Abstract
Background
Our recent genome-wide association study identified a novel breast cancer susceptibility locus at 9q31.2 (rs865686).
Methods
To further investigate the rs865686–breast cancer association, we conducted a replication study within the Breast Cancer Association Consortium, which comprises 37 case–control studies (48,394 cases, 50,836 controls).
Results
This replication study provides additional strong evidence of an inverse association between rs865686 and breast cancer risk [study-adjusted per G-allele OR, 0.90; 95% confidence interval (CI), 0.88; 0.91, P = 2.01 × 10–29] among women of European ancestry. There were ethnic differences in the estimated minor (G)-allele frequency among controls [0.09, 0.30, and 0.38 among, respectively, Asians, Eastern Europeans, and other Europeans; P for heterogeneity (Phet) = 1.3 × 10–143], but no evidence of ethnic differences in per allele OR (Phet = 0.43). rs865686 was associated with estrogen receptor–positive (ER+) disease (per G-allele OR, 0.89; 95% CI, 0.86–0.91; P = 3.13 × 10–22) but less strongly, if at all, with ER-negative (ER–) disease (OR, 0.98; 95% CI, 0.94–1.02; P = 0.26; Phet = 1.16 × 10–6), with no evidence of independent heterogeneity by progesterone receptor or HER2 status. The strength of the breast cancer association decreased with increasing age at diagnosis, with case-only analysis showing a trend in the number of copies of the G allele with increasing age at diagnosis (P for linear trend = 0.0095), but only among women with ER+ tumors.
Conclusions
This study is the first to show that rs865686 is a susceptibility marker for ER+ breast cancer.
Impact
The findings further support the view that genetic susceptibility varies according to tumor subtype.
Introduction
Several genome-wide association studies (GWAS) have examined, since 2007, the role of common genetic variation in breast cancer risk leading to the identification of more than 20 risk loci (1–10). We recently conducted a multi-stage GWAS, involving a total of 11,781 cases and 12,378 controls that identified a novel breast cancer locus at 9q31.2 (rs865686) with an estimated per G-allele OR of 0.89 [95% confidence interval (CI) 0.85–0.92, P = 1.75 × 10–10 (11; Fig. 1)]. Although this result is statistically significant by the usual criteria, rs865686 lies over 600 kb from the nearest gene and is not in linkage disequilibrium with any genomic elements that suggest a possible causal mechanism. Statistical replication of this association is therefore important to establish this region as a risk locus for breast cancer. To provide a more precise estimate of the magnitude of the rs865686 association with breast cancer risk, we conducted a replication study within the Breast Cancer Association Consortium (BCAC), a large international consortium comprising more than 95,000 breast cancer cases and controls, which has been used to confirm most other GWAS-identified breast cancer susceptibility loci (e.g., refs. 12–14). This large case–control series also provided an opportunity to investigate whether the strength of the SNP–breast cancer association varies by ethnicity, age at diagnosis, or tumor subtype.
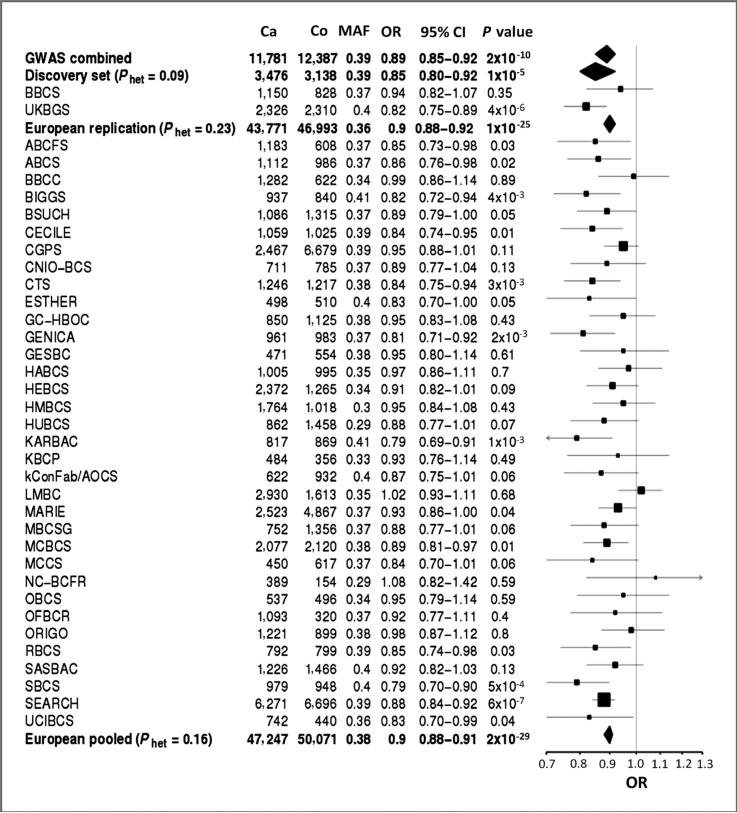
Figure 1.
Forest plot of the association of 9q31.2-rs865686 with breast cancer risk among women of European ancestry in the original GWAS, all stages combined (11), and replication within the Breast Cancer Association Consortium (BCAC). Ca, no. cases; Co, no. controls; MAF, minor allele frequency. Study abbreviations as in Supplementary Table S1. * The discovery set (see Results) comprises data from 2 BCAC studies that had contributed data to the GWAS analysis [BBCS contributed data on 1,711 (out of the 1,978 subjects examined here) to stages 1 and 2 of the GWAS; UKBGS contributed data on 4,621 (out of the 4,661 subjects examined here) to stage 3 of the GWAS].
