Summary
Circadian clocks generate daily rhythms in molecular, cellular, and physiological functions providing temporal dimension to organismal homeostasis. Recent evidence suggests two-way relationship between circadian clocks and aging. While disruption of the circadian clock leads to premature aging in animals, there is also age-related dampening of output rhythms such as sleep/wake cycles and hormonal fluctuations. Decay in the oscillations of several clock genes was recently reported in aged fruit flies, but mechanisms underlying these age-related changes are not understood. We report that the circadian light-sensitive protein CRYPTOCHROME (CRY), is significantly reduced at both mRNA and protein levels in heads of old Drosophila melanogaster. Restoration of CRY using the binary GAL4/UAS system in old flies significantly enhanced the mRNA oscillatory amplitude of several genes involved in the clock mechanism. Flies with CRY overexpressed in all clock cells maintained strong rest/activity rhythms in constant darkness late in life when rhythms were disrupted in most control flies. WE also observed a remarkable extension of healthspan in flies with elevated CRY. Conversely, CRY deficient mutants showed accelerated functional decline and accumulated greater oxidative damage. Interestingly, overexpression of CRY in central clock neurons alone was not sufficient to restore rest/activity rhythms or extend healthspan. Together, these data suggest novel anti-aging functions of CRY and indicate that peripheral clocks play an active role in delaying behavioral and physiological aging.
Keywords: cryptochrome, circadian clock, aging, healthspan, Drosophila
Introduction
Circadian clocks are intrinsic mechanisms that generate daily rhythms in behavior, physiology, and cellular processes, ensuring temporal homeostasis coordinated with day/night cycles (Reddy & O’Neill 2010). Increasing evidence suggests bidirectional relationships between circadian clocks and aging. On one hand, genetic or environmental disruptions of the clock function accelerate physiological aging, onset of late life diseases, and mortality risk in mammals (Davidson et al. 2006; Kondratov et al. 2006; Antoch et al. 2008; Yu & Weaver 2011). On the other hand, aging is also known to impair circadian rhythms as evidenced by disruption of sleep/wake cycles, dampening of hormonal rhythms, and weakening of clock gene oscillations (Valentinuzzi et al. 1997; Huang et al. 2002; Hofman & Swaab 2006; Kondratova & Kondratov 2012). Age-related decline in temporal coordination of metabolic, physiological and neurological functions have profound effects on health and disease susceptibility, yet the mechanisms underlying the decay of the circadian system are not understood.
Bidirectional relationships between circadian clocks and aging have also been established in Drosophila melanogaster. Disruption of the circadian clock increases the susceptibility of aging fruit flies to oxidative stress and neurodegeneration (Krishnan et al. 2009; Krishnan et al. 2012). Conversely, aging flies show fragmented sleep/activity patterns and dampened clock gene oscillations (Koh et al. 2006; Luo et al. 2012; Rakshit et al. 2012; Umezaki et al. 2012). In the current study, we used Drosophila to address mechanisms underlying the decay of the circadian system, and investigate whether this decay could be reversed in aging flies.
The molecular mechanism of the circadian clock is based on transcription-translation feedback loops that are evolutionarily conserved from flies to mammals (Stanewsky 2003; Yu & Hardin 2006). In fruit flies, Clock (Clk) and cycle (cyc) genes encode transcription factors that form CLK-CYC activator complexes and stimulate the expression of genes period (per) and timeless (tim) early at night. Subsequently, PER and TIM proteins form heterodimers and accumulate in the cell nuclei repressing CLK-CYC transcriptional activity, and thus suppressing their own transcription (Hardin 2011). In another negative feedback loop, CLK-CYC complexes induce the expression of transcription factors Par domain protein 1ε (Pdp1ε) and vrille (vri) that act as an activator and repressor of Clk (Cyran et al. 2003). This basic clock mechanism is cell-autonomous and operates in the central and peripheral clock cells. The central clock is formed by a network of 150 pacemaker neurons in the fly brain, which regulate rest/activity rhythms (Nitabach & Taghert 2008). Peripheral clocks are found in many cells of the nervous system, such as retinal photoreceptors, glia, sensory neurons, and in non-neural tissues in the head and body (Hardin 2011; Xu et al. 2011).
High-amplitude oscillations of per, tim, Pdp1ε, and vri mRNA as well as PER and TIM proteins observed in young flies, are significantly reduced in heads of aging flies (Luo et al. 2012; Rakshit et al. 2012). Oscillations of PER and TIM proteins were also evaluated in central clock neurons (Luo et al. 2012; Umezaki et al. 2012); however, these neurons constitute only a small fraction of clock cells compared to peripheral oscillators, which are therefore responsible for reduced clock gene oscillations observed in the heads. The causes of dampening of clock gene oscillations are not clear. In young flies, the blue light photoreceptive flavin-binding protein CRYPTOCHROME (CRY), is critical for the synchronization of individual oscillator cells (Emery et al. 1998; Stanewsky et al. 1998). CRY targets TIM for degradation after lights on and synchronizes the circadian oscillations with external day/night cycles (Busza et al. 2004). In addition to acting as circadian photoreceptor that mediates light input into the clock, CRY appears to function as the central clockwork component in peripheral clocks. Indeed, in hypomorphic cryb mutants, rhythmic expression of per and tim at mRNA and protein levels is abolished in the peripheral clock cells but not in the central clock neurons (Stanewsky et al. 1998). Furthermore, CRY is necessary for circadian clock functions in other peripheral oscillators under constant darkness (Ivanchenko et al. 2001; Krishnan et al. 2001), but its role is not yet understood. Mammalian mCry1 and mCry2 are not photoreceptive and act as circadian transcriptional regulators in the clock negative feedback loop (Kume et al. 1999). Interestingly, a recent report shows that human HsCRY-1 protein confers light-independent biological activity in transgenic Drosophila (Vieira et al. 2012), suggesting functional similarities between fly and human CRY. More unexpectedly, it was shown that endogenous fly CRY also regulates considerable light-independent transcriptional activity in Drosophila (Vieira et al. 2012). An increased repertoire of CRY functions in flies including effects on metabolism (Fogle et al. 2011; Seay & Thummel 2011; Kumar et al. 2012), suggest that CRY may act via multiple mechanisms that remain to be understood.
