Abstract
The chromatin remodeling complex Isw2 from S. cerevisiae (yIsw2) mobilizes nucleosomes through an ATP-dependent reaction that is coupled to the translocation of the helicase domain of the enzyme along intranucleosomal DNA. In this study we demonstrate that yIsw2 is capable of translocating along single-stranded DNA in a reaction that is coupled to ATP hydrolysis. We propose that single-stranded DNA translocation by yIsw2 occurs through a series of repeating uniform steps with an overall macroscopic processivity of P = (0.90 ± 0.02), corresponding to an average translocation distance of (20 ± 2) nucleotides before dissociation. This processivity corresponds well to the processivity of nucleosome sliding by yIsw2 thus arguing that single-stranded DNA translocation or tracking may be fundamental to the double-stranded DNA translocation required for effective nucleosome mobilization. Furthermore, we find evidence that a slow initiation process, following DNA binding, may be required to make yIsw2 competent for DNA translocation. We also provide evidence that this slow initiation process may correspond to the second step of a two-step DNA binding mechanism by yIsw2 and a quantitative description of the kinetics of this DNA binding mechanism.
Keywords: chromatin remodeling, motor protein, translocase, kinetics, ATPase, ISWI
Members of the ISWI family of chromatin remodeling enzymes (termed remodelers) hydrolyze ATP to reposition nucleosomes along DNA (1–3). The ATP-hydrolyzing subunits possessed by members of this and other families of remodelers have significant homology to those of the helicase family of proteins (4–6). In fact, remodelers are classified as members of the SF2 superfamily of helicases based upon amino acid sequence comparison (7), although remodelers lack helicase activity (8). However, the ATP-dependent DNA translocation activity within the helicase derivative domains of remodelers is similar to that which is observed in helicases (9–18). This DNA translocation property plays a central role in certain models for ATP-dependent chromatin remodeling by members of both the ISWI and the SWI/SNF subfamilies of chromatin remodeling enzymes (2, 4, 14, 15, 19–21). In one such model, the ATPase subunit of the remodeling enzyme translocates directionally along the DNA at an internal site on the nucleosome. This ATP-dependent DNA translocation results in the movement of the DNA around the nucleosome by drawing in DNA from one side of the nucleosome while simultaneously pumping it out the other side. More specifically, the translocation results in the formation of a loop or bulge of DNA that then propagates around the surface of the octamer (4, 14, 19, 22, 23), thus resulting in a translation of the octamer relative to the DNA. The size of the propagated loop of DNA is believed to be related to the kinetic step size of DNA translocation by the remodeler and estimates of it vary between enzymes: 1 bp for S. cerevisiae RSC (19), 2 bp for S. cerevisiae Isw2 (2, 20), and ∼50 bp for S. cerevisiae SWI/SNF (20).
yIsw2 comprises a heterodimer of the Itc1p (145 kDa) and Isw2p (130 kDa) proteins and is a representative member of the ISWI subfamily of the SWI2/SNF2 superfamily of chromatin remodeling proteins (21, 24); both Itc1 and Isw2p are required for the chromatin remodeling activity of the holocomplex (25). In addition, two small histone-fold containing proteins, Dpb4p and Dls1p, are present in at least a fraction of Isw2 complexes purified from yeast (26). yIsw2 has been demonstrated to affect in vivo repression of transcription of several genes (3, 27–29) and, together with Ino80, to promote replication fork progression (30). It has been suggested that the yISW2 accomplishes these activities by modifying the spacing of sequential mononucleosomes along short, contiguous stretches of chromatin through nucleosome sliding (24, 31). Interestingly, yIsw2 is able to accomplish this sliding of nucleosomes without disrupting the integrity of the association of the DNA and the octamer (1, 32).
The ATPase activity of yIsw2 is stimulated in the presence of double-stranded DNA (21, 24); this is consistent with the hypothesis that yIsw2 is a double-stranded DNA translocase. As mentioned above, double-stranded DNA translocation by yIsw2 is also required for models of nucleosome mobilization by the enzyme (20, 21, 33). The ability of yIsw2 to slide nucleosomes with a specific directional bias has lead to the suggestion that DNA translocation by yIsw2 is also directionally biased (2, 20, 32, 33). Furthermore, the ability of yIsw2 to slide nucleosomes when either single- or doublestranded DNA is present in the extranucleosomal linker suggests that yIsw2 is both a single- and double-stranded DNA translocase (33). Interestingly, the genetically related D. melanogaster ISWI enzyme has been shown to be a 3’ to 5’ single-stranded DNA translocase. Taken together, this suggests that it is the single-stranded, directionally-biased DNA translocation activity which is fundamental to the double-stranded DNA translocation activity of yIsw2 and, by extension, to its nucleosome mobilization activity.
In spite of this, there has not yet been a quantitative description of the single- or double-stranded DNA translocation mechanism of yISW2. It is worth noting, however, that although kinetic studies of the of the single-stranded DNA translocation activity of the D. melanogaster ISWI enzyme have been published, an associated quantitative description of the mechanism of translocation was not presented (15). The results presented here thus represent the first quantitative characterization of the kinetic mechanism of single-stranded DNA translocation by a chromatin remodeling enzyme and, furthermore, offer insight not only into the relationship between single-stranded DNA binding and DNA translocation by yIsw2, but also into the possible mechanistic similarities between subfamilies of chromatin remodeling enzymes.
Materials and Methods
Buffers
Reaction buffer were prepared with reagent grade chemicals using twice distilled water that was subsequently deionized with a Milli-Q purification system (Millipore Corp., Bedford, MA). All ATPase and DNA binding reactions were carried out in 10 mM HEPES (pH 7.0), 5% glycerol, 20 mM potassium acetate, 5 mM MgCl2, 0.5 mM dithiothreitol, 0.1 mg/ml BSA at 37 °C.
ISW2 protein and DNA
yISW2 was expressed and purified as described (20). Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) dialyzed extensively into Milli-Q water before use. Poly(dT) was purchased from Midland Reagent company (Midland, TX) and dialyzed extensively into Milli-Q water before use.
Radioactive ATPase Experiments
ATPase activity was determined by measuring the rate of conversion of ATP to ADP using [α-32P]ATP. All reactions were conducted at 37 °C. 140 nM ISW2 and 100 µM (nucleotide) single-stranded DNA were incubated together on ice for 5 minutes prior to the initiation of the translocation reaction. DNA translocation was initiated through the addition of ATP to the solution containing the preformed complexes of yISW2 and DNA; the final concentration of ATP was 1 mM and the final reaction volume was 30 µL. 4 µL aliquots of the resulting solution were removed every 6 minutes and quenched by adding an equal volume of 0.5 M EDTA. The extent of product formation at each time point was monitoring by spotting 2 µL samples onto polyethyleneimine(PEI)-cellulose TLC plates (Merck KGaA, Darmstadt, Germany) which were developed using 0.6 M KPO4 (pH 3.4) and quantitatively analyzed using a BioRad Molecular Imager FX (BioRad, Hercules, CA). Spots corresponding to the radiolabeled ATP and ADP were quantified using the Quantity One software supplied by the manufacturer.