Materials and Methods
Study subjects
Thirty-seven BCAC case–control studies contributed to this analysis. A brief description of the participating studies is given in Supplementary Table S1. The large majority of women (98%) were of self-reported European descent; those who participated in studies conducted in Eastern European countries (HMBCS and HUBCS) and those in OFBCR, who self-reported themselves as being Eastern European, were further classified as Eastern Europeans. In all, 2% were of self-reported Asian ethnicity; the latter included all the participants in the only study (ACP) conducted in Asia as well as Asians who participated in other studies (restricted to studies with at least 20 Asians). Women of other self-reported ethnicities were excluded because of small numbers. Data on morphology (invasive vs. in situ) and receptor status [estrogen receptor (ER), progesterone receptor (PR), and HER2], obtained mainly from clinical notes, were available for certain subsets of cases (Supplementary Table S2).
Each study was approved by the relevant Institutional Review Boards/Ethics Committees. Written informed consent was obtained from each subject. Only anon-ymised data were made available to BCAC.
Genotyping
9q31.2-rs865686 was genotyped by MALDI-TOF MS (Sequenom), TaqMan (Applied Biosystems) or Fluidigm technologies (Fluidigm) using standard protocols (Supplementary Table S3). Strict quality control criteria, as implemented by BCAC (15), were applied. Briefly, the genotyping concordance was verified by including at least 2% of samples in duplicate and a common set of 93 CEPH DNAs used by the HapMap Consortium (HAPMAPPT01, Coriell Institute for Medical Research, Cambden, NJ). All samples from any study with more than 2 discordant genotypes on the CEU plate were excluded. Overall single nucleotide polymorphism (SNP) call rates were more than 97% for all studies (Supplementary Table S3). There was no evidence of departure from Hardy–Weinberg equilibrium (HWE) among controls in any individual participating study (P > 0.01 for all studies; Supplementary Table S3).
Statistical methods
The SNP association with breast cancer risk was assessed by estimating per-allele ORs with 95% CI. Two European BCAC studies (BBCS and UKBGS) had contributed data to the original GWAS and were therefore analysed separately as the “discovery” (hypothesis-generating) set; cases and controls of self-reported European ancestry from the remaining BCAC studies constituted the independent “replication” set (Fig. 1).
In secondary analyses, data from the discovery and replication sets were combined to maximize power. There is a possibility of “winner's curse” bias in this approach, which we assessed by comparing the primary SNP association in the combined data to the unbiased estimate in the replication set and also to the more accurate unbiased uniformly minimum variance conditional unbiased estimator (UMVCUE) proposed by Bowden and Dudbridge (16), which corrects for a “winner's curse” effect in stage I of the GWAS. The 95% CI and P value for the UMVCUE OR were calculated using the variance from 1,000,000 bootstrap samples. The 9q31.2-rs865686 SNP in stage I of the discovery GWAS had P = 2.28 × 10–4, whereas all other SNPs from the GWAS had P > 5 × 10–4 (11), hence the latter was taken as the P value threshold within the UMVCUE calculations. We assumed that a lack of detectable selection bias in the primary SNP association with breast cancer would justify combining the discovery and replication sets in the secondary analyses.
Between-study heterogeneity in ORs was assessed using the Breslow–Day test. Pooled results were adjusted for study using Cochran–Mantel–Haenszel tests. Dominant and recessive models were also considered and departure from log-additivity was tested. Heterogeneity in ORs between different ethnic groups was assessed by a Wald test of the coefficients of the ethnicity covariates obtained from a logistic regression including interactions between OR and ethnicity.
Per-allele ORs specific to each breast cancer subtype (as defined by ER, PR, and HER2 status) were estimated by taking all available controls as each reference outcome and adjusting for study. Heterogeneity in the OR by subtypes was tested using case-only logistic regression, with each binary receptor status as the predictor, allele (major/minor) as the response, and study as a covariate. This model was then extended to include all receptor statuses in a combined analysis of association and heterogeneity.
Heterogeneity in the OR by age at diagnosis (<40, 40–49, 50–59, 60–69, 70 years) was evaluated by conducting 5 separate case–control analyses among Europeans, taking all the controls as the reference outcome for each subgroup, with adjustment for study. In addition, a case-only allelic logistic regression, adjusted for study, was used to test for a linear trend between number of copies of the G-allele and age at diagnosis.
In some studies, cases had been selected to have an increased genetic susceptibility to improve power. To assess whether there was heterogeneity in the OR by level of genetic enrichment of the cases, the combined OR estimate for the 10 European studies (BBCS, CNIO-BCS, GC-HBOC, GC-HBOC, HEBCS, KARBAC, kConFab/AOCS, MBCSG, NC-BCFR, OFBCR, and RBCS) that selected all, or a subset, of cases with increased genetic susceptibility (e.g., those with 2 independent primaries and/or with at least one affected first-degree relative) was compared with the combined OR estimate for the remaining 26 European studies of unselected cases, with adjustment for study within each group.
In addition, a quantitative “family history” score was assigned to each individual woman reflecting her number of affected first-degree relatives and whether she was a unilateral or a bilateral/ipsilateral case (a value of 2 was added to the score for bilateral/ipsilateral cases) and included as a covariate within a logistic regression model, adjusting for study. A significant interaction between this score and the SNP effect provided a test of effect modification by family history.
To assess whether rs865686 is associated with age at menarche, independently of disease status, we used linear regression with age at menarche as response and genotype as predictor, with covariate adjustment for both case–control status and study.
All statistical tests were 2-sided. All analyses were carried out using PLINK (17, 18) and R (19).
