Abstract
Kirkwood-Buff analysis was performed on aqueous solutions of N-methylacetamide and acetamide using the Chemistry at HARvard Molecular Mechanics additive and Drude polarizable all-atom force fields. Comparison of a range of properties with experimental results, including Kirkwood-Buff integrals, excess coordination numbers, solution densities, partial molar values, molar enthalpy of mixing, showed both models to be well behaved at higher solute concentrations with the Drude model showing systematic improvement at lower solution concentrations. However, both models showed difficulties reproducing experimental activity derivatives and the excess Gibbs energy, with the Drude model performing slightly better. At the molecular level, the improved agreement of the Drude model at low solute concentrations is due to increased structure in the solute-solute and solute-solvent interactions. The present results indicate that the explicit inclusion of electronic polarization leads to improved modeling of dilute solutions even when those properties are not included as target data during force field optimization.
INTRODUCTION
The properties of a solute in its bulk or neat state, such as viscosity, diffusion constant, dielectric constant, and surface tension, will typically differ from those in liquid mixtures comprised of the solute in an aqueous solution or any other solvent, with those differences being concentration dependent.1, 2 The macroscopic differences arise from microscopic changes in the interactions among the components of the solutions at different concentrations on the molecular level. In the past decade, interest in understanding the properties of liquid mixtures has been steadily growing, leading to a number of computer simulations of the properties of liquid mixtures.3, 4, 5, 6, 7, 8, 9, 10, 11
Two ingredients are required to clarify the relationship between the molecular details of liquid mixtures and the macroscopic properties using computer simulations. The first is an exact molecular theory of liquid mixtures to formally link the macroscopic properties to the underlying microscopic details of the system. There are essentially two theories of solutions that can be considered exact: the McMillan–Mayer theory12 and Fluctuation Solution Theory (FST).13 The McMillan–Mayer theory is mostly limited to solutes at low concentrations, while FST has no such limitations.14 It is an exact theory applicable to any stable solution that consists of any types of molecules of any sizes, any number of components and concentrations, that was initially derived by Kirkwood and Buff.13 Hence it is also widely referred to as Kirkwood-Buff (KB) theory in the literature. It quantifies the molecular distributions of liquid mixtures in terms of so-called Kirkwood-Buff integrals (KBIs) that can be expressed as a function of fluctuations in the number of the particles in the open system. The second ingredient is an accurate potential energy function coupled with a set of optimized force field (FF) parameters. Current empirical force fields are generally developed to reproduce experimental data for pure liquids.15 Ideally, force fields so developed have captured the essential characteristics of the molecules and should be able to describe interactions among different species of molecules adequately. In practice, however, it has been shown that such a set of force field parameters may work very well for pure liquids but not necessarily for liquid mixtures without modifications.4, 16 One possible reason could be that in many classical models the nonbond terms in the potential functions are treated in a pairwise additive fashion and induced polarization is incorporated implicitly by optimizing the fixed partial atomic charges to yield average polarizable induction in the bulk phase.17 Explicit inclusion of polarizability, or nonadditivitiy, into force fields is anticipated to alleviate such limitations.
There are three main classes of polarizable empirical force fields:18 the point dipole, charge transfer (or fluctuating charge), and classical Drude oscillator (or shell or charge-on-spring) models. In point dipole models, atoms are assigned a fixed partial atomic charge and an inducible dipole. The dipoles are then self-consistently adjusted in response to the surrounding electric field for any configuration of the system.19, 20 Charge transfer models treat the partial atomic charges as variables that are allowed to change according to a self-consistent atomic electronegativity equalization scheme.21 Polarizable force field development in our laboratory has primarily been based on the classical Drude oscillator model.22, 23, 24, 25 In the model a point charge, called the Drude oscillator or particle, is attached to each non-hydrogen atom via a harmonic spring. Induced polarization is modeled by Drude particles relaxing in response to their surrounding electrostatic environment and seeking their local energy minima for any given configuration. The approach was proposed by Paul Drude in 190026 as a simple way to describe the dispersive properties of materials and introduced into CHARMM (Chemistry at HARvard Molecular Mechanics) back in 2003.22, 27 Ever since, force field parameters based on Drude oscillator models have been developed for water,22, 28 alkanes,29 alcohols,30 ethers,31 amides,32 aromatics,33 nitrogen-containing heteroaromatics,34 sulfur-containing compounds,35, 36 and ions.37, 38 Those for biomolecules, such as proteins, nucleic acids, lipids, and carbohydrates,39 are under active development. While these models have shown improvements in many properties as compared with their additive counterparts, it is of interest to see how they will perform in describing the behavior of liquid mixtures when compared to the additive force field.
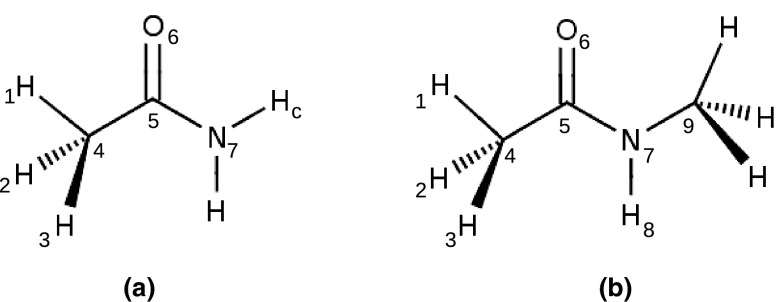
Small amide molecules are representative of several functional groups in proteins and nucleic acids. For instance, they represent models for the peptide group, asparagine/glutamine side chains, and amide-like functionality present in the nucleic acid bases. Hence, many simulations have been carried out to investigate the properties of these molecules in solutions.40, 41, 42, 43, 44, 45 In aqueous solutions, if the solvation of the amides is not favorable enough self-aggregation of the solute molecules may occur while too favorable solvation may lead to disruption of the native structures of peptides, proteins, and nucleic acids. Therefore, it is of great importance to ensure that the force field achieves a correct balance of hydrogen bonding between the amides and their surrounding water molecules. The present study accomplished the objective by performing Kirkwood-Buff analysis on computer simulations of aqueous amide solutions using the polarizable CHARMM Drude force field,27 as well as the non-polarizable CHARMM additive force field.15, 46 This effort includes optimization of parameters for acetamide (ACEM) and an update of the previously parametrized N-methylacetamide (NMA) model (see Fig. 1).32 As described below the optimization was performed following standard protocols used for the Drude model in our laboratory.23, 24, 29, 30, 31 The final parameter set was then used in the KB analysis.
Figure 1.
Structures of (a) acetamide (ACEM) and (b) N-methylacetamide (NMA).
THEORY
The Kirkwood-Buff theory of solutions involves relationships between molecular distribution functions and thermodynamic quantities for multicomponent systems in the grand canonical ensemble (V, T, μ).13 Thermodynamic properties such as compressibility (κT), partial molar volumes (PMVs), and derivatives of the chemical potential can be expressed in terms of the Kirkwood-Buff integrals (KBI), defined by
| (1) |
where gij(r) is the radial distribution function (RDF) in the grand canonical ensemble between species i and j, and Gij is the corresponding KB integral.
We focus on two-component systems in the present study. They consist of a cosolvent (c) that is either NMA, or ACEM, and water (w). The following thermodynamic properties are involved: the partial molar volumes of the two components, and , and the isothermal compressibility of the solution, κT. These terms can be expressed as functions of the integrals Gcc, Gcw, and Gww, and the number densities, ρc and ρw of cosolvent and water, respectively, as follows:13, 47
| (2) |
| (3) |
| (4) |
where kB is the Boltzmann constant,
| (5) |
and
| (6) |
Given the experimental measurements of the partial molar volumes, the isothermal compressibility of the solution, and density, KB theory described above can be reversed by an inversion procedure to obtain KBIs,48, 49 which can be directly compared to those computed based on RDFs obtained from integral equations models of solution or molecular dynamics simulations, as in the present study,
| (7) |
| (8) |
| (9) |
where
| (10) |
and
| (11) |
xc and xw are the mole fraction of cosolvent and water, is the excess molar Gibbs free energy of the solution. The second term in the definition of D is often denoted as the activity derivative, fcc, in literature,50
| (12) |
where
| (13) |
COMPUTATIONAL METHODS
QM calculations were performed with NWChem51 and Gaussian 03.52 Geometry optimizations were performed at the MP2(fc)/6-31G(d) level of theory. This level of theory provides molecular geometries consistent with available gas-phase experimental data and it has been previously utilized in the optimization of other small molecules with the Drude force field.30, 32, 33, 34, 53 QM calculations of the molecular electrostatic potentials (ESP) were performed on MP2-optimized geometries using the B3LYP hybrid functional54, 55, 56 and the correlation-consistent double-zeta Dunning aug-cc-pVDZ basis set.57 Single-point energy B3LYP calculations were performed with the tight convergence criteria producing the target QM ESP maps. QM calculations on complexes between the model compounds and the rare gas atoms were performed at the MP3/6-311++G(3d,3p) level.58
Empirical force field calculations were performed with the program CHARMM59 utilizing both the additive CHARMM general force field60 and Drude polarizable force fields27 at 313 K and 1 atm in the NPT ensemble to match the experimental conditions for the aqueous amide solutions.61, 62 The TIP3P water model63 was used for the additive FF calculations and the SWM4-NDP water model28 was used in all calculations involving the polarizable model.
