disease. Biomarkers for smoking cessation, as considered herein, are those that may be useful for the development of drugs as aids to smoking cessation and that are delineated across a range of methodologies including metabolism, genetics, and neuroimaging. Analytical and regulatory perspectives are also described. Finally, it is important to note that more than 4,000 compounds can be created when a cigarette is burned. Of these myriad compounds, nicotine—even though it is generally recognized as the most addictive substance in smoke—represents only one. erefore, although this review focuses on biomarkers for nicotine dependence and their translational potential for enhancing the development of smoking-cessation medications, biomarkers not involving nicotine should also be considered for future development.
Box 1. The three major types of biomarkers considered herein to enhance therapeutic development of smoking-cessation medications.
1. Diagnostic biomarker for patient selection
Definition: a diagnostic biomarker described herein is defined as a measure of an underlying (patho-) physiological process as it relates to the course of a specific clinical outcome. A diagnostic “patient selection” biomarker is used to identify/qualify key biological aspects of a disease and optimally stratify patient populations into the most appropriate phenotype for treatment.
No therapeutic intervention is involved.
This type of marker could be identified via either retrospective or prospective analyses.
Some diagnostic biomarkers for smoking currently exist; one example of a diagnostic biomarker for smoking status is a measure of carbon monoxide in exhaled air.
2. A pharmacodynamic (PD) biomarker for optimal dosing
Definition: a PD biomarker described herein is defined as a “response indicator” for a pharmaco- therapy and is used to assess its potential dose– response in vivo.
If there were strong modulation of the target by the pharmacotherapy, there would be a noted associa- tion with the clinical outcome, probably due to the drug interacting with its biologically intended tar- get; however, if there were minimal/no engagement of a pharmacotherapy with its biologically intended target, there would be little/no noted association with the clinical outcome, probably due to other factors (e.g., unfavorable properties of the drug, insufficient dosing).
A PD biomarker of smoking cessation, for example, could be used to define a clear qualitative physio- logical drug response and thereby improve decision- making processes (e.g., go/no-go decision points for a drug development project).
3. Predictive biomarker for therapeutic efficacy
Definition: a predictive biomarker described herein is defined as a measure used to identify before treatment those patients who might respond to a particular therapeutic.
Predictive biomarkers correlate the response to a therapeutic intervention, positive (efficacy) and/or negative (safety/toxicity), with a clinical outcome before administration of the intervention but are not necessarily associated with disease progression.
Molecular features in particular are likely to identify specific features of a disease that may differentiate/target an efficacious (or harmful) therapy a priori.
One example of a predictive biomarker for smoking cessation may be the nicotine metabolic ratio– predicted quit success with transdermal nicotine-replacement therapy (refs. 22–24).
METABOLIC BIOMARKERS
The nicotine metabolic ratio as a diagnostic biomarker
e substantial variability in smoking behaviors and cessation is attributable, in part, to heritable differences in the clearance rates of nicotine. Nicotine, the primary addictive compound in tobacco, is metabolized primarily by the hepatic enzyme CYP2A6 to cotinine (COT) and then exclusively by CYP2A6 to 3′-hydroxycotinine (3HC).9 the gene encoding this enzyme is highly polymorphic, resulting in large variation in rates of nicotine metabolic inactivation. Because of the major role that CYP2A6 plays in nicotine’s metabolism, genetic variation in CYP2A6 alters total nicotine clearance, non-renal plasma clearance, and nicotine plasma half-life.10 Genetically slow metabolizers are less likely to be smokers; those who are smokers are less dependent and smoke fewer cigarettes per day.11,12 Dependence scores differ significantly by CYP2A6 genotype12 and by nicotine metabolic ratio (NMR)13; for example, the individuals in the first quartile (i.e., slow metabolizers) had Fagerström Test for Nicotine Dependence (FTND) scores of 4.6, whereas those in the upper three quartiles had scores of 5.7–6.1. the slow-metabolizer genotype is also found more often among shorter-duration smokers,11 former smokers,14 and those responding to placebo or patch treatment.15 ese smokers also have a lower risk for tobacco-related illnesses, including lung cancer.12 One difficulty in genotyping individuals, however, is the complexity of the CYP2A6 gene loci; there are numerous characterized and uncharacterized alleles that vary in frequency across world populations. Whereas genotypic variation accounts for a substantial portion of the variation in CYP2A6 activity, other influences not captured by genotype include gender, body mass index, and some drugs. Accordingly, a non-invasive assessment of CYP2A6 activity that could be measured in smokers would expedite translation of data to the clinic.
A biomarker for CYP2A6 activity has been proposed on the basis of the exclusive role that CYP2A6 plays in COT metabolism to 3HC and the high correlation of this to total nicotine clearance. the ratio of 3HC/COT, referred to as the NMR, reflects CYP2A6 genetic variation and environmental factors influencing nicotine clearance in vivo. COT has a long half-life (averaging 16 h), making its assessment a relatively robust indicator of the “time to last cigarette.” By comparison, 3HC has a shorter half-life (averaging 5 h). Among regular smokers, because levels of COT reach steadystate, the elimination of 3HC is CYP2A6 formation-dependent, and the NMR ratio is stable over time.16 the reliability coefficient for stability tests of log-transformed NMR was 0.92. Assessments of NMR in two different laboratories were consistent (ratio: 1.06), and observed measures were within the outside limit of agreement of mean bias ±2 SDs. the reliability coefficient for repeated measurements of NMR over 44 weeks (i.e., repeated sampling of individuals over time) was 0.85.16 the NMR can be measured in the saliva, urine, or plasma of smokers and has been validated as a biomarker of nicotine clearance.17 the NMR is also closely related to CYP2A6 genotype and more recently has been used to characterize novel CYP2A6 alleles.18
New data on the reproducibility and reliability of the NMR are emerging. the saliva NMR does not vary with time of day.19 Moreover, recent experiments have documented the concordance of NMR values across two analytical laboratories, stability of the NMR assays using plasma and saliva stored at room temperature and in blood at 4 °C, concordance of blood NMR with saliva and plasma NMRs, and excellent reliability for repeated plasma NMR measurements in smokers over 44 weeks of sampling.16 erefore, genetically distinguishing fast vs. slow metabolizers using the NMR diagnostic biomarker may be an efficient approach to differentiate less dependent smokers from more dependent smokers and personalize their smoking cessation treatment accordingly.
The NMR as a predictive biomarker
The NMR also has value as a predictive biomarker. the NMR has been utilized in several clinical trials as a genetically informed marker of CYP2A6 activity.20,21 In the first study of the clinical utility of the NMR, treatment-seeking smokers were randomized to transdermal nicotine or nicotine nasal spray. the NMR predicted quit success with transdermal NRT; however, the NMR did not predict quit success for those prescribed nicotine nasal spray, presumably because smokers were able to titrate the dose of nasal spray on the basis of differential metabolism.22,23 the NMR was validated as a predictor of quit success in an independent sample of more than 500 smokers after 8 weeks of transdermal nicotine treatment, for which the NMR was a better predictor of quitting than cigarettes per day, FTND, or baseline COT.24 On the basis of these results, the efficacy of extended (6-month) transdermal nicotine therapy vs. the standard 8-week therapy was examined. ose with reduced rates of metabolism were found to benefit more from extended therapy than from the standard 8-week therapy as compared with normal metabolizers.15
To determine whether the predictive validity of the NMR is specific for transdermal nicotine therapy, another study evaluated this biomarker in a 10-week placebo-controlled randomized trial of bupropion, a drug not metabolized by CYP2A6 (ref. 25). Fast metabolizers, as determined by the NMR, had significantly lower quit rates than slow metabolizers if treated with placebo (i.e., counseling only) but had quit rates equivalent to those of slow metabolizers if treated with bupropion.25 In a clinical trial of African-American light smokers, slow metabolizers, as determined on the basis of the nicotine metabolite ratio, had higher quit rates than fast metabolizers treated with nicotine gum or placebo.26 ese retrospective data suggest that slow metabolizers, as defined by the NMR, have a greater success with transdermal nicotine, nicotine gum, or with counseling only, whereas fast metabolizers would not be ideal candidates for these treatments. For fast metabolizers, a nonnicotine therapy such as bupropion may be most efficacious. Evidence for the efficacy of higher-dose patch therapy for fast metabolizers is modest. However, these hypotheses remain to be tested in a prospective head-to-head clinical trial before establishing NMR cutoffs and metrics that can be used for clinical integration.