Stopped-Flow Experiments
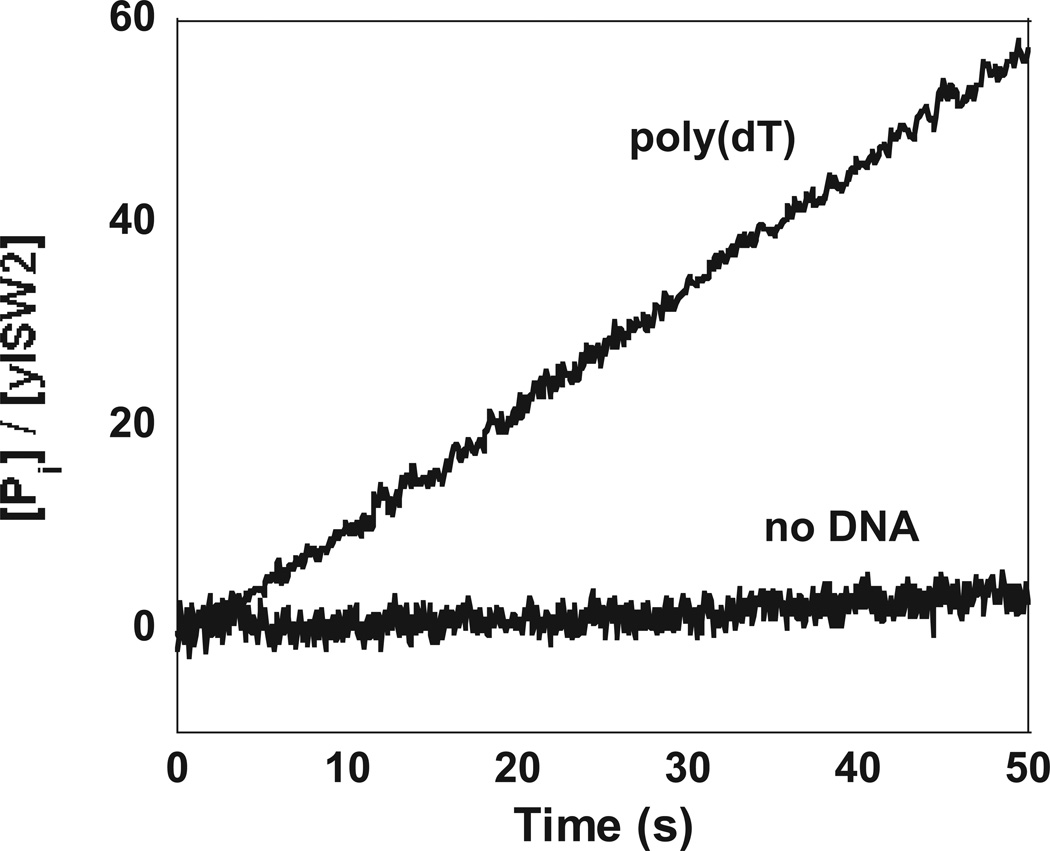
Measurements of the production of Pi in the stopped-flow spectrometer were performed using the EnzChek® phosphate assay kit (Invitrogen, Carlsbad, CA); this assay was first described by Webb (42). In the presence of Pi, the substrate 2-amino-6-mercapto-7-methylpurine riboside (MESG) is converted enzymatically by purine nucleoside phosphorylase (PNP) to ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine. The enzymatic conversion of MESG results in a spectrophotometric shift in maximum absorbance from 330 nm for the substrate to 360 nm for the product which is monitored in the stopped-flow spectrometer. The kcat for the reaction is 40 s−1, which is sufficiently fast to monitor the kinetics of ATPase by the translocating yISW2 complex. For the experiments shown in Figure 4 the concentration of yISW2 was 25 nM and the concentration of the poly(dT) was 100 µM (nucleotide).
Figure 4.
Time courses of Pi production during single-stranded DNA translocation along poly(dT) monitored using the EnzChek® phosphate assay as described in Materials and Methods. A linear analysis of the time course resulted in an estimate of Vmax = (63 ± 2) [Pi]/[yISW2]/min, consistent with the estimate of (61 ± 4) [Pi]/[yISW2]/min determined from our radioactivity-based assay, and an intercept on the ordinate of (−1 ± 0.5) [Pi]/[yISW2]. This negative intercept on the ordinate is consistent with the kinetic model depicted in Scheme 4. A control experiment in which no DNA was included with the reaction is also shown. A linear analysis of this time course resulted in an estimate of Vmax = (2 ± 1) [Pi]/[yISW2]/min, consistent with the results from our radioactivity-based assay, and an intercept on the ordinate of (0.0 ± 0.1).
Measurements of the kinetics of yISW2 binding to fluorescein or Cy3 labeled oligo(dT) were performed in the stopped-flow spectrometer by monitoring the change in the fluorescence of these fluorophores. Fluorescein was excited at 492 nm and its fluorescence emission was monitored at wavelengths >520 nM using a long-pass filter. Cy3 was excited at 515 nm and its fluorescence emission monitored at wavelengths >570 nm.
Measurements of the kinetics of yISW2 binding to dT70 we performed in the stopped-flow spectrometer by monitoring the change in the intrinsic tryptophan fluorescence of yISW2 that occurs upon DNA binding. In these experiments the tryptophan fluorescence was excited at 280 nm and emission monitored at wavelengths greater than 305 nm using a WG 305 Schott glass filter. BSA was omitted from these reactions since its tryptophan fluorescence created too large a background signal. The final reaction concentration of yISW2 was 10 nM.
Analysis of Kinetic Data
The ATPase rate for each length of single-stranded DNA was determined from LLS analysis of the corresponding ATPase time courses. Four independent measurements of Vmax were made for each DNA length.
All LLS and NLLS analyses using Equations (2), (5) and (6) were performed using Conlin, kindly provided by Dr. Jeremy Williams. Fitting models and Monte Carlo simulation programs were written in the C++ computer language and compiled with the Microsoft C++ 6.0 compiler. The software library CNL50 (Visual Numerics Incorporated, Houston, TX) was used for the numerical calculation of the inverse Laplace transform in Equation (5). The uncertainties in all fitted parameters reported in this manuscript represent 68% confidence limits (1 standard deviation) and were determined by performing a 1000 cycle Monte Carlo simulation using the routine built into Conlin.
Results
yISW2 is an ATP-dependent single-stranded DNA translocase
We studied the kinetics of yIsw2 translocation along single-stranded DNA by analyzing the stimulation of yIsw2’s ATPase activity in the presence of single-stranded DNA of varying lengths. ATPase experiments were performed as described in Materials and Methods under solution conditions identical to those previously used to study the DNA- and nucleosome-stimulate ATPase activity of yIsw2 (21). In order to minimize complications arising from secondary structure in the DNA we performed our experiments using oligodeoxythmidylates. To ensure that our experiments were performed at DNA concentrations sufficient to elicit maximum steady-state ATPase stimulation, we first determined the KM for the single-stranded DNA stimulated ATPase activity of yIsw2. We estimate KM = (5 ± 2) µM (mononucleotide concentration), independent of the length of the DNA (data not shown).
We subsequently measured the stimulation of yIsw2’s ATPase activity in the presence of DNA of varying lengths, but at constant mononucleotide concentration (14, 18) of 100 µM (20 fold larger than KM). As shown in Figure 1 and Figure 2, we observe that the steady state ATPase rate of yIsw2 increases with increasing DNA length. These results are consistent with the previously published hypothesis that yIsw2 is a singlestranded DNA translocase (33). Furthermore, since these experiments were conducted under conditions of excess DNA concentration the binding of single complexes of yIsw2 to the DNA is favored. This is further supported by equilibrium studies of DNA binding by yISW2 experiments which demonstrated that the occluded site size of yISW2 binding to DNA is larger than 100 bp (Fitzgerald et al., unpublished). Therefore, taken together the results suggest that monomeric yIsw2 is functional as a single-stranded DNA translocase.