As mentioned above, rhythmic expression of per and tim is abolished in peripheral clock cells such as photoreceptors and glia of hypomorphic cryb mutants (Stanewsky et al. 1998). Age-related dampening of cry mRNA oscillations was recently reported in heads of flies, along with significantly reduced mRNA oscillations of per, tim, Pdp1ε, and vri (Luo et al. 2012; Rakshit et al. 2012). We reasoned that reduced levels of cry in heads of old flies may be responsible for diminished cycling of per and tim mRNA, similar as in young cryb mutants (Stanewsky et al. 1998).
In this study, we observed that CRY is reduced with age at both mRNA and protein levels, and attempted to replenish CRY in old flies using the binary GAL4/UAS system. We report that overexpression of CRY in all clock expressing cells improves the mRNA oscillatory amplitude of several genes involved in the clock mechanism, and restores strong circadian rest/activity rhythms in old flies. We further show that flies with elevated CRY levels have significantly improved healthspan during aging. Conversely, cry-null mutants show accelerated functional decline and accumulate greater oxidative damage, together suggesting novel anti-aging role of CRY. Interestingly, restored rest/activity rhythms and healthspan benefiting effects are only observed when cry is overexpressed in all clock cells, but not when cry overexpression is restricted to the central clock neurons, suggesting that peripheral clocks play an active role in maintaining organismal health during aging.
Results
CRY is reduced at both mRNA and protein levels in heads of old flies
We obtained daily mRNA expression profiles of cry in the heads and bodies of adult Canton S (CS) males on day 5 (young), 35 (middle aged), and 50 (old). In heads of young flies, cry mRNA showed expected daily oscillations, with peak at Zeitgeber time (ZT) 4–8 and trough at ZT 16 (Figure 1A). The levels and amplitude of cry mRNA oscillations were significantly dampened in 35 day-old flies and further reduced on day 50 (Figure 1A, Table S1), similar to a recent report (Luo et al. 2012). In contrast to heads, there was no statistically significant effect of age on cry oscillations in bodies (Table S1).
Fig. 1. CRY is reduced with age at both mRNA and protein levels.
A) Daily mRNA profiles of cry on days 5, 35, and 50 in heads of CS males, normalized to the trough (ZT 16) values set at 1 for day 5. White and black horizontal bars mark periods of light and dark respectively. Each data point represents mean ± SEM of three independent RNA samples. Statistical significance between day 5 vs. 35 and day 5 vs. 50 values was determined by two-way ANOVA with Bonferonni’s post hoc test (***p< 0.001, **p<0.01, and *p<0.05). B) Western blot showing CRY protein profile in head extracts of 5 and 50 day-old CS males with cryb and tim>CRY serving as negative and positive controls. Bar graph (below) indicates the relative band intensity for different ages and time points, with signal intensity at peak (ZT 0) in 5 day-old flies set as 100. Values are mean ± SEM of three independent bioreplicates. Statistical significance between day 5 vs. 50 values was determined using two-way ANOVA with Bonferonni’s post hoc test, and is denoted by ***p< 0.001 and *p<0.05 (LC-Loading control).
Given the age-related decline in cry mRNA levels, we compared CRY protein profiles in head extracts of 5 and 50 day-old CS flies. In young flies, the highest levels of CRY protein were detected near the end of the dark phase at ZT 0/24. CRY declined during the day to a trough at ZT 12. By comparison, CRY levels were significantly lower in old flies across all time points except ZT 16 (Figure 1B, Table S1).
Old flies with elevated CRY have increased oscillatory amplitude of several genes involved in the clock mechanism
We and others recently reported that the oscillatory amplitude of per, tim, Pdp1ε, and vri is dampened during aging (Luo et al. 2012; Rakshit et al. 2012); however, the underlying mechanisms are not fully understood. Since per and tim transcription is reduced and non-rhythmic in young cryb mutants (Stanewsky et al. 1998), we investigated whether CRY deficiency may contribute to the dampened oscillations of clock genes in old flies. We increased wild type CRY levels in all central and peripheral clock cells by combining the timGAL4 driver with the UAS-cry responder (tim>CRY). As a control, timGAL4 flies were crossed to UAS-cryb (tim>CRYB), which carries a missense mutation at the putative flavin-binding residue, rendering CRYB protein light-insensitive and unstable (Stanewsky et al. 1998; Emery et al. 2000; Busza et al. 2004). As a second control, timGAL4 was crossed to w1118 (timGAL4/+).
We first determined that CRY protein was significantly elevated in heads of both young and old tim>CRY flies, compared to the unstable mutant protein in tim>CRYB (Figure S1). At both ages, CRY levels in heads of tim>CRY flies were lower during the day (ZT 4) than at night (ZT 16) suggesting that the ectopic CRY undergoes light-dependent degradation similar as the endogenous CRY. To determine whether higher CRY levels affected the expression of clock genes, we obtained daily expression profiles of per and tim in heads of old flies. The control timGAL4/+ flies showed considerably dampened daily oscillations of per mRNA with trough at ZT 4 and a shallow peak between ZT 12–16 on day 50, consistent with recent reports (Luo et al. 2012; Rakshit et al. 2012). In contrast, age-matched tim>CRY flies had significantly higher amplitude of per mRNA expression with a well-defined peak at ZT 16 (Figure 2, Table S2), when a peak is usually observed in young flies (Rakshit et al. 2012). Expression of mutant CRYB protein did not increase per levels in old tim>CRYB flies, and they had a very similar mRNA profile as the timGAL4/+ control. We next measured the expression profile of tim, which encodes TIM protein that forms heterodimers with PER. Flies overexpressing CRY had higher amplitude of tim cycling due to significantly higher levels at the peak time point (ZT 16), compared to the age-matched controls (Figure 2, Table S2).