As part of the Pharmacogenomics Research Network, an ongoing prospective stratified clinical trial of more than 1,300 smokers is currently evaluating the efficacy and clinical utility of the NMR as a biomarker of response to placebo, transdermal nicotine, and varenicline. Estimates of sensitivity, specificity, predictive value, and optimized cutoff points will be established from this trial. If favorable, the NMR test could begin to be utilized within clinical practice as a point-of-care predictor to tailor an individual’s smoking-cessation treatment.
PHARMACOGENOMIC BIOMARKERS
Twin studies have consistently indicated that there are genetic contributions to an individual’s ability to quit smoking (e.g., ref. 27). ere have been a number of pharmacogenomic investigations of quit success in individuals who quit in community settings or in clinical trials with FDA-approved medications (e.g., refs. 20, 28–30). No major gene effects have been found in this work, although findings with 10−2 > P > 10−8 have been obtained. Variants in candidate genes that appear to influence the numbers of cigarettes smoked per day provide only modest effects on an individual’s overall ability to quit when their influences on smoking quantity itself are statistically corrected for.31,32 Variants in the CHRNα5-α3-β4 gene cluster may predict a therapeutic response for smoking-cessation treatments, but the proportion of variance accounted for is small.31,33 Although pathway- or systems-based genomic analysis may provide a biologically comprehensive approach for pharmacogenomic research on smoking cessation,34,35 the required sample sizes to detect gene–gene–drug interactions can be rate-limiting. A recent genome-wide association study meta-analysis of a retrospective smoking-cessation phenotype identified a number of genes of interest; however, only rs3025343 near the dopamine β-hydroxylase (DBH) gene reached genome-wide significance.36 is finding has been replicated in at least one candidate-gene case–control study.37 Other DBH variants have been examined with mixed results in some clinical trials (for review, see ref. 38). To our knowledge, the genome-wide significant association of rs3025343 with cessation has yet to be evaluated in prospective clinical trial samples. is should be pursued in future investigations.
The “v1.0 quit success score” as a predictive biomarker
Genome-wide association studies for smoking-cessation clinical trials using 500,000 to 1 million single-nucleotide polymorphism (SNP) markers30,39–41 have provided groups of clustered SNPs that display 10−2 > P > 10−8 differences between successful and unsuccessful quitters treated with either NRT or bupropion. the chromosomal regions and genes identified by these clustered, nominally significant SNPs from different samples overlap much more than would be expected by chance. ese studies identify several genes, including, for example, those that encode cell adhesion molecule cadherin 13, that have been reported from multiple, independent studies. More than 12,000 SNPs displayed 10−2 > P > 10−8 associations with the ability to quit in at least one of three initial clinical trials.30 ese SNPs were weighted based on their P value and number of samples in which the association occurred to form a “v1.0 quit success score.” Modeling studies supported substantially increased power of phase II–sized clinical trials if participants were genomically stratified based on the v1.0 quit success score that identified even half of the genetic influences on quit success.40 Indeed, this marker has been found to predict quit success in two independent, prospective, double-blind clinical trials39 as well as in a recently reported trial in which NRT doses were matched to a level of dependence; the v1.0 quit success scores estimate an area under the receiver–operator curve of 0.67.42
An NRT-induced reduction in smoking (i.e., as measured by a reduction in exhaled carbon monoxide concentration) before a targeted quit date also provides robust ability to predict quit success that is not well captured by the v1.0 quit success score. When combined with the v1.0 quit success score, the precessation smoking reduction associated with a higher rate of short-term smoking abstinence provides a more robust “biosignature,” or combination of biomarkers. Together, this combination of physiological and genetic biomarkers provides an example by which two distinct biomarkers capture a “more comprehensive biological snapshot,” leading to a somewhat enhanced clinical predictive validity with an area under the receiver–operator curve of 0.73.42 As this genotype-based quit-success score is tested in populations of disparate genetic backgrounds, we will learn whether the application of this biosignature is generalizable.
At this time, the drug products investigated most widely in pharmacogenetic and pharmacogenomic studies, including many cited above, are bupropion and NRTs. Feasibility and diffusion studies are needed to explore the challenges, logistics, and patient-centered outcomes germane to implementation in real-world health-care organizations. BEACON (Behaviorally Enhanced Counseling on Nicotine Dependence), a US-based feasibility study of a treatment protocol, used a single-marker approach as a simple proof of concept to genetically tailor smoking-cessation treatment to patients in a health-care setting to either bupropion (rs1800497 CC genotype) or a nicotine patch (rs1800497 TT/TC genotype). is study randomized patients in the Group Health Cooperative integrated delivery system to receive theoretically grounded genetic feedback or standard care.43 the results of the BEACON study suggested that telephone-based counseling combined with genetically matched treatment is feasible, with high patient satisfaction and no evidence of adverse psychological impact. Similar results were found in the much larger, UK-based, Personalised Extra Treatment (PET) trial. is trial randomized smokers to increased dosing of NRT by OPRM1 rs1799971 AA genotype ((6 mg)/GA; AA genotype (12 mg)) vs. dosing NRT by FTND score (score < 8 (6 mg); ≥8 (12 mg)) but found no evidence of adverse psychological or behavioral impact of genetic feedback in the context of genetically tailored smoking-cessation treatment.44
NEUROIMAGING BIOMARKERS
Although it has become increasingly appreciated that drug dependence is a CNS disease of dysregulated learning and memory, the modifications that have occurred as a consequence of addiction remain poorly understood. Which observed neuroadaptations are specific to nicotine dependence? Are they transient or long-lasting? Why might neuroimaging measures serve as potential biomarkers of such drug-induced neuroplasticity and addiction? ere are at least three key roles for neuroimaging biomarkers. First, given that they can index brain structure and function, imaging biomarkers can serve as putative endo-phenotypic measures that intermediate distal biomarkers (e.g., DNA, gene expression) with more proximal biomarkers (e.g., behavior, self-report) and their clinical outcomes, ideally correlating meaningfully with the phenotypes on the causal path of a disease or a biological response to a therapy. As such, whereas the outcome of a particular treatment may be related to genetic factors or the effects of the treatment on self-reported craving, there may be multiple pathways that underlie the influence of a gene or genes on outcome, or multiple pathways that mediate the subjective experience of craving (e.g., salience of cues, self-awareness). Measures of brain function may provide assessment of the relative contribution of these pathways to clinical course and/or outcome. Second, again reflecting their position as a potential endophenotype, neuroimaging markers can potentially provide a simultaneous assessment of both dynamic, acute-state (e.g., withdrawal, craving) and more chronic trait-like (e.g., addiction severity) markers relevant to addiction outcomes. ird, based on brain regions identified as involved in clinical phenotypes, such as withdrawal symptoms or cue-elicited craving, modification of activity within these regions can subsequently serve as directed mechanistic interventional targets and predictive surrogates of smoking behavior.
