Summary
Aging and age-related pathology is a result of a still incompletely-understood intricate web of molecular and cellular processes. We present a C57BL/6J female mice in vivo aging study of five organs (liver, kidney, spleen, lung and brain), in which we compare genome-wide gene expression profiles during chronological aging with pathological changes throughout the entire murine lifespan (13, 26, 52, 78, 104 and 130 weeks). Relating gene expression changes to chronological aging revealed many differentially expressed genes (DEGs) and altered gene-sets (AGSs) were found in most organs, indicative of intra-organ generic aging processes. However, only ≤ 1% of these DEGs are found in all organs. For each organ, at least one of 18 tested pathological parameters showed a good age-predictive value, albeit with much inter- and intra-individual (organ) variation. Relating gene expression changes to pathology-related aging revealed correlated genes and gene-sets, which made it possible to characterize the difference between biological and chronological aging. In liver, kidney and brain, a limited number of overlapping pathology-related AGSs were found. Immune responses appeared to be common, yet the changes were specific in most organs. Furthermore, changes were observed in energy homeostasis, reactive oxygen species, cell cycle, cell motility and DNA damage. Comparison of chronological and pathology-related AGSs revealed substantial overlap and interesting differences. For example, the presence of immune processes in liver pathology-related AGSs which were not detected in chronological aging. The many cellular processes that are only found employing aging–related pathology could provide important new insights into the progress of aging.
Keywords: aging dynamics, in vivo, gene expression, pathology, database
Introduction
Aging is a complex process comprising a wide variety of interconnected features and effects, like progressive functional decline, gradual deterioration of physiological function, decrease in fertility and viability. (Maslov and Vijg, 2009). Deterioration of physical health is the principal factor associated with aging, but has proven difficult to mechanistically dissect or translate into consistent biomarkers. As gene expression is involved in or affected by most cellular processes, whole-transcriptome analysis offers a unique opportunity to better define the aging process at a molecular level. Numerous transcriptome studies thus have provided new insights into aging mechanisms in multiple species, genotypes and organs (de Magalhaes et al., 2009; Park et al., 2009; Swindell, 2007; Swindell, 2009; Swindell et al., 2012; Zahn et al., 2007). Fairly consistent changes in the immune system and in metabolic rate were observed during aging (Park et al., 2009; Swindell, 2009). Also, several mechanisms have been postulated that are involved in aging at the cellular level, such as the prevailing theory: the free radical or oxidative damage theory of aging. The latter proposes that macromolecular damage, either due to normal toxic by-products of metabolism or inefficient repair/defensive systems, accumulates during lifespan and causes aging (Garinis et al., 2008; Parkes et al., 1998; Treiber et al., 2011). In addition, molecular pathways involving the IGF-1/GH axis (Bluher et al., 2003; Holzenberger et al., 2003) and mTOR (Harrison et al., 2009; Anisimov et al., 2010) have been implicated in the aging process.
A next step could be to correlate the commonly used patho-physiological aging end points to molecular and cellular phenotypes to investigate general health deterioration and loss of homeostasis in aging. For this, whole-transcriptome changes could serve as a surrogate for the molecular phenotype of aging, as the state of the transcriptome is generally considered to determine the cellular phenotype (Kim and Eberwine, 2010). Correlation studies however demand extensive time series over the entire lifespan of an organism in multiple organs. Unfortunately, aging transcriptome studies often: compare just two age groups (young versus old) (Barger et al., 2008; Southworth et al., 2009; Park et al., 2009); use model organisms with prolonged or reduced longevity (Schumacher et al., 2008; Swindell, 2007); use samples from a (human) population (Grondahl et al., 2010; Zahn et al., 2006), adding unwanted genetic and environmental variation; or do not use a full genome platform (Zahn et al., 2007).
Here, we measured the whole transcriptome in female C57BL/6J mice in a controlled in vivo aging study and incorporated temporal as well as physical-health aspects into our analysis. Biological samples were taken at regular time intervals during the entire murine lifespan. Next to whole transcriptome analysis of five different organs, also pathological analyses were performed to address health and physiological deterioration. This allows for the conventional temporal monitoring of aging during lifespan (‘chronological aging’), but also link gene expression to age-related pathology providing novel insights into ‘biological aging’. For example, our study suggests a correlation between immune response and the level of oxidative stress during aging in liver, whereas these processes were not found in the temporal analysis. Chronological aging in liver predominantly show energy-related metabolic and mitochondrial gene expression changes. This exemplifies the added value of the additional pathology-related aging analysis performed in this study.
Results
Survival and age-related pathology
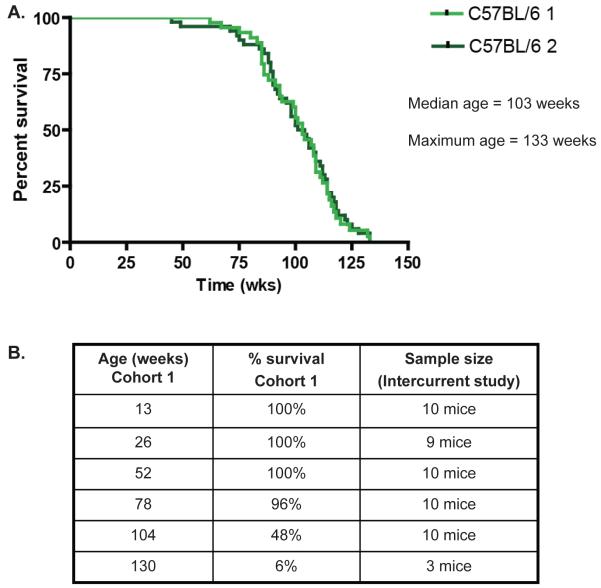
Using whole-transcriptome data during aging, we investigated six cross-sectional age groups spanning the adult lifespan of C57BL6/J female mice (Figure 1). Given the differences in aging as well as gene usage per organ, we investigated liver, kidney, spleen, lung and brain for age-related pathology and gene expression changes. Survival and cause-of-death pathology has been reported previously in studies on a concurrent longevity cohort of female mice (n=50, (Wijnhoven et al., 2005; Melis et al., 2008). The most prevalent causes of death were neoplasms, inflammation and general conditional decline. To show that the stringent standardized conditions, under which the survival cohort was kept and made them representative for murine aging, we performed an identical survival study several years later. The survival curves of both cohorts were quite similar, with a median lifespan of 103 weeks of age and a maximum lifespan of 133 weeks (Figure 1A).