Fig. 2. Old flies with elevated CRY have increased clock gene oscillations.
Daily mRNA profiles of clock genes in heads of timGAL4/+, tim>CRY, and tim>CRYB males on day 50, normalized to the trough (ZT 4/ZT 16 for Clk) values set at 1 for timGAL4/+ control. White and black horizontal bars mark periods of light and dark respectively. Each data point represents mean ± SEM of three independent RNA samples. Statistical significance between the genotypes was determined by two-way ANOVA with Bonferonni’s post hoc test, and is denoted by ***p<0.001 and **p<0.01.
To determine whether other components of the clock mechanism are enhanced by the overexpression of CRY, we examined the mRNA profiles of Pdp1ε and vri that are also activated by CLK/CYC complexes and oscillate in phase with per and tim. We observed that mRNA oscillations for the two genes were significantly higher in heads of 50 day-old tim>CRY flies than in both controls (Figure 2, Table S2). Together, these data suggested that flies overexpressing CRY could have increased levels of Clk and cyc genes leading to increased CLK/CYC mediated activation of per, tim, Pdp1ε and vri. To begin addressing this question, we examined the expression levels of Clk and cyc mRNA in heads of old males. Control flies showed expected Clk mRNA oscillations, with peak levels at ZT 4 and trough at ZT 16. There was no significant difference in Clk mRNA profiles in heads of tim>CRY flies compared to both controls (Figure 2, Table S2). The expression of cyc is non-rhythmic (Bae et al. 2000), and does not significantly change with age in flies (Rakshit et al. 2012). Similar as for Clk, there was no difference in cyc mRNA levels in heads of cry overexpressing flies and controls (Figure 2, Table S2).
CRY overexpression prevents age-related weakening of rest/activity rhythms
Higher oscillations of several clock genes in heads of old CRY overexpressing flies prompted us to examine the strength of behavioral rhythms in these flies. We recorded locomotor activity rhythms in tim>CRY and control flies on days 5–15, 35–45, and 50–60, to assess age-related changes in rhythm strength and other circadian parameters. In addition, we also monitored activity across lifespan in two lines of cry-null flies (cry01 and cry02) that show normal rest/activity rhythms when young (Dolezelova et al. 2007). Males were monitored for 3 d in LD 12:12 followed by 7 d in DD. Young control flies showed bimodal activity distribution in LD with morning and evening peaks and anticipation of lights on and off (Fig. S2, Table S3). There was a general age-related reduction in average LD activity counts accompanied by the loss of morning anticipation. However, 50 day-old tim>CRY flies showed pronounced morning and evening peaks while one or both of these peaks were attenuated in control or cry-null flies at this age (Figure S2), suggesting that the well-known role of CRY in circadian photoreception persists in old flies overexpressing CRY.
Our analysis of free-running rhythms in DD revealed that 50–60 day-old tim>CRY flies maintained remarkably robust rest/activity rhythms while those rhythms decayed in most control flies (Figure 3). The average rhythm strength (FFT) was significantly higher in young tim>CRY flies than in controls and remained higher across lifespan (Figure 3B). Young cry-null flies had strong activity rhythms as expected (Dolezelova et al. 2007), and interestingly, the average FFT for cry01 was significantly higher than for cry02 (Figure 3B, S3A). To better understand senescence in context of rhythm strength, we calculated the time required for this function to decline to 50% and 75% of its peak value as described (Martin et al. 2005). This analysis suggests that aging tim>CRY flies have longer FFT decline time than cry01, while both showed similar FFT at young age (Figure S3A).
Fig. 3. CRY overexpression prevents disruption of locomotor activity rhythms.
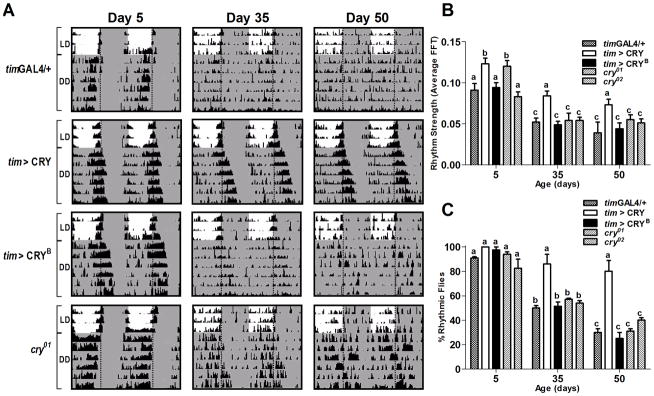
A) Locomotor activity profiles of representative 5, 35, and 50 day-old males of the indicated genotypes. Flies of each age were monitored in LD (12:12) for 3 d, followed by 7 d in DD at 25°C. Shaded areas represent periods of darkness. Vertical dotted lines indicate time of lights-off (ZT/CT 12). B) Average rhythm strength, and C) Percentage of rhythmic flies on days 5, 35, and 50. Flies with FFT values >0.04 were considered rhythmic. Values are mean ± SEM of two independent activity analyses. Total number of flies analyzed for each genotype is indicated in Table S3. Statistical significance was determined using two-way ANOVA with Bonferroni’s post hoc test, and bars with different letters are significantly different at p<0.05.
Overall calculation of the percentage of rhythmic flies pooled from two independent experiments demonstrated that tim>CRY flies were still 82% rhythmic on days 50–60 when the proportion of rhythmic flies decreased to below 40% in control and cry-null lines (Figure 3C, Table S3). Additional analysis of rhythmicity as a function of age for all genotypes confirmed that tim>CRY flies had reduced rates of rhythm loss and extended rhythmicity decline times compared to all other genotypes examined (Figure S3B). Persistence of behavioral rhythms in DD suggests that a durable intrinsic circadian system is maintained in old tim>CRY flies.