Acute prequit reductions in smoking-cue brain reactivity as predictive biomarkers
e relatively few studies using neuroimaging as a potential biomarker have focused on imaging as a predictive marker for smoking cessation. For example, acute pre-quit reductions in smoking-cue reactivity within the thalamus and ventral striatum are associated with improved cessation outcomes;45 pre-quit activation in the insula and anterior cingulate in response to smoking cues during a distraction-based task, predicted postquit slips;46 and larger hippocampal volumes are predictive of poorer cessation outcomes.47 Although few in number and of limited sample size, these studies provide preliminary evidence that neuroimaging measures may be useful predictive biomarkers for smoking cessation.
Other studies have focused on behavioral and/or pharmacological manipulations. For example, studies have examined the effects of pharmacological and behavioral interventions on brain reactivity to visual smoking cues.45,48–50 ree of these studies48–50 showed that interventions with putative effects on reactivity to smoking cues decreased brain responses to these cues in ventral aspects of the anterior cingulate cortex and/or medial orbitofrontal cortex. ese studies suggest that changes in braincue reactivity, particularly in ventromedial structures, may serve as a possible pharmacodynamic biomarker and, if further qualified, may be used to define a clear, qualitative drug response.
Large-scale, brain-network interactions as a diagnostic biomarker
Assessment of functional magnetic resonance imaging signals in the absence of a task reveals patterns of coherent, low-frequency, spontaneous fluctuations that may more clearly indicate underlying pathophysiological changes of a substance use disorder such as nicotine dependence. the patterns of large-scale networks observed at “rest” are largely consistent with, and constrained by, anatomical connections and appear to comprise the full set of networks activated when an individual performs specific cognitive tasks. One such network, the so-called default mode network (DMN), which includes the medial prefrontal cortex, medial temporal lobes, and posterior cingulate cortex, is deactivated during task performance and is thought to reflect episodic memory, ruminations, or “mind wandering.”
Although used to investigate a wide range of psychiatric disorders including depression and attention-deficit hyperactivity disorder, resting-state functional connectivity is increasingly being utilized to assess nicotine dependence and its possible applications to smoking-cessation treatment. For example, it has been demonstrated that anterior cingulate cortex–ventral striatal functional connectivity strength is negatively correlated with nicotine usage and is unaltered by acute NRT, although other more “cognitively” related cingulate-cortical circuits are enhanced by nicotine, suggesting the existence of a potential “addiction circuit” diagnostic biomarker for nicotine dependence. A very similar circuit is moderated by variation in the α5 nicotine receptor subunit gene.51 Furthermore, improvement in cognitive withdrawal symptoms in abstinent smokers is related to the degree of negative correlation between executive control and DMNs.52 Among nonsmokers, nicotine simultaneously decreases activity within the DMN while increasing visual attention networks.53 Taken together, these findings are consistent with the hypothesis that nicotine acts by enhancing executive control and focusing attention toward external stimuli.
A model-based approach has recently been proposed for the use of resting-state functional connectivity. is model, which incorporates the interactions between large-scale networks and alterations in the addiction trajectory, might profitably serve as a diagnostic biomarker of addiction. is model centers on the interoceptive monitoring role of the insula, which, together with the anterior cingulate cortex, is thought to direct attention toward either internal or external stimuli by mediating dynamic activity between two large-scale brain networks, the DMN and the executive control network. ese networks interact competitively during task performance, whereby suppression of DMN activity is often associated with optimal behavioral outcome. During nicotine abstinence, the insula may track withdrawal-induced bodily sensations (e.g., perceived as cravings) and, in turn, direct attention toward the homeostatically salient internal state via increased interactions with the DMN at the expense of decreased exogenously directed attention mediated by executive control network. is resting-state functional connectivity circuitry model warrants further empirical testing and validation.
A number of practical issues need to be considered in assessing neuroimaging as a potential drug development tool. First, short- and intermediate-term longitudinal imaging studies should be performed, with multiple measures of cessation-treatment outcome, to determine whether specific brain circuits and networks change as a function of alterations in addiction severity that occur with extended abstinence during treatment. Second, future studies that evaluate putative biomarkers need to include former smokers. is cohort is understudied and needs to be examined in order to answer questions of possible long-term recovery of function. ird, genetic markers should be used to complement the power of, and individual differences in, imaging biomarkers. Fourth, negative results need to be reported/shared among investigators. Fifth, there must be a novel, multimodal, composite set of imaging and behavioral testing batteries, standardized for use across multiple sites. For example, whether task-based or task-independent, most neuroimaging studies to date have relied on group analysis, mostly due to the intrinsic low sensitivity of the functional magnetic resonance imaging signal. Low signal-to-noise ratios have generally precluded the use of individual subject analysis, something required for an analytical tool to be clinically useful as a biomarker. It remains to be seen whether this biophysical limitation will preclude its routine application. Finally, small sample sizes, cost, and reliability and reproducibility issues remain concerns for neuroimaging, as they do for any potential biomarker assay. the incorporation of standard research methods across laboratories and development of large-scale databases may help overcome sample-size issues. Whereas MRI-based neuroimaging measures are currently expensive to obtain, their potential usefulness in predicting treatment response may outweigh these costs when considered against the cost of addiction at the individual and societal level. In addition, increased knowledge of the mechanisms that underlie nicotine dependence can lead to new and refined, and relatively less expensive, behavioral or electroencephalogram-based markers that may be more easily transferred into clinical practice.
ANALYTICAL CONSIDERATIONS
Development of biomarkers requires defining optimal methods for data analysis. For instance, characterization of heterogeneity in treatment response for smoking cessation, using biomarkers, presents specific statistical challenges. “Subgroup analyses” that evaluate outcome as a function of the interaction of a baseline covariate (subgroup indicator) and treatment are not without controversy. Evaluating the degree to which biomarkers account for heterogeneity in treatment response raises concerns regarding type I and II errors, leading to recommendations favoring a Bayesian approach. the Bayesian method permits estimation of a “posterior distribution” that summarizes evidence regarding the existence of a subgroup effect. is posterior distribution is a function of investigators’ prior information and the data observed in the current study. Inspection of the posterior distribution for the interaction term permits statements regarding the probability that an interaction effect of some magnitude exists given the data. Two points are salient: (i) inspection of the posterior distribution permits direct evaluation of the alternative hypothesis, a feature that is not available via conventional frequentist statistical reasoning, and (ii) the posterior distribution, the Bayesian method, permits the estimation of the probability that a given treatment confers benefit or harm as a function of a specified subgroup variable. Recognizing that investigators may disagree regarding the state of the prior evidence, specification of multiple priors representing distinct perspectives (e.g., optimistic, neutral, or skeptical) is possible. is formalization, termed a “community” of priors, permits evaluation of the degree to which the data should result in revision of differing levels of preexisting skepticism.54,55 Such properties permit evaluation of the degree to which convincing evidence for a biomarker effect exists.