Figure 1. Survival curves longevity cohorts.
(A) Survival curves of the concurrent female wild type longevity cohort (C57BL/6J 1, n=50) of this study. A control female cohort (C57BL/6J 2, n=50) from several years later. (B) Survival percentages for the intercurrent age groups used in this study as deducted from cohort C57BL/6J 1.
Age-predictive value of pathological parameters
There are several known organ-specific pathological parameters that are predictive of chronological aging in mice. We analyzed in total 18 different pathological parameters. Significant aging effects were observed for almost all parameters (Table 1, SI01). However, their dynamics during lifespan vary considerably and the profiles over age differed considerably (SI01). A generally accepted marker of aging at organ level is accumulation of lipofuscin (Gray and Woulfe, 2005) and also levels of karyomegaly have been proposed as a putative aging marker (Thoolen et al., 2010). We have scored lipofuscin accumulation during aging in a mitotic organ (liver) as well as a post-mitotic organ (brain). We observed a promising relationship with chronological aging in both organs. Karyomegaly appeared to be a promising marker in liver, but the correlation with aging in kidney appeared to be low and non-significant. Glomerular membrane thickening in the kidney however can be used as a good predictor for chronological aging in this tissue.
Table 1. Relationship chronological aging and pathological markers.
The pathological markers are either quantified on a continuous scale, an ordinal scale using four or five levels of severity, or on occurrence (absent - present). The values indicate the mean (continuous), median (ordinal) or percentage (binary) for each age class with the number of analyzed mice between brackets. The lipofuscin index in liver was calculated combining the abundance, intensity (ordinal) and size (ordinal) of lipofuscin spots. The last column indicates age prediction as indicated by the significance of a difference between age classes (SI02).
| Chronological age in weeks | correlation chronological age1 |
PPCG 2 |
pathology AGS 3 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Organ | Pathological parameter | Scale | 13 | 26 | 52 | 78 | 104 | 130 | |||
|
| |||||||||||
| Liver | Focal lymphoid proliferation | A/P | 33 (9) | 33 (9) | 17 (6) | 100 (7) | 80 (10) | 100 (2) | 0.0020 | 3 | 26 |
| Scattered extramedullary hematopoiesis | 0-5 | 1 (9) | 1 (9) | 0 (9) | 1 (10) | 1 (10) | 4 (3) | 0.1000 | 67 | 9 | |
| Karyomegaly | 0-5 | 3 (9) | 3 (9) | 3 (9) | 3.5 (10) | 4 (9) | 5 (3) | 0.0020 | 330 | 45 | |
| Intranuclear droplets | % | 0 (9) | 0 (9) | 0 (9) | 30 (10) | 89 (9) | 33 (3) | 0.0050 | 18 | 4 | |
| Hepatocellular vacuolization | 0-5 | 1 (9) | 1 (8) | 2 (9) | 3 (10) | 4 (10) | 1 (3) | 0.0005 | 96 | 16 | |
| Ito cell vacuolization | 0-5 | 0 (9) | 0 (9) | 3 (8) | 2 (8) | 3 (7) | 3 (3) | 0.0005 | 29 | 12 | |
| Lipofuscin index4 | # | 0.6 (9) | 5.4 (9) | 39 (10) | 87 (10) | 112 (10) | 231 (3) | 2×10−7 | 626 | 125 | |
| Kidney | Karyomegaly | 0-5 | 1 (9) | 0 (9) | 1 (10) | 1 (10) | 1 (10) | 2 (3) | 0.3067 | 18 | 4 |
| Lymphoid proliferation | A/P | 0 (10) | 0 (9) | 56 (9) | 30 (10) | 40 (10) | 33 (3) | 0.0255 | 10 | 25 | |
| Glomerular membrane thickening4 | 0-5 | 0 (10) | 0 (8) | 0 (9) | 2 (10) | 2.5 (10) | 4 (3) | 3×10−6 | 279 | 238 | |
| Tubular degeneration | 0-5 | 0 (10) | 0 (9) | 0 (8) | 1.5 (10) | 1.5 (10) | 2 (3) | 0.0008 | 398 | 61 | |
| Spleen | Lymphocytolysis4 | 0-5 | 3 (10) | 2 (8) | 1.5 (8) | 1.5 (10) | 0 (10) | 0 (3) | 0.0004 | 65 | 8 |
| Iron laden macrophages | 0-5 | 0 (10) | 0 (8) | 2.5 (8) | 3 (10) | 2 (10) | 1 (3) | 0.0050 | 31 | 11 | |
| Lung | Peribronchiolar lymphoid proliferation4 | A/P | 10 (10) | 0 (8) | 50 (10) | 89 ( 9) | 89 (9) | 50 (2) | 0.0005 | 33 | 5 |
| Brain | Vacuolisation white matter | 0-5 | 0 (9) | 0 (7) | 1 (10) | 2 (10) | 3 (10) | 5 (3) | 1×10−5 | 173 | 83 |
| Lipofuscinosis in neurons4 | 0-5 | 0 (9) | 0 (7) | 0 (8) | 2 (10) | 2 (10) | 2.5 (2) | 7×10−6 | 148 | 1 | |
| Periventricular GFAP5 | 0-4 | 1 (9) | 0 (6) | 2 (10) | 2 (10) | 1 (9) | 3 (3) | 0.0050 | 77 | 29 | |
| Hippocampus GFAP5 | 0-4 | 1 (9) | 0 (6) | 2 (10) | 2 (10) | 2 (9) | 4 (3) | 0.0020 | 67 | 33 | |
P-value, red values are not-significant
PPCG; Pathological parameter correlated genes
AGS: Overrepresented altered gene sets
Pathological parameter with the best correlation to chronological aging
GFAP: Glial Fibrillary Acidic Protein IHC staining
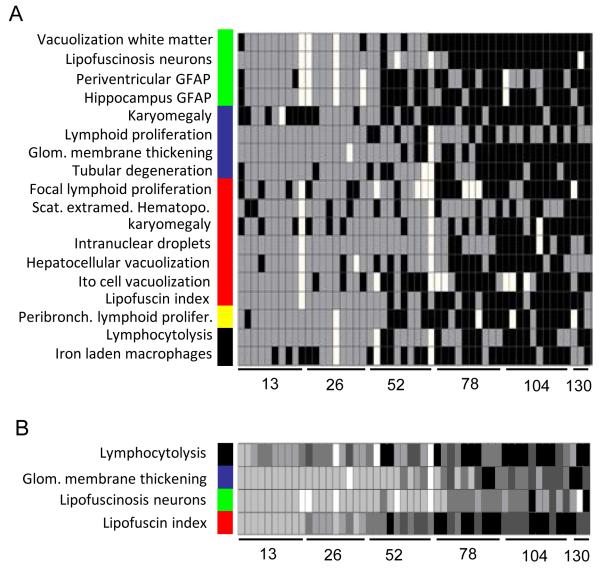
Unfortunately, good correlation of predictive parameters does not mean that they are good biomarkers for chronological aging for each individual mouse. Hence, we analyzed how individual organ samples could be age-identified by these pathological parameters. Figure 2A illustrates to what extent the pathology was indicative for chronological age. Virtually none of the parameters resulted in a perfect separation between “young” (grey) and “old” (black) in this analysis. To estimate the bias, we looked for their performance i.e. correctly identified samples, only from the two youngest time points and the two oldest time point (SI02). Based on all age class predictions (SI02), the overall best correlating pathological parameters per organ for chronological aging were: lipofuscin accumulation in brain and liver, glomerular membrane thickening in kidney, decrease in lymphocytolysis in spleen and increased peribronchiolar lymphoid proliferation in lung.