Results
A total of 48,394 breast cancer cases and 50,836 controls from 37 BCAC case–control studies contributed to the analysis. Overall, 98% of the subjects were of self-reported European ancestry and 2% were of self-reported Asian ancestry. The mean (±SD) age at diagnosis for European cases was 55 (±14) years. The estimated minor (G)-allele frequency (MAF) of rs865686 was significantly higher among European (MAF = 0.38) than Asian controls (MAF = 0.09; P = 1.11 × 10–114). There was no clear evidence of subethnic differences in the estimated MAF among Asians [0.12 for controls from the Indian subcontinent (N = 99); 0.09 for controls from South East Asia (N = 660); P = 0.21] but, among Europeans, the estimated MAF was lower among controls from Eastern European populations (MAF = 0.30) than among those from other European populations (MAF = 0.38, P = 1.08 × 10–31).
The replication set (comprising Europeans from all studies except BBCS and UKBGS) provided strong independent support for an association between rs865686 and overall breast cancer risk among women of European ancestry (Fig. 1), with an estimated OR of 0.90 (95% CI, 0.88–0.92; P = 1.23 × 10–25) per G-allele and no evidence of between-study heterogeneity (Phet = 0.23). Genotype-specific ORs were 0.89 (95% CI, 0.87–0.92) for GT vs. TT and 0.81 (95% CI, 0.78–0.85) for GG vs. TT. None of the estimates departed from those expected under a log-additive model (P = 0.34) with the OR being 0.87 (95% CI 0.85, 0.90) and 0.87 (95% CI 0.83, 0.90), respectively, for dominant (GG and GT vs. TT) and recessive (GG vs. GT and TT) models.
There was no evidence of heterogeneity in the per-allele OR between the discovery and the replication sets (Phet = 0.16; Fig. 1), with their combined data yielding a pooled per-allele OR of 0.90 (95% CI, 0.88–0.91; P = 2.01 × 10–29) for women of European descent. This pooled estimate is potentially biased by the “winner's curse” effect, but in fact is almost identical to the unbiased UMVCUE estimator of Bowden and Dudbridge (ref. 16; OR 0.8999, 95% CI, 0.8823–0.9178; P = 1.07 × 10–25) and the replication estimate (OR, 0.8996; 95% CI, 0.8819–0.9176; P = 1.23 × 10–25). Therefore, any selection bias in the discovery set is minimal, and in the following analyses, we combined the data from all women of European ancestry.
There was no evidence of ethnic variation in the per-allele OR estimate (Phet = 0.43), but the power of the study to detect this was low due to the relatively small sample size (N = 1,882; 1,123 cases and 759 controls) and the low MAF (0.09) among Asians. The per-allele OR estimates were similar for non-Eastern Europeans (OR, 0.90; 95% CI, 0.88–0.91) and Eastern Europeans (OR, 0.92; 95% CI, 0.84–1.0), and the higher estimate for Asians had a wide CI (OR, 1.02; 95% CI, 0.80–1.31), which overlapped the other 2 CIs.
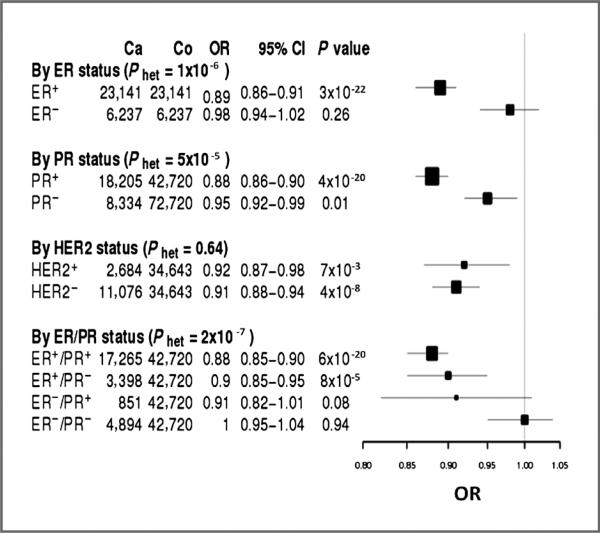
Among women of European ancestry, the allelic ORs were similar for invasive and in situ tumors (Phet = 0.62), but differed according to receptor status (Fig. 2). The SNP was more strongly associated with ER-positive (ER+) than ER-negative (ER–) tumors (Phet = 1.16 × 10–6) and with PR-positive (PR+) than PR-negative (PR–) tumors (Phet = 5.14 × 10–5). There was no evidence, albeit based on a smaller sample size, of effect modification by HER2 status (Phet = 0.64). Stratification according to the 4 different possible combinations of ER/PR status (Fig. 2) showed that the rs865686 was strongly associated with ER+/PR+ and ER+/PR– disease (P = 5.78 × 10–20 and P = 8.21 × 10–5, respectively) but not with ER–/PR+ or ER–/PR– disease (P = 0.08 and P = 0.94, respectively, Phet = 1.75 × 10–7). When both ER and PR status were modeled jointly in a case-only analysis, only the association with ER status remained significant (P = 0.0012 for ER, P = 0.21 for PR), with no evidence of interaction between ER and PR status (Phet = 0.27). Together, these findings are consistent with the SNP being strongly associated with ER+ disease (per-allele OR, 0.89; 95% CI, 0.86–0.91; P = 3.13 × 10–22) but less strongly, if at all, with ER– disease (OR, 0.98; 95% CI, 0.94–1.02; P = 0.26; Fig. 2). There was no evidence of heterogeneity in ORs by tumor histology, or receptor status, among women of Asian descent, but these analyses were based on a much smaller sample size than for women of European ancestry (data not shown).
Figure 2.