For the KB analysis a range of concentrations in terms of mole fraction were considered from pure water through mixed solutions to pure cosolvent. The initial configuration for each composition was prepared by randomly placing a certain number of amide molecules in a cubic box then adding water molecules to match the concentration as listed in Table 1. The length of the cubic box is approximately 50 Å. Such a large simulation box was suggested previously to allow the tails of RDFs to converge properly therefore minimizing possible long-range effects when calculating KBIs.10 The systems, using the additive model, were then simulated to allow for extensive equilibration. RDFs were monitored to make sure that they became stable with increased simulation time, indicating that the solutions were fully mixed at the end of equilibration (usually about 1 ns). Then 10 ns production simulations were run at each concentration for data collection. For the additive simulations pressure and temperature were maintained by extended system algorithms64 with the mass of the pressure piston set to 1000 amu and Langevin piston collision frequency set to 25 per ps. Equation of motion was integrated by Leap-Frog algorithm at a time step of 2 fs.
Table 1.
Summary of MD simulations of amide solutions. All simulations were performed at 313 K and 1 atm in the NPT ensemble for 10 ns. Symbols are Nc, number of amide molecules; Nw, number of water molecules; xc, amide mole fraction. NA, not applicable.
| Molarity | Box length (Å) |
Volume (nm3) |
Molality (mol/l) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nc | Nw | xc | (mol/kg) | Additive | Drude | Additive | Drude | Additive | Drude | |
| NMA | 343 | 3087 | 0.10 | 6.17 | 51.51 ± 0.06 | 51.26 ± 0.04 | 136.631 ± 0.486 | 134.659 ± 0.303 | 4.17 | 4.23 |
| 512 | 2048 | 0.20 | 13.88 | 50.24 ± 0.06 | 49.81 ± 0.04 | 126.809 ± 0.461 | 123.580 ± 0.323 | 6.70 | 6.88 | |
| 729 | 1354 | 0.35 | 29.89 | 51.08 ± 0.06 | 50.63 ± 0.05 | 133.237 ± 0.476 | 129.754 ± 0.361 | 9.09 | 9.33 | |
| 805 | 805 | 0.50 | 55.51 | 50.17 ± 0.06 | 49.82 ± 0.05 | 126.249 ± 0.458 | 123.685 ± 0.366 | 10.59 | 10.81 | |
| 875 | 472 | 0.65 | 102.90 | 50.03 ± 0.06 | 49.81 ± 0.06 | 125.225 ± 0.465 | 123.551 ± 0.415 | 11.60 | 11.76 | |
| 925 | 232 | 0.80 | 221.32 | 49.93 ± 0.06 | 49.81 ± 0.06 | 124.461 ± 0.461 | 123.573 ± 0.428 | 12.34 | 12.43 | |
| 985 | 0 | 1.00 | NA | 50.03 ± 0.06 | 50.04 ± 0.06 | 125.248 ± 0.473 | 125.293 ± 0.474 | 13.06 | 13.05 | |
| ACEM | 200 | 3800 | 0.05 | 2.92 | 51.08 ± 0.06 | 51.14 ± 0.04 | 133.272 ± 0.466 | 133.735 ± 0.311 | 2.49 | 2.48 |
| 350 | 3150 | 0.10 | 6.17 | 50.43 ± 0.06 | 50.36 ± 0.04 | 128.287 ± 0.459 | 127.739 ± 0.312 | 4.53 | 4.55 | |
| 650 | 0 | 1.00 | NA | 39.46 ± 0.06 | 39.16 ± 0.06 | 61.433 ± 0.29 | 60.064 ± 0.268 | 17.57 | 17.97 | |
| Water | 0 | 2058 | 0.00 | 0.00 | 39.49 ± 0.07 | 39.68 ± 0.05 | 61.595 ± 0.328 | 62.465 ± 0.216 | 0.00 | 0.00 |
The final solution configurations from the additive production runs were used as starting configurations for the Drude simulations. Force field parameters for Drude models of amides were originally from Harder et al.32 with additional adjustments as described below. A Nose-Hoover thermostat65, 66 with a relaxation time of 0.1 ps was applied to all the atoms to control the global temperature of the system at 313 K. A modified Andersen-Hoover barostat67 with a relaxation time of 0.1 ps was used to maintain the system at constant pressure.27 The extended Lagrangian double-thermostat formalism68 was used in all polarizable MD simulations where a mass of 0.4 amu was transferred from parent atoms to the corresponding Drude particles.27 The amplitude of their oscillation was controlled with a separate low-temperature thermostat (at T = 1 K) to ensure that their time course approximates the self-consistent field (SCF) regime. A relaxation time of 5 fs was used for the Drude oscillator thermostat. The force constant was set at 500 kcal/(mol Å2) for the Drude particle-parent atom harmonic terms. Simulations were run for 11 ns using the velocity Verlet integrator67 at a time step of 1 fs, with the last 10 ns used for analyses.
In both the additive and Drude simulations, electrostatic interactions were treated by the Particle Mesh Ewald method69 to model the long-range electrostatics with a real space cutoff of 12 Å, a coupling parameter of 0.34, and a sixth-order spline for mesh interpolation. All nonbonded electrostatic interactions involving Drude particles are treated in the same way as electrostatic interactions between parent atoms while the nonbonded interactions are modified to allow 1-2 and 1-3 screened dipole-dipole interactions, as proposed by Thole.70 Lennard-Jones (LJ) interactions were switched off between 10 Å and 12 Å. Nonbonded pair lists were maintained up to 14 Å and updated heuristically. Isotropic long-range corrections to the LJ terms were applied.71 All bonds involving hydrogens were constrained using SHAKE72 for additive simulations and using the SHAKE/Roll and RATTLE/Roll procedures for Drude simulations.67 Coordinates were saved every 0.2 ps for bulk property calculations and RDF computations.
Additional solution properties were calculated as follows. Solution density () is the total mass divided by average volume of the simulation box ⟨V⟩:
| (14) |
The excess coordination number Nij was computed by
| (15) |
where ρj is the component density and Gij is the Kirkwood-Buff integral. The molar enthalpy of mixing ΔHm was calculated from the potential energy via73
| (16) |
where Um is the potential energy of the mixture, Uc and Uw the potential energies of the pure cosolvent and water, respectively. The self-diffusion constant (D0) was evaluated from the mean squared displacement of the center-of-mass (COM) of all molecules under the Stokes-Einstein relation,74 with a system size correction as follows:75
| (17) |
| (18) |
where η is shear viscosity using Green-Kubo relation76, 77 from a separate NVT simulation, L is the box length of the cubic simulation box. The dielectric constant was calculated from the dipole moment fluctuations using the following equation:78, 79
| (19) |
where M is the total dipole moment of the simulation box. The high-frequency optical dielectric constant, ɛ∞, was estimated from the Clausius-Mossotti equation80 for the Drude model. For the additive simulations ɛ∞ was set to 1 since no electronic degrees of freedom were explicitly modeled. The isothermal compressibility could be evaluated from KBIs using Eq. 4 in theory but it is typically statistically unreliable.81 Instead it was calculated from30, 82
| (20) |
where ⟨V⟩ is the ensemble average of volume and ⟨V2⟩ is the volume fluctuations. Errors were estimated by using five block averages.
RESULTS AND DISCUSSION
Drude parameter optimization and validation
The Drude force field parameters of NMA represent an update of the previously published parameters32 and were optimized following a modified version of the protocol presented in that work. This involves an iterative approach in which changes in any parameter are followed by re-evaluation of the reproduction of all target data and additional optimization of parameters performed as required. Results presented below are for the final parameter set. Internal parameters, including bond length, valence angle, dihedral angle, and improper dihedral terms, in the updated model represent a compromise of those for polypeptides, for example, the alanine dipeptide, as the presently studied compounds share common atom types and chemical groups with the protein backbone. Target data included gas-phase geometries from electron diffraction,83, 84, 85 surveys of the Cambridge Crystallographic Data bank86 and the Protein Data Bank87 and from quantum mechanical optimized structures of the model compounds. Force constants were optimized against QM vibrational spectra and potential energy surfaces for rotation around selected dihedrals. Presented in Table 2 are the Drude optimized geometries along with the different target data. The optimized values for the three models generally fall in the range of the various target data. During the fitting certain terms common to the model compounds as well as polypeptides (e.g., the bond lengths in the peptide bond (C5-N7 and C5-O6) were biased towards the crystallographic survey data as they will ultimately be used in proteins. We note that the use of individual atom types for the different compounds would lead to better agreement with the target data for the specific compounds, but this was not performed in order to obtain a more general force field. QM vibrational frequencies and assignments, calculated using the Molvib module in CHARMM were used to optimize the force constants along with QM dihedral potential energy scans (PES). Comparison of the vibrational data for the QM and Drude models shows the FF to satisfactorily reproduce the general pattern of the frequencies and the largest normal mode contributing to the frequencies (see Table S1 and S2 in the supplementary material88 for details). The NMA O6=C5-N7-C9 PES for the Drude and QM levels of theory shows the overall agreement to be satisfactory (see Figures S1 in the supplementary material88 for details). The quality of the level agreement for these energy surfaces along with the level of agreement for the lowest frequency modes indicates that the empirical model will correctly treat out-of-plane distortions of the ring systems that occur in MD simulations.
