Abstract
In pediatric sickle cell disease (SCD) patients, it has been reported that higher systolic blood pressure (SBP) is associated with increased risk of a silent cerebral infarction (SCI). SCI is a major cause of neurologic morbidity in children with SCD, and blood pressure is a potential modulator of clinical manifestations of SCD; however, the risk factors underlying these complications are not well characterized. The aim of this study was to identify genetic variants that influence SBP in an African American population in the setting of SCD, and explore the use of SBP as an endo-phenotype for SCI. We conducted a genome-wide meta-analysis for SBP using two SCD cohorts, as well as a candidate screen based on published SBP loci. A total of 1,617 patients were analyzed, and while no SNP reached genome-wide significance (P-value<5.0x10-8), a number of suggestive candidate loci were identified. The most significant SNP, rs7952106 (P-value=8.57x10-7), was in the DRD2 locus on chromosome 11. In a gene-based association analysis, MIR4301 (micro-RNA4301), which resides in an intron of DRD2, was the most significant gene (P-value=5.2x10-5). Examining 27 of the previously reported SBP associated SNPs, 4 SNPs were nominally significant. A genetic risk score was constructed to assess the aggregated genetic effect of the published SBP variants, demonstrating a significant association (P=0.05). In addition, we also assessed whether these variants are associated with SCI, validating the use of SBP as an endo-phenotype for SCI. Three SNPs were nominally associated, and only rs2357790 (5’ CACNB2) was significant for both SBP and SCI. None of these SNPs retained significance after Bonferroni correction. Taken together, our results suggest the importance of DRD2 genetic variation in the modulation of SBP, and extend the aggregated importance of previously reported SNPs in the modulation of SBP in an African American cohort, more specifically in children with SCD.
Introduction
Sickle cell disease (SCD) is an inherited hemoglobin disorder affecting approximately 1 in 600 individuals of African American ancestry in the United States [1]. The clinical manifestations of SCD begin early in life [2] and continue with an increasing incidence of adverse events, involving genetic as well as environmental factors [3]. Previous studies have demonstrated that the arterial blood pressure in steady state patients with SCD is significantly lower than that of age, sex and race matched controls [4-9]. These findings are counterintuitive in view of the well