Figure 2. Age prediction by pathological characteristics.
(A) For each pathological parameter a clustering was performed in which each sample was assigned to one of the two main clusters, that were identified as “young” (grey) or “old” (black) based on the average age of the samples in each cluster (SI02).
(B) To enable detailed comparison within individuals, the most evidential pathological marker for each organ is shown at an ordinal scale from one (light gray) to four (black). White indicates missing values. The color bar indicates organ: brain (green), kidney (blue), liver (red), lung (yellow) and spleen (black).
The observation that almost no individual mouse showed a consistently “young” or “old” phenotype across the multiple organs (Figure 2), feeds into the increasing awareness that chronological age and biological age are different features. Our results indicate that there is ample inter- and intra-individual variation, despite the fact that some pathological parameters give a good general indication of young and old age. Biological aging was not consistently reflected by the various pathological parameters over the multiple organs and hardly any of the individuals showed an identical ranking for all these parameters, especially in older animals (Figure 2B). Given that several pathological parameters (≈ biological aging) were highly correlated with chronological aging based on average scores per age group, we assume that despite inter- and intra-individual (organ) variation, it is still valid to analyze chronological aging in the context of the “average aging process” in this population.
Gene expression related to chronological aging
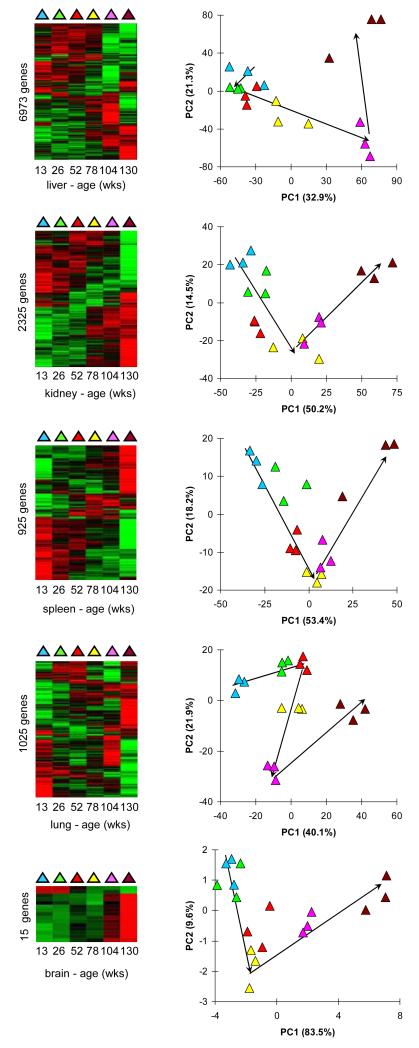
As next step we investigated the changing molecular phenotype of aging as embodied by whole transcriptome gene expression of three randomly selected samples from each time point. For this, whole-transcriptome gene expression profiling (35,283 transcripts) was performed on all age groups in all five organs. We first investigated all gene expression profiles together in a principal component analysis (PCA) (SI03, SI04). The PCA plot revealed that the strongest variance in gene expression could be found between organs, so much that all samples from each organ clustered together (SI03). Therefore, we continued our analyses per organ. Firstly, we looked at several established longevity-related genes based on literature: mTOR, p16, GhH, IGF, Pten, Sirt1, TgfB, Tert. Interestingly, none these genes showed an evenly graded gene expression profile during lifespan (SI05) and we conclude that their involvement in aging could not unambiguously be confirmed from the results from this experiment. To still identify genes affected during lifespan, we determined differentially-expressed genes (DEGs, FDR < 0.05): 6,973 in liver; 2,325 in kidney; 925 in spleen; 1,025 in lung; and 15 in brain (SI04). This implies that liver here appears to be the organ with the most consistent transcriptome changes during aging. Organizing the DEGs in heatmaps revealed marked gene expression differences in mice of 130 weeks illustrated by clusters of genes that are exclusively highly expressed in these mice (Figure 3). In the associated PCAs, the samples showed quite a distinct order that largely agrees with chronological aging (Figure 3). Altogether, there are many genes that show regulated gene expression during aging.
Figure 3. Age-related gene expression.
Differentially expressed genes (DEGs) in chronological aging. The first column shows heatmaps after clustering of the DEGs. The second column shows PCA of the gene expression values from the DEGs. The colors indicate chronological age according to the first column.
We compared the DEGs from different organs for overlapping genes (SI04, SI06). As expected, most DEGs (88%) are exclusively found in one organ. The number of overlapping DEGs rapidly decreased when more tissues were compared at the same time. Only one gene was found to be differentially expressed in all organs tested: Lilrb4 (Affymetrix probe-set identifier 1420394_s_at) (SI04A). Expression of Lilrb4 increases during aging and the produced protein is an immunoglobulin-like receptor that is involved in regulation of immune tolerance (SI04B). Five of the 11 other DEGs that are found in at least four organs, are immunoglobulin lambda and kappa complex related (SI04A) and all DEGs showed increased gene expression during aging.