Table 2.
Geometric data on NMA and ACEM. See Fig. 1 for the atom numbering.
| Drude | Gas1 | Crystal2 | Crystal survey | MP2/6–31G* | |
|---|---|---|---|---|---|
| NMA | |||||
| C4-C5 | 1.545 | 1.520 | 1.515 | 1.529 ± 0.041 | 1.514 |
| C5-N7 | 1.328 | 1.386 | 1.326 | 1.300 ± 0.032 | 1.365 |
| C5=O6 | 1.228 | 1.224 | 1.246 | 1.248 ± 0.026 | 1.232 |
| N7-H8 | 1.019 | NA | 1.026 | 0.945 ± 0.095 | 1.010 |
| N7-C9 | 1.448 | 1.469 | 1.455 | 1.432 ± 0.039 | 1.448 |
| C4-C5-N7 | 114.0 | 114.1 | 116.3 | 116.1 ± 3.2 | 115.0 |
| C5-N7-H8 | 117.3 | NA | 118.8 | 118.9 ± 3.2 | 118.8 |
| C5-N7-C9 | 122.7 | 119.6 | 121.3 | 123.1 ± 3.8 | 122.1 |
| O6=C5-C4 | 118.0 | 124.13 | 121.9 | 120.6 ± 1.9 | 122.0 |
| ACEM | |||||
| C4-C5 | 1.528 | 1.519 | 1.503 | 1.503 ± 0.006 | 1.515 |
| C5-N7 | 1.318 | 1.380 | 1.325 | 1.323 ± 0.006 | 1.375 |
| C5=O6 | 1.220 | 1.220 | 1.243 | 1.247 ± 0.004 | 1.228 |
| N7-H8 | 1.015 | 1.022 | 0.862 | 0.891 ± 0.067 | 1.012 |
| N7-H9 | 1.018 | 1.022 | 0.946 | 0.911 ± 0.083 | 1.010 |
| C4-C5-N7 | 116.5 | 115.1 | 117.2 | 117.5 ± 0.7 | 114.6 |
| C5-N7-Hc | 122.0 | NA4 | 116.3 | 121.2 ± 4.0 | 116.2 |
| C5-N7-Ht | 119.9 | NA | 117.7 | 119.8 ± 3.0 | 120.6 |
| O6=C5-C4 | 120.0 | 123.0 | 120.6 | 120.6 ± 0.4 | 123.3 |
Electrostatic parameters, including the partial atomic charges, atomic polarizabilities, and the atom-atom Thole scale factors, were developed for each model compound following the methodology described before.23 Initial values of the partial atomic charges were either taken from the C22 additive all-atom force field46 or, in the case of ACEM, the initial charges were from NMA. Starting values of the polarizabilities were adjusted Miller's atomic hybrid polarizabilities (ahp) values,23, 89, 90 and the Thole factors were set to 1.3, the value originally determined for benzene. The fitted electrostatic models yielded dipole moments in good agreement with the reference values as shown in Table 3.
Table 3.
Molecular dipole moments (μ) and polarizabilities (αi) of NMA and ACEM. is the trace of the polarizability tensor. Δ1α and Δ2α are invariant measurements of polarizability. Dipole moments are in Debye and polarizabilities in Å3. QM values were determined at the MP2/aug-cc-pVDZ level.
| μ | α1 | α2 | α3 | Δ1α | Δ2α | ||
|---|---|---|---|---|---|---|---|
| NMA | |||||||
| Drude | 3.72 | 7.16 | 6.46 | 4.98 | 6.20 | 1.44 | 1.93 |
| QM | 3.91 | 9.45 | 7.89 | 5.96 | 7.77 | 2.53 | 3.03 |
| ACEM | |||||||
| Drude | 3.77 | 6.01 | 5.53 | 3.81 | 5.12 | 1.34 | 2.00 |
| QM | 3.89 | 6.45 | 6.78 | 4.52 | 5.92 | 0.80 | 2.10 |
Polarizabilities of all the model compounds were compared to QM values. Comparisons included the mean polarizability, given by the trace of the polarization tensor,
| (21) |
in which a1 = azz ⩽ a2 ⩽ a3 are the eigenvalues or principal values of the polarization tensor. The polarizability anisotropy, which is the difference between the in-plane and out-of-plane components, as given by Eq. 22 was also calculated and compared to QM data as was the Kerr effect as calculated using Eq. 23,
| (22) |
| (23) |
Comparison of QM reference values and those from the final Drude model is presented in Table 3. Overall, the discrepancies between the polarizable Drude model and the QM data are small, showing the ability of the polarizable model to reproduce subtle aspects of the electrostatic behavior of the molecules under study. The systematically lower values for NMA and ACEM are consistent with the scaling of the polarizabilities in the Drude force field as previously discussed.17, 28
LJ parameters were optimized targeting pure solvent properties and QM interactions with rare gases, as previously described.15 Heats of vaporization and molecular volumes for the final Drude models are shown in Table 4. For ACEM and NMA the Drude model reproduce the experimental target data within 2%, the targeted limit. The use of different temperatures for the molecular volumes and heats of vaporization calculations on NMA was performed as previously discussed.30 Free energies of hydration were also computed for the model compounds (Table 4). The free energies of hydration for NMA and ACEM are comparable to the experimental measurements. The final set of parameters (see Table S3–S6 in the supplementary material88 for details) was then subjected to the Kirkwood-Buff analysis without further adjustments.
Table 4.
Comparison of experimental and calculated condensed phase properties for NMA and ACEM. Molecular volumes, Vm, in Å3 and heats of vaporization, ΔHvap, and free energies of hydration, ΔGhyd, in kcal/mol. Experimental data: molecular volumes from Ref. 105, heat of vaporization from Refs. 106 and 107, free energy of hydration from Refs. 44 and 108.
| Drude |
Experimental data |
||||||
|---|---|---|---|---|---|---|---|
| T (K) | Vm | ΔHvap | ΔGhyd | Vm | ΔHvap | ΔGhyd | |
| NMA | 298 | NA | NA | −9.9 ± 0.1 | NA | NA | −10.1 |
| 373 | 133.7 ± 0.2 | NA | NA | 135.9 | NA | NA | |
| 410 | NA | 13.3 ± 0.1 | NA | NA | 13.0 | NA | |
| ACEM | 298 | NA | NA | −10.1 ± 0.1 | NA | NA | −9.7 |
| 358 | 94.9 ± 0.2 | 15.6 ± 0.5 | NA | 98.2 | 14.8 | NA | |
Kirkwood-Buff solution analysis
A summary of the simulations carried out is presented in Table 1. They cover the entire concentration range for NMA, as this molecule is the main focus of the present study. ACEM was studied at two low concentrations because it has relatively low solubility in water at the temperature studied. All mixtures were simulated for 10 ns to ensure adequate precision in the data.