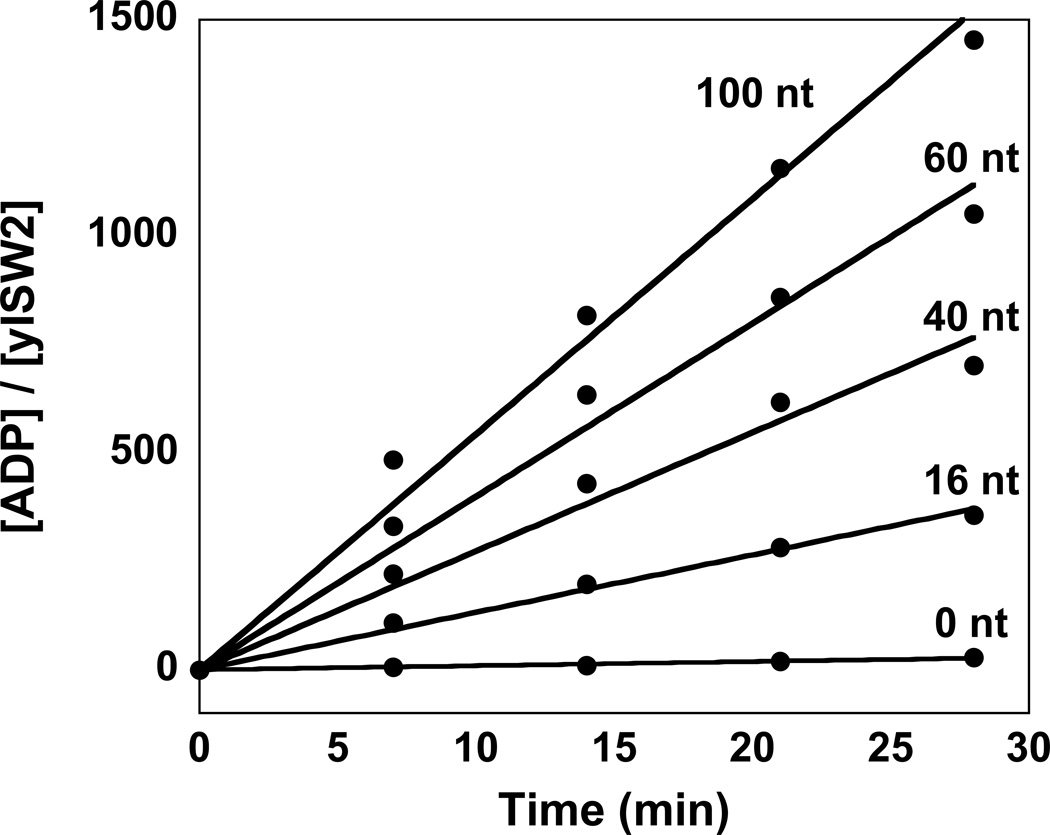
Figure 1.
Time courses of ATP hydrolysis for yIsw2 in the presence of a saturating concentration of single-stranded DNA of varying length. In these experiments yISW2 and single-stranded DNA were incubated together before the addition of ATP. The lengths of the DNA are 16 nt, 40 nt, 60 nt, and 100 nt. A control experiment in which no DNA is included in the reaction is also shown.
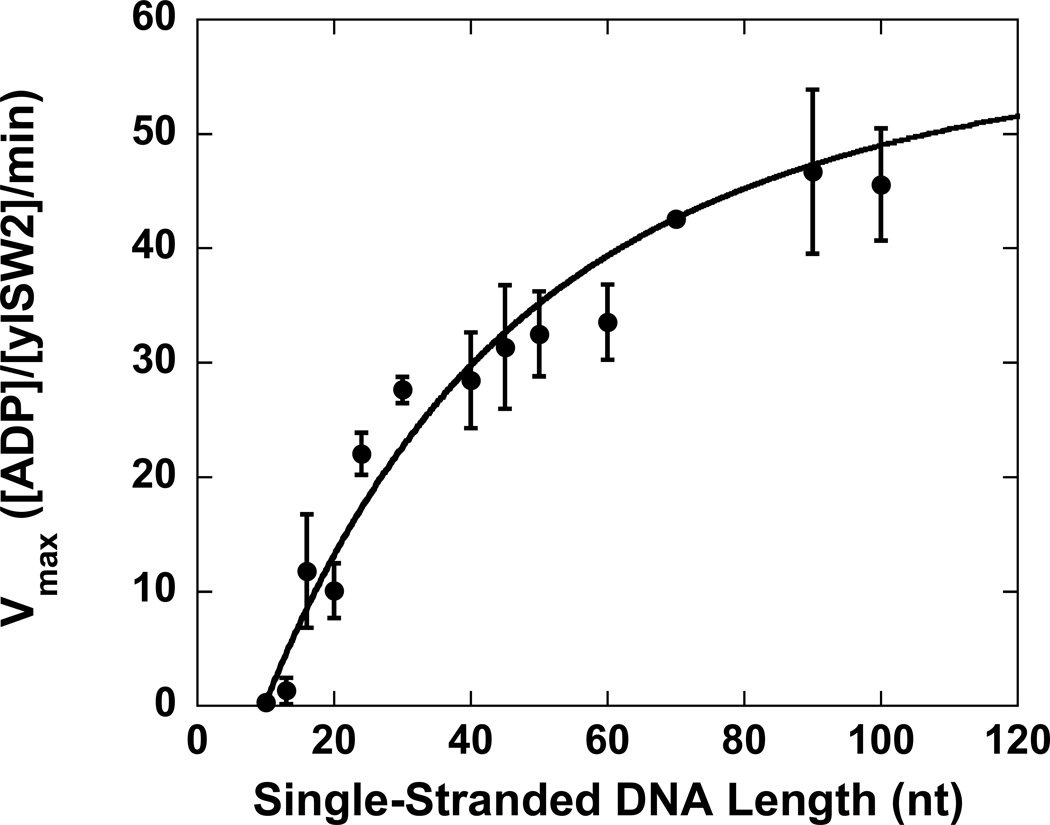
Figure 2.
The dependence of Vmax upon single-stranded DNA length. Each data point is the average of 4 separate measurements of Vmax and the error bars represent the standard deviation of those measurements. The solid line is a NLLS fit of the data to Equation (2) (Scheme 1). The NLLS fits using Equations (10) and (12) are identical. The parameter estimates associated with these fits are in Tables 1 and 2.