Cellular processes related to chronological aging
Identifying DEGs is just a first step in discovering functional processes involved in aging. We connected gene expression to cellular processes by testing the top 10% genes with the most significantly changed gene expression per organ for overrepresentation of genes that are functionally related, such as in a pathway or cellular process (Tomlins et al., 2007). We observed many altered gene sets (AGSs): 122 in liver; 203 in kidney; 307 in spleen; 59 in lung; and 82 in brain (SI07). We would like to issue a word of caution with regard to results in brain, because the low amount of brain DEGs (15) makes overrepresentation of gene sets prone to false discovery. Nevertheless, the observed AGSs in these different organs encompass a pleiotropic collection of pathways and cellular processes. Plotting the gene expression of selected AGSs (as described in the M&M) revealed that several of them showed a continuously increasing or decreasing profile during aging (SI08). This means that the genes of associated pathways and cellular processes are collectively regulated during aging.
We compared AGSs from different organs for overlap (SI07, SI06). Similar to the DEGs, most AGSs (79%) are exclusively found in one organ. The number of overlapping AGSs rapidly decreased when more tissues were compared at the same time. No common AGS was found in the four remaining organs tested. In all organs, except liver, many gene sets involved in immunological processes are changed during aging. Despite the observed overlapping AGSs, plotting the gene expression profiles revealed a decrease of immune-related gene expression in the spleen during aging as compared to an increase of immune-related gene expression in kidney and lung for several exemplarily AGSs (SI09). This stresses that each organ generally has a specific aging course, which corroborates the pathology data.
Gene expression related to pathological aging parameters
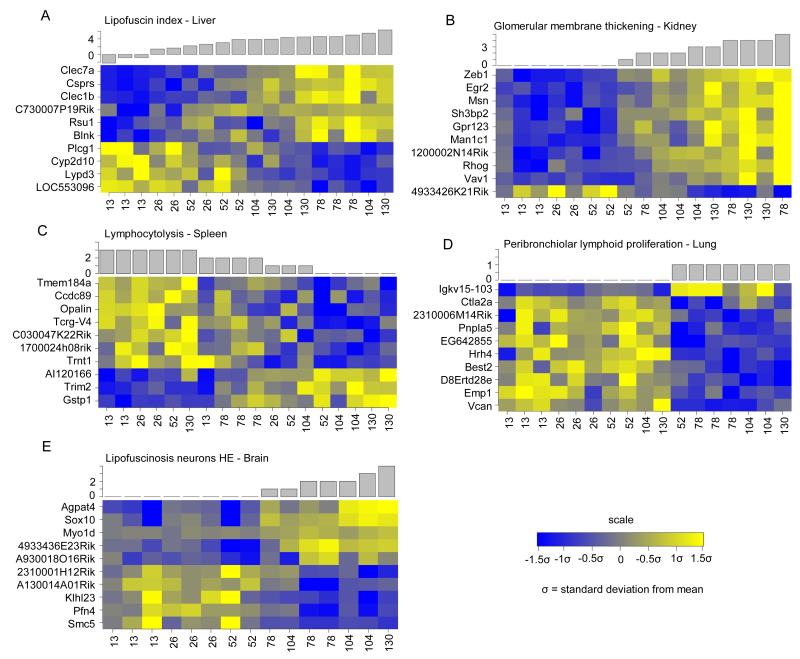
We investigated if it is possible to better define the specific progression of aging in organs by employing organ specific pathological markers rather than chronological aging time. As a first step towards discovery of the molecular mechanisms underlying the changing pathology during aging we correlated all pathological parameters with gene expression profiles for each organ. This resulted in a wide range of pathological parameter correlated genes (PPCGs), from 3 (focal lymphoid proliferation, liver) to 626 (lipofuscin index, liver) (Table 1, SI10). As an example, we plotted the gene expression (Figure 4) of the top ten annotated genes that are most strongly co-expressed (either positive or negative) with the pathological parameters, which in each organ correlated the best with chronological aging (Table 1). Notably, even though these parameters were selected because they correlated per organ the best with chronological aging, the heatmaps ordered by each parameter showed that in all cases the chronological age order is reshuffled in a specific way (Figure 4). This indicates that the biological aging of each organ is different than the chronological aging.
Figure 4. Examples of genes which expression correlates with pathological parameters.
Heatmaps depicting the top 10 annotated genes which expression correlates with pathological aging parameter: (A) lipofuscin index in the liver; (B) glomerular membrane thickening in the kidney; (C) lymphocytolysis in the spleen; (D) peribronchiolar lymphoid proliferation in lung; and (E) lipofuscinosis neurons in the brain.
Cellular processes related to pathological aging and comparison to chronological aging
For each pathological parameter in each organ we used the changes in expression of correlated genes (using the top 10% correlated genes per parameter as input) to find biological processes related to pathological aging. Supplemental information 11 gives a full overview of the correlated biological processes for all parameters and tissues. The number of altered gene sets (AGS) per pathological aging parameter is also indicated in the last column of Table 1.
To indicate how pathological aging-related biological processes can be compared to chronological aging processes, we used the most significantly correlated parameters lipofuscin (liver) and glomerular membrane thickening (kidney) as an example. SI11 shows which biological pathways and/or processes do overlap between chronological aging and pathological aging and which are specific for pathological aging. In liver, we find overlapping processes and components related to deterioration of mitochondrial function and lipid metabolic processes. Interestingly, this mitochondrial dysfunction was also correlated with karyomegaly severity in this tissue (SI11). The large majority of the lipofuscin-related AGSs, not overlapping with chronological aging, is involved in immunological cellular responses. This immune response was not shown by the chronological aging generated AGSs (SI07, SI08). A comparable analysis for the parameter glomerular membrane thickening (kidney) shows that overlapping functional pathways with chronological aging are amongst others processes involved in cell migration, adhesion, motility, angiogenesis and extracellular matrix components (compare responses shown in SI08 and SI11). Also several immune responses and EMT-related processes apparent in chronological aging are corroborated in pathological aging analyses (SI11). Our results allow comparable analyses for the investigated pathological parameters and tissues indicated in Table 1. This database thereby creates a good starting point for further aging research and can provide important new insights into the processes of aging.