Statistical analysis of multiple, diverse biomarkers
Recent advances in technology now make it feasible to obtain numerous biomarkers, ranging from genetics to imaging to behavior, for the more complete evaluation of subgroup effects. In this context, the discovery of noteworthy subgroups and the investigation of potential interactive effects dramatically increase the number of statistical tests evaluated. Often these tests are viewed solely in the context of the total number of tests or investigators rely on post hoc justification of preconceived ideas.
For high-dimensional data analysis, formal Bayesian approaches ofter a statistical approach that formalizes the prior knowledge and balances the trade-off between that knowledge and the data. With regard to genome-wide association studies, rankings may be expanded upon using methods that incorporate prior information into the determination of the false-discovery rate—a weighted false-discovery rate56 or a Bayesian false-discovery probability.57 For effect estimation and model building, hierarchical models and Bayesian stochastic search approaches may be used.58 Hierarchical modeling has the advantage of formalizing prior knowledge into predictor variables that describe potential relationships between the factors. In genetics, this could define simple variables systematically extracted from public databases and ontologies, such as gene annotation and the pathway in which each gene is thought to be active. In addition, more experimentally derived information can be used, such as eQTLs or ChIP-seq,36,59 in silico predictions based on evolutionary conservation or predicted effect on protein conformation60 or predictions from simulations of the pathway.61 Moreover, because there may also exist a hierarchy across all factors, separate sets of priors can be constructed for SNPs within a linkage disequilibrium block,62 genes and genes within pathways,63 or for main effects and interactions.64 Finally, hypothesized biological pathways may be used to construct a “prior topology” or combination of factors that then informs the model search and evaluation by giving greater prior probability to combinations of factors in the data that are more closely related to the prior structure or biological mechanism.65
The Bayesian analytical approach to characterizing heterogeneity in treatment effects as a function of biomarkers confers a number of benefits. Bayesian analyses permit direct estimates of the probability that subgroup effects exist and of the degree to which treatment may incur benefit or harm as a function of a subgroup variable, and it may do so in the context of sample sizes too small to register an effect using conventional frequentist methods. The Bayesian approach permits evaluation of the degree to which the data might sway investigators articulating reasonable differences of opinion. Empirically derived prior information can improve the precision of resulting subgroup estimates. For subsequent trials, evaluating the probability of obtaining a meaningful outcome conditional on the estimate of the subgroup effect and its corresponding uncertainty should provide a more robust prediction of the decisiveness of a planned trial than conventional power analysis.
PRECLINICAL TRANSLATION
As a tool for drug development (Figure 2), animal models may be used at three stages of the drug discovery process: first, animals may be utilized early in the drug development process, during the “target validation” stage. is sometimes sophisticated and multifaceted process serves to increase the understanding of a particular drug target and may help investigators determine whether a particular drug target meets criteria for initiation into a drug-screening campaign. Second, when pharmacological agent(s) that target a system of interest are uncovered, they can be examined in an animal model of nicotine dependence to obtain “proof of concept” that the compounds show some degree of efficacy. Positive (or negative) results may then be taken into consideration when deciding whether a potential project is worth pursuing further. ird, animal models of nicotine dependence may also be used later in the drug development process. ese types of models can be used to advance efforts when several agents within one class or different classes of compounds have been developed. Likewise, animal models may play an important, multifaceted role in guiding iteratively the development of compounds into the clinic for testing.
Figure 2.
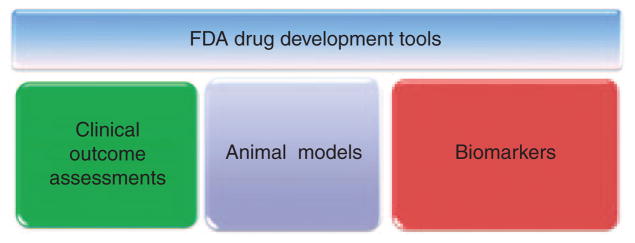
US Food and Drug Administration (FDA) drug development tools (DDTs). The DDT qualification program, including biomarkers, was created by the Center for Drug Evaluation and Research to guide and prepare drug development efforts for rigorous safety and efficacy testing and eventual regulatory evaluation.
Although not without potential drawbacks, it is reassuring to note that varenicline decreases intravenous nicotine self-administration behavior in rodent models.66 the utility of the self-administration procedure, as well as other animal models of nicotine dependence that may be used to support drug development efforts, would be further validated by the identification of reliable biomarkers that, optimally, predict the efficacy of smoking-cessation agents in human smokers.
Translational utility
Biomarkers may prove useful in the evolution of preclinical animal models to better encapsulate various aspects of nicotine dependence and, in the long-term, enhance decision making during drug development. Indeed, it can be envisioned that modifications of an animal model based on the response of a qualified biomarker in human smokers may help drive animal model development closer to the human condition, possibly improving predictive power for drug development. Such bio-markers could also be used to stratify/prioritize animal models and potentially identify those animal tests that specifically model aspects of nicotine dependence that have the most predictive power for identifying efficacious smoking-cessation medications in human smokers. us, cross-species biomarkers may help guide and validate the development of animal models for human nicotine dependence.
Animal models of nicotine dependence are critical for the development of novel smoking-cessation medications. the translatability of available animal models could be substantially increased if reliable, cross-species biomarkers of nicotine dependence that have diagnostic, pharmacodynamic, and/or predictive uses could be identified in human smokers. is is especially relevant for CNS diseases such as nicotine dependence, in which the direct assessment of brain tissue is costly and impractical. Translatable biomarkers may capture key aspects of nicotine dependence that help characterize the underlying mechanism(s) and can be used to validate measures for more efficient drug development. Biomarkers may also prove useful in the conception of improved animal models, which better reflect the human condition, and/or be used to stratify/rank pre-clinical models, potentially identifying those animal tests that specifically model causal features of nicotine dependence with the most predictive power for identifying efficacious smoking-cessation medications in human smokers. us, confirmation of biomarkers across species may not only help advance their qualification but also serve to guide the development and prioritization of superior animal models.
FUTURE DIRECTIONS
Biomarker discovery
Readily measurable biomarkers from peripheral fluids (e.g., plasma or urine) that could reproducibly reflect pathologically and/or pharmacologically induced processes in the brain would provide a significant step toward elucidating further underpinnings of the disease, identifying putative targets, confirming pharmacological target effect, and so on, all from a “surrogate” tissue. One promising discovery approach for novel biomarkers may arise from the proteomic or transcriptomic analysis of circulating proteins/nucleic acids in the blood of cigarette smokers. A recent proteomic analysis of serum-bound proteins in the blood of humans with a history of alcohol abuse revealed a discrete protein expression profile relative to control subjects, including dysregulated expression of gelsolin, selenoprotein P, serotransferrin, tetranectin, hemopexin, histidinerich glycoprotein, plasma kallikrein, and vitronectin.67
Another innovative and important area of research that is likely to yield useful biomarkers for nicotine dependence is that of circulating microRNAs (miRNAs). miRNAs are small (~21–25 nucleotides) regulatory RNAs that are emerging as powerful biomarkers of disease. Recent evidence suggests that miRNAs can be readily detected in blood, urine, saliva, amniotic fluid, and pleural fluid, with expression levels similar across different individuals matched for health status.26 As an example of the potential utility of miRNAs as sensitive biosignatures of physiological processes, the placenta-associated miRNAs miR-527, miR-526a, miR-141, and miR-149 were significantly elevated in the sera of pregnant women as compared with non-pregnant subjects,26,29 with the magnitude by which these miR-NAs were upregulated being correlated with pregnancy stage.26 the identification and validation of blood miRNA may offer a viable approach to develop biomarkers for tobacco dependence. Importantly, however, alterations in circulating miRNAs in response to a pathophysiological state may not only represent diagnostic tools but also provide insight into disease processes that may co-occur in more distal tissues. is could potentially occur through at least two mechanisms: first, miRNAs whose expression is dysregulated in diseased tissues (e.g., miRNAs in prostate cancer tissues; miRNAs in hepatocytes during drug-induced hepatic toxicity) may be “released” from the site of damage directly into circulation. is may be particularly important in the case of lung damage. Indeed, genome-wide association study profiling has revealed miRNA diagnostic signatures of non–small cell lung cancer; 12 specific miRNAs have been shown to be robustly overexpressed in diseased lung tissue and to have prognostic utility for estimating life expectancy.68 Specifically, miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-214, and miR-212 were upregulated in diseased lung tissue as compared with normal lung tissue. miR-21 levels in particular were closely correlated with a positive prognosis and overall survival of lung cancer patients.68 Hence, serum-based miRNAs can serve as biomarkers for tobacco dependence, but the same miRNAs may also reveal insights into the progression of tobacco-associated diseases.