9q31.2-rs865686 and breast cancer risk amongst women of European ancestry stratifying by ER, PR, and HER2 receptor status*. Ca, no. cases; Co, no. controls; * P for heterogeneity (Phet) in the OR by ER status: 6.9 × 10–4 amongst PR– tumors and 0.52 amongst PR+ tumors. Phet in the OR by PR status: 0.13 amongst ER– cases and 0.52 amongst ER+ cases.
In women of European ancestry, case–control analyses suggest a positive trend in the magnitude of the per-allele OR (i.e., ORs became closer to unity) with increasing age at diagnosis of breast cancer (Table 1). This result was confirmed by a case-only analysis, but only for ER+ tumors, which showed a significant trend (P = 0.0095) in the number of copies of the G-allele with increasing age of onset of ER+ disease (Table 1), but did not detect any association among either all cases, or ER– cases (P = 0.37 and P = 0.35, respectively). Note that the estimated OR for cases diagnosed at age <40 years, albeit based on a small sample size, did not follow the overall trend with its magnitude being as high as the OR for the oldest age group (≥70 years). This may be due to chance or a truly different effect among very young cases.
Table 1.
Association of 9q31.2-rs865686 and breast cancer risk among women of European ancestry, by ER status of the tumor and age at diagnosis
| Class | Age at diagnosis | Cases | Controls | OR (95% CI)* | P |
|---|---|---|---|---|---|
| All cases | |||||
| Case-control analysis: | |||||
| 1 | <40 | 4,934 | 50,216 | 0.93 (0.89, 0.97) | 0.001 |
| 2 | 40–49 | 10,643 | 50,216 | 0.88 (0.85, 0.91) | 4.21 × 10–15 |
| 3 | 50–59 | 14,703 | 50,216 | 0.88 (0.86, 0.91) | 3.59 × 10–18 |
| 4 | 60–69 | 11,298 | 50,216 | 0.91 (0.88, 0.94) | 5.44 × 10–10 |
| 5 | ≥70 | 4,978 | 50,216 | 0.93 (0.89, 0.97) | 0.001 |
| NK | 836 | – | – | – | |
| Case-only analysis: | |||||
| All | Exact Age | 46,556 | – | – | P for linear trend = 0.37 |
| ER-positive cases | |||||
| Case-control analysis: | |||||
| <40 | 1,749 | 43,706 | 0.93 (0.87, 1.01) | 0.066 | |
| 40—49 | 4,593 | 43,706 | 0.84 (0.80, 0.88) | 7.84 × 10–13 | |
| 50—59 | 7,160 | 43,706 | 0.87 (0.83, 0.90) | 4.53 × 10–13 | |
| 60–69 | 6,335 | 43,706 | 0.91 (0.87, 0.95) | 4.07 × 10–6 | |
| ≥70 | 3,255 | 43,706 | 0.93 (0.88, 0.98) | 0.011 | |
| NK | 29 | ||||
| Case-only analysis: | |||||
| All | Exact Age | 23,112 | – | – | P for linear trend = 0.0095 |
| ER-negative cases | |||||
| Case-control analysis: | |||||
| <40 | 1,017 | 43,706 | 0.99 (0.90, 1.09) | 0.81 | |
| 40-49 | 1,443 | 43,706 | 0.96 (0.88, 1.04) | 0.28 | |
| 50-59 | 1,923 | 43,706 | 0.98 (0.91, 1.05) | 0.52 | |
| 60-69 | 1,278 | 43,706 | 1.02 (0.94, 1.10) | 0.71 | |
| ≥70 | 567 | 43,706 | 0.95 (0.84, 1.07) | 0.40 | |
| NK | 9 | – | – | – | |
| Case-only analysis: | |||||
| All | Exact Age | 6,228 | – | – | P for linear trend = 0.35 |
Abbreviation: NK, not known.
Study-adjusted OR and 95% CI.
There was no evidence among Europeans that the per-allele OR differed (Phet = 0.97) between the 10 case–control studies based on “genetically enriched” cases (combined per-allele OR, 0.90; 95% CI, 0.86–0.94; P = 1.48 × 10–6) and the 26 studies based on unselected cases (combined per-allele OR 0.90, 95% CI 0.88, 0.92, P = 2.38 × 10–24). Analyses by family history score, which reflects number of affected first-degree relatives and whether the woman had one (unilateral case) or 2 (ipsilateral and bilateral cases) independent primary tumors (see Methods section), gave similar results with no evidence that the SNP-associated effect was modified by this variable (Phet = 0.47). The OR for bilateral disease may be somewhat weaker than expected (20), but redefining the family history score as simply the number of affected relatives, regardless of whether the woman was a unilateral or a bilateral/ipsilateral case, did not affect the findings (Phet = 0.45).
Two recent GWAS (21, 22) reported associations between 9q31.2 SNPs (rs7861820, rs12684013, rs4452860, rs7028916, and rs2090409) and age at menarche, a factor known to affect breast cancer risk. These loci map of more than 2 Mb from rs865686 are not correlated with it (r2 < 0.01, D′ <0.09 in CEU HapMap phase 2). We found no evidence that rs865686 was associated with age at menarche (per-allele difference in age at menarche: 0.0017 years; 95% CI, –0.0169–0.0203; P = 0.86), within the BCAC subset of 58,983 European women (31,522 cases and 27,461 controls) with information available on age at menarche and rs865686 genotype.