Discussion
In this comprehensive study, we systematically analyzed in vivo aging of C57BL6/J mice, based on regular samples taken during their lifespan (13, 26, 52, 78, 104 and 130 weeks of age) from five organs for pathology and gene expression analyses. The average survivorship of the female mice in these studies (see concurrent survival cohort data in Figure 1) is comparable to previous findings (Kunstyr and Leuenberger, 1975). We identified pathological hallmarks that are correlated with chronological aging and employed these to assess individual pathology-related (biological) aging. Besides the generally accepted aging marker lipofuscin accumulation, several other hallmarks of aging were revealed in five different tissues (e.g. glomerular membrane thickening, lymphocytolysis in spleen and lymphoid proliferation in kidney and lung). The majority of our findings was supported by the few large scale studies available that specifically report on non-neoplastic or degenerative lesions in relation to aged C57BL/6J or BL/6-related mice (Bronson and Lipman, 1991; Lipman et al., 1999; Haines et al., 2001). However, the dynamics of these aging hallmarks as described in our study is novel. In addition, ito cell vacuolisation can be considered a novel hallmark of aging.
Chronological and biological aging, as defined by pathological parameters, were functionally characterized by gene expression profiling, with the aim to confirm or discover underlying mechanisms for aging and to investigate their temporal progression during lifespan. The pathological parameters in our current study demonstrated that signs of aging are predominantly organ-specific. The gene expression profiles confirmed this organ-specific regulation both in chronological and biological aging. However, analyses based on individual genes as well as on functionally related gene sets, gave one recurrent result: the genes commonly changing in multiple organs during aging were related to immune processes. The pronounced involvement of the immune system was found previously in other large-scale age-related gene expression studies (Swindell, 2009; Park et al., 2009; Zahn et al., 2007). In the comprehensive meta-analysis study of Swindell, immune responses were similarly the most commonly regulated processes over all organs examined, but also biological processes like cellular respiration and other mitochondrial-related processes in liver were significantly regulated. Our study allows for monitoring of temporal dynamics of these and other possible age-related processes during the entire murine lifespan, which is an attractive extension for the aging field. We have chosen to analyze the aging processes in a temporal chronological and pathology-related (biological) manner.
Functional characterization of gene expression pinpointed immune responses which steadily increase or decrease with age, depending on the organ type, for instance a decreasing expression of immune-related gene sets in spleen, but an increasing expression in kidney and lung. The temporal analysis showed that the decreasing expression in spleen occurred quite sudden after 104 weeks, but increasing expression in the other organs was more gradual. This may indicate increased immune cell infiltration upon cell death or cellular senescence in the organs with age, but an overall decline in functionality of the immune system in spleen. The intercurrent pathological observation of lymphoid proliferation (in lung, kidney and liver) and lymphocytolysis in spleen could partially reflect these transcriptome changes.
Our study demonstrated that the most apparent gene expression changes observed in the liver were related to electron transport chain, metabolic processes and the mitochondrial membrane, which can reflect increasing mitochondrial and cellular dysfunction over time. The consequences of aging for mitochondria and oxidative phosphorylation have extensively been reviewed (Lesnefsky and Hoppel, 2006) and decreased levels in electron transport chain have been shown in aging human organ and other species (Zahn et al., 2007). Previous data support the finding that the rate of oxidative phosphorylation decreases during aging (Okatani et al., 2002). Our results indicate that mitochondrial processes and oxidative phosphorylation increased moderately from very young adulthood to mature adulthood, remained constant until 78 weeks of age and then decreased considerably during the remainder of the lifespan (104 and 130 weeks). The mTOR and PTEN signaling pathways had similar dynamics in liver. Deregulation in both pathways has been associated with metabolic changes during aging and can affect cancer susceptibility (Zoncu et al., 2011; Keniry and Parsons, 2008). In other organs too, we found gene expression changes that have been associated with cancer. In kidney the gene expression profiles revealed an up-regulation during aging of processes like cell motility, cell migration and angiogenesis. Several cancer associated pathways were likewise up-regulated in spleen: increased cell cycle and DNA damage responses were apparent, especially during the final stages of the lifespan.
Our study furthermore showed that the pathological biomarker for ROS, lipofuscin accumulation (Jung et al., 2007), gradually increased over time, in mitotic as well as post-mitotic organs. This supports the hypothesis that ROS and free radicals contribute to protein, lipid, RNA and DNA damage accumulation and homeostatic imbalance during aging in our study. Interestingly, the gene most strongly co-expressed with lipofuscin accumulation in liver was Clec7a (Figure 4A), which is an innate immune receptor and can mediate production of ROS in the cell (Goodridge et al., 2011). Part of the correlated pathways to biological aging in liver revealed additional processes that were linked to ROS, such as angiotensin II-induced production of ROS (SI11). Another interesting result linking lipofuscin accumulation to increased ROS was the fact that biological aging analyses in liver yielded pathways related to reactive oxygen species and numerous immune-related responses, while these responses were not identified by chronological aging analyses. These results suggest a correlation between immune response and the level of oxidative stress during aging in liver and moreover exemplify the added value of pathology-related aging analyses. Combining temporal responses spanning the murine C57BL/6J lifespan and gene expression patterns linked to aging pathology our study provides more information on the intricate processes involved in aging.
Processes identified being differentially regulated during aging, like mitochondrial dysfunction and the regulation of immune-related processes, are able to greatly increase ROS in cells (Cui et al., 2012; West et al., 2011), thereby potentially causing collateral DNA and other macromolecular damage. Since our data did not show a substantial regulation of DNA repair pathways over time or over biological aging and considering DNA repair responses are mostly post-transcriptionally regulated in mice, we expect this damage to accumulate slowly over time and influence age-related disease in most organs, be it at different rates. We have previously shown that mutations, caused by intrinsic DNA damage, increased at different rates during aging in several organs in mice from the same intercurrent aging cohorts that were used in our current study (Melis et al., 2008). Reversely, in vivo defects in DNA damage repair machinery have demonstrated to promote (segmental) accelerated aging phenotypes (Wijnhoven et al., 2005; Wijnhoven et al., 2007).
In this extensive study we analyzed the process of aging on multiple levels: biological and chronological aging were assessed, combining age-related pathology and gene expression profiling. We proposed several distinguishable and potentially novel pathological hallmarks that are highly correlated to chronological aging in different organs. But because of inter- and intra-individual variation the pathological hallmarks are additionally useful to study pathology-related, biological aging. In this study it was evident that the immune responses played the most distinguished role in both chronological and biological aging, but manifested itself with highly organ-specific dynamics. Our results furthermore support several aging hypotheses at the cellular level, like processes that can cause increased levels of ROS, an imbalanced metabolic or energy homeostasis or increased mutational load. Our study enables studying temporal dynamics of genes and processes spanning the entire lifespan over multiple organs.