A second mechanism through which serum-based miRNAs may provide information concerning molecular events in distal diseased tissues is if the same signaling events that occur in diseased tissue are conserved in nucleated blood cells such that the same complement of miRNAs is altered in both tissues. Indeed, patterns of gene expression in nucleated blood cells are remarkably consistent across healthy individuals,36 and many of the same signal transduction cascades are conserved in nucleated blood cells that are present in other cells, including neurons. erefore, it is possible that exposure to nicotine and other components of tobacco smoke may initiate the same signal transduction cascades in nucleated blood cells as those that are engaged in neurons localized within addiction-relevant brain circuits, resulting in the sample complement of miRNAs dysregulated in brain and blood. erefore, in addition to their clinical benefits, identification of circulating miRNA biomarkers could reveal those that are mechanistically important to addiction.
Finally, a characteristic of miRNAs that makes them particularly attractive as putative biomarkers is their remarkable stability within a peripheral (i.e., within a non-CNS, “surrogate”) tissue. It has been shown that serum-expressed miRNAs are resistant to endogenous ribonuclease activity, stable at room temperature, and resistant to degradation by freeze–thaw cycles.69,70 Importantly, it has been shown that the expression levels of miRNAs in serum, which are easy to collect and more readily available from archived samples, are highly correlated with levels in plasma samples from the same human subjects.70 erefore, the stability of miRNAs, their relatively simple separation from other nucleic acids in blood serum, and their easy accessibility from within a “surrogate,” non-CNS peripheral tissue, all suggest that proteomic and/or transcriptomic approaches to analyzing circulating blood-based protein or nucleic acids, particularly miRNAs, may yield reliable biomarkers for nicotine dependence.
Biosignatures
A surrogate biomarker has been defined as a single biomarker “intended to substitute for a clinical endpoint…and predict clinical benefit (or harm).”8 In terms of the development of smoking-cessation medications, a surrogate marker is already in use; clinical benefit is inferred when assessed through 4 weeks of self-reported abstinence, confirmed with periodic, exhaled carbon monoxide breath levels. What is likely to be of additional value in future drug development will be the application of a combination of biomarkers, or biosignature, which can more accurately capture the heterogeneous biologic and/or pharmacologic responses associated with a clinical end point. is may be especially true if the putative biosignature incorporates nonoverlapping approaches combining proximal and distal biomarkers (Figure 3). Together, these data would increase the sensitivity (true positive) and/or specificity (true negative) of the assay. A biosignature such as this is more likely to assess risks (i.e., off-target, nonpathway effects) and benefits (i.e., intended targeted pathway activation/repression) and more accurately reflect a clinical end point (Figure 4). For example, genetic and neuroimaging data, when combined, can be used to more clearly suggest which CNS circuits are affected, better elucidate underlying pathophysiological mechanisms, and characterize which neurochemical processes might contribute causally to nicotine dependence (see “Neuroimaging Biomarkers” above; refs. 51,71). A second example would be the combination of a physiological measure (NRT-induced reduction in carbon monoxide before a targeted quit date) with the “quit success” score with its ability to predict robustly the quit success for populations across diverse clinical settings (see “Pharmacogenomic Biomarkers” above; ref. 39). As robust assays for biosignatures like these are prioritized, validated, and developed (Table 1, Figure 1), their application as drug development tools (Figure 2, Box 1) will become more generally applicable to drug development studies.
Figure 3.
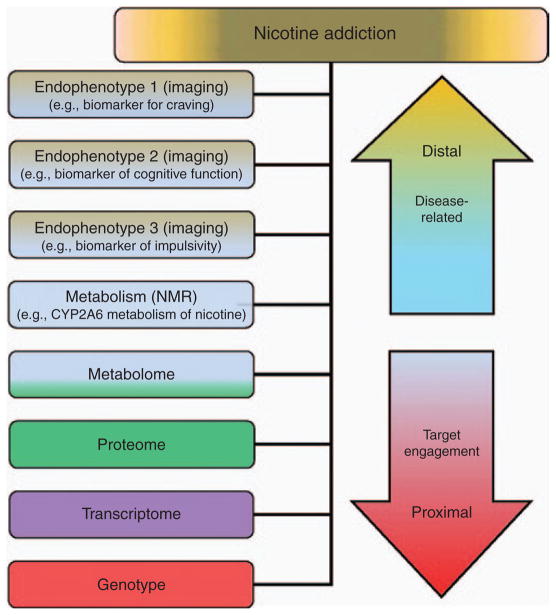
Proximal vs. distal biomarkers. Proximal markers are those biomarkers closely associated with the underlying molecular mechanisms of a disease; proximal biomarkers are not likely to capture the disease phenotype in its entirety (e.g., nicotine dependence). Distal markers, by comparison, are those biomarkers that reflect the endophenotypes further up the phenotypic “tree”; they are more likely to capture more features of an underlying pathophysiology or a biological response to a therapeutic intervention. NMR, nicotine metabolic ratio.
Figure 4.
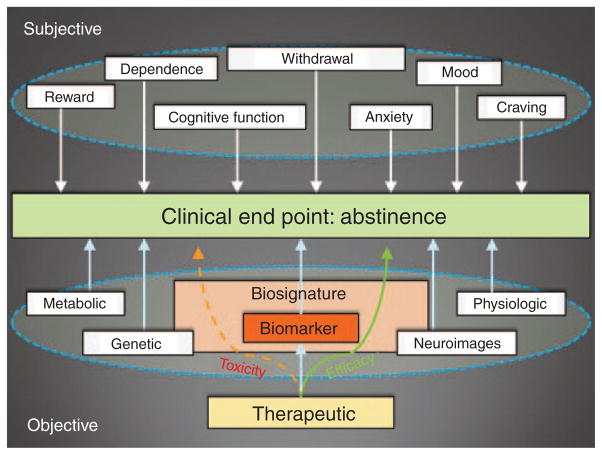
A biomarker is an objective measure of a clinical outcome. A biomarker is an objectively measured indicator of a normal biological process, pathogenic process, or biological response to a therapeutic intervention. As opposed to other more subjective measures related to nicotine dependence (e.g., craving, mood) (top circle), an objective biomarker for nicotine dependence might include a measure collected via a metabolic (e.g., NMR), genetic (e.g., quit-success score), physiologic (e.g., precessation response to NRT), or neuroimaging approach (e.g., rsFC) (see text for details). However, these data may reflect only a single aspect of a disease phenotype or pharmacotherapeutic response. For example, if combined as a “biosignature,” two or more biomarkers (e.g., neuroimages + genetics; see text) are more likely to capture key processes underlying efficacy and/or toxicity along the causal pathway(s) of a pharmacological process (large “Biosignature” box) that might have otherwise been missed if a single measure were applied (small “Biomarker” box). In terms of drug development, this type of data could help assess the overall risk (i.e., off-target, nonpathway effects) to benefit (i.e., intended targeted pathway activation/repression) ratio and improve the efficiency by which clinical trials are conducted. NMR, nicotine metabolic ratio; NRT, nicotine-replacement therapy; rsFC, resting-state functional connectivity.