Replenishment of CRY in all clock-expressing cells improves the healthspan of aging flies
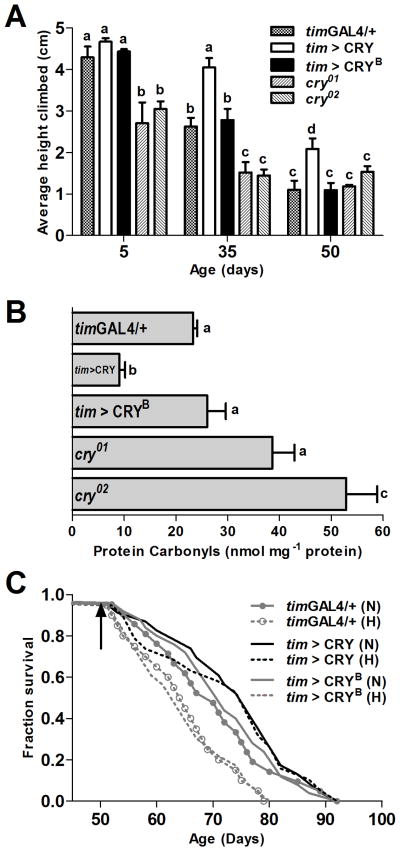
Correlational data suggest links between degradation of circadian rhythms and accelerated aging; however, it is not known whether rejuvenation of the circadian system is associated with extended healthspan. Our finding that replenishment of CRY restores molecular and behavioral rhythms in aging flies provided a unique opportunity to examine the relationship between strength of circadian rhythms and the rate of aging. One of the important biomarkers of aging in Drosophila is the decline of climbing ability, which can be measured via the RING assay that utilizes negative geotaxis to assess vertical mobility (Rhodenizer et al. 2008). We used the RING assay to compare climbing ability in flies with altered CRY levels across lifespan. On day 5, the climbing ability of tim>CRY flies was not different from the timGAL4/+ or tim>CRYB controls; however, both cry01 and cry02 mutants had significantly lower climbing ability even at this young age (Figure 4A, Table S4). The climbing ability declined significantly in both controls and cry-null flies on day 35. In contrast, tim>CRY flies maintained significantly higher vertical mobility that was not different from day 5 (p>0.05). Flies of all genotypes further lost their climbing ability with progressing age; however, tim>CRY flies performed significantly better than controls even on day 50 (Figure 4A, Table S4). This is confirmed by additional calculations showing that tim>CRY flies had the longest decline times for climbing ability (Figure S3C).
Fig. 4. Flies overexpressing CRY have improved healthspan.
A) Vertical mobility was measured by the RING assay in 5, 35, and 50 day-old males of the indicated genotypes. Bars represent mean height climbed (±SEM), based on testing 4 vials per genotype, each containing 25 flies. Statistical significance was determined using two-way ANOVA with Bonferroni’s post hoc test, and bars with different letters are significantly different at p<0.05. B) Oxidative damage in the form of protein carbonyls (PC) were measured in heads of 50 day-old males of the indicated genotypes at ZT 8. tim>CRY flies had significantly lower PC compared to the timGAL4/+ and tim>CRYB controls, and cry01. There was significantly higher PC accumulation in heads of cry02 flies compared to timGAL4/+ control. Bars represent mean carbonyl levels (±SEM), based on testing 3 independent sets of flies, each containing 75 flies in 3 technical repeats of 25 flies each. Statistical significance was determined using one-way ANOVA with Tukey’s post hoc test, and bars with different letters are significantly different at p<0.05. C) Lifespan of timGAL4/+, tim>CRY, and tim>CRYB flies in normoxia (solid lines) and following 24 h hyperoxia (dotted lines) on day 50 (marked by arrow). In normoxia, there was no significant difference in mean survival curves among the genotypes. Hyperoxia on day 50 significantly shortened the lifespan of timGAL4/+, and tim>CRYB controls (p<0.05) but not tim>CRY flies.
Another biomarker of aging is the accumulation of oxidatively damaged proteins, which can be biochemically measured as levels of protein carbonyls (PC) in the total protein extract (Krishnan et al. 2009). Since alterations in CRY levels affected climbing ability, we tested whether the accumulation of oxidative damage in old age was also affected. We quantified PC in heads of 50 day-old flies at ZT 8, which is the peak of PC accumulation (K. Rakshit, unpublished). Indeed, tim>CRY flies had significantly lower PC accumulation in heads, compared to the controls (Figure 4B). On the other hand, both cry01 and cry02 flies had higher PC levels than the controls and this difference reached statistical significance in cry02 flies. Together, these data suggest that CRY may have novel anti-aging functions related to proteostasis.
An important aspect of healthy aging is the ability to withstand homeostatic insults, such as oxidative stress. We reported that flies with disrupted clocks showed significantly increased mortality risk after short-term oxidative challenge (24 h of 100% hyperoxia) in the middle or old age (Krishnan et al. 2009). We used this assay here to test whether flies with elevated CRY levels are capable of buffering short-term oxidative challenge later in life, by exposing 50 day-old flies to 24 h hyperoxia. This treatment significantly shortened the lifespan of timGAL4/+ and tim>CRYB flies. In contrast, tim>CRY flies were resilient to this stress and had very similar survival curves as their normoxia controls, despite a few initial deaths (Figure 4C, Table S5). The lifespan of tim>CRY flies under normoxia was ~7% longer than control flies, albeit this difference was not statistically significant.