Discussion
This combined analysis of data from a large international consortium confirms 9q31.2-rs865686 to be a breast cancer susceptibility locus in women of European ancestry, yielding a very precise estimate of the per-allele OR among European women (OR, 0.90; 95% CI, 0.88–0.91). Replication studies have shown that genetic susceptibility to breast cancer varies by expression levels of ER in breast tumors, with certain variants being associated with both ER+ and ER– disease, whereas others are more strongly associated with ER+ or triple-negative (ER–, PR–, HER2–) disease (23). Our study is the first to show that the 9q31.2-rs865686 is associated with ER+ breast cancer but less so, if at all, with ER– disease. These results are not affected by the “winner's curse” bias as data on receptor status were available only for studies in the replication set. Interestingly, a recent study of African-American women (24) found no evidence of heterogeneity by ER status (Phet = 0.17), but with a significant association being observed among ER– (per-allele OR, 0.87; 95% CI, 0.78–0.97) but not ER+ disease (OR, 0.94; 95% CI, 0.86–1.03; ref. 24). However, this other study had low power to detect differences by receptor status (n = 1520 ER+ cases, 988 ER– cases, and 2,745 controls).
Our original GWAS (11) found no evidence of an association in the per-allele ORs with either age at menarche or age at diagnosis. The present study did not reveal any association of rs865686 with age at menarche or age at diagnosis overall among women of European ancestry, but it found a trend in the number of copies of the G-allele with increasing age at diagnosis among ER+ breast cancer cases. No strong trends in risk with age at diagnosis for the other known common breast cancer SNPs have been reported despite the fact that the familial relative risk of breast cancer is much higher at younger ages, particularly in relatives of young cases (25).
Our study was well-powered to identify ethnic variations in MAF, with this being much lower among women of Asian descent (MAF = 0.09) than among those of European ancestry (MAF = 0.38), and among the latter lower among Eastern European (MAF = 0.30) than among other European women (MAF = 0.38). However, the magnitude of the per-allele OR for the breast cancer association was estimated less precisely in Eastern European and Asian women. The Asian sample was markedly under powered for detecting an association between the SNP and breast cancer risk (only 15% power at P < 0.05 as estimated by the Genetic Power Calculator (ref. 26) assuming the same OR as estimated in European women). Assuming a similar case–control ratio as in the present BCAC dataset in approximately 11,000 Asian cases would be required to attain 80% power. The Eastern European sample had a greater power (~70%), with approximately 1,000 additional cases required to attain 80% power. Overall, these ethnic differences are consistent with those from the study of African-American women mentioned above (24) showing that they had a higher MAF (0.48) than European women but a similar per-allele OR (0.93; 95% CI, 0.85–0.99; P = 0.034).
Further genetic and functional studies will be required to identify the causal variant (or variants) and the mechanisms underlying the 9q31.2-rs865686 association with breast cancer risk. This SNP maps to a 17 kb region of LD (109927817-109944558 bp) on 9q31.2 with no known genes. The nearest genes are Kruppel-like factor 4 (KLF4, 636 kb centromeric), RAD23B (799 kb centromeric), and actin-like 7A (ACTL7A, 736 kb telomeric). Interrogation of the Oncomine database (27) showed a decrease in KLF4 gene transcripts in breast cancers and a correlation between KLF4 expression and ERα positivity. However, a causal link remains to be established between these functional mechanisms and sequence variation at or near rs865686. To motivate such functional studies, indisputable epidemiologic evidence is needed for association between rs865686 and breast cancer, which we now report in this study. With such evidence in place, this region now presents a strong example of a noncoding SNP whose causal mechanism on disease is unclear. Determination of the mechanism is a considerable challenge but will eventually shed further light on breast cancer oncogenesis and, potentially, noncoding mechanisms in other complex diseases.
In conclusion, this large replication study found strong evidence for an association between the 9q31.2-rs865686 SNP and ER+ breast cancer among women of European ancestry. The findings are consistent with breast cancer being a biologically heterogeneous disease and highlight the need for subtype-specific studies to be conducted in different ethnic populations.
Supplementary Material
Acknowledgments
The authors thank all the women who participated in the 37 contributing studies and all the researchers, clinicians, technicians, and administrative staff who supported this work. Special thanks to - ABCFS: The many women and their families that generously participated in this study and consented to us accessing their pathology material. ABCS: Laura Van't Veer, Richard van Hien, Senno Verhoef, Frans Hogervorst and Bas Buenode-Mesquita (the latter for the release of control samples). ACP: The women who participated in the study; the Thai Ministry of Public Health (MOPH), the doctors and the nurses who helped with the data collection, and Dr. Prat Boonyawongviroj, the former Permanent Secretary of MOPH, and Dr Pornthep Siriwanarungsan, the Department Director-General of Disease Control, who have supported the study throughout. BBCC: Sonja Öser and Silke Landrith. BBCS: Lorna Gibson, Eileen Williams, Elaine Ryder-Mills, and Kara Sargus for their support. BIGGS: Niall McInerney, Gabrielle Colleran, Andrew Rowan, and Nicola Miller for their support. BSUCH: Anne Langheinz for MassArray analysis. CGPS: Staff and participants of the Copenhagen General Population Study. CNIO-BCS: Charo Alonso, Tais Moreno, Guillermo Pita, Primitiva Menendez, and Anna González-Neira. GC-HBOC: Anne Langheinz. GENICA: Ute Hamann, Sylvia Rabstein, Anne Spickenheuer, Hans-Peter Fischer, Beate Pesch, Volker Harth, and Christian Baisch. HABCS: Johann H. Karstens. HEBCS: Dr. Kirsimari Aaltonen, Dr. Karl von Smitten, and RN Irja Erkkilä for help with the patient data and samples. KBCP: Helena Kemiläinen, Aija Parkkinen and Eija Myöhänen for their skillful technical assistance. KConFab-AOCS: Heather Thorne, Eveline Niedermayr, the AOCS management group (D Bowtell, G Chenevix-Trench, A deFazio, D Gertig, A Green, P Webb), the ACS management group (A Green, P Parsons, N Hayward, P Webb, D Whiteman). MBCSG: Marco Pierotti, Bernard Peissel and Daniela Zaffaroni of the Fondazione IRCCS Istituto Nazionale Tumori; Bernardo Bonanni and Monica Barile of the Istituto Europeo di Oncologia; and the personnel of the Cancer Genetics Testing laboratory at the IFOM-IEO campus. OBCS: Taina Turpeenniemi-Hujanen, Saila Kauppila, Kari Mononen, Meeri Otsukka. OFBCR: The participants; and Teresa Selander and Gord Glendon for their contributions to the study. ORIGO: E. Krol-Warmerdam and J. Blom. RBCS: Ans van den Ouweland, Elisabeth Huijskens, Ellen Crepin, Petra Bos, Jannet Blom and Anja Nieuwlaat. SBCS: Sue Higham, Helen Cramp, and Dan Connley. SEARCH: The SEARCH and EPIC teams.