Table 1.
Putative biomarkers for smoking cessation
| Biomarker | Biomarker use | Methodological assay | Context of use (critical parameters) | Identification and correlative development | Analytical validation | Clinical validation | Qualification (FDA) | Utilization (FDA) | Clinical valuation | Market- effect valuation | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lab/clinical | Lab/clinical | Clinical | Clinical/drug development | Clinical/drug development | Clinical/health care | Clinical/health care | |||||
| Nicotine metabolic ratio (NMR) | Predictive | Pharmacogenetic | Genotyping from blood sample; Homo sapiens; nicotine (drug) metabolism |
|
19–21 | ||||||
| NMR | Diagnostic | Pharmacogenetic | Genotyping from blood sample; Homo sapiens; nicotine (drug) metabolism |
|
14 | ||||||
| v1.0“Quit success” score | Predictive | Physiological (CO) + genetic panel of biomarkers | ↑ Quit-success score is correlated with ↑ pharmacological treatment success for smoking cessation; exhaled [CO]; 2-week success treatment with NRT; GWAS from blood sample; Homo sapiens |
|
31 | ||||||
| Gray matter volume (bilateral hippocampi) | Predictive | Voxel-based morphometry (high-resolution MRI) | ↑ Bilateral hippocampi volume are associated with ↓ smoking- cessation outcomes; Homo sapiens |
|
39 | ||||||
| Task-based neuroimages of thalamus | Predictive | fMRI | ↓ Reactivity to smoking cues in the thalamus is associated with ↑ smoking cessation; Homo sapiens |
|
37 | ||||||
| Task-based neuroimages of ventral striatum | Pharmaco- dynamic (PD) | fMRI | ↓ Reactivity to smoking cues in the ventral striatum is associated with ↑ smoking cessation; Homo sapiens |
|
37 | ||||||
| Task-based neuroimages of insula | Predictive | fMRI | ↑ Prequit insular activation during smoking cue–based distractors is associated with ↓ smoking- cessation outcomes; Homo sapiens |
|
38 | ||||||
| Task-based neuroimages of anterior cingulate cortex (ACC) | Predictive | fMRI | ↑ Prequit ACC activation during smoking cue–based distractors is associated with ↓ smoking- cessation outcomes; Homo sapiens |
|
38 | ||||||
| Nontasked neuroimages of dorsal ACC striatal connectivity | PD | Resting-state fMRI | ↓ Dorsal ACC-striatal connectivity is associated with a ↑ nicotine addiction severity; Homo sapiens |
|
73 | ||||||
| Cell adhesion molecule-13 gene | Diagnostic | Pharmacogenomic | Genotyping from blood sample; Homo sapiens |
|
34 | ||||||
References:http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM267449.pdf. As defined by the Critical Path Institute (http://www.c-path.org), one of the key next steps will be to determine feasibility; that is, to determine whether the biomarker DDT (Figure 2) possesses a significant enough impact to both industry and regulatory agencies that it justifies future efforts toward a regulatory qualification submission (Table 2).
ACC, anterior cingulate cortex; CO, carbon monoxide; DDT, drug development tool; FDA, US Food and Drug Administration; fMRI, functional magnetic resonance imaging; GWAS, genome-wide association study; NRT, nicotine-replacement therapy; ↑, increase; ↓, decrease.
CONCLUSION
It is estimated that by 2030, tobacco will kill more than 8 million people worldwide each year.1 the vast majority of smokers want to quit, yet many are unable to remain abstinent using existing therapeutic options. More effective antismoking treatments are needed to address this large medical need. To get there, however, the translational efficiency by which pharmaceutical development can be achieved needs to be improved. Biomarkers, as a drug development tool, hold significant promise in this regard.
To enhance our potential for translating these potential biomarkers, some of the approaches described herein might also be used to define more clearly various aspects of addiction. If, for example, various aspects of nicotine dependence were broken down into (i) reward, (ii) withdrawal, and (iii) relapse, the qualification of a specific smoking-cessation biomarker/biosignature could perhaps be honed and correlated with a risk-of-relapse clinical outcome measure that the putative biomarker most accurately reflects.72 is would theoretically help to circumvent complications that may arise due to the diverse influences on smoking cessation as a clinical outcome and thereby boost the efficiency by which biomarkers are identified and accepted.
Although some of the approaches described herein outline the first step(s) in biomarker development (Figure 1; Table 1), the discovery and development of putative biomarkers in general are not without issues. First, the term “biomarker” in general is overused; because evidence collection in the past has not been assembled in a standardized manner, there are diverse methodological approaches (e.g., imaging, genetics, physiological, and “omics”) and varied applications (e.g., diagnosis, prognosis, prediction), and the term has frequently been applied haphazardly. Second, biomarkers are frequently considered individually; there is a need for future studies to validate findings in both the clinic and the laboratory. ird, the pathway for biomarker evaluation is still evolving.73 Collectively, these issues have worked to further complicate the already multifaceted evidentiary process of linking biomarker data to a clinical outcome. A plan for the development for smoking-cessation biomarkers that circumvents many of these issues is presented in Table 2.
Table 2.
A suggested plan for biomarker development for smoking cessation
| Developmental steps | Collective steps |
|---|---|
| 1 | Identification: Diagnostic, pharmacodynamic, and/or predictive biomarkers for smoking cessationa Purported biomarker biological summary/association → begin to define its context of use
|
| 2 | Prioritization:
|
| 3 | Assay development: Analytical process is defined and optimized into a robust, reproducible assay/device
|
| 4 | Regulatory:
|
| 5 | Analytical validation:
|
| 6 | Public comment:
|
| 7 | Clinical validation:
|
| 8 | Regulatory review: Submit results of analytical validation and clinical study(s) to FDA to
|
Biomarkers represent a key translational bridge between an improved understanding of the mechanistic underpinnings of nicotine dependence, preclinical development, and clinical evaluation. ere is an expectation that biomarkers/biosignatures for smoking cessation will be able to help guide future therapeutic development processes as well as tailor medical practice to the individual smoker. the further development of the biomarkers presented here—diagnostic biomarkers for patient selection, pharmacodynamic biomarkers for optimal dosing, and predictive biomarkers, which can be used to select patients likely to respond therapy—represent the crucial first steps toward more efficient development of therapeutic interventions. the translation and application of smoking-cessation biomarkers should increase the number of personalized treatment options for smokers who want to quit and, as such, enhance appreciably general public health.