Overexpression of CRY in central pacemaker neurons does not improve rest/activity rhythms during aging
In young flies, locomotor activity rhythms are controlled by ~150 central pacemaker neurons in the brain (Nitabach & Taghert 2008). The timGAL4 driver increases CRY in both central and peripheral clock cells. Therefore, we next examined whether overexpression of CRY in central pacemaker neurons alone is sufficient to restore locomotor activity rhythms during aging. We initially used the PdfGAL4 driver to overexpress CRY or CRYB in PDF-positive lateral ventral neurons (LNvs), which are crucial for DD activity rhythms in young flies (Nitabach & Taghert 2008). Also, PdfGAL4 driven UAS-cry can rescue behavioral photo responses in cryb mutants (Emery et al. 2000). Surprisingly, we found that Pdf driven expression of CRY in LNvs is not sufficient to prevent age-related decay of behavioral rhythms (activity of PdfGAL4 in PDF-positive clock neurons was confirmed by crossing with UAS-gfp - not shown). Pdf>CRY flies showed weakening and age-related fragmentation of rest/activity rhythms similar as the Pdf>CRYB controls (Figure 5A). There was no significant difference in the average daily activity in LD between Pdf>CRY and Pdf>CRYB flies (Figure S4, Table S3). Analysis of fly activity in DD revealed very similar age-related decline in the rhythm strength (Figure 5B) and the percentage of rhythmic flies (Figure 5C) in both Pdf>CRY and Pdf>CRYB flies, with no statistical difference at any tested age (Table S3). Additional analysis of rhythm strength and % rhythmicity functions with age confirmed similar rates of decline in Pdf>CRY and Pdf>CRYB flies (Figure S5A–B).
Fig. 5. Rescue of CRY in central pacemaker neurons alone does not improve locomotor activity rhythms.
A) Locomotor activity profiles of representative Pdf>CRY and Pdf>CRYB males on days 5, 35, and 50. Flies of each age were monitored in LD (12:12) for 3 d, followed by 7 d in DD at 25°C. Shaded areas represent periods of darkness. Vertical dotted lines indicate time of lights-off (ZT/CT 12). B) Average rhythm strength, and C) Percentage of rhythmic Pdf>CRY and Pdf>CRYB flies on days 5, 35, and 50. D) Representative locomotor activity profiles, E) Average rhythm strength, and F) Percentage of rhythmic cry39>CRY and cry39>CRYB flies on days 5, 35, and 50. (B, C, D, F - Flies with FFT values >0.04 were considered rhythmic. Values are mean ± SEM of two independent activity analyses. Total number of flies analyzed for each genotype is indicated in Table S3. Statistical significance was determined using two-way ANOVA with Bonferroni’s post hoc test, and bars with different letters are significantly different at p<0.05.)
We then overexpressed CRY or CRYB using another driver line cryGAL4-39, which is active in additional groups of central pacemaker neurons regulating rest/activity rhythm (Klarsfeld et al. 2004). We confirmed that the cryGAL4-39 driver drives expression in several groups of pacemaker neurons but not in peripheral clocks by crossing with UAS-gfp (not shown). Despite that a larger subset of central pacemaker neurons was targeted, rhythms of locomotor activity decayed at a similar rate in cry39>CRY flies as in cry39>CRYB controls (Figures 5D–F, S4, S5A–B, Table S3).
Rescue of CRY in central pacemaker neurons alone does not improve healthspan
We tested whether rescue of CRY in the central pacemaker neurons affects biomarkers of aging that were improved by CRY overexpression in both central and peripheral clocks. There was no statistically significant difference in climbing ability of Pdf>CRY or cry39>CRY flies compared to their respective controls at any physiological age (Figure 6A, S5C). Neither was there any statistically significant difference in PC accumulation in heads of old Pdf>CRY or cry39>CRY flies compared to their respective Pdf>CRYB and cry39>CRYB controls (Figure 6B).
Fig. 6. CRY-restoration in central pacemaker neurons alone does not improve healthspan.
A) Vertical mobility was measured by the RING assay in 5, 35, and 50 day-old males of the indicated genotypes. Bars represent mean height climbed (±SEM), based on testing 4 vials per genotype, each containing 25 flies. Statistical significance was determined using two-way ANOVA with Bonferroni’s post hoc test, and bars with different letters are significantly different at p<0.05. B) Oxidative damage in the form of protein carbonyls (PC) were measured in heads of 50 day-old males of the indicated genotypes at ZT 8. Bars represent mean carbonyl levels (±SEM), based on testing 3 independent sets of flies, each containing 75 flies in 3 technical repeats of 25 flies each. Statistical significance was determined using one-way ANOVA with Tukey’s post hoc test.
Discussion
The decline of circadian output rhythms is a common signature of aging in animals including humans. It was recently reported in Drosophila that aging is associated with dampened molecular oscillations of core clock genes in peripheral but not central oscillators (Luo et al. 2012; Rakshit et al. 2012). Given the persistence of molecular oscillations in the central clock, there is currently no explanation as to why rest/activity rhythms lose their robustness during aging. The results of our study suggest that enhancement of peripheral clocks may prevent degradation of the circadian output. Furthermore, our data suggest a cause and effect relationship between the molecular and behavioral decay of circadian rhythms and fly healthspan.
We observed reduction in cry mRNA levels in heads of old flies, consistent with a previous study (Luo et al. 2012). Interestingly, cry decline was observed in the heads but not in bodies similar to our results on other clock genes (Rakshit et al. 2012), suggesting that peripheral clocks in the fly head are more susceptible to aging than clocks in the body tissues. We also report that CRY protein is significantly reduced in heads of 50 day-old CS flies at most time points, although it still oscillates in a phase similar to young flies. This suggests that the circadian system is severely attenuated yet functional in old flies.