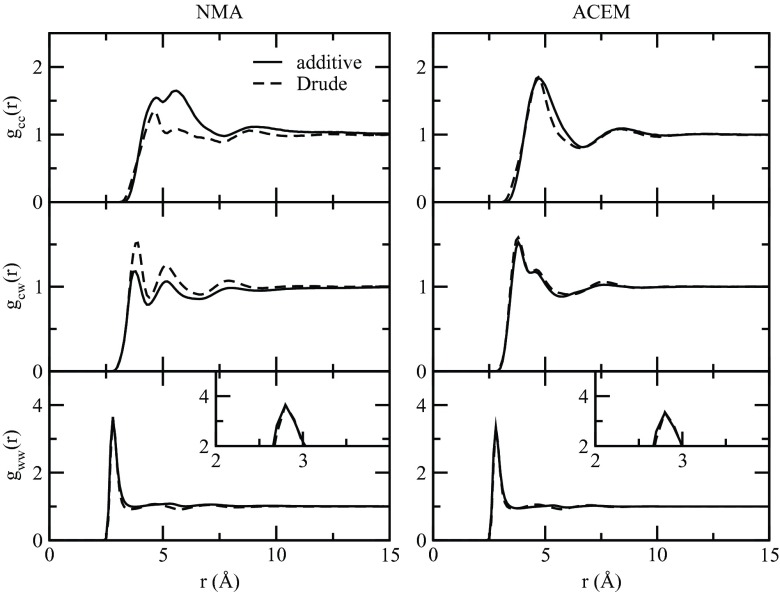
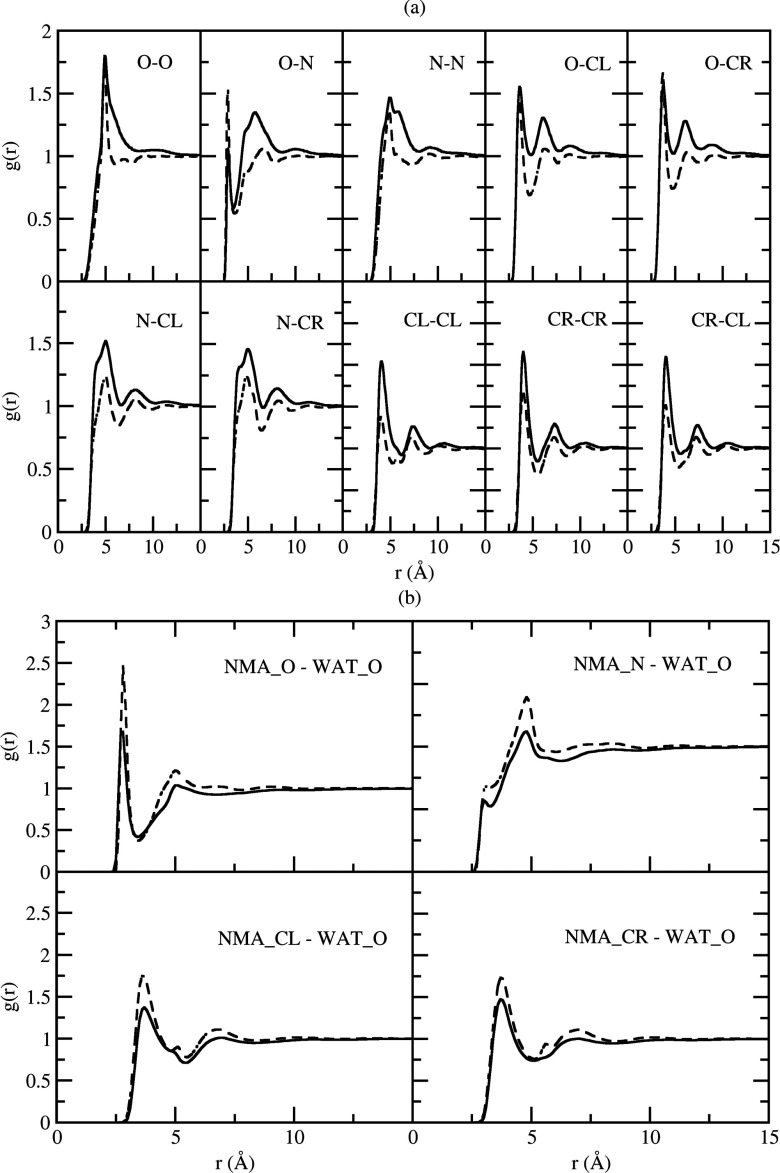
Examples of the center of mass based RDFs calculated for all three amide mixtures at xc = 0.1 are displayed in Fig. 2. As indicated by the gcc plot, solute-solute interactions modeled by the CHARMM additive and Drude force fields in the NMA solutions were largely different in terms of the shape and the maxima/minima for the first shell. The additive FF predicted stronger solute-solute interactions and displayed two consecutive local maxima with the second slightly higher. In contrast, the Drude FF showed a smaller maximum at about the same position as the first additive peak with the second maximum much smaller than the first as well as its additive counterpart. Both FFs displayed a visible third shell, with the Drude FF displaying a fourth one. For solute-solvent interactions in the NMA solutions, gcw, the Drude FFs had a higher first peak as compared to the additive. This is to be expected given the anticorrelated nature of the gcc and gcw RDFs due to the limited space surrounding each solute molecule. Such differences almost disappeared for the ACEM solutions. The degree of both solute-solute and solute-solvent interactions increased in ACEM versus NMA for the Drude models. This is due to the increase in the number of potential hydrogen bonding sites in the former. The additive model gww RDFs show a small decrease in the first peak in ACEM versus NMA, again consistent with their hydrogen bonding capacity. The effect was present in both the additive and polarizable FFs, with the effect slightly larger in the Drude model. The overall feature of the RDFs at xc = 0.1 in Fig. 2 is reminiscent of those published by Smith and co-workers, based on the KBFF which they developed by targeting experimental KB data.91 Small variations existing in these RDFs have significant implications on the performance of the KB analysis, as shown below.
Figure 2.
Center of mass based RDFs for xc = 0.1 mixtures of NMA and ACEM. The solid line is for the CHARMM additive force field and the dashed line is for Drude polarizable force field.
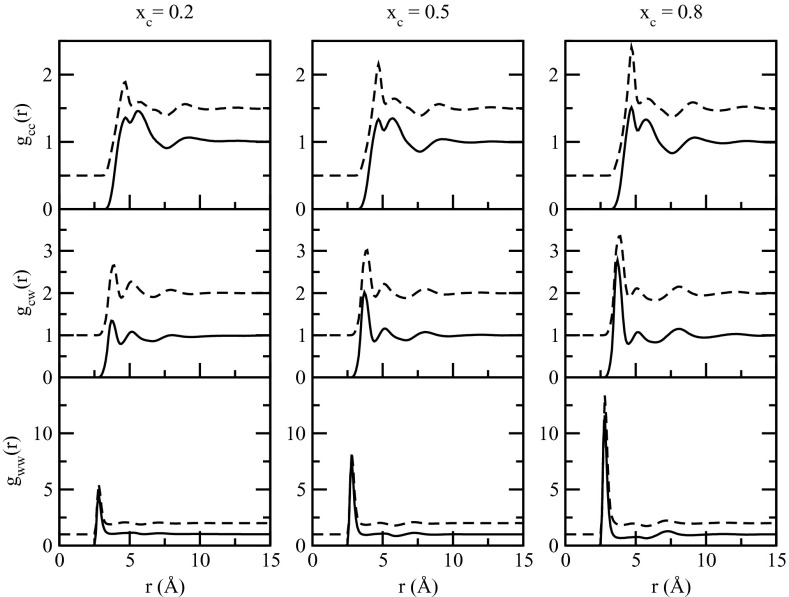
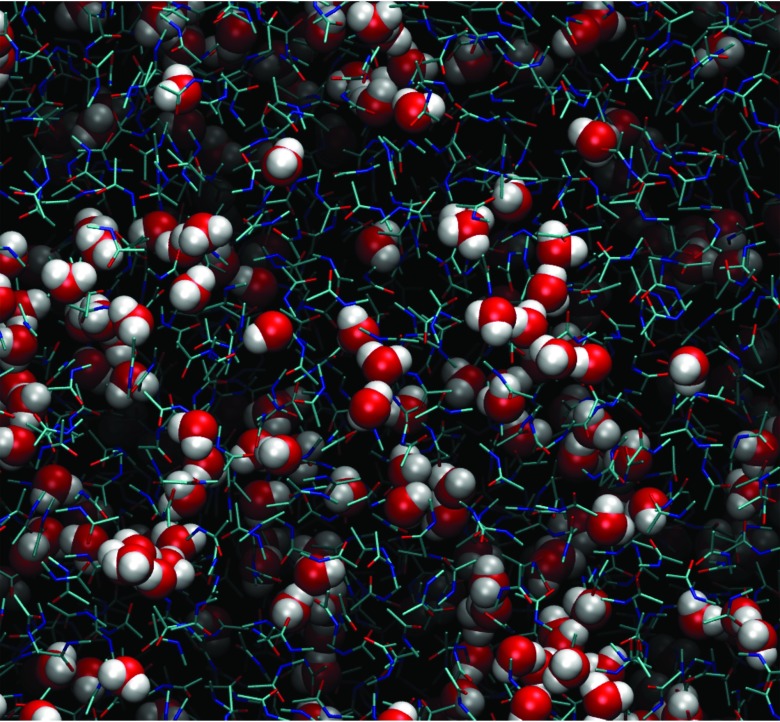
We further examined RDFs of NMA solutions as a function of concentration. They are presented in Fig. 3 for xc = 0.2, 0.5, and 0.8. Similar to those at xc = 0.1, the RDFs displayed very similar positions for maxima and minima for both FFs. Differences between the FFs are still evident for the solute-solute RDFs, though the differences are less at higher concentrations. The solute-solvent and solvent-solvent RDFs are similar for the two models. With respect to concentration effects the most prominent feature was that the first shells in all RDFs increased as a function of mole fraction of the solute, indicative of enhanced solute-solute, solute-solvent, and solvent-solvent interactions. This has been observed in other types of aqueous solutions as well.81 The phenomenon is essentially a natural consequence of crowding as well as the normalization of the RDFs to unity at infinite separation. As the number of solute molecules increases the number of solute molecules with which to interact increases leading to increases in gcc. In the case of solute-solvent and solvent-solvent interactions, the increased crowding by the solute molecules limits that space accessible to the solvent so they interact to a relatively higher extent with solute molecules or with other solvent molecules when normalized to unity at long distances. This effect leads to the large first peak in gww RDFs. An image of the NMA xc = 0.8 solution is shown in Fig. 4, where it can be seen that the water molecules hydrogen bond to adjacent water molecules, leading to the large first peak in Fig. 3. In some cases chains of multiple water molecules are present, corresponding to the small second and third peaks in the gww RDFs.