Acknowledgments
We thank Susan Kerns, who provided excellent editorial assistance during the preparation of the manuscript. We also thank Celia Winchell (US Food and Drug Administration (FDA)/Center for Drug Evaluation and Research) and Phil Skolnick (National Institutes of Health (NIH)/National Institute on Drug Abuse (NIDA)), who provided helpful review of the manuscript. This work was supported in part through grants funded by the NIH (Pharmacogenomics Research Network U01 DA020830, P50 CA143187, P50 DA027840, P50 DA009262, R01DA025983, R01 DA017441, R01 DA025876, R01 CA140561, R01 ES019876, and R21DA027331), the NIDA Intramural Research Program, Canadian Institutes of Health Research grants (MOP-86471 and TMH-109787), and the Centre for Addiction and Mental Health.
Footnotes
All listed authors contributed to the composition of this review. The views expressed in this presentation are those of the authors and may not necessarily reflect the position of the FDA (S.A.) or NIH (K.J.B.).
CONFLICT OF INTEREST
C.L. has received funding and/or served as a consultant to GlaxoSmithKline, Pfizer, and AstraZeneca. R.F.T. has participated in 1-day advisory meetings for Novartis and McNeil. Associate Editor R.F.T. was not involved in the review or decision process for this article. F.J.M. has received research funding from Pfizer (primary investigator: Munafo). J.E.R. and G.R.U. are listed as co-inventors on a patent application filed by Duke University based on genomic markers that distinguish successful quitters from unsuccessful quitters. S.P.D. has consulted for a 1-day Pfizer-sponsored conference on behavioral treatments for smoking cessation and is a scientific advisor to Genophen. D.V.C. has served as a consultant to Pfizer. The other authors declared no conflict of interest.
References
- 1.World Health Organization. < http://www.who.int/tobacco/enpower/tobacco_facts/en/>.
- 2.Centers for Disease Control and Prevention. Smoking-attributable mortality, years of potential life lost, and productivity losses—United States, 2000–2004. MMWR Morb Mortal Wkly Rep. 2008;57:1226–1228. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Morbidity and Mortality Weekly Report (MMWR) < http://www.cdc.gov/mmwr/preview/mmwrhtml/SH6004a12/htm>.
- 4.Centers for Disease Control and Prevention. National Health Interview Survey, United States. 2005–2010 < http://www.cdc.gov/mmwr/pdf/wk/mm6035.pdf#page=21>.
- 5.Bauld L, Bell K, McCullough L, Richardson L, Greaves L. The effectiveness of NHS smoking cessation services: a systematic review. J Public Health (Oxf) 2010;32:71–82. doi: 10.1093/pubmed/fdp074. [DOI] [PubMed] [Google Scholar]
- 6.Ray R, Schnoll RA, Lerman C. Nicotine dependence: biology, behavior, and treatment. Annu Rev Med. 2009;60:247–260. doi: 10.1146/annurev.med.60.041707.160511. [DOI] [PubMed] [Google Scholar]
- 7.Hurko O. The uses of biomarkers in drug development. Ann NY Acad Sci. 2009;1180:1–10. doi: 10.1111/j.1749-6632.2009.04946.x. [DOI] [PubMed] [Google Scholar]
- 8.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 9.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;282:1608–1614. [PubMed] [Google Scholar]
- 10.Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther. 2006;80:457–467. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sofuoglu M, Herman AI, Nadim H, Jatlow P. Rapid nicotine clearance is associated with greater reward and heart rate increases from intravenous nicotine. Neuropsychopharmacology. 2012;37:1509–1516. doi: 10.1038/npp.2011.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu DF, Hinks LJ, Morton NE, Day IN. The use of long PCR to confirm three common alleles at the CYP2A6 locus and the relationship between genotype and smoking habit. Ann Hum Genet. 2000;64:383–390. doi: 10.1046/j.1469-1809.2000.6450383.x. [DOI] [PubMed] [Google Scholar]
- 15.Lerman C, et al. Genetic variation in nicotine metabolism predicts the efficacy of extended-duration transdermal nicotine therapy. Clin Pharmacol Ther. 2010;87:553–557. doi: 10.1038/clpt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.St Helen G, et al. Reproducibility of the nicotine metabolite ratio in cigarette smokers. Cancer Epidemiol Biomarkers Prev. 2012;21:1105–1114. doi: 10.1158/1055-9965.EPI-12-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dempsey D, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Mwenifumbo JC, et al. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat. 2008;29:679–688. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 19.Lea RA, Dickson S, Benowitz NL. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J Anal Toxicol. 2006;30:386–389. doi: 10.1093/jat/30.6.386. [DOI] [PubMed] [Google Scholar]
- 20.Gold AB, Lerman C. Pharmacogenetics of smoking cessation: role of nicotine target and metabolism genes. Hum Genet. 2012 doi: 10.1007/s00439-012-1143-9. e-pub ahead of print 31 January 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray R, Tyndale RF, Lerman C. Nicotine dependence pharmacogenetics: role of genetic variation in nicotine-metabolizing enzymes. J Neurogenet. 2009;23:252–261. doi: 10.1080/01677060802572887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerman C, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin Pharmacol Ther. 2006;79:600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11:400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 24.Schnoll RA, Patterson F, Wileyto EP, Tyndale RF, Benowitz N, Lerman C. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: a validation study. Pharmacol Biochem Behav. 2009;92:6–11. doi: 10.1016/j.pbb.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson F, et al. Toward personalized therapy for smoking cessation: a randomized placebo-controlled trial of bupropion. Clin Pharmacol Ther. 2008;84:320–325. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 26.Ho MK, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin Pharmacol Ther. 2009;85:635–643. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmelli D, Swan GE, Robinette D, Fabsitz R. Genetic influence on smoking–a study of male twins. N Engl J Med. 1992;327:829–833. doi: 10.1056/NEJM199209173271201. [DOI] [PubMed] [Google Scholar]
- 28.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David SP, Johnstone EC, Churchman M, Aveyard P, Murphy MF, Munafò MR. Pharmacogenetics of smoking cessation in general practice: results from the patch II and patch in practice trials. Nicotine Tob Res. 2011;13:157–167. doi: 10.1093/ntr/ntq246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhl GR, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen LS, et al. Interplay of genetic risk factors (CHRNA5-CHRNA3-CHRNB4) and cessation treatments in smoking cessation success. Am J Psychiatry. 2012;169:735–742. doi: 10.1176/appi.ajp.2012.11101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munafò MR, Johnstone EC, Walther D, Uhl GR, Murphy MF, Aveyard P. CHRNA3 rs1051730 genotype and short-term smoking cessation. Nicotine Tob Res. 2011;13:982–988. doi: 10.1093/ntr/ntr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarginson JE, et al. Markers in the 15q24 nicotinic receptor subunit gene cluster (CHRNA5-A3-B4) predict severity of nicotine addiction and response to smoking cessation therapy. Am J Hum Genet B Neuropsych Genet. 2011;156B:275–284. doi: 10.1002/ajmg.b.31155. [DOI] [PubMed] [Google Scholar]
- 34.Conti DV, et al. Pharmacogenetics of Nicotine Addiction and Treatment Consortium. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King DP, et al. Smoking cessation pharmacogenetics: analysis of varenicline and bupropion in placebo-controlled clinical trials. Neuropsychopharmacology. 2012;37:641–650. doi: 10.1038/npp.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siedlinski M, et al. COPDGene Investigators; ECLIPSE Investigators. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66:894–902. doi: 10.1136/thoraxjnl-2011-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David SP, Munafò MR. Genetic variation in the dopamine pathway and smoking cessation. Pharmacogenomics. 2008;9:1307–1321. doi: 10.2217/14622416.9.9.1307. [DOI] [PubMed] [Google Scholar]