Our data shows that age-related CRY decline is functionally linked to decay of the circadian system, as overexpression of wild type CRY significantly improved the amplitude of per, tim, Pdp1ε, and vri, genes in heads of old flies. These results may seem surprising as it was reported that overexpression of CRY together with PER using the eye-specific gmrGAL4 driver suppressed tim mRNA expression (Collins et al. 2006). However, this study also reported that overexpression of CRY alone significantly increased the amplitude of tim and vri in young flies (Collins et al. 2006), consistent with our current data in old flies. Further studies are required to determine whether replenishment of CRY in peripheral clock cells may enhance the transcriptional activity of CLK-CYC complexes in the heads. Although we show that neither aging nor CRY overexpression altered Clk and cyc mRNA levels, downstream post-translational mechanisms or epigenetic modifications could be conceivably affected in old flies, and restored by CRY overexpression. In support of this notion, it was reported that CLK protein levels are constitutively low in heads of cryb flies (Collins et al. 2006). Further experiments would be required to understand the mode of CRY action in maintaining molecular circadian rhythms. Interestingly, age-related decay of the circadian clock at molecular and behavioral levels was reported in mPer1 and mCry2 knockout mice (Oster et al. 2003).
Our study reveals that restoration of CRY in all clock expressing cells prevented the decay of locomotor activity rhythms with age. While rhythms in LD were somewhat improved in tim>CRY flies consistent with its role as a photoreceptor, remarkable differences were observed in DD where 35 and 50 day-old flies with overexpressed CRY showed almost young-like rest/activity rhythms suggesting that the free-running circadian system was intact in those flies. We also found that augmentation of CRY in central clock neurons alone was not sufficient to restore rest/activity rhythms. While only central clocks are required for behavioral rhythms in young flies (Hardin 2011), our data suggest that peripheral clocks may additionally be necessary for maintaining neuronal homeostasis and preventing the degradation of circadian output pathways in aging individuals. A recent study suggests that circadian rhythms may be also strengthened by Pdf overexpression (Umezaki et al. 2012), suggesting that circadian rejuvenation may be achieved in more than one way. Interestingly, decay of clock output pathways despite strong mPer2 oscillations in the central clock (SCN) has been recently reported in mouse (Nakamura et al. 2012).
Our data show that CRY-mediated strengthening of circadian rhythms is associated with deceleration of functional decline normally observed in aging flies. Namely, CRY overexpressing flies maintained significantly higher climbing ability than their controls at every age tested, suggesting that these flies had a slower rate of aging. Furthermore, these flies had significantly lower oxidative damage in old age. Consistent with these findings, they were also able to recover from exposure to short-term hyperoxia. In contrast, cry-null flies showed accelerated functional decline and higher accumulation of oxidatively damaged proteins. Together, these results suggest that strong circadian rhythms are important for maintaining the health of an organism during aging, and also point toward novel anti-aging functions of CRY. While several studies demonstrated that genetic or environmental disruptions of the clock function accelerate physiological aging and age-related diseases, it is not clear whether rejuvenation of the circadian system can delay aging. Here, we achieved circadian rejuvenation by ectopic increase in age-reduced protein CRY tha, and demonstrate that this treatment extends healthspan in flies. CRY supplementation and conversely, CRY deficiency at the same chronological age, shifted the readouts of aging in opposite directions. Namely, tim>CRY flies showed better climbing vigor and reduced oxidative damage than age-matched controls, while climbing was impaired even in the young cry-null flies and they accumulated greater oxidative damage during aging. A limitation of our study is that the timGAL4 driver causes supra-physiological increase in the levels of CRY and the associated phenotype may be a result of excess of CRY instead of a mere replenishment with age. However, tim>CRY flies did not show aberrant phenotypes that could suggest off-target effects in molecular or behavioral parameters measured. Moreover, tim>CRYB controls show results very similar to wild type controls despite supra-physiological levels of cry mRNA (not shown).
Taken together, our data suggest that CRY-mediated improvement of circadian rhythms may delay functional aging. This could be linked to the enhanced expression of clock-controlled effector genes that are involved in many pathways maintaining temporal homeostasis (Wijnen & Young 2006; Krishnan et al. 2008). On the other hand, we cannot exclude that healthspan benefiting effects of CRY overexpression may be uncoupled from circadian rhythms and instead linked to its pleiotropic effects. Several novel non-circadian functions of CRY have been recently suggested in Drosophila (Fogle et al. 2011; Seay & Thummel 2011; Kumar et al. 2012). Intriguingly, recent in vivo analysis comparing the transcriptional activity of human and fly cryptochromes in transgenic Drosophila suggests that these proteins may share common signaling pathways in DD, regulating genes implicated in stress response (Vieira et al. 2012).
In summary, we demonstrate that age-related dampening of clock gene oscillations and daily activity rhythms can be significantly improved by the genetic manipulation of a protein that is best known for its role in circadian photo-transduction. We provide evidence that overexpression of CRY helps slow down the aging process and reverse age-associated phenotypes. Our study suggests novel anti-aging role for the gene cryptochrome in Drosophila, as it has profound effects on the health and fitness most likely by improving clock-controlled effector genes. However, the mode of CRY action as an anti-aging factor will require future studies.
Experimental Procedures
Fly rearing
D. melanogaster were reared on 1% agar, 6.25% cornmeal, 6.25% molasses, and 3.5% Red Star yeast at 25°C in 12 h light:12 h dark (LD) cycles (with an average light intensity of 2000 lux). By convention, lights-off is denoted as Zeitgeber time (ZT) 12. For experiments on aging flies, cohorts of 50–75 mated males, were housed in 8 oz round bottom polypropylene bottles (Genesee Scientific, San Diego, CA) inverted over 60 mm Falcon Primaria Tissue culture dishes (Becton Dickinson Labware, Franklin Lakes, NJ) containing 15 mL of diet. Diet dishes were replaced daily without CO2, after tapping flies to the bottom of the bottle. For other assays, cohorts of 25 mated males were reared in narrow vials (Genesee Scientific).