Figure 3.
Center of mass based RDFs for NMA at xc = 0.2, 0.5, and 0.8, respectively. The solid line is for the CHARMM additive force field and the dashed line is for Drude polarizable force field. Drude RDFs were offset for clarity.
Figure 4.
Snapshot of the NMA-water mixture at xc = 0.8. Water is in VDW representation and NMA is in atom-colored licorice representation. Image is generated using VMD.102
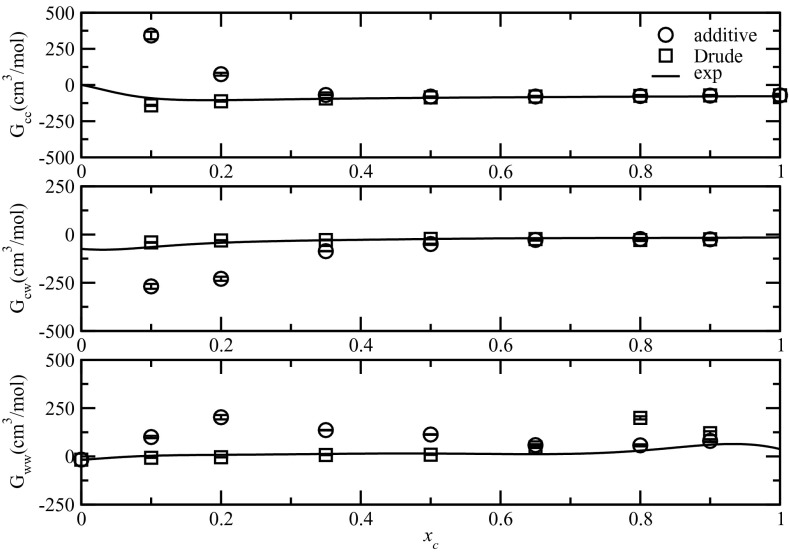
The simulated KBIs from the RDFs of NMA shown above are displayed in Fig. 5 along with experimental KBIs of aqueous NMA solutions computed from experimental data61, 62 from inverse KB procedure.48, 49 Simulated KBIs were computed via Eq. 1 by integrating the RDFs via the trapezoidal rule and taking the average between 10 and 15 Å.81 Previous KB studies used averages from 9.5 to 12 Å81 or from 15 to 20 Å.81, 92 As shown in Fig. 5, KBIs computed from Drude-based RDFs compared very favorably to the experimental data at all concentrations. The largest deviation was a small underestimation for Gcc at low concentrations. In contrast, while the additive FF performed satisfactorily at high concentrations (xc ≥ 0.5) it showed large deviations at low concentrations. There were significant overestimations for Gcc and Gww as well as underestimations for Gcw. Considering that the positions of maxima and minima were almost the same in RDFs for both additive and Drude FFs, the deviations must arise from the differences in the magnitudes of those RDFs. The additive RDFs overestimated the solute-solute, solvent-solvent interactions and underestimated the solute-solvent interactions, leading to overestimations in Gcc and Gww and underestimations in Gcw. This may be an outcome of the requirement that the charges in the additive model be enhanced to mimic the experimental pure solvent properties during FF development, leading to a small imbalance with respect to the interactions between the solutes and water.46 The Drude FF, despite not being explicitly optimized by targeting KB data, achieved a better balance among those interactions hence better agreement with experimental KBIs.
Figure 5.
KBIs as a function of mole fraction for the NMA solutions.
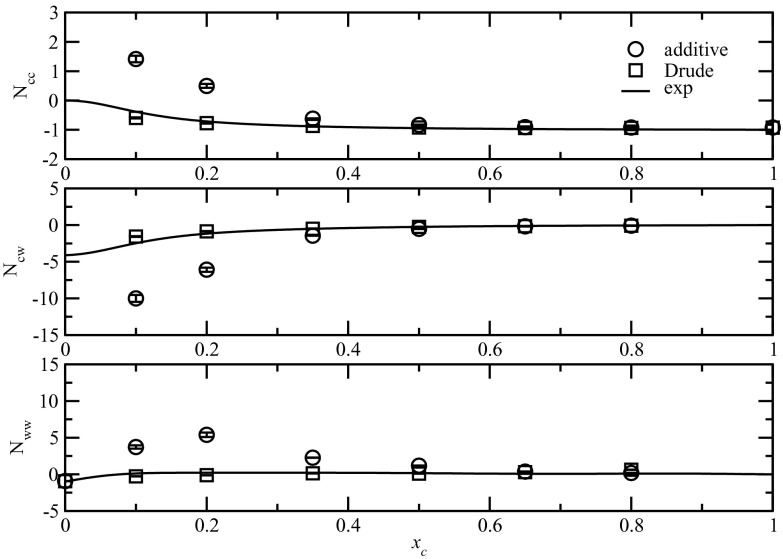
Excess coordination numbers obtained as Nij = ρjGij from both simulations of NMA solutions and the experimental data are shown in Fig. 6. The experimental data were nicely reproduced by the Drude model at all but the low concentrations (xc = 0.1 and 0.2). At these concentrations, Ncc and Nww were slightly overestimated while Ncw was slightly underestimated. In sharp contrast, large deviations were observed for the additive model at low concentrations up to xc = 0.35. Ncc and Nww were significantly overestimated while Ncw were significantly underestimated, consistent with the results that Kang et al. reported using the additive NMA model of the CHARMM force field.91 It indicates a large degree of self-aggregation of NMA and water molecules, consistent with the RDFs and KBIs in Figs. 25. Given that water yields the proper coordination number in the pure liquid for the additive FF, this further suggests that the solute-solute interactions are too strong and solute-solvent interactions are too weak. With the introduction of polarization in the Drude FF, a more realistic balance of those interactions is achieved. It is worth noting that correct excess coordination numbers can be obtained in the framework of additive FF, as reported by Smith and co-workers by specifically targeting KB experimental data.91 Excess coordination numbers for ACEM modeled by both the additive and Drude FFs are presented in Figures S2 in the supplementary material.88 The trend was the same as for aqueous NMA, though the deviations were much less for the ACEM solutions.
Figure 6.
Excess coordination numbers as a function of mole fraction for the NMA solutions.
Solution density and partial molar volumes as a function of concentrations for aqueous NMA mixtures are presented in Fig. 7. The additive model consistently underestimated the solution density, while there was a slight overestimation by the Drude model in the range from xc = 0.3 to 0.8. For the pure NMA solutions, the additive FF actually yields more accurate solution densities than the Drude FF. While this is a direct outcome of the Drude model as it was parametrized, in part, targeting the pure NMA data, the improved reproduction of the densities of the mixed solutions further indicates the ability of the polarizable model to properly balance the solute-solute, solute-solvent, and solvent-solvent interactions.
Figure 7.
Density (g/cm3) and partial molar volumes, and , (cm3/mol) as a function of mole fraction for the NMA solutions.
The overall trend of the partial molar volumes, computed via Eq. 2, 3, were reasonably well reproduced by both FFs. At high concentrations (xc ≥ 0.5), the additive FF reproduced even slightly better PMV for NMA, , than the Drude FF. At low concentrations, the additive FF overestimated the PMV while the Drude FF showed a somewhat small underestimation, which was present throughout the range of xc values. For the water PMV, , the Drude model was generally in good agreement with the experimental data. In contrast, noticeable deviations occurred at high concentrations (xc ≥ 0.35) for the additive FF. Overall, the Drude FF performed better than the additive model in reproducing the PMVs of the studied NMA solutions. It is worth noting that the fact that PMVs computed from simulated KBIs are close to the experimental data as shown in Fig. 7 is further validation that the proper KBIs were obtained from averaging the integrals in Eq. 1 over the range of 10 and 15 Å.
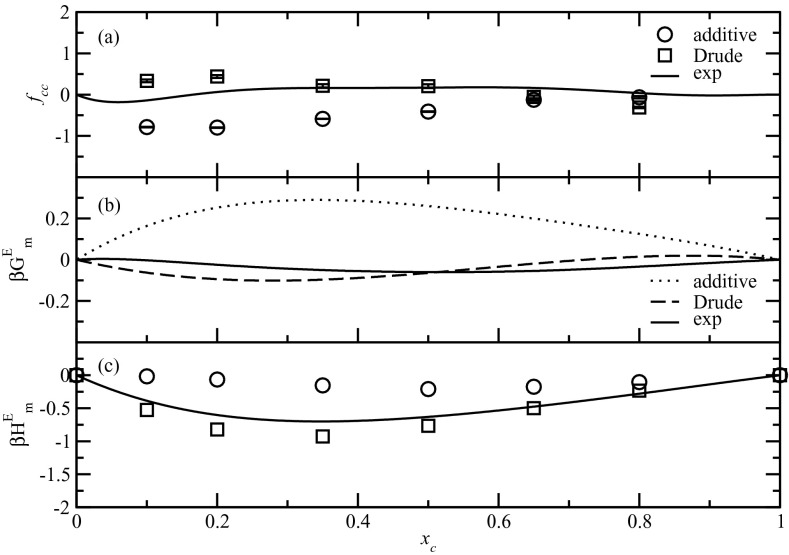
Activity derivatives, fcc, were obtained from simulated KBIs using Eq. 12 as shown in Fig. 8a. Then by fitting fcc with Redlich-Kister equation,93 one can obtain the excess Gibbs energy. The results are displayed in Fig. 8b. For these properties, both the additive and Drude FFs showed some deviations from experimental data, with the additive FF being larger. The deviations were small at high concentrations but started to increase in opposite directions as the concentrations become lower. In those regions, the additive FF underestimated the activity derivative while it overestimated the excess Gibbs energy. The Drude FF also showed deviations but to a much lesser degree. The Drude FF showed much better agreement with the experimental data for the mixing enthalpies (Fig. 8c), while the additive FF predicted almost no enthalpy change during the mixing, although a slight overestimation still exists at low concentrations. The low enthalpy of mixing could be a reason for the enhanced self-aggregation in the additive model, as previously shown in aqueous sodium chloride studies by Smith and co-workers.94 The excess enthalpy with the Drude model was in satisfactory agreement with the experiment results. The fact that Drude model did not reproduce the excess Gibbs energy indicates that problems are possibly present with the excess entropy of mixing, which will be taken into account in future force field optimizations.