Experimental procedures
Experimental Design
Female C57BL6/J mice were randomized in different groups; i.e. longevity cohorts (described in Wijnhoven et al. (Wijnhoven et al., 2005) (n=50), or cross-sectional cohorts (n=10-20). In cross-sectional cohorts, mice were sacrificed at a fixed age of 13, 26, 52, 78, 104 or 130 weeks. The microbiological status was monitored every three months during the entire studies. Mice were bred in-house and accommodated under pathogen-free conditions, strict standardized day/night (12:12hr) regime, consistent temperature (20°C), and controlled air pressure. Moreover, to minimize confounding effects on health status, mice were housed in small groups of 4-5 in Macrolon II type cages with sufficient cage-enrichment. Standard lab chow (Hope Farms, the Netherlands) and water were supplied ad libitum. Complete autopsy was performed on the mice; organs were isolated from each animal and stored for further histopathological analysis. Organ from liver, kidney, spleen, lung and brain (frontal lobe of cerebellum) were snap-frozen for gene expression profiling.
Histopathology
Organ samples of each animal were preserved in a neutral aqueous phosphate-buffered 4% solution of formaldehyde. Organs required for microscopic examination were embedded in paraffin wax, sectioned at 4 μm and stained with haematoxylin and eosin. Detailed microscopic examination was performed on the intercurrent cohort samples and on all gross lesions suspected of being tumors or representing major pathological conditions. For each animal, histopathological abnormalities, tumors, and non-neoplastic lesions were recorded.
The difference between the age groups of binomial data was tested using a chi-squared contingency table test. The difference between the age groups of ordinal data was tested using Kruskal-Wallis rank sum test. To determine to which extent the pathology was indicating aging, the following analysis was performed. For each pathological variable, the samples were clustered based on severity, using hierarchical clustering and the complete agglomeration method (with binary distance if required). The average age of each of the two main clusters was calculated. The allocation of each sample to either a “young” cluster or an “old” age group was recorded (Figure 2) and was afterwards compared to the chronological age of this sample. A pathological parameter was considered aging related if samples of 13 and 26 weeks old were allocated to the young age groups and if samples from 104 and 130 weeks old were allocated to the old age group.
Microarray analysis
Total RNA was isolated from liver, kidney, spleen, lung and brain, using the RNeasy Midi Kit (Qiagen, Valencia, CA, USA;). RNA quality was tested using automated gel electrophoresis (Bioanalyzer 2100; Agilent technologies, Amstelveen, the Netherlands (RIN > 7)). RNA samples (n = 3 per time point per organ, 90 samples in total) were labeled and hybridized to Mouse GeneChip 430 2.0 (Affymetrix, Santa Clara, CA, USA) arrays according to the manufacturer’s protocol. All raw data passed the quality criteria, but relevant effects of labeling batches were detected. The raw data from each organ was normalized using the robust multi-array average algorithm (Irizarry et al., 2003) and annotated according de Leeuw (de Leeuw et al., 2008). The data were corrected for labeling-batch effects using a linear model with group-means-parametrization and coefficients for age (fixed) and labeling-batch (random). The resulting normalized expression values were analyzed for differentially expressed genes (DEGs) by fitting natural cubic splines as a function of chronological age, as described by Storey et al. (Storey et al., 2005), using a false discovery rate corrected p-value cut-off of < 0.05 (Storey and Tibshirani, 2003). The temporal profiles of the DEGs were explored using agglomerative hierarchical clustering and principal components analysis. Altered gene sets (AGSs) were determined using overrepresentation analysis and gene set analysis. For overrepresentation analysis, the top 10% of most significant differentially expressed genes (Tomlins et al., 2007) in each organ were tested for disproportionally high numbers of functionally related genes, using the hypergeometric test (p < 10−3). For gene set analysis, a priori defined functionally related sets of genes were tested for concordant aging effects, using the Wilcoxon rank test (p < 10−5 (Michaud et al., 2008)). Gene sets were defined by the Gene Ontology, Metacore (http://www.genego.com/metacore.php) and a predefined list of gene-sets (SI12). The profile of an overrepresented gene set was determined based on the genes in the top 10% (yi) by calculating the eigengene x (Alter et al., 2000), and the direction by Σicor(x,yi): a negative score resulted in reversion of the eigengene.
Co-expression was quantified using ρx,y and ρx,y*z, where ρ indicates Spearman rank correlation, x = a phenotypic variable, y = a gene and z = age. P-values for ρx,y were determined using a permutation based Spearman rank correlation test (ordinal data), or using a chi-squared contingency test (binomial data) and for ρx,y*z a conditional Spearman correlation test (ordinal data) or a Cochran-Mantel-Haenszel test (binomial data). A score for co-expression was calculated as the negative sum of the logs of the p-values of the correlation and the conditional correlation test. This methodology corrects for correlations with age. In order to define AGSs, the co-expression scores were subjected to gene set analysis and overrepresentation analysis (using the top 3% (1000) highest co-expression scores). Gene expression data has been submitted to the public Gene Expression Omnibus, number GSE34378.
Supplementary Material
SI01
Dynamics of the average values of the age-related pathological parameters (cf. Table 1) during murine lifespan.
SI02
For each pathological parameter a clustering was performed in which each sample was assigned to one of the two main clusters, that were identified as predicted “young” or “old” based on the average age of the samples in each cluster. The accuracy of each pathological parameter for chronological age prediction was tested. For the chronologically young group (here defined as 13 and 26 weeks, average in weeks), individuals should be predicted as “young” in the cluster analysis (Figure 2, grey). For the chronologically old group (here defined as 104 and 130 weeks, average in weeks), individuals should be predicted as “old” in the cluster analysis (Figure 2, black). Per organ, one parameter (bold) is indicated as the most predictive and most correlated to chronological aging.
SI03
Principal component analysis (PCA) plot depicting the samples plotted against the first two principal components calculated from the gene expression values of the perfect-match probes. The colors indicate liver (yellow), kidney (blue), spleen (green), lung (grey), and brain (red). The numbers indicate the chronological age of each sample (in weeks).