Kinetic Models for Single-stranded DNA Translocation
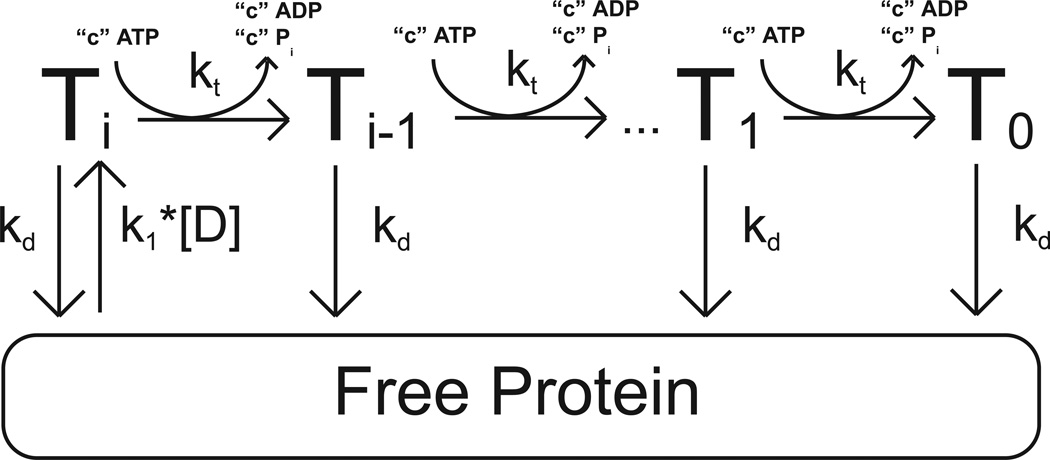
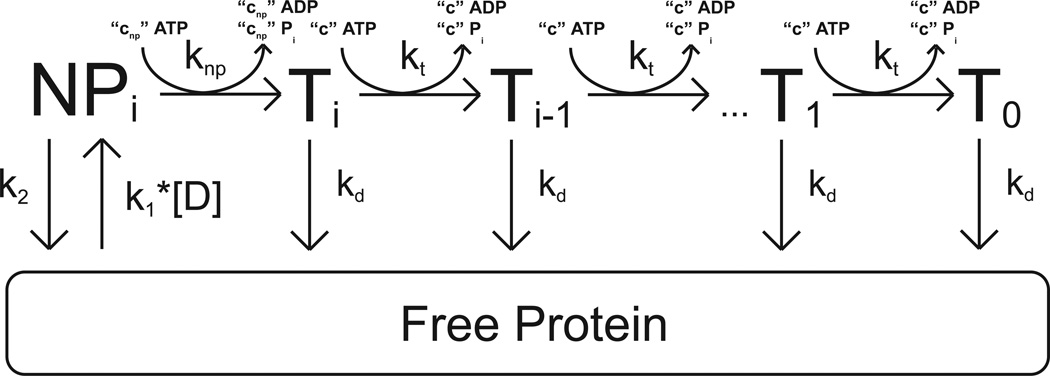
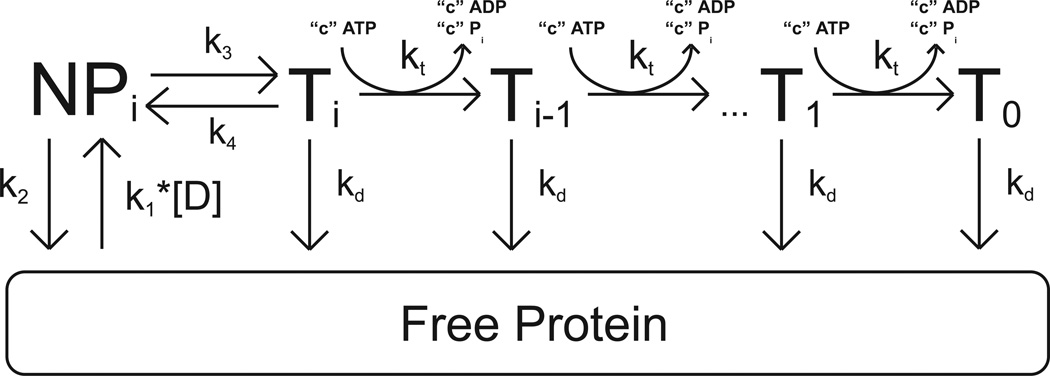
The dependence of the Vmax of the single-stranded DNA stimulated ATPase activity of yISW2 on DNA length shown in Figure 2 is consistent with yISW2 acting as a single-stranded DNA translocase. The simplest model for single-stranded DNA translocation by yIsw2 which is consistent with the data in Figures 1 and 2 is shown in Scheme 1. In this model, we assume that the yIsw2 complex binds randomly to one of its possible binding sites on the single-stranded DNA. Upon binding, the yISW2 complex is i translocation steps away from the end of the DNA (the Ti state in Scheme 1). Upon subsequent repeating cycles of ATP binding, ATP hydrolysis and release of ADP and inorganic phosphate, the yIsw2 complex translocates along the single-stranded DNA with directional bias (33) and finite processivity, . This translocation occurs in discrete steps, of m nucleotides per step, with an associated translocation rate constant kt and c molecules of ATP hydrolyzed per step; the constant c is thus the thermodynamic coupling efficiency for the process associated with the kt rate constant and m is the kinetic step size (34, 35). The rate constant for dissociation during translocation is kd. The maximum number of translocation steps, n, for a given length of DNA, L, is related to the kinetic step size of translocation, m, through the approximate relation , where d is the interaction site size of the protein (18, 34). Upon reaching the end of the DNA (the T0 state in Scheme 1), the yIsw2 complex can no longer translocate and so dissociates with rate constant kd. We further assume that since our experiments are performed under conditions of excess DNA concentration there will be only one yIsw2 complex bound per DNA and thus we have ignored any potential protein-protein interactions.
Scheme 1.
All dissociated protein (and protein that was initially free in solution at the start of the reaction) will bind the DNA randomly at any of available binding sites. The second order rate constant for yIsw2 binding single-stranded DNA (and forming the complex Ti) is k1. It is worth noting that we assume the binding affinity is the same for all binding sites and thus we ignore electrostatic effects associated with the ends of the DNA. Furthermore, in our experiments the concentration of DNA is much larger than the concentration of protein so we can treat the binding of yIsw2 to DNA as a pseudo-first order process with associated rate constant k1*[D], where [D] is the nt concentration of DNA in solution.
It is also important to note that we have neglected any contributions from the potential futile hydrolysis of ATP by yIsw2 bound at the ends of the DNA. Such futile hydrolysis has been observed for helicase translocation along single-stranded DNA (35), but we cannot determine if yIsw2 is exhibiting similar behavior based upon our data. It is worth noting, however, that the contribution of this futile hydrolysis (if occurring) would decrease with increasing DNA length (18) making its contribution to the dependence of the total observed steady-state ATPase rate on DNA length negligible.
The differential equations describing the concentration of protein bound at each position (Ti and free protein) in Scheme 1 can be solved to obtain time-dependent equations for these protein concentrations that are functions of the maximum number of translocation steps, n, associated with a given DNA length; this is equivalent to finding expressions for these populations as a function of the length of the DNA and the kinetic step size of translocation (11, 18, 34). In this derivation we assume an initial equilibrium between the bound and free complexes of yISW2, as determined by k1, [D], and kd, exists before the addition of ATP in order to match our experimental conditions (see Materials and Methods). These expressions can then be combined to form a time-dependent expression (Equation (1)) for the concentration of ATP hydrolyzed (or the concentration of ADP or Pi produced) during the translocation of yIsw2 along single-stranded DNA.
| (1) |
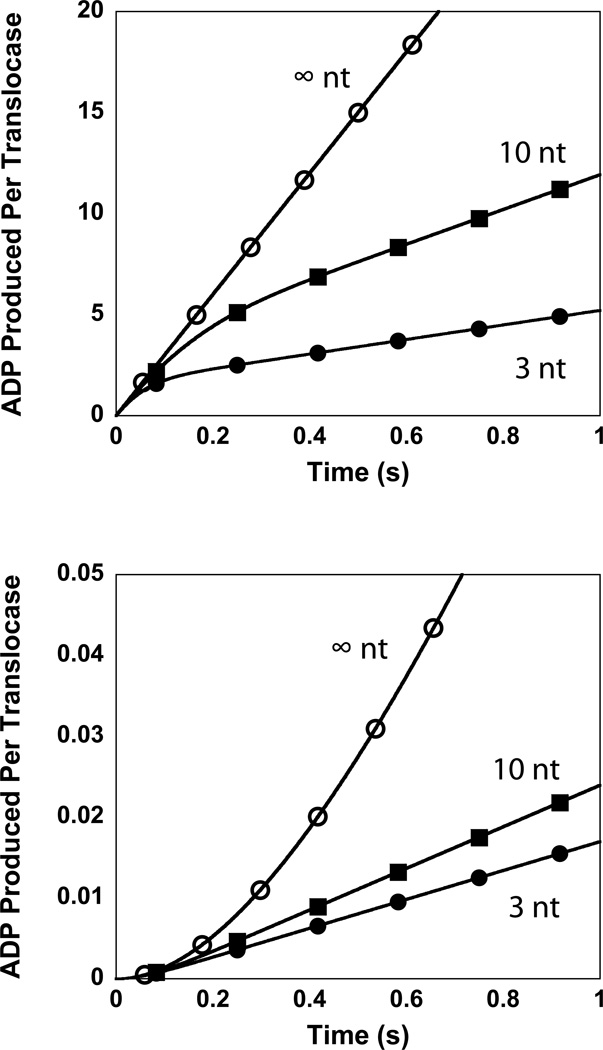
Simulated time courses based upon this model are shown in Figure 3. As seen in Figure 3, these time courses consist of an initial rapid exponential increase in the ADP produced (ATP hydrolyzed) by the translocating proteins, associated with their pre-steady state translocation on the single-stranded DNA, followed by a linear phase corresponding to their steady-state translocation activity. The rate of the steady-state ATP hydrolysis by these translocating enzymes obeys Michaelis-Menten kinetics with Vmax and KM shown in Equations (2) and (3), respectively.