Figure 8.
Activity derivative ( fcc), excess Gibbs energy, and molar enthalpy of mixing for aqueous the NMA solutions. Experimental data for mixing enthalpy is at 308 K.91
A summary of selected bulk properties of the pure liquids of NMA and water are presented in Table 5. Results based on the KBFF models developed by Kang et al., who used the SPC/E water model, were also included for comparison.91 The diffusion constants for pure water by the Drude and SPC/E water models were close to the experimental value while additive TIP3P water model overestimated the values by a factor of two, as previously reported.95 All models overestimate the diffusion constant for NMA, with the Drude FF producing the smallest difference. It should be noted that the system size correction for diffusion constant calculations suggested by Yeh et al.75 was adopted in the present study. The correction was relatively small (a few percent) because relatively large simulation boxes were used as in the present study as well as in most other KB studies. More accurate pure solvent dielectric constants have been shown to be one of the strengths of the polarizable force fields.22, 28, 79 For pure water, both Drude and SPC/E predicted the correct dielectric constant compared to the experiment, while TIP3P model overestimated it by a large margin, due to the intrinsic limitation of this model.22 NMA represents a difficult case for dielectric constant prediction. The results calculated by all three models were only one third of the experimental value, though the Drude model gave the largest value, consistent with the explicit inclusion of polarizability. We note that the value of the dielectric constant is significantly smaller than a previously reported value for the Drude model.32 This is due to changes in the Lennard-Jones parameters as part of the present study, performed to obtain a better balance of quantum mechanical interactions with water, hydrogen bonding between peptide groups on model peptides as well as pure solvent and free energies of aqueous solvation properties. These differences illustrate the subtle and complex dependence of the dielectric constant on factors other than the molecular charge distribution and polarizability. For the isothermal compressibility, results for the Drude FF were also superior to those from the additive FF and well as the KBFF model.
Table 5.
Properties of pure liquids. Experimental data: densities from Refs. 61, 109, and 110, diffusion constant from Refs. 111 and 112, dielectric constant from Refs. 113 and 114, isothermal compressibilities from Refs. 114 and 115.
| Density | Diffusion constant | Dielectric constant | Isothermal compressibility | ||
|---|---|---|---|---|---|
| ρ g/cm3 | D × 10−5 cm2 s−1 | ɛ | κт × 10−5 atm−1 | ||
| NMA | Additive | 0.940 | 0.97 ± 0.04 | 45 ± 5 | 5.5 ± 0.2 |
| Drude | 0.954 | 0.66 ± 0.05 | 67 ± 4 | 6.7 ± 0.1 | |
| KBFF | 0.935 | 0.70 | 52 | 7.5 | |
| Expt. | 0.942 | 0.46 | 191, 166 | 6.3 | |
| Water | TIP3P | 0.994 | 7.54 ± 0.05 | 93 ± 6 | 5.7 ± 0.2 |
| SWM4 | 0.986 | 3.81 ± 0.06 | 72 ± 2 | 4.5 ± 0.1 | |
| SPC/E | 0.987 | 3.5 | 69 | 4.6 | |
| Expt. | 0.992 | 3.7 | 70 | 4.4 |
The improved performance by the Drude FF over the additive FF in the KB analysis indicates that the polarizable model yields a more accurate description of the molecular interactions occurring in the NMA solutions, especially at low concentrations where significant differences occur (Figs. 56). To better understand the differences at a molecular level atom-based RDFs were computed between NMA and NMA and between NMA and water. They are shown in Figs. 9a, 9b, respectively. It is evident that all the atom-atom RDFs contribute to the differences in the sharper first peaks in the center-of-mass based NMA-NMA Drude RDFs (Figs. 23) while the smaller second peak mainly comes from N-O RDF though most of the other RDFs also show a smaller peak in the Drude FF. The dominance of the first peak in the Drude indicates that the interactions with NMA are better defined at low concentrations with contributions from all atoms in the molecule. For NMA-water interactions, all Drude atom-based RDFs showed a more pronounced first peak, contributing to the larger first peak in the center-of-mass based NMA-water RDFs (Figs. 23). Thus, in dilute solutions the Drude model yields more structured NMA-NMA interactions as well as NMA-water interactions as compared to the additive model.
Figure 9.
Atom-based RDFs from NMA to NMA (a) and RDFs from NMA to water (b) at xc = 0.1. The atom names in NMA are (CL)C=ONH(CR) where CL is the methyl carbon connected to the carbonyl carbon and CR is the methyl carbon connected to the nitrogen atom. Additive FF (solid line) and Drude FF (dashed line) results are shown.
CONCLUSIONS
Kirkwood-Buff analysis was performed on two aqueous amide solutions modeled by both the CHARMM additive and Drude polarizable force fields. It was shown that the Drude model reproduced experimental results adequately well at all concentrations while the additive model tended to have significant deviations at low concentrations in terms of KBIs, excess coordination numbers, solution densities, partial molar values, molar enthalpy of mixing. These results indicate a better balance of the solute-solute, solvent-solute, and solvent-solvent interactions in the Drude FF at low solute concentrations. However, both models did not optimally reproduce activity derivative and the excess Gibbs energy. Given that the excess enthalpy with the Drude model was in satisfactory agreement with the experiment results, this indicates that problems are present with the excess entropy of mixing. This suggests an area that could be improved in future force field optimization efforts, though addressing this issue will be a considerable challenge.
In the framework of nonpolarizable force fields, Smith and co-workers developed KBFF force field parameters for amides by specifically targeting experimental data analyzed by KB theory.81, 91, 94, 96, 97, 98, 99, 100, 101 The results showed that without introducing polarization good agreement with experimental KB data can be obtained. The present study demonstrated that the polarizable Drude force field, which was not developed by targeting experimental KB data, can also achieve almost the same degree of accuracy compared to KBFF, except for activity derivative and excess Gibbs free energy, which will be the focus in our future force field optimization efforts.
The molecular interactions in aqueous amide solutions revealed by the RDFs were similar for the water-water interactions, but largely different for the solute-solute and solute-solvent interactions. Analysis of selected atom-atom RDFs for NMA solutions suggested that this is associated with more structured solute-solute and solute-solvent interactions at low solute concentrations. The improved agreement with experiment as judged by the KB analysis in the Drude model indicated that these more structured interactions are more representative of the experimental regime. The improvements may be attributed to the introduction of polarizability into both amides and water in the Drude model and it showed that polarizability plays an important role in the behavior of liquid mixtures.
ACKNOWLEDGMENTS
Financial support from the NIH (GM051501 and GM072558) is acknowledged.