SI04
List of genes that were differentially expressed (FDR corrected p-value ≤ 0.05) in any of the organs The p-values and q-values (FDR adjusted p-values) are listed, indicating the degree of significance of differential expression of all DEGs during aging. A: DEGs are ordered based on q-value, and qvalues < 0.05 are marked green. The visible overlap is summarized in SI06. B: Expression profilesduring aging in all organs of the only gene that is significantly differentially expressed in all five organs examined: Lilrb4 (Affymetrix probe-set identifier 1420394_s_at).
SI05
Gene expression dynamics of several literature based age-related genes during murine lifespan. Liver (red), kidney (blue), spleen (black), lung (yellow), and brain (green).
SI06
The organ intersections in differentially expressed genes (DEGs) (SI04) and functionally related altered gene sets (AGSs) (SI07).
SI07
Lists of altered gene sets (AGSs) that were found to be involved in aging in the different organs after gene set analysis (GSA), overrepresentation analysis (ORA) and Metacore overrepresentation analysis (M-ORA).
SI08
Dynamics of average, age-related gene expression changes of selected organ-specific, altered gene sets (AGSs).
SI09
Organ-specific gene expression dynamics of common immune related altered gene sets (AGSs) in kidney, spleen and lung.
SI10
List of genes that showed significant correlation (p-value ≤ 0.001) with any pathological parameter in any of the organs. The p-values of the correlation (cor) and partial correlation, which includes a correction for chronological age, (pcor) are shown. The union of the correlated and partial correlated genes per pathological parameter is presented in Table 1.
SI11
Lists of altered gene sets (AGSs) correlated to the pathological parameters in five organs after gene set analysis (GSA), overrepresentation analysis (ORA) and Metacore overrepresentation analysis (M-ORA). If present, overlap with chronological aging AGSs is indicated. 1= Found as an AGS (green). Grey cells indicate processes also present as AGS in chronological aging
SI12
Predefined list of aging related gene-sets based on literature.
Acknowledgements
We thank the Animal Facilities of the Netherlands Vaccine Institute (NVI) for their skilful (bio)technical support. The work presented here was in part financially supported by IOP Genomics IGE03009, NIH/NIA (3PO1 AG017242), STW Grant STW-LGC.6935 and Netherlands Bioinformatics Center (NBIC) BioRange II – BR4.1. Support was also obtained from Markage (FP7-Health-2008-200880), LifeSpan (LSHG-CT-2007-036894), European Research Council (ERC advanced scientist grant JHJH).
References
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, Yurova MN, Antoch MP, Blagosklonny MV. Rapamycin extends maximal lifespan in cancer-prone mice. Am J Pathol. 2010;176:2092–2097. doi: 10.2353/ajpath.2010.091050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger JL, Kayo T, Vann JM, Arias EB, Wang J, Hacker TA, Wang Y, Raederstorff D, Morrow JD, Leeuwenburgh C, Allison DB, Saupe KW, Cartee GD, Weindruch R, Prolla TA. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Bronson RT, Lipman RD. Reduction in rate of occurrence of age related lesions in dietary restricted laboratory mice. Growth Dev Aging. 1991;55:169–184. [PubMed] [Google Scholar]
- Cui H, Kong Y, Zhang H. Oxidative stress, mitochondrial dysfunction, and aging. J Signal Transduct. 2012;2012:646354. doi: 10.1155/2012/646354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw WC, Rauwerda H, Jonker MJ, Breit TM. Salvaging Affymetrix probes after probe-level re-annotation. BMC Res Notes. 2008;1:66. doi: 10.1186/1756-0500-1-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, Weiss A, Vasilakos JP, Underhill DM. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature. 2011;472:471–475. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DA, Woulfe J. Lipofuscin and aging: a matter of toxic waste. Sci Aging Knowledge Environ. 2005;2005:re1. doi: 10.1126/sageke.2005.5.re1. [DOI] [PubMed] [Google Scholar]
- Grondahl ML, Yding AC, Bogstad J, Nielsen FC, Meinertz H, Borup R. Gene expression profiles of single human mature oocytes in relation to age. Hum Reprod. 2010;25:957–968. doi: 10.1093/humrep/deq014. [DOI] [PubMed] [Google Scholar]
- Haines DC, Chattopadhyay S, Ward JM. Pathology of aging B6;129 mice. Toxicol Pathol. 2001;29:653–661. doi: 10.1080/019262301753385988. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jung T, Bader N, Grune T. Lipofuscin: formation, distribution, and metabolic consequences. Ann N Y Acad Sci. 2007;1119:97–111. doi: 10.1196/annals.1404.008. [DOI] [PubMed] [Google Scholar]
- Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- Kim J, Eberwine J. RNA: state memory and mediator of cellular phenotype. Trends Cell Biol. 2010;20:311–318. doi: 10.1016/j.tcb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunstyr I, Leuenberger HG. Gerontological data of C57BL/6J mice. I. Sex differences in survival curves. J Gerontol. 1975;30:157–162. doi: 10.1093/geronj/30.2.157. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, Hoppel CL. Oxidative phosphorylation and aging. Ageing Res Rev. 2006;5:402–433. doi: 10.1016/j.arr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Lipman RD, Dallal GE, Bronson RT. Lesion biomarkers of aging in B6C3F1 hybrid mice. J Gerontol A Biol Sci Med Sci. 1999;54:B466–B477. doi: 10.1093/gerona/54.11.b466. [DOI] [PubMed] [Google Scholar]
- Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta. 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis JP, Wijnhoven SW, Beems RB, Roodbergen M, van den BJ, Moon H, Friedberg E, van der Horst GT, Hoeijmakers JH, Vijg J, van Steeg H. Mouse models for xeroderma pigmentosum group A and group C show divergent cancer phenotypes. Cancer Res. 2008;68:1347–1353. doi: 10.1158/0008-5472.CAN-07-6067. [DOI] [PubMed] [Google Scholar]
- Michaud J, Simpson KM, Escher R, Buchet-Poyau K, Beissbarth T, Carmichael C, Ritchie ME, Schutz F, Cannon P, Liu M, Shen X, Ito Y, Raskind WH, Horwitz MS, Osato M, Turner DR, Speed TP, Kavallaris M, Smyth GK, Scott HS. Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genomics. 2008;9:363. doi: 10.1186/1471-2164-9-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatani Y, Wakatsuki A, Reiter RJ, Miyahara Y. Hepatic mitochondrial dysfunction in senescence-accelerated mice: correction by long-term, orally administered physiological levels of melatonin. J Pineal Res. 2002;33:127–133. doi: 10.1034/j.1600-079x.2002.02109.x. [DOI] [PubMed] [Google Scholar]