Figure 3.
Simulated ATPase time courses for protein translocating along single-stranded DNA of varying lengths (3 nt, 10 nt and ∞ nt) under the conditions of a saturating concentration of DNA. In these simulations kt = 30 steps/s, kd = 2 s−1, m = 1 nt/step, and c = 1 ATP/step. (A) Simulations according to Scheme 1. (B) Simulations according to Scheme 2 with knp = 0.01 s−1.
| (2) |
| (3) |
The variable P in Equation (2) is the previously defined processivity of translocation. It is interesting to note that this model (and corresponding Equation (3)) predicts that KM is independent of the maximum number of steps (n), or equivalently independent of the length of the DNA. This is consistent with measurements of KM of yIsw2 translocating along single-stranded DNA.
In order to obtain a better estimate of the kinetic parameters from our NLLS analysis of the data in Figure 2 we also measured the ATPase time course for the translocation of yIsw2 along poly(dT). Poly(dT) can effectively mimic infinitely long DNA and thus the value of Vmax determined for the translocation of yIsw2 along poly(dT) can serve as a constraint in the analysis of the data in Figure 2 using Equation (2) (18). Specifically, in the limit of infinitely long DNA, Equation (2) simplifies to Equation (4).
| (4) |
We determined Vmax,∞ = (61 ± 4) [ADP]/[yISW2]/min from the analysis of the ATPase time courses obtained in the presence of poly(dT). We then determined estimates of P and d through NLLS analysis of the dependence of Vmax on DNA length (Figure 2) using Equation (2) with our estimate of Vmax,∞ as a fixed constraint. In this analysis we further assumed that m = 2 nt since an estimate of 2 nt for the kinetic step size of double-stranded DNA translocation by yIsw2 was recently reported (20). The parameter estimates obtained from this analysis are presented in Table 1 and the solid line overlaying the data in Figure 2 is a simulation using Equation (2) and these parameter estimates.
Table 1.
Estimates of kinetic parameters for yIsw2 translocation along single-stranded DNA according to Scheme 1 (Equation (2)).
| P | (0.90 ± 0.02) | |
| ckt | (61 ± 4) [ADP]/[yISW2]/min (fixed) |
|
| d | (9.85 ± 0.09) nt | |
| (20 ± 2) nt | ||
| Variance of fit | 0.65 |
As described above and shown in Figure 3, the ATPase time courses associated with Scheme 1 contain an initial exponential phase of activity associated with the pre-steady state translocation of the proteins along the DNA. It is worth noting that within the resolution of our measurements we could not detect this phase; rather, we determine a zero intercept, within error, on the ordinate of the time courses shown in Figure 1. A simple explanation for this discrepancy would be that the rate of dissociation from the end of the DNA (from the T0) state is much faster than kt and not equal to kd. Such a kinetic mechanism would be consistent with the lack of any observed pre-steady state translocation (and corresponding ATPase) activity of the yISW2 complex, however this mechanism would also suggest that there would be no dependence of Vmax upon DNA length. Since we do see a dependence of Vmax on DNA length we can reject this hypothesis.
In order to improve the time resolution of our measurements of the ATPase activity of translocating yISW2 and thus also specifically improve the resolution of the pre-steady state kinetics of translocation, we next performed single-stranded DNA translocation experiments using a stopped-flow technique which monitors the production of Pi produced by yISW2 during translocation. Pi in solution was measured spectrophotometrically using the commercially available EnzChek® phosphate assay (see Materials and Methods).
Since the rate of the single-stranded DNA stimulated steady-state ATPase activity of yISW2 is small we measured the kinetics of Pi release in the presence of a saturating concentration of poly(dT) to maximize the observed fluorescence signal; representative time courses from these experiments are shown in Figure 4. Linear analysis of the data in Figure determined a hydrolysis rate of (63 ± 2) [Pi]/[yISW2]/min, consistent with the estimate of (61 ± 4) [ADP]/[yISW2]/min determined from our radioactivity-based assay, and an intercept on the ordinate of (−2 ± 1) [Pi]/[yISW2]. As shown in Figure 3, the time course of Pi release should be linear with a zero intercept on the ordinate if Scheme 1 is correct.
Alternative Model for DNA Translocation
The deviation from a zero intercept for the time course of Pi release by yISW2 translocating along poly(dT) is inconsistent with the kinetic model outlined in Scheme 1. Although the deviation is small, we nevertheless consider it important to consider alternative models for single-stranded DNA translocation by yISW2. For example, it is worth noting that a previous study of the double-stranded DNA translocation by RSC (18) also detected no initial exponential phase of ATPase activity that could be associated with pre-steady state kinetics. On the basis of this observation a kinetic model for double-stranded DNA translocation was proposed that included a slow process, following DNA binding by the RSC, which was required to make RSC competent for double-stranded DNA translocation. In this model, shown in Scheme 2, the protein is i translocation steps away from the end of the DNA upon binding the DNA, but is unable to initiate translocation (it is in the NPi state in Scheme 2). The rate constant for dissociation from the NPi state is k2 and may be different from kd. Upon the addition of ATP, the protein undergoes a slow process, described by the rate constant knp, before becoming competent for DNA translocation (the Ti state in Scheme 2). The thermodynamic coupling efficiency for the knp rate constant is given by cnp.
Scheme 2.
The differential equations describing the concentration of protein bound at each position (NPi, Ti, and free protein) in Scheme 2 can be solved to obtain time-dependent equations for these protein concentrations that are functions of the maximum number of translocation steps (n) associated with a given DNA length. As before, these expressions can then be combined to form a time-dependent expression (Equation (5)) for the concentration of ATP hydrolyzed (or ADP or Pi produced) during yISW2 translocation along single-stranded DNA.
| (5) |
Simulated time courses based upon this model are shown in Figure 3. As seen in Figure 3, these time courses consist of an initial slow exponential increase in the ADP produced (ATP hydrolyzed) by the translocating proteins, associated with their pre-steady state translocation along the single-stranded DNA, followed by a linear phase corresponding to their steady-state translocation activity. It is worth noting that the presence of this slow initial phase is consistent with the linear analysis of time course for Pi release shown in Figure 4. Specifically, the presence of this slow phase would result in an estimate of a negative intercept on the ordinate from a linear analysis of the time course.