References
- Hill T. L., J. Am. Chem. Soc. 79, 4885 (1957). 10.1021/ja01575a016 [DOI] [Google Scholar]
- Prigogine I., The Molecular Theory of Solutions (North-Holland Publishing Company, New York, 1957). [Google Scholar]
- Smith P. E., J. Phys. Chem. B 103, 525 (1999). 10.1021/jp983303c [DOI] [Google Scholar]
- Chitra R. and Smith P. E., J. Phys. Chem. B 104, 5854 (2000). 10.1021/jp000558t [DOI] [Google Scholar]
- Chitra R. and Smith P. E., J. Chem. Phys. 115, 5521 (2001). 10.1063/1.1396676 [DOI] [Google Scholar]
- Perera A. and Sokolic F., J. Chem. Phys. 121, 11272 (2004). 10.1063/1.1817970 [DOI] [PubMed] [Google Scholar]
- Lopez-Rendon R., Mora M. A., Alejandre J., and Tuckerman M. E., J. Phys. Chem. B 110, 14652 (2006). 10.1021/jp0619540 [DOI] [PubMed] [Google Scholar]
- Zielkiewicz J. and Mazerski J., J. Phys. Chem. B 106, 861 (2002). 10.1021/jp012833x [DOI] [Google Scholar]
- Kokubo H., Rosgen J., Bolen D. W., and Pettitt B. M., Biophys. J. 93, 3392 (2007). 10.1529/biophysj.107.114181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo H. and Pettitt B. M., J. Phys. Chem. B 111, 5233 (2007). 10.1021/jp067659x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo H., Roesgen J., Bolen D. W., and Pettitt B. M., Biophys. J. 93, 3392 (2007). 10.1529/biophysj.107.114181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J. W. G. and Mayer J. E., J. Chem. Phys. 13, 276 (1945). 10.1063/1.1724036 [DOI] [Google Scholar]
- Kirkwood J. G. and Buff F. P., J. Chem. Phys. 19, 774 (1951). 10.1063/1.1748352 [DOI] [Google Scholar]
- Smith P. E., Matteoli E., and O’Connell J. P., Fluctuation Theory of Solutions: Applications in Chemistry, Chemical Engineering, and Biophysics (CRC Press, Boca Raton, FL, 2013). [Google Scholar]
- A. D.MackerellJr., J. Comput. Chem. 25, 1584 (2004). 10.1002/jcc.20082 [DOI] [PubMed] [Google Scholar]
- Chitra R. and Smith P. E., J. Chem. Phys. 114, 426 (2001). 10.1063/1.1330577 [DOI] [Google Scholar]
- Lopes P. E. M., Roux B., and A. D.MacKerellJr., Theor. Chem. Acc. 124, 11 (2009). 10.1007/s00214-009-0617-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T. A. and Damm W., Curr. Opin. Struct. Biol. 11, 236 (2001). 10.1016/S0959-440X(00)00196-2 [DOI] [PubMed] [Google Scholar]
- Onsager L., J. Am. Chem. Soc. 58, 1486 (1936). 10.1021/ja01299a050 [DOI] [Google Scholar]
- Kirkwood J. G., J. Chem. Phys. 7, 911 (1939). 10.1063/1.1750343 [DOI] [Google Scholar]
- Nalewajski R. F., J. Phys. Chem. 89, 2831 (1985). 10.1021/j100259a025 [DOI] [Google Scholar]
- Lamoureux G., A. D.MacKerellJr., and Roux B., J. Chem. Phys. 119, 5185 (2003). 10.1063/1.1598191 [DOI] [Google Scholar]
- Anisimov V. M., Lamoureux G., Vorobyov I. V., Huang N., Roux B., and A. D.MackerellJr., J. Chem. Theory Comput. 1, 153 (2005). 10.1021/ct049930p [DOI] [PubMed] [Google Scholar]
- Harder E., Anisimov V. M., Vorobyov I. V., Lopes P. E. M., Noskov S. Y., A. D.MacKerellJr., and Roux B., J. Chem. Theory Comput. 2, 1587 (2006). 10.1021/ct600180x [DOI] [PubMed] [Google Scholar]
- Zhu X., Lopes P. E. M., and A. D.MacKerellJr., WIREs Comput. Mol. Sci. 2, 167 (2012). 10.1002/wcms.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drude P., The Theory of Optics, translated from the German by Mann C. R. and Millikan R. A. (Longmans, Green and Co., London, 1901). [Google Scholar]
- Lamoureux G. and Roux B., J. Chem. Phys. 119, 3025 (2003). 10.1063/1.1589749 [DOI] [Google Scholar]
- Lamoureux G., Harder E., Vorobyov I. V., Roux B., and A. D.MacKerellJr., Chem. Phys. Lett. 418, 245 (2006). 10.1016/j.cplett.2005.10.135 [DOI] [Google Scholar]
- Vorobyov I. V., Anisimov V. M., and A. D.MacKerellJr., J. Phys. Chem. B 109, 18988 (2005). 10.1021/jp053182y [DOI] [PubMed] [Google Scholar]
- Anisimov V. M., Vorobyov I. V., Roux B., and A. D.MacKerellJr., J. Chem. Theory Comput. 3, 1927 (2007). 10.1021/ct700100a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobyov I., Anisimov V. M., Greene S., Venable R. M., Moser A., Pastor R. W., and A. D.MacKerellJr., J. Chem. Theory Comput. 3, 1120 (2007). 10.1021/ct600350s [DOI] [PubMed] [Google Scholar]
- Harder E., Anisimov V. M., Whitfield T. W., A. D.MacKerellJr., and Roux B., J. Phys. Chem. B 112, 3509 (2008). 10.1021/jp709729d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes P. E. M., Lamoureux G., Roux B., and A. D.MacKerellJr., J. Phys. Chem. B 111, 2873 (2007). 10.1021/jp0663614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes P. E. M., Lamoureux G., and A. D.MacKerellJr., J. Comput. Chem. 30, 1821 (2009). 10.1002/jcc.21183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. and A. D.MacKerellJr., J. Comput. Chem. 31, 2330 (2010). 10.1002/jcc.21527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riahi S. and Rowley C. N., J. Phys. Chem. B 117, 5222 (2013). 10.1021/jp401847s [DOI] [PubMed] [Google Scholar]
- Yu H. B., Whitfield T. W., Harder E., Lamoureux G., Vorobyov I., Anisimov V. M., A. D.MacKerellJr., and Roux B., J. Chem. Theory Comput. 6, 774 (2010). 10.1021/ct900576a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Jiang W., Yu H. B., A. D.MacKerellJr., and Roux B., Faraday Discuss. 160, 135 (2013). 10.1039/c2fd20068f [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lopes P. E. M., and A. D.MacKerellJr., Biopolymers 99, 724 (2013). 10.1002/bip.22286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J. W. and Kollman P. A., J. Phys. Chem. 99, 6208 (1995). 10.1021/j100016a067 [DOI] [Google Scholar]
- Rizzo R. C. and Jorgensen W. L., J. Am. Chem. Soc. 121, 4827 (1999). 10.1021/ja984106u [DOI] [Google Scholar]
- Rick S. W. and Berne B. J., J. Am. Chem. Soc. 118, 672 (1996). 10.1021/ja952535b [DOI] [Google Scholar]
- Patel S. and Brooks C. L., J. Comput. Chem. 25, 1 (2004). 10.1002/jcc.10355 [DOI] [PubMed] [Google Scholar]
- Ding Y. B., Bernardo D. N., Kroghjespersen K., and Levy R. M., J. Phys. Chem. 99, 11575 (1995). 10.1021/j100029a040 [DOI] [Google Scholar]
- Nutt D. R. and Smith J. C., J. Chem. Theory Comput. 3, 1550 (2007). 10.1021/ct700053u [DOI] [PubMed] [Google Scholar]
- A. D.MacKerellJr., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., and Karplus M., J. Phys. Chem. B 102, 3586 (1998). 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Ben-Naim A., Molecular Theory of Solutions (Oxford University Press, Oxford, 2006). [Google Scholar]
- Ben-Naim A., J. Chem. Phys. 67, 4884 (1977). 10.1063/1.434669 [DOI] [Google Scholar]
- Smith P. E., J. Chem. Phys. 129, 124509 (2008). 10.1063/1.2982171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitra R. and Smith P. E., J. Phys. Chem. B 105, 11513 (2001). 10.1021/jp012354y [DOI] [Google Scholar]
- Valiev M., Bylaska E. J., Govind N., Kowalski K., Straatsma T. P., Van Dam H. J. J., Wang D., Nieplocha J., Apra E., Windus T. L., and de Jong W. A., Comput. Phys. Commun. 181, 1477 (2010). 10.1016/j.cpc.2010.04.018 [DOI] [Google Scholar]
- Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Montgomery J. A., Vreven T., Kudin K. N., Burant J. C., Millam J. M., Iyengar S. S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G. A., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J. E., Hratchian H. P., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Ayala P. Y., Morokuma K., Voth G. A., Salvador P., Dannenberg J. J., Zakrzewski V. G., Dapprich S., Daniels A. D., Strain M. C., Farkas O., Malick D. K., Rabuck A. D., Raghavachari K., Foresman J. B., Ortiz J. V., Cui Q., Baboul A. G., Clifford S., Cioslowski J., Stefanov B. B., Liu G., Liashenko A., Piskorz P., Komaromi I., Martin R. L., Fox D. J., Keith T., Laham A., Peng C. Y., Nanayakkara A., Challacombe M., Gill P. M. W., Johnson B., Chen W., Wong M. W., Gonzalez C., and Pople J. A., Gaussian 03, Revision C.02 (2003).