- 39.Rose JE, Behm FM, Drgon T, Johnson C, Uhl GR. Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score. Mol Med. 2010;16:247–253. doi: 10.2119/molmed.2009.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhl GR, Drgon T, Johnson C, Rose JE. Nicotine abstinence genotyping: assessing the impact on smoking cessation clinical trials. Pharmacogenomics J. 2009;9:111–115. doi: 10.1038/tpj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhl GR, et al. Genome-wide association for smoking cessation success: participants in the Patch in Practice trial of nicotine replacement. Pharmacogenomics. 2010;11:357–367. doi: 10.2217/pgs.09.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhl GR, et al. Smoking quit success genotype score predicts quit success and distinct patterns of developmental involvement with common addictive substances. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.155. e-pub ahead of print 6 November 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClure JB, et al. Pharmacogenetic smoking cessation intervention in a health care setting: a pilot feasibility study. Nicotine Tob Res. 2013;15:518–526. doi: 10.1093/ntr/nts173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marteau TM, et al. Effect on adherence to nicotine replacement therapy of informing smokers their dose is determined by their genotype: a randomised controlled trial. PLoS ONE. 2012;7:e35249. doi: 10.1371/journal.pone.0035249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClernon FJ, Hiott FB, Liu J, Salley AN, Behm FM, Rose JE. Selectively reduced responses to smoking cues in amygdala following extinction-based smoking cessation: results of a preliminary functional magnetic resonance imaging study. Addict Biol. 2007;12:503–512. doi: 10.1111/j.1369-1600.2007.00075.x. [DOI] [PubMed] [Google Scholar]
- 46.Janes AC, et al. Brain reactivity to smoking cues prior to smoking cessation predicts ability to maintain tobacco abstinence. Biol Psychiatry. 2010;67:722–729. doi: 10.1016/j.biopsych.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Froeliger B, Kozink RV, Rose JE, Behm FM, Salley AN, McClernon FJ. Hippocampal and striatal gray matter volume are associated with a smoking cessation treatment outcome: results of an exploratory voxel-based morphometric analysis. Psychopharmacology (Berl) 2010;210:577–583. doi: 10.1007/s00213-010-1862-3. [DOI] [PubMed] [Google Scholar]
- 48.Culbertson CS, et al. Effect of bupropion treatment on brain activation induced by cigarette-related cues in smokers. Arch Gen Psychiatry. 2011;68:505–515. doi: 10.1001/archgenpsychiatry.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franklin T, et al. Effects of varenicline on smoking cue–triggered neural and craving responses. Arch Gen Psychiatry. 2011;68:516–526. doi: 10.1001/archgenpsychiatry.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westbrook C, Creswell JD, Tabibnia G, Julson E, Kober H, Tindle HA. Mindful attention reduces neural and self-reported cue-induced craving in smokers. Soc Cogn Affect Neurosci. 2013;8:73–84. doi: 10.1093/scan/nsr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong LE, et al. A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc Natl Acad Sci USA. 2010;107:13509–13514. doi: 10.1073/pnas.1004745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cole DM, Beckmann CF, Long CJ, Matthews PM, Durcan MJ, Beaver JD. Nicotine replacement in abstinent smokers improves cognitive withdrawal symptoms with modulation of resting brain network dynamics. Neuroimage. 2010;52:590–599. doi: 10.1016/j.neuroimage.2010.04.251. [DOI] [PubMed] [Google Scholar]
- 53.Tanabe J, et al. Nicotine effects on default mode network during resting state. Psychopharmacology (Berl) 2011;216:287–295. doi: 10.1007/s00213-011-2221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parmar MK, Ungerleider RS, Simon R. Assessing whether to perform a confirmatory randomized clinical trial. J Natl Cancer Inst. 1996;88:1645–1651. doi: 10.1093/jnci/88.22.1645. [DOI] [PubMed] [Google Scholar]
- 55.Spiegelhalter DJ, et al. Bayesian Approaches to Clinical Trials and Health-Care Evaluation. Wiley; West Sussex, UK: 2004. [Google Scholar]
- 56.Roeder K, Bacanu SA, Wasserman L, Devlin B. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenland S. Bayesian perspectives for epidemiological research. II. Regression analysis. Int J Epidemiol. 2007;36:195–202. doi: 10.1093/ije/dyl289. [DOI] [PubMed] [Google Scholar]
- 59.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rebbeck TR, Spitz M, Wu X. Assessing the function of genetic variants in candidate gene association studies. Nat Rev Genet. 2004;5:589–597. doi: 10.1038/nrg1403. [DOI] [PubMed] [Google Scholar]
- 61.Thomas DC, Conti DV, Baurley J, Nijhout F, Reed M, Ulrich CM. Use of pathway information in molecular epidemiology. Hum Genomics. 2009;4:21–42. doi: 10.1186/1479-7364-4-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Conti DV, Witte JS. Hierarchical modeling of linkage disequilibrium: genetic structure and spatial relations. Am J Hum Genet. 2003;72:351–363. doi: 10.1086/346117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conti DV, Gauderman WJ. SNPs, haplotypes, and model selection in a candidate gene region: the SIMPle analysis for multilocus data. Genet Epidemiol. 2004;27:429–441. doi: 10.1002/gepi.20039. [DOI] [PubMed] [Google Scholar]
- 64.Conti DV, Cortessis V, Molitor J, Thomas DC. Bayesian modeling of complex metabolic pathways. Hum Hered. 2003;56:83–93. doi: 10.1159/000073736. [DOI] [PubMed] [Google Scholar]
- 65.Baurley JW, Conti DV, Gauderman WJ, Thomas DC. Discovery of complex pathways from observational data. Stat Med. 2010;29:1998–2011. doi: 10.1002/sim.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Le Foll B, et al. Varenicline decreases nicotine self-administration and cue-induced reinstatement of nicotine-seeking behaviour in rats when a long pretreatment time is used. Int J Neuropsychopharmacol. 2012;15:1265–1274. doi: 10.1017/S1461145711001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai X, Liangpunsakul S, Crabb DW, Ringham HN, Witzmann FA. A proteomic workflow for discovery of serum carrier protein-bound biomarker candidates of alcohol abuse using LC-MS/MS. Electrophoresis. 2009;30:2207–2214. doi: 10.1002/elps.200800775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–1704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 69.Gilad S, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ray R, et al. Human Mu Opioid Receptor (OPRM1 A118G) polymorphism is associated with brain muopioid receptor binding potential in smokers. Proc Natl Acad Sci USA. 2011;108:9268–9273. doi: 10.1073/pnas.1018699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Institute of Medicine. Neuroscience Biomarkers And Biosignatures: Converging Technologies, Emerging Partnerships, Workshop Summary. National Academies Press; Washington, DC: 2008. [PubMed] [Google Scholar]
- 73.Institute of Medicine. Perspectives on Biomarker and Surrogate Endpoint Evaluation: Discussion Forum Summary. National Academies Press; Washington, DC: 2011. [PubMed] [Google Scholar]
- 74.Patterson SD, Van Eyk JE, Banks RE. Report from the Wellcome Trust/EBA “Perspectives in Clinical Proteomics” retreat–a strategy to implement next-generation proteomic analyses to the clinic for patient benefit: pathway to translation. Proteomics Clin Appl. 2010;4:883–887. doi: 10.1002/prca.201000113. [DOI] [PubMed] [Google Scholar]