The rate of steady-state ATP hydrolysis by these translocating enzymes obeys Michaelis-Menten kinetics with Vmax and KM shown in Equations (6) and (7), respectively.
| (6) |
| (7) |
As expected, Equations (6) and (7) are equal to Equations (2) and (3), respectively, in the limit of large knp. If ATP hydrolysis is associated with the knp step then an additional term (Equation (8)) must be included in our expression for ADP produced by the translocating protein.
| (8) |
This expression is dominated by a linear term which obeys Michaelis-Menten kinetics with a value of KM identical to that shown in Equation (7) and a value of Vmax given in Equation (9)
| (9) |
Therefore, we can write the expression for the total ADP produced during translocation in Equation (10)
| (10) |
The parameter estimates obtained from analysis of the data in Figure 2 using Equation (10) are presented in Table 2. In this analysis it is not possible to simultaneous determine estimates of cnp and d since these variables are strongly correlated in Equation (10) (18). Thus we first analyzed the data in Figure 2 assuming that cnp = 0 and then reanalyzed the data using d = 10 nt since this corresponded to the smallest length of single-stranded DNA that stimulated the ATPase activity of yISW2. It is important to note that since equations (2) and (10) predict an identical dependence of Vmax on n analysis of the dependence of Vmax on DNA length in Figure 2 using either model yields an identical estimate of P = (0.90 ± 0.02) and describe the data equally well.
Table 2.
Estimates of kinetic parameters for yIsw2 translocation along single-stranded DNA according to Scheme 2 (Equation (10)).
| P | (0.90 ± 0.02) | (0.90 ± 0.02) | |
| (61 ± 4) [ADP]/[yISW2]/min (fixed) | |||
|
|
0 [ADP]/[yISW2]/min (fixed) | (0.2 ± 0.2) [ADP]/[yISW2]/min | |
| d | (9.85 ± 0.09) nt | 10 nt (fixed) | |
|
|
(20 ± 2) nt | (20 ± 2) nt | |
| Variance of fit | 0.65 | 0.65 | |
Kinetics of single-stranded DNA binding by yIsw2
It is worth noting that the estimate of cnp obtained in the analysis of the data in Figure 2 using Equation (10) is, within error, equal to zero (Table 2). This suggests that the knp step in Scheme 2 may not be associated with significant hydrolysis of ATP. Since this step follows single-stranded DNA binding in Scheme 2 we decided to investigate whether it might correspond to the second step of a two step single-stranded DNA binding mechanism by yIsw2.
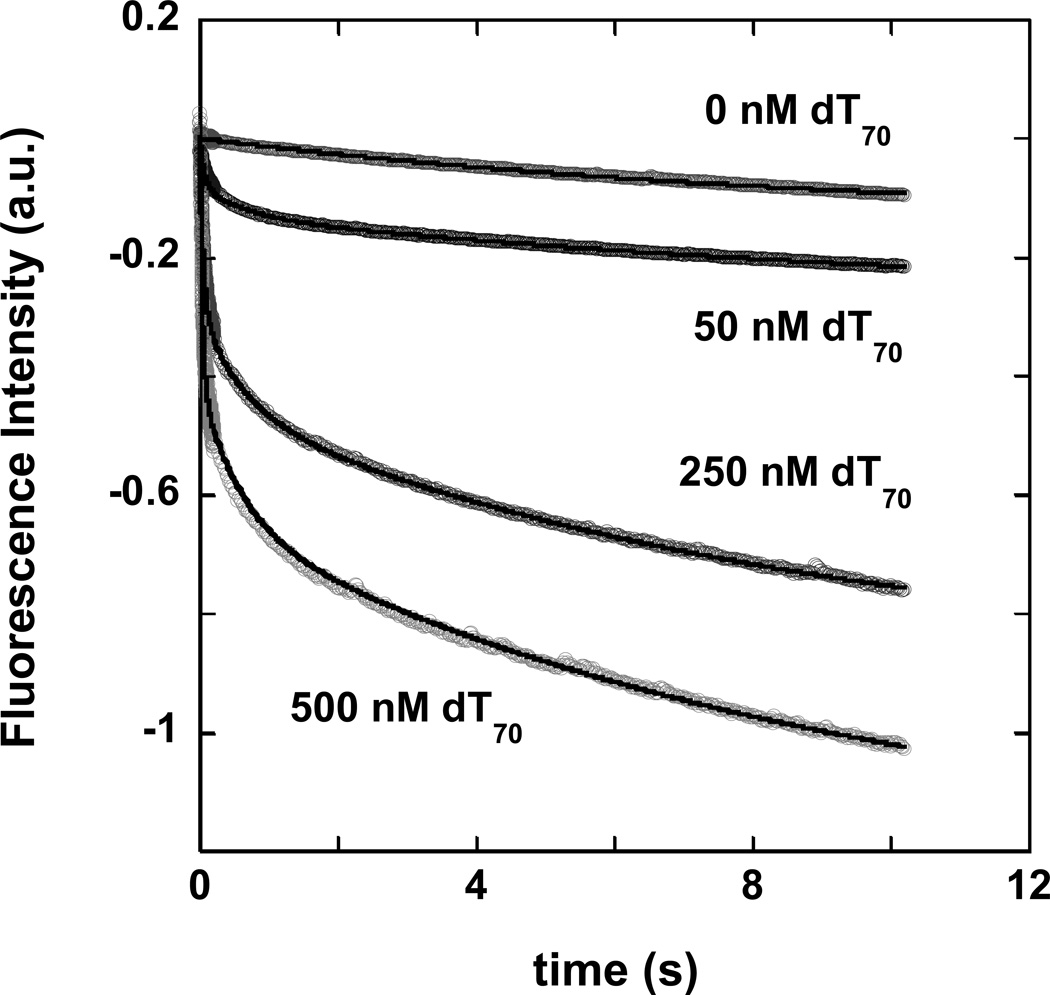
In our single-stranded DNA binding experiments we monitored the binding of yIsw2 to excess concentrations of dT70 (Figure 5). In the absence of DNA the observed fluorescence time course decayed monoexponentially. The rate of this signal change was independent of the fluorescence excitation wavelength or the bandwidth of the excitation monochromator, suggesting that it was not associated with photobleaching of the tryptophan residues, but rather may result from the non-specific binding of yISW2 to the walls of the spectrometer flow-cell. This slow signal change was also observed in the presence of the DNA and the magnitude of the rate constant associated with this signalchange was independent of the concentration of the DNA (data not shown); this suggests that this process is not associated with the binding of yISW2 to the single-stranded DNA. In the presence of DNA, however, two additional exponential phases are observed. The first observed rate constant had a linear dependence on DNA concentration, whereas the second observed rate constant had a hyperbolic dependence on DNA concentration (data not shown).
Figure 5.
Kinetics of yISW2 binding to an excess concentration of dT70, monitored by the quenching of the intrinsic tryptophan fluorescence of yIsw2. The time courses were observed in the stopped-flow upon mixing 10 nM yIsw2 (final concentration) with 0, 50, 250, and 500 nM dT70. The solid lines overlaying each time course are NLLS squares fits obtained from global NLLS analysis of the time courses using Equation (11).
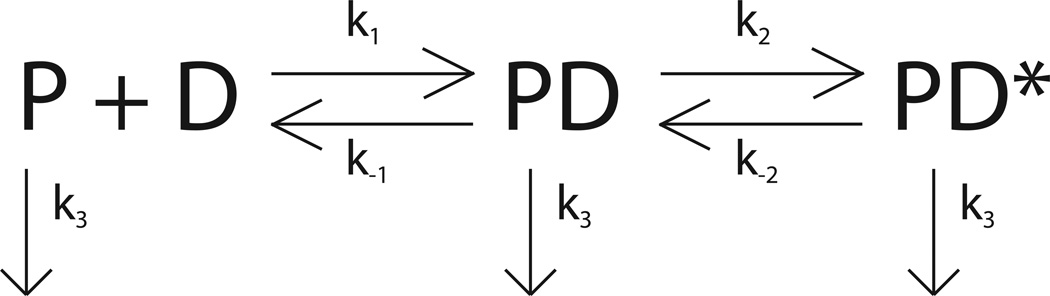
The minimal mechanism associated with these time courses and the associated dependence of these three observed rate constants on DNA concentration is shown in Scheme 3. The equation that describes the time dependence of the intrinsic tryptophan fluorescence associated with the binding mechanism shown in Scheme 3 is given in Equation (11).