- Park SK, Kim K, Page GP, Allison DB, Weindruch R, Prolla TA. Gene expression profiling of aging in multiple mouse strains: identification of aging biomarkers and impact of dietary antioxidants. Aging Cell. 2009;8:484–495. doi: 10.1111/j.1474-9726.2009.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- Schumacher B, van dP I, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, Breit TM, van Steeg H, Niedernhofer LJ, van Ijcken W, Bartke A, Spindler SR, Hoeijmakers JH, van der Horst GT, Garinis GA. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth LK, Owen AB, Kim SK. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 2009;5:e1000776. doi: 10.1371/journal.pgen.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. Gene expression profiling of long-lived dwarf mice: longevity-associated genes and relationships with diet, gender and aging. BMC Genomics. 2007;8:353. doi: 10.1186/1471-2164-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585. doi: 10.1186/1471-2164-10-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Sun L, Xing X, Fisher GJ, Bulyk ML, Elder JT, Gudjonsson JE. Meta-profiles of gene expression during aging: limited similarities between mouse and human and an unexpectedly decreased inflammatory signature. PLoS One. 2012;7:e33204. doi: 10.1371/journal.pone.0033204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoolen B, Maronpot RR, Harada T, Nyska A, Rousseaux C, Nolte T, Malarkey DE, Kaufmann W, Kuttler K, Deschl U, Nakae D, Gregson R, Vinlove MP, Brix AE, Singh B, Belpoggi F, Ward JM. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol. 2010;38:5S–81S. doi: 10.1177/0192623310386499. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- Treiber N, Maity P, Singh K, Kohn M, Keist AF, Ferchiu F, Sante L, Frese S, Bloch W, Kreppel F, Kochanek S, Sindrilaru A, Iben S, Hogel J, Ohnmacht M, Claes LE, Ignatius A, Chung JH, Lee MJ, Kamenisch Y, Berneburg M, Nikolaus T, Braunstein K, Sperfeld AD, Ludolph AC, Briviba K, Wlaschek M, Florin L, Angel P, Scharffetter-Kochanek K. Accelerated aging phenotype in mice with conditional deficiency for mitochondrial superoxide dismutase in the connective tissue. Aging Cell. 2011;10:239–254. doi: 10.1111/j.1474-9726.2010.00658.x. [DOI] [PubMed] [Google Scholar]
- West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnhoven SW, Beems RB, Roodbergen M, van den BJ, Lohman PH, Diderich K, van der Horst GT, Vijg J, Hoeijmakers JH, van Steeg H. Accelerated aging pathology in ad libitum fed Xpd(TTD) mice is accompanied by features suggestive of caloric restriction. DNA Repair (Amst) 2005;4:1314–1324. doi: 10.1016/j.dnarep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Wijnhoven SW, Hoogervorst EM, de Waard H, van der Horst GT, van Steeg H. Tissue specific mutagenic and carcinogenic responses in NER defective mouse models. Mutat Res. 2007;614:77–94. doi: 10.1016/j.mrfmmm.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, Lakatta EG, Boheler KR, Xu X, Mattson MP, Falco G, Ko MS, Schlessinger D, Firman J, Kummerfeld SK, Wood WH, III, Zonderman AB, Kim SK, Becker KG. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2006;2:e115. doi: 10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI01
Dynamics of the average values of the age-related pathological parameters (cf. Table 1) during murine lifespan.
SI02
For each pathological parameter a clustering was performed in which each sample was assigned to one of the two main clusters, that were identified as predicted “young” or “old” based on the average age of the samples in each cluster. The accuracy of each pathological parameter for chronological age prediction was tested. For the chronologically young group (here defined as 13 and 26 weeks, average in weeks), individuals should be predicted as “young” in the cluster analysis (Figure 2, grey). For the chronologically old group (here defined as 104 and 130 weeks, average in weeks), individuals should be predicted as “old” in the cluster analysis (Figure 2, black). Per organ, one parameter (bold) is indicated as the most predictive and most correlated to chronological aging.
SI03
Principal component analysis (PCA) plot depicting the samples plotted against the first two principal components calculated from the gene expression values of the perfect-match probes. The colors indicate liver (yellow), kidney (blue), spleen (green), lung (grey), and brain (red). The numbers indicate the chronological age of each sample (in weeks).
SI04
List of genes that were differentially expressed (FDR corrected p-value ≤ 0.05) in any of the organs The p-values and q-values (FDR adjusted p-values) are listed, indicating the degree of significance of differential expression of all DEGs during aging. A: DEGs are ordered based on q-value, and qvalues < 0.05 are marked green. The visible overlap is summarized in SI06. B: Expression profilesduring aging in all organs of the only gene that is significantly differentially expressed in all five organs examined: Lilrb4 (Affymetrix probe-set identifier 1420394_s_at).
SI05
Gene expression dynamics of several literature based age-related genes during murine lifespan. Liver (red), kidney (blue), spleen (black), lung (yellow), and brain (green).
SI06
The organ intersections in differentially expressed genes (DEGs) (SI04) and functionally related altered gene sets (AGSs) (SI07).
SI07
Lists of altered gene sets (AGSs) that were found to be involved in aging in the different organs after gene set analysis (GSA), overrepresentation analysis (ORA) and Metacore overrepresentation analysis (M-ORA).
SI08
Dynamics of average, age-related gene expression changes of selected organ-specific, altered gene sets (AGSs).
SI09
Organ-specific gene expression dynamics of common immune related altered gene sets (AGSs) in kidney, spleen and lung.
SI10
List of genes that showed significant correlation (p-value ≤ 0.001) with any pathological parameter in any of the organs. The p-values of the correlation (cor) and partial correlation, which includes a correction for chronological age, (pcor) are shown. The union of the correlated and partial correlated genes per pathological parameter is presented in Table 1.
SI11
Lists of altered gene sets (AGSs) correlated to the pathological parameters in five organs after gene set analysis (GSA), overrepresentation analysis (ORA) and Metacore overrepresentation analysis (M-ORA). If present, overlap with chronological aging AGSs is indicated. 1= Found as an AGS (green). Grey cells indicate processes also present as AGS in chronological aging
SI12
Predefined list of aging related gene-sets based on literature.