- Baker C. M. and A. D.MacKerellJr., J. Mol. Mod. 16, 567 (2010). 10.1007/s00894-009-0572-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becke A. D., Phys. Rev. A 38, 3098 (1988). 10.1103/PhysRevA.38.3098 [DOI] [PubMed] [Google Scholar]
- Becke A. D., J. Chem. Phys. 98, 5648 (1993). 10.1063/1.464913 [DOI] [Google Scholar]
- Lee C., Yang W., and Parr R. G., J. Mol. Struct.: THEOCHEM 163, 305 (1988). 10.1016/0166-1280(88)80397-X [DOI] [Google Scholar]
- Dunning J. T. H., J. Chem. Phys. 90, 1007 (1989). 10.1063/1.456153 [DOI] [Google Scholar]
- Yin D. and A. D.MacKerellJr., J. Comput. Chem. 19, 334 (1998). [DOI] [Google Scholar]
- Brooks B. R., C. L.BrooksIII, A. D.MacKerellJr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., and Karplus M., J. Comput. Chem. 30, 1545 (2009). 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., and A. D.MacKerellJr., J. Comput. Chem. 31, 671 (2010). 10.1002/jcc.21367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielkiewicz J., J. Chem. Thermodyn. 31, 1597 (1999). 10.1006/jcht.1999.0559 [DOI] [Google Scholar]
- Zielkiewicz J., J. Chem. Thermodyn. 31, 819 (1999). 10.1006/jcht.1999.0502 [DOI] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L., J. Chem. Phys. 79, 926 (1983). 10.1063/1.445869 [DOI] [Google Scholar]
- Feller S. E., Zhang Y. H., Pastor R. W., and Brooks B. R., J. Chem. Phys. 103, 4613 (1995). 10.1063/1.470648 [DOI] [Google Scholar]
- Nosé S., Mol. Phys. 52, 255 (1984). 10.1080/00268978400101201 [DOI] [Google Scholar]
- Hoover W. G., Phys. Rev. A 31, 1695 (1985). 10.1103/PhysRevA.31.1695 [DOI] [PubMed] [Google Scholar]
- Martyna G. J., Tuckerman M. E., Tobias D. J., and Klein M. L., Mol. Phys. 87, 1117 (1996). 10.1080/00268979600100761 [DOI] [Google Scholar]
- Rick S. W., Stuart S. J., and Berne B. J., J. Chem. Phys. 101, 6141 (1994). 10.1063/1.468398 [DOI] [Google Scholar]
- Darden T., York D., and Pedersen L., J. Chem. Phys. 98, 10089 (1993). 10.1063/1.464397 [DOI] [Google Scholar]
- Thole B. T., Chem. Phys. 59, 341 (1981). 10.1016/0301-0104(81)85176-2 [DOI] [Google Scholar]
- Allen M. P. and Tildesley D. J., Computer Simulation of Liquids (Oxford University Press, 1989). [Google Scholar]
- Ryckaert J.-P., Ciccotti G., and Berendsen H. J. C., J. Comp. Phys. 23, 327 (1977). 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- Walser R., Mark A. E., van Gunsteren W. F., Lauterbach M., and Wipff G., J. Chem. Phys. 112, 10450 (2000). 10.1063/1.481680 [DOI] [Google Scholar]
- Einstein A., Ann. Phys. 322, 549 (1905). 10.1002/andp.19053220806 [DOI] [Google Scholar]
- Yeh I. C. and Hummer G., J. Phys. Chem. B 108, 15873 (2004). 10.1021/jp0477147 [DOI] [Google Scholar]
- Green M. S., J. Chem. Phys. 22, 398 (1954). 10.1063/1.1740082 [DOI] [Google Scholar]
- Kubo R., J. Phys. Soc. Jpn. 12, 570 (1957). 10.1143/JPSJ.12.570 [DOI] [Google Scholar]
- Gereben O. and Pusztai L., Chem. Phys. Lett. 507, 80 (2011). 10.1016/j.cplett.2011.02.064 [DOI] [PubMed] [Google Scholar]
- Yu W., Lopes P. E. M., Roux B., and A. D.MacKerellJr., J. Chem. Phys. 138, 034508 (2013). 10.1063/1.4774577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rysselberghe P. V., J. Phys. Chem. 36, 1152 (1932). 10.1021/j150334a007 [DOI] [Google Scholar]
- Weerasinghe S. and Smith P. E., J. Phys. Chem. B 109, 15080 (2005). 10.1021/jp051773i [DOI] [PubMed] [Google Scholar]
- Mulero A., Faundez C. A., Parra M. I., and Cuadros F., Thermochim. Acta 334, 1 (1999). 10.1016/S0040-6031(99)00132-X [DOI] [Google Scholar]
- Kitano M., Fukuyama T., and Kuchitsu K., Bull. Chem. Soc. Jpn. 46, 384 (1973). 10.1246/bcsj.46.384 [DOI] [Google Scholar]
- Kitano M. and Kuchitsu K., Bull. Chem. Soc. Jpn. 46, 3048 (1973). 10.1246/bcsj.46.3048 [DOI] [Google Scholar]
- Mack H.-G. and Oberhammer H., J. Am. Chem. Soc. 119, 3567 (1997). 10.1021/ja964374x [DOI] [Google Scholar]
- Allen F. H., Acta Cryst. B 58, 380 (2002). 10.1107/S0108768102003890 [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J. B., E. F.MeyerJr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., and Tasumi M., Arch. Biochem. Biophys. 185, 584 (1978). 10.1016/0003-9861(78)90204-7 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4818731 for Figures S1 and S2 and Tables S1–S6.
- Huang L. and Roux B., J. Chem. Theory Comput. 9, 3543 (2013). 10.1021/ct4003477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. J., J. Am. Chem. Soc. 112, 8533 (1990). 10.1021/ja00179a044 [DOI] [Google Scholar]
- Kang M. and Smith P. E., J. Comput. Chem. 27, 1477 (2006). 10.1002/jcc.20441 [DOI] [PubMed] [Google Scholar]
- Zhong Y. and Patel S., J. Phys. Chem. B 114, 11076 (2010). 10.1021/jp101597r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlich O. and Kister A. T., Ind. Eng. Chem. 40, 345 (1948). 10.1021/ie50458a036 [DOI] [Google Scholar]
- Weerasinghe S. and Smith P. E., J. Chem. Phys. 119, 11342 (2003). 10.1063/1.1622372 [DOI] [Google Scholar]
- Mark P. and Nilsson L., J. Comput. Chem. 23, 1211 (2002). 10.1002/jcc.10117 [DOI] [PubMed] [Google Scholar]
- Bentenitis N., Cox N. R., and Smith P. E., J. Phys. Chem. B 113, 12306 (2009). 10.1021/jp904806f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee M. B., Cox N. R., Jiao Y. F., Bentenitis N., Weerasinghe S., and Smith P. E., J. Chem. Theory Comput. 7, 1369 (2011). 10.1021/ct100517z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploetz E. A. and Smith P. E., Phys. Chem. Chem. Phys. 13, 18154 (2011). 10.1039/c1cp21883b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe S. and Smith P. E., J. Chem. Phys. 118, 10663 (2003). 10.1063/1.1574773 [DOI] [Google Scholar]
- Weerasinghe S. and Smith P. E., J. Phys. Chem. B 107, 3891 (2003). 10.1021/jp022049s [DOI] [Google Scholar]
- Weerasinghe S. and Smith P. E., J. Chem. Phys. 121, 2180 (2004). 10.1063/1.1768938 [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., and Schulten K., J. Mol. Graphics Modell. 14, 33 (1996). 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Hamzaoui F. and Baert F., Acta Crystallogr. 50, 757 (1994). 10.1107/S0108270193010832 [DOI] [Google Scholar]
- Bats J. W., Haberecht M. C., and Wagner M., Acta Crystallogr. 59, o1483 (2003). 10.1107/S1600536803019494 [DOI] [Google Scholar]
- Bøje L. and Hvidt A., J. Chem. Thermodyn. 3, 663 (1971). 10.1016/S0021-9614(71)80088-5 [DOI] [Google Scholar]
- Chickos J. S. and Acree J. W. E., J. Phys. Chem. Ref. Data 32, 519 (2003). 10.1063/1.1529214 [DOI] [Google Scholar]
- A. D.MacKerellJr., Shim J. H., and Anisimov V. M., J. Chem. Theory Comput. 4, 1307 (2008). 10.1021/ct8000969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R., Biochemistry 17, 201 (1978). 10.1021/bi00594a030 [DOI] [PubMed] [Google Scholar]
- Jelinska-Kazimierczuk M. and Szydlowski J., J. Mol. Liq. 30, 623 (2001). [Google Scholar]
- Scharlin P. and Steinby K., J. Chem. Thermodyn. 35, 279 (2003). 10.1016/S0021-9614(02)00359-2 [DOI] [Google Scholar]
- Williams W. D., Ellard J. A., and Dawson L. R., J. Am. Chem. Soc. 79, 4652 (1957). 10.1021/ja01574a020 [DOI] [Google Scholar]
- Pruppacher H. R., J. Chem. Phys. 56, 101 (1972). 10.1063/1.1676831 [DOI] [Google Scholar]
- Whitfield T. W., Martyna G. J., Allison S., Bates S. P., Vass H., and Crain J., J. Phys. Chem. B 110, 3624 (2006). 10.1021/jp053140+ [DOI] [PubMed] [Google Scholar]
- Haynes W. M., Lide D. R., and Bruno T. J., Handbook of Chemistry and Physics, 93rd ed. (CRC press, 2012). [Google Scholar]
- Marcus Y. and Hefter G. T., J. Mol. Liq. 73-74, 61 (1997). 10.1016/S0167-7322(97)00057-3 [DOI] [Google Scholar]