Scheme 3.
| (11) |
In this equation, the parameter A is the fluorescence signal change associated with the first step of the binding, the parameter B is the fluorescence signal change associated with the second step of the binding, the parameter C is the fluorescence signal change associated with the non-specific binding of the protein to the walls of the flow cell, and the rate constants k1, k−1, k2, k−2 and k3 are as shown in Scheme 3. Since the slowest signal change occurs at a rate that is independent of the DNA concentration we have assumed that an identical process affects all three protein species in Scheme 3. The parameter estimates obtained from the NLLS analysis of the time courses in Figure 5 using Equation (11) are shown in Table 3.
Table 3.
Estimates of the kinetic parameters for single-stranded DNA binding by yIsw2 according to Scheme 3 (Equation (11).
| k1 (M−1 s−1) | (2.8 ± 0.5) × 106 |
| k−1 (s−1) | 1.40 ± 0.02 |
| k2 (s−1) | 0.013 ± 0.002 |
| k−2 (s−1) | 0.157 ± 0.001 |
| k3 (s−1) | 0.009 ± 0.002 s−1 |
| k−1/k1 (M) | (4.9 ± 0.2 ) × 10−7 |
| k−2/k2 | 0.08 ± 0.01 |
| BA | 6.0 ± 0.8 |
Model of DNA Translocation Combined With DNA Binding
It is interesting to note that the equilibrium constant for the second step in the binding mechanism is highly unfavorable (k2/k−2 = 0.08 ± 0.01). This is consistent with the requirement that the slow conformation change in Scheme 2 is actually the second, and unfavorable, step of the two step binding mechanism for yIsw2 to single-stranded DNA (see also (18)). Naturally the rate constants for single-stranded DNA binding (k1, k− 1, k2 and k−2) will be affected by the presence of nucleotide, but the presence of this second step in the single-stranded DNA binding mechanism by yISW2 should nevertheless be incorporated into our kinetic model for single-stranded DNA translocation by yISW2.
A revised kinetic model for single-stranded DNA translocation by yIsw2 is shown in Scheme 4. In this scheme, the initial conformational change required to make yIsw2 competent for DNA translocation (the knp step in Scheme 2) has been replaced by a second reversible process associated with single-stranded DNA binding by yIsw2. As before for Scheme 2, the differential equations describing the concentration of protein bound at each position (NPi, Ti, and free protein) in Scheme 4 can be solved and subsequently combined to form a time-dependent expression for the concentration of ATP hydrolyzed (or ADP or Pi produced) during the translocation of yIsw2 along singlestranded DNA. This equation is too cumbersome to reproduce here, however it does predict a linear term associated with the steady-state single-stranded DNA translocation activity of the enzyme. The magnitude of the rate constant associated with this steady-state ATPase activity follows Michaelis-Menten kinetics with Vmax and KM as defined in Equations (12) and (13), respectively.
Scheme 4.
| (12) |
| (13) |
Not surprisingly Equation (2), (6) and (12) display an identical dependence of P on n. Therefore, analysis of data in Figure 2 using Equation (12) also yields an estimate of P = (0.90 ± 0.02) and is equally as good at describing the data as the results of the analysis of the data using Equations (2) or (6).
Discussion
The chromatin remodeling activity of yIsw2 is manifest in the ability of the enzyme to directionally slide nucleosomes along DNA without disrupting the integrity of the nucleosome itself (1, 2, 31, 32). The current model for nucleosome mobilization by yISW2 involves the directionally biased translocation of the DNA on the surface of the nucleosome by yISW2 in an ATP-dependent reaction (15, 20, 31, 33). In this paper we provide the first quantitative characterization of mechanisms of both single-stranded DNA translocation and the single-stranded DNA binding by yISW2 and show how these two activities may be interrelated. Our direct observation of the single-stranded DNA translocation activity of yISW2 confirms previous indirect measurements (33) and offers a mechanism by which both double-stranded DNA translocation and nucleosome sliding can be directionally biased.
yISW2 translocase activity for single-stranded DNA
The dependence on single-stranded DNA length of the single-stranded DNA stimulated ATPase activity of yIsw2 is consistent with yIsw2 being a single-stranded DNA translocase (Figure 2). Furthermore, this single-stranded DNA translocation activity may form the basis for double-stranded DNA translocation and nucleosome sliding by yIsw2 (2, 20, 33) and, perhaps, other remodelers as well (14, 18, 19).
Our estimate of the processivity of single-stranded DNA translocation by yIsw2,P = (0.90 ± 0.02), is consistent with enzyme moving approximately nt on average before dissociation. This processivity is quite low compared to what is observed for helicase translocation along single-stranded DNA (11, 13, 35, 36), but similar to the processivity of double-stranded DNA translocation by RSC chromatin remodeling complex from S. cerevisiae (18) and similar to the processivities of single- and double-stranded DNA translocation by D. melanogaster ISWI (15).
It is worth noting that D. melanogaster ISWI is proposed to translocate along single-stranded DNA with a 3’ to 5’ directional bias, whereas either a 5’ to 3’ directional bias, based upon modifications to the extranucelosomal linker DNA (33), or no directional bias, based upon modifications to the internucleosomal DNA (20), has been proposed for yIsw2. As mentioned previously, the ability of yISW2 to slide nucleosomes with directional bias implies that the enzyme also possess directionally-biased DNA translocation activity. Thus, it’s possible that the interaction of one domain of yIsw2 with the extranucleosomal DNA dictate the directional bias to nucleosome sliding and the translocating motor domain moves either 3’ to 5’ or 5’ to 3’, accordingly, along a singlestrand of the internucleosomal DNA in order to accomplish the directionally-biased sliding. In light of this, an important future experiment would be to directly test the directional bias of both single- and double-stranded DNA translocation by yIsw2 using a spectrophotometric assay (11, 34). Since either directional bias (3’ to 5’ or 5’ to 3’) of the single-stranded DNA translocation by yIsw2 would consistent with the assumptions of our model, the resolution of this issue is not necessary to support the conclusions we draw here.
Translocation initiation is likely coupled to DNA binding
The linear analysis of the ATPase time course shown in Figure 4 suggests that a slow initiation step might be present, following single-stranded DNA binding by yISW2, to make the enzyme competent for single-stranded DNA translocation. We propose that the slow initiation process may actually be the second step of a two-step mechanism for single-stranded DNA binding by yIsw2. Although yIsw2 is able to bind single-stranded DNA with reasonable affinity (Table 3), only a small fraction of the total population of bound protein has completed the second process in the binding reaction and thus is competent for DNA translocation. This hypothesis is consistent with previously published results which also demonstrated that although yIsw2 could bind DNA efficiently (25), it’s ATPase activity was not strongly stimulated by DNA binding (21).