Fly Stocks
Canton S mated males were used for RNA and protein measurements of cry during aging. For the cry overexpression studies, w1118 served as the wild type control to which driver and responder lines were backcrossed for 8 generations. Driver lines w1118;+;BsR-B (PdfGAL4) (Park et al. 2000), w;cryGal4#39;+ (cry39GAL4) (Klarsfeld et al. 2004), w;timGal4(62);+ (timGAL4) (Kaneko & Hall 2000) were crossed to either responder line w1118;UAS-cry24;+ (UAS-cry) (Emery et al. 1998), or w1118;w+ UAScryb31;+ (UAS-cryb) (Emery et al. 2000) as a control. Additionally, cry01 and cry02 mutants (Dolezelova et al. 2007) were backcrossed to w1118 and used in some experiments.
Quantitative Real-Time Polymerase Chain Reaction
Three independent bio-replicates of flies were collected at 4 h intervals around the clock on days 5, 35, and 50. Total RNA was extracted from fly heads and bodies separately using TriReagent (Sigma, St. Louis, MO). The samples were purified and treated with Takara Recombinant DNase I (Clontech Laboratories Inc., Mountain View, CA). Synthesis of cDNA was achieved with the iScript cDNA synthesis kit (BioRad, Hercules, CA). Quantitative real-time PCR (qRT-PCR) was performed on the StepOnePlus Real-Time machine (Applied Biosystems, Carlsbad, CA) under default thermal cycling conditions, with a dissociation curve step. Every reaction contained Power SYBR Green (Applied Biosystems), 10 ng cDNA, and 400 nM primers. Primer sequences are available upon request. Data were analyzed using the 2−ΔΔCT method with mRNA levels normalized to the gene rp49. Relative mRNA amplitude was calculated with respect to the trough levels on day 5 set as 1 (for aging study) or trough levels of timGAL4/+ (control) set as 1 on day 50 (for cry overexpression studies).
Western Blotting
Three independent bioreplicates of 5 and 50 day-old males of different genotypes were collected at specific ZT points. About 5–10 fly heads/time point were homogenized on ice in Laemmli buffer, sonicated, boiled at 100°C for 5 min, and centrifuged at 12000 g at 4°C. A constant ratio of the buffer (7 μL/head) was used to ensure equal protein loading and separation on 10% acrylamide gel. Proteins were transferred to the 0.45 μm polyvinylidene fluoride (PVDF) Immobilon-FL membrane (Millipore Billerica, MA) and incubated in 1X TBST (10mM Tris, 0.15M NaCL, 0.1% Tween-20, pH 7.5) + 5% milk for 2 h, then overnight at 4°C with 1:2,000 anti-CRY (Rush et al. 2006) in blocking buffer. Membranes were treated for 2 h with 1:20,000 goat anti-rabbit IRDye680 (LI-COR Biosciences, Lincoln, NE). Proteins were quantified using the LI-COR Odyssey Infrared Imaging System software (v. 3.0).
Locomotor Activity Analysis
Flies were entrained in LD 12:12 at 25°C. Locomotor activity of 5, 35, and 50 day-old males was recorded or 3 d in LD 12:12, followed by 7 d in constant darkness (DD) using the Trikinetics locomotor activity monitor (Waltham, MA). For a quantitative measure of circadian rhythmicity in DD, Fast Fourier Transform (FFT) analysis was conducted using ClockLab software (Actimetrics; Coulbourn Instruments, Whitehall, PA). Flies with FFT values <0.04 were classified as arrhythmic, ones with values of 0.04–0.08 were classified as weakly rhythmic, whereas flies with FFT values >0.08 were considered strongly rhythmic. Flies with both weak and strong rhythms that showed a single peak in the periodogram were included in the calculation of the free running period using the ClockLab software (Actimetrics, Wilmette, IL).
Rapid iterative negative geotaxis (RING) assay
Vertical mobility was tested using the RING assay as described (Rhodenizer et al. 2008; Krishnan et al. 2012). Briefly, 4 groups of 25 mated males of each age and genotype were transferred into empty vials without anesthesia, and the vials were loaded into the RING apparatus. The apparatus was rapped three times in rapid succession to initiate a negative geotaxis response. The flies’ movements in tubes were videotaped and digital images captured 4 s after initiating the behavior. The climbed distance was calculated for each fly and expressed as average height climbed in the 4 s interval. The performance of flies in a single vial was calculated as the average of 5 consecutive trials (interspersed with a 30 s rest).
Protein Carbonyl (PC) Assay
To assess oxidative damage, protein carbonyls were measured in head homogenates of 50 day-old males of various genotypes at 370 nm after reaction with 2,4-dinitrophenylhydrazine (DNPH), using a Synergy 2 plate reader (BioTek, Winooski, VT), as described previously (Krishnan et al. 2008). Results were expressed as nmol mg−1 protein using an extinction coefficient of 22,000 M−1 cm−1.
Hyperoxia treatment and lifespan analysis
For hyperoxia exposure, 4 cohorts of 25 males in narrow vials with diet were placed in a Plexiglass chamber filled with oxygen (100% medical grade) flowing at a constant rate (300ml/min) for 24 h, as described (Krishnan et al. 2009). Control flies were transferred to narrow vials with diet and kept under normoxia. After the treatment, flies were transferred from narrow vials to bottles as described in the fly rearing section. Diet was replaced on alternate days without anesthesia, and mortality was recorded daily.
Statistical Analysis
Data were statistically analyzed with GraphPad Prism (v.5.0) and GraphPad Instat (v.3.0; San Diego, CA). The qRT-PCR, locomotor activity analysis and RING data were evaluated by two-way analysis of variance (ANOVA) with Bonferroni’s post hoc test. Additionally, we plotted FFT, % rhythmic, and RING data as points to better visualize the slope of functional decline with age for each genotype, and calculated the time required for each function to decline to 50% and 75% of its peak value, interpolated from second-order polynomial curves as described (Martin et al. 2005). PC data were evaluated by one-way ANOVA with Tukey’s post hoc test. For Western data, the relative strength of the signals was quantified using LI-COR Image analysis software (v.3.0) and subjected to two-way ANOVA with Bonferroni’s post hoc test. Life span and survival curves were plotted following Kaplan Meier survival analysis and statistical significance of curves assessed using the Log-Rank (Mantel-Cox) test.