Grant Support
This work was supported by the following sources - ABCFS: National Health and Medical Research Council of Australia, the New South Wales Cancer Council; the Victorian Health Promotion Foundation (Australia); the United States (US) National Cancer Institute, NIH [RFA CA 06 503]; and through cooperative agreements with members of the Breast Cancer Family Registry (CFR) and principal investigators. The University of Melbourne [U01 CA69638] contributed data to this study. ABCS: Dutch Cancer Society [NKI 2001-2423, 2007-3839] and Dutch National Genomics Initiative. ACP: Breast Cancer Research Trust, UK. BBCC: ELAN program of the University Hospital of Erlangen. BBCS: Cancer Research UK [C150/A5660 and C1178/A3947]; Breakthrough Breast Cancer; and NHS funding to the NIHR Biomedical Research Centre and the National Cancer Research Network (NCRN). H. Warren is funded by the UK Medical Research Council [G0801414]. BCAC: Cancer Research UK [C1287/A10118, C1287/A12014] and the European Community's Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F2-2009-223175) (COGS). BCAC meetings have been funded by the European Union COST program [BM0606]. BIGGS: NIHR Comprehensive Biomedical Research Centre, Guy's & St. Thomas’ NHS Foundation Trust in partnership with King's College London [to E.J. Sawyer]; Oxford Biomedical Research Centre (to I. Tomlinson]. BSUCH: Helmholtz Society; German Cancer Research Center; and Dietmar-Hopp Foundation. CECILE: Fondation de France [2004012618, 2007005156]; Institut National du Cancer (INCa) [2007-1/SPC2, 2008-1-CP-4, 2009-1-SHS/SP-04]; Association pour la Recherche contre le Cancer (ARC) [2008-1-CP-4]; Agence Française de Sécurité Sanitaire de l'Environnement et du Travail (AFSSET - ANSES) [ST-2005-003, EST2008/1/26, and VS-2009-21]; Ligue contre le Cancer Grand Ouest; and Agence Nationale de la Recherche (ANR) [00047-05]. CGPS: Herlev Hospital; Chief Physician Johan Boserup; and Lise Boserup's Fund. CNIO-BCS: Genome Spain Foundation; the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer; and the Fondo de Investigación Sanitario [PI081583, PI081120]. CTS: California Breast Cancer Act of 1993; NIH [R01 CA77398]; the Lon V Smith Foundation [LVS39420]; and the California Breast Cancer Research Fund [contract 97-10500]. Collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer-reporting program mandated by California Health and Safety Code Section 103885. ESTHER: Baden-Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study that was supported by a grant from the Deutsche Krebshilfe (German Cancer Aid). GESBC: Deutsche Krebshilfe e.V. [70492]; and genotyping in part by the state of Baden-Württemberg through the Medical Faculty of the University of Ulm [P.685]. GC-HBOC: Deutsche Krebshilfe [107054]; the Dietmar-Hopp Foundation; the Helmholtz society; and the German Cancer Research Centre (DKFZ). GENICA: Federal Ministry of Education and Research (BMBF) Germany [01KW9975/5, 01KW9976/8, 01KW9977/0, 01KW0114]; Robert Bosch Foundation, Stuttgart; Deutsches Krebsforschungszentrum (DKFZ), Heidelberg; Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn; Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, Germany. HABCS: Rudolf Bartling Foundation. HEBCS: Helsinki University Central Hospital Research Fund, Academy of Finland [132473]; Finnish Cancer Society; Nordic Cancer Union; and Sigrid Juselius Foundation. HMBCS: Hannelore Munke Stipend [to N. Bogdanova]. HUBCS: German Federal Ministry for Research and Education [RUS 08/017]. KARBAC: Swedish Cancer Society; Stockholm Cancer Society; and Gustav V Jubilee Foundation. KBCP: Finnish Cancer Society; Academy of Finland [127220], special Government Funding (EVO) of Kuopio University Hospital [5654113, 5501]; and strategic funding from the University of Eastern Finland. KConFab-AOCS: National Breast Cancer Foundation; National Health and Medical Research Council (NHMRC); Queensland Cancer Fund; Cancer Councils of New South Wales, Victoria, Tasmania and South Australia; and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC [145684, 288704, 454508]. Financial support for the AOCS was provided by the US Army Medical Research and Materiel Command [DAMD17-01-1-0729], the Cancer Council of Tasmania and Cancer Foundation of Western Australia and the NHMRC [199600]. G. Chenevix-Trench and P. Webb are supported by the NHMRC. LMBC: ‘Stichting tegen Kanker’ [232-2008, 196-2010]. MARIE: Deutsche Krebshilfe e.V. [70-2892-BR I]; Hamburg Society; German Cancer Research Center; and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MBCSG: Ministero della Salute (Extraordinary National Cancer Program 2006 “Alleanza contro il Cancro”, and “Progetto Tumori Femminili”) (to P. Radice); Ministero dell'Universita’ e Ricerca (RBLAO3-BETH; to P. Radice); Fondazione Italiana per la Ricerca sul Cancro (Special Project “Hereditary tumors”); Associazione Italiana per la Ricerca sul Cancro (4017; to P. Peterlongo) and by funds from Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5 1000”). MCBCS: NIH [CA122340]; Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201]; the Komen Foundation for the Cure; and the Breast Cancer Research Foundation (BCRF). MCCS: Australian National Health and Medical Research Council [209057, 251533, 396414, 504711], with infrastructure support provided by the Cancer Council Victoria, and cohort recruitment partly funded by VicHealth. NC-BCFR: US National Cancer Institute, NIH [RFA-CA-06-503] and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario [U01 CA69467], the Cancer Prevention Institute of California [U01 CA69417], and The University of Melbourne [U01 CA69638]. Samples from the NC-BCFR were processed and distributed by the Coriell Institute for Medical Research. OBCS: Finnish Cancer Foundation; Academy of Finland; University of Oulu; and Oulu University Hospital. OFBCR: US National Cancer Institute, NIH [RFA-CA-06-503] and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario [U01 CA69467], Northern California Cancer Center [U01 CA69417], and University of Melbourne [U01 CA69638] and by Cancer Care Ontario. ORIGO: Dutch Cancer Society [RUL 1997-1505]; and Biobanking and Biomolecular Resources Research Infrastructure [BBMRI-NL CP16]. RBCS: Dutch Cancer Society [DDHK 2004-3124, DDHK 2009-4318]. SASBAC: NIH [RO1 CA58427] [to P. Hillemanns and J. Liu]; and the Agency for Science, Technology and Research (A* STAR; Singapore). SBCS: Yorkshire Cancer Research; and Breast Cancer Campaign. SEARCH: Programme grant from Cancer Research UK [C490/A10124]. UCIBCS: US NIH [CA58860, CA92044] and the Lon V Smith Foundation [LVS39420]. UKBGS: Breakthrough Breast Cancer.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Authors’ Contributions
Conception and design: H. Warren, N. Orr, J.L. Hopper, M.W. Beckmann, R.L. Milne, A. Ziogas, H. Brenner, T. Bruning, A. Mannermaa, G. Chenevix-Trench, G.G. Giles, G. Severi, P. Hall, P.D.P. Pharoah, A.M. Dunning, M. Ghoussaini, I. dos Santos Silva
Development of methodology: J.L. Hopper, M.J. Kerin, A. Ziogas, T. Bruning, A. Miron, M. Ghoussaini
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): N. Orr, N. Johnson, J.L. Hopper, C. Apicella, M.C. Southey, M.K. Schmidt, K. Muir, A. Lophatananon, A. Chaiwerawattana, S. Wiangnon, P.A. Fasching, M.W. Beckmann, A.B. Ekici, E.J. Sawyer, I. Tomlinson, M.J. Kerin, B. Burwinkel, F. Marme, A. Schneeweiss, C. Sohn, P. Guénel, T. Truong, P. Laurent-Puig, S.E. Bojesen, S.F. Nielsen, H. Flyger, B.G. Nordestgaard, R.L. Milne, J.-I. Arias-Pérez, M.P. Zamora, H. Anton-Culver, A. Ziogas, L. Bernstein, C.A. Clarke, H. Brenner, H. Muller, V. Arndt, A. Langheinz, A. Meindl, M. Golatta, C.R. Bartram, R.K. Schmutzler, H. Brauch, C. Justenhoven, T. Bruning, J. Chang-Claude, S. Wang-Gohrke, U. Eilber, T. Dork, P. Schurmann, M. Bremer, P. Hillemanns, H. Nevanlinna, T.A. Muranen, K. Aittomäaki, C. Bromqvist, N. Bogdanova, N. Antonenkova, Y. Rogov, M. Bermisheva, D. Prokofyeva, G. Zinnatullina, E. Khusnutdinova, A. Lindblom, S. Margolin, A. Mannermaa, J.M. Hartikainen, V. Kataja, G. Chenevix-Trench, J. Beesley, D. Lambrechts, A. Smeets, R. Paridaens, C. Weltens, D. Flesch-Janys, S. Behrens, P. Peterlongo, L. Bernard, S. Manoukian, P. Radice, F.J. Couch, C.M. Vachon, X. Wang, J.E. Olsen, G.G. Giles, L. Baglietto, C.A. McLean, G. Severi, E.M. John, A. Miron, R. Winqvist, K. Pylkäs, A. Jukkola-Vuorinen, M. Grip, I.L. Andrulis, J.A. Knight, A.M. Mulligan, P. Devilee, R.A.E.M. Tollenaar, J.W. M. Martens, C.M. Seynaeve, M.J. Hooning, A. Hollestelle, A. Jager, M.M.A. Tilanus-Linthorst, P. Hall, K. Czene, L. Jianjun, A. Cox, S.S. Cross, M.W.R. Reed, P.D.P. Pharoah, A.M. Dunning, A. Ashworth, A. Swerdlow, M. Jones, M.J. Schoemaker, D.F. Easton, I. dos Santos Silva
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): H. Warren, F. Dudbridge, O. Fletcher, M.C. Southey, P.A. Fasching, A. Langheinz, T. Bruning, C.A. McLean, A. Miron, P. Hall, D.F. Easton, I. dos Santos Silva
Writing, review, and/or revision of the manuscript: H. Warren, F. Dud-bridge, O. Fletcher, N. Orr, J.L. Hopper, M.C. Southey, M.K. Schmidt, A. Broeks, K. Muir, A. Lophatananon, A. Chaiwerawattana, S. Wiangnon, P. A. Fasching, M.W. Beckmann, A.B. Ekici, R. Schulz-Wendtland, B. Burwinkel, A. Schneeweiss, P. Guénel, T. Truong, S.E. Bojesen, H. Flyger, B.G. Nordestgaard, R.L. Milne, Javier Benítez, M.P. Zamora, H. Anton-Culver, L. Bernstein, V. Arndt, A. Langheinz, R.K. Schmutzler, H. Brauch, C. Justenhoven, T. Bruning, J. Chang-Claude, S. Wang-Gohrke, T. Dork, P. Schurmann, P. Hillemanns, H. Nevanlinna, K. Aittomäki, C. Bromqvist, N. Bogdanova, E. Khusnutdinova, S. Margolin, A. Mannermaa, V.-M. Kosma, V. Kataja, G. Chenevix-Trench, J. Beesley, R. Paridaens, C. Weltens, K. Buck, P. Peterlongo, F.J. Couch, C.M. Vachon, X. Wang, G.G. Giles, G. Severi, E.M. John, R. Winqvist, A. Jukkola-Vuorinen, I.L. Andrulis, J.A. Knight, R.A.E.M. Tollenaar, J.W.M. Martens, C.M. Seynaeve, M.J. Hooning, A. Hollestelle, A. Jager, M.M.A. Tilanus-Linthorst, K. Czene, L. Jingmei, A. Cox, P.D.P. Pharoah, A. Swerdlow, D.F. Easton, M.K. Humphreys, I. dos Santos Silva
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.L. Hopper, C. Apicella, M.K. Schmidt, A. Broeks, S. Cornelissen, L.M. Braaf, A. Lophatananon, A.B. Ekici, B. Burwinkel, S.F. Nielsen, B.G. Nordestgaard, A. Ziogas, L. Bernstein, A. Langheinz, C.R. Bartram, H. Brauch, T. Bruning, S. Wang-Gohrke, U. Eilber, P. Schurmann, P. Hillemanns, N. Antonenkova, S. Margolin, J.M. Hartikainen, V. Kataja, X. Chen, D. Lambrechts, S. Behrens, J.E. Olsen, R. Winqvist, N. Weerasooriya, M.J. Hooning, K. Czene, L. Jianjun, I.W. Brock, F.M. Blows, M.K. Humphreys, Q. Wang
Study supervision: J.L. Hopper, C. Apicella, M.C. Southey, M. Mahmoodi, P. Guénel, B.G. Nordestgaard, A. Meindl, T. Bruning, T. Dork, G. Zinnatullina, I. dos Santos Silva
Primary analyst of combined data: H. Warren
Biocollections: C. Mulot
Designed and managed the California Teachers Study which has contributed data and DNA to BCAC for this manuscript: L. Bernstein
Contribution done as PI of the GENICA Network: T. Bruning
Initiator of the Sheffield Breast Cancer Study: M.W.R. Reed
Supplementary data for this article are available at Cancer Epidemiology, Biomarkers & Prevention Online (http://cebp.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
J.L. Hopper is a National Health and Medical Research Council Australia Fellow. M.C. Southey is a National Health and Medical Research Council Senior Research Fellow. J.L. Hopper and M.C. Southey are both group leaders of the Victoria Breast Cancer Research Consortium. The content of this manuscript does not necessarily reflect the views or the policies of the National Cancer Institute or any of the collaborating centers in the CFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the CFR.No potential conflicts of interest were disclosed.
References
- 1.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 4.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng W, Long J, Gao Y-T, Li C, Zheng Y, Xiang Y-B, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43:1210–4. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103:425–35. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 12.Milne RL, Benitez J, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, et al. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J Natl Cancer Inst. 2009;101:1012–8. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueroa JD, Garcia-Closas M, Humphreys M, Platte R, Hopper JL, Southey MC, et al. Associations of common variants at 1p11.2 and 14q24.1 (RAD51L1) with breast cancer risk and heterogeneity by tumor subtype: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milne RL, Goode EL, Garcia-Closas M, Couch FJ, Severi G, Hein R, et al. Confirmation of 5p12 as a susceptibility locus for progesterone-receptor-positive, lower grade breast cancer. Cancer Epidemiol Bio-markers Prev. 2011;20:2222–31. doi: 10.1158/1055-9965.EPI-11-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Riches-son DA, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowden J, Dudbridge F. Unbiased estimation of odds ratios: combining genomewide association scans with replication studies. Genet Epidemiol. 2009;33:406–18. doi: 10.1002/gepi.20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S. [2009 Oct 10];PLINK, version 1.07. Available from: http://pngu.mgh.harvard.edu/~purcell/plink/
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. [2011 Jul 8];The R statistical programming language. Version 2.13.1. Available from: http://www.r-project.org/
- 20.Dudbridge F, Fletcher O, Walker K, Johnson N, Orr N, dos-Santos-Silva I, et al. Estimating causal effects of genetic risk variants for breast cancer using marker data from bilateral and familial cases. Cancer Epidemiol Biomarkers Prev. 2012;21:262–72. doi: 10.1158/1055-9965.EPI-11-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–8. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41:648–50. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:3289–303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F, Chen GK, Millikan RC, John EM, Ambrosone CB, Bernstein L, et al. Fine-mapping of breast cancer susceptibility loci characterizes genetic risk in African Americans. Hum Mol Genet. 2011;20:4491–503. doi: 10.1093/hmg/ddr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beral V, Bull D, Doll R, Peto R, Reeves G. Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358:1389–99. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.