Similarities to other motors
The kinetic mechanism for single-stranded DNA translocation proposed here for yIsw2 is similar to the mechanism recently proposed for the mechanism of doublestranded DNA translocation by the S. cerevisiae RSC chromatin remodeling complex (18). In both cases a slow initiation step is required after DNA binding to make the enzyme competent for DNA translocation. In addition, slow initiation steps have also been proposed for mechanisms of single-stranded DNA translocation by the B. stearothermophilus PcrA helicase (37, 38) and the T7 DNA helicase (36). The initiation steps for the PcrA and T7 DNA helicases were proposed to be ATP-dependent conformational changes (36–38), whereas no conclusion regarding the ATP dependence of the initiation step for RSC was proposed (18). However, it is worth noting that quantitative descriptions of the kinetic mechanisms for single-stranded DNA binding by these helicases have yet to be determined. Thus, it is not possible to further resolve the dependence of single-stranded DNA translocation by these enzymes on their single-stranded DNA binding characteristics.
In contrast, the kinetic mechanism of single-stranded DNA binding by monomers of the E. coli Rep helicase has been determined (39). E. coli Rep is a member of the SF1 superfamily of helicases and is a known single-stranded DNA translocase (9). Interestingly, the second step in the two-step mechanism for single-stranded DNA binding by Rep is associated with a favorable equilibrium constant (39) and the kinetic mechanism for single-stranded DNA translocation by Rep does not include or require the presence of a slow initiation process to make the monomer competent for single-stranded DNA translocation (9). This correlation between the differences in both the mechanisms of single-stranded DNA binding and translocation for E coli Rep and yIsw2 may further support our hypothesis that these two processes are linked; i.e., that the slow initiation process required to make yIsw2 competent for single-stranded DNA translocation is actually the second step of a two-step mechanism for single-stranded DNA binding by yIsw2. Similarly, the lower affinity of single-stranded DNA binding by yISW2 when compared to that of Rep likely correlates with the lower processivity of single-stranded DNA translocation by yISW2 when compared to Rep.
Implications for nucleosome mobilization
yIsw2 is able to slide nucleosomes without disrupting the integrity of the nucleosome (1, 2, 32). This suggests that the loops (or bulges) of DNA formed on the surface of the nucleosome as a result of the translocation of yIsw2 are likely small in size (2). The low processivity of single-stranded DNA translocation by yIsw2 reported here is consistent with the ability of yIsw2 to form small loops of DNA during its translocation (18). Although our studies here can provide no direct measurement of the average loop size or kinetic step size of translocation, we can safely assume that both are well below the processivity limit of the enzyme (∼ 20 nt); as mentioned above, a kinetic step size of 2 nt has been proposed for single-stranded DNA translocation by yIsw2 based upon the enzyme’s nucleosome sliding ability (20). As with the question of directional bias to single-stranded DNA translocation by yIsw2, the determination of the kinetic step size could likely be determined in future work using a spectrophotometric assay for single-stranded DNA translocation (11, 34).
Perhaps more interesting is the similarity between our estimate of the processivity of single-stranded DNA translocation and reported step-sizes of nucleosome sliding by yIsw2 (20). The reported step size of 9–11 bp for nucleosome sliding (20) is smaller than our estimate of yISW2 translocating (20 ± 2) nt on average before dissociation. One could therefore propose that the processivity of single-stranded DNA translocation may be ultimately limiting for the double-stranded DNA translocation and associated nucleosome sliding activity of the enzyme. Of course the processivity of translocation for double-stranded DNA translocation may well be different from the processivity of single-stranded DNA translocation and could also be affected by the presence of and binding to the nucleosome. Nevertheless, this model is also consistent with recent in vivo studies of the chromatin remodeling activity of yIsw2 suggest that the enzyme is bound to the chromatin only transiently during the specific period of nucleosome mobilization (1, 40 ). Furthermore, these studies also suggest that chromatin remodeling by yIsw2 involves the directional sliding of a few nucleosomes and, contrary to what is observed in vitro (32, 41), does not result in regularly spaced arrays of nucleosomes on the chromatin. Although in vitro studies of mononucleosome sliding by yIsw2 confirm that sliding is directionally biased from the ends of the DNA toward its center (31, 40), the differences between the results obtained in vitro and in vivo suggest that other cellular factors play a role in regulating the nucleosome mobilization activity of yIsw2, and also determining its directionality. Naturally, it would be interesting to determine what these factors are and how they also affect the single- and double-stranded DNA translocation activity of yIsw2.
Acknowledgements
The authors would like to thank Dr. Timothy J. Richmond (ETH-Hönggerberg HPK, Zürich) for providing samples of yISW2 for this work and for their critical review of this manuscript. This research was supported, in part, by startup funding from the University of Kansas (to C.J.F.) and by NIH grant P20 RR17708 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources (to C.J.F.).
Abbreviations
- NLLS
non-linear least squares
- LLS
linear least squares
- ATP
adenosine triphosphate
References
- 1.Fazzio TG, Tsukiyama T. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol Cell. 2003;12:1333–1340. doi: 10.1016/s1097-2765(03)00436-2. [DOI] [PubMed] [Google Scholar]
- 2.Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat Res. 2007;618:3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J Biol Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 6.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Bio. 1993;3:419–429. [Google Scholar]
- 8.Cote J, Peterson CL, Workman JL. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci U S A. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci U S A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillingham MS, Soultanas P, Wiley P, Webb MR, Wigley DB. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc Natl Acad Sci U S A. 2001;98:8381–8387. doi: 10.1073/pnas.131009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. Embo J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. [DOI] [PubMed] [Google Scholar]
- 14.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns B, Peterson CL, Bustamante C. DNA Translocation and Loop Formation Mechanism of Chromatin Remodeling by SWI/SNF and RSC. Molecular Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer CJ, Saha A, Cairns BR. Kinetic model for the ATP-dependent translocation of Saccharomyces cerevisiae RSC along double-stranded DNA. Biochemistry. 2007;46:12416–12426. doi: 10.1021/bi700930n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 20.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald DJ, DeLuca C, Berger I, Gaillard H, Sigrist R, Schimmele K, Richmond TJ. Reaction cycle of the yeast Isw2 chromatin remodeling complex. Embo J. 2004;23:3836–3843. doi: 10.1038/sj.emboj.7600364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 23.Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci U S A. 2007;104:4913–4918. doi: 10.1073/pnas.0700706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatinremodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConnell AD, Gelbart ME, Tsukiyama T. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol Cell Biol. 2004;24:2605–2613. doi: 10.1128/MCB.24.7.2605-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 29.Sherriff JA, Kent NA, Mellor J. The Isw2 chromatin-remodeling ATPase cooperates with the Fkh2 transcription factor to repress transcription of the B-type cyclin gene CLB2. Mol Cell Biol. 2007;27:2848–2860. doi: 10.1128/MCB.01798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nature structural & molecular biology. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. Embo J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer CJ, Lohman TM. ATP-dependent translocation of proteins along single-stranded DNA: models and methods of analysis of pre-steady state kinetics. J Mol Biol. 2004;344:1265–1286. doi: 10.1016/j.jmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A Nonuniform Stepping Mechanism for E. coli UvrD Monomer Translocation along Single-Stranded DNA. Mol Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DE, Narayan M, Patel SS. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J Mol Biol. 2002;321:807–819. doi: 10.1016/s0022-2836(02)00733-7. [DOI] [PubMed] [Google Scholar]
- 37.Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 38.Dillingham MS, Wigley DB, Webb MR. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 39.Bjornson KP, Moore KJ, Lohman TM. Kinetic mechanism of DNA binding and DNA-induced dimerization of the Escherichia coli Rep helicase. Biochemistry. 1996;35:2268–2282. doi: 10.1021/bi9522763. [DOI] [PubMed] [Google Scholar]
- 40.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langst G, Becker PB. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci. 2001;114:2561–2568. doi: 10.1242/jcs.114.14.2561. [DOI] [PubMed] [Google Scholar]
- 42.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci U S A. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]