Supplementary Material
Acknowledgments
We are grateful to Eileen Chow for help with fly rearing. We thank Patrick Emery for anti-CRY antibody and fly stocks. Other fly lines were kindly shared by Paul Hardin, Paul Taghert, and Jeff Hall. We are also grateful to anonymous reviewers for their constructive criticism and insightful comments. This research was supported by NIH R21-AG038989 and R21-NS075500 grants to JMG. KR is supported by NSF IGERT in Aging Sciences Fellowship at Oregon State University (DGE 0965820).
Footnotes
Author Contributions
KR and JMG designed the study. KR conducted all the experiments and wrote the manuscript. JMG provided critical feedback and revised the first draft. The authors declare no conflict of interest.
Contributor Information
Kuntol Rakshit, Email: rakshitk@science.oregonstate.edu.
Jadwiga M. Giebultowicz, Email: giebultj@science.oregonstate.edu.
References
- Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, Lee C, Nikitin AY. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–1204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Lee C, Hardin PE, Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER- TIM complex. J Neurosci. 2000;20:1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Collins B, Mazzoni EO, Stanewsky R, Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr Biol. 2006;16:441–449. doi: 10.1016/j.cub.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol. 2006;16:R914–916. doi: 10.1016/j.cub.2006.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics. 2007;177:329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, So V, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Foster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman MA, Swaab DF. Living by the clock: the circadian pacemaker in older people. Ageing Res Rev. 2006;5:33–51. doi: 10.1016/j.arr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Huang YL, Liu RY, Wang QS, Van Someren EJ, Xu H, Zhou JN. Age-associated difference in circadian sleep-wake and rest-activity rhythms. Physiol Behav. 2002;76:597–603. doi: 10.1016/s0031-9384(02)00733-3. [DOI] [PubMed] [Google Scholar]
- Ivanchenko M, Stanewsky R, Giebultowicz JM. Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J Biol Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Malpel S, Michard-Vanhee C, Picot M, Chelot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103:13843–13847. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, Hall JC, Hardin PE, Dryer SE. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Krishnan N, Davis AJ, Giebultowicz JM. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;374:299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Kretzschmar D, Rakshit K, Chow E, Giebultowicz J. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging. 2009;1:937–948. doi: 10.18632/aging.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Rakshit K, Chow ES, Wentzell JS, Kretzschmar D, Giebultowicz JM. Loss of circadian clock accelerates aging in neurodegeneration-prone mutants. Neurobiol Dis. 2012;45:1129–1135. doi: 10.1016/j.nbd.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Chen D, Sehgal A. Dopamine acts through Cryptochrome to promote acute arousal in Drosophila. Genes Dev. 2012;26:1224–1234. doi: 10.1101/gad.186338.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Luo W, Chen WF, Yue Z, Chen D, Sowcik M, Sehgal A, Zheng X. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell. 2012;11:428–438. doi: 10.1111/j.1474-9726.2012.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Gargano JW, Grotewiel M. A proposed set of descriptors for functional senescence data. Aging Cell. 2005;4:161–164. doi: 10.1111/j.1474-9726.2005.00155.x. [DOI] [PubMed] [Google Scholar]
- Nakamura TJ, Nakamura W, Yamazaki S, Kudo T, Cutler T, Colwell CS, Block GD. Age-related decline in circadian output. J Neurosci. 2012;31:10201–10205. doi: 10.1523/JNEUROSCI.0451-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Oster H, Baeriswyl S, Van Der Horst GT, Albrecht U. Loss of circadian rhythmicity in aging mPer1−/−mCry2−/− mutant mice. Genes Dev. 2003;17:1366–1379. doi: 10.1101/gad.256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit K, Krishnan N, Guzik EM, Pyza E, Giebultowicz JM. Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol Int. 2012;29:1–10. doi: 10.3109/07420528.2011.635237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, O’Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodenizer D, Martin I, Bhandari P, Pletcher SD, Grotewiel M. Genetic and environmental factors impact age-related impairment of negative geotaxis in Drosophila by altering age-dependent climbing speed. Exp Gerontol. 2008;43:739–748. doi: 10.1016/j.exger.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush BL, Murad A, Emery P, Giebultowicz JM. Ectopic CRYPTOCHROME renders TIM light sensitive in the Drosophila ovary. J Biol Rhythms. 2006;21:272–278. doi: 10.1177/0748730406290416. [DOI] [PubMed] [Google Scholar]
- Seay DJ, Thummel CS. The circadian clock, light, and cryptochrome regulate feeding and metabolism in Drosophila. J Biol Rhythms. 2011;26:497–506. doi: 10.1177/0748730411420080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in. Drosophila Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Umezaki Y, Yoshii T, Kawaguchi T, Helfrich-Forster C, Tomioka K. Pigment-Dispersing Factor Is Involved in Age-Dependent Rhythm Changes in Drosophila melanogaster. J Biol Rhythms. 2012;27:423–432. doi: 10.1177/0748730412462206. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Scarbrough K, Takahashi JS, Turek FW. Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am J Physiol. 1997;273:R1957–1964. doi: 10.1152/ajpregu.1997.273.6.R1957. [DOI] [PubMed] [Google Scholar]
- Vieira J, Jones AR, Danon A, Sakuma M, Hoang N, Robles D, Tait S, Heyes DJ, Picot M, Yoshii T, Helfrich-Forster C, Soubigou G, Coppee JY, Klarsfeld A, Rouyer F, Scrutton NS, Ahmad M. Human cryptochrome-1 confers light independent biological activity in transgenic Drosophila correlated with flavin radical stability. PLoS One. 2012;7:e31867. doi: 10.1371/journal.pone.0031867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13:639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EA, Weaver DR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 2011;3:479–493. doi: 10.18632/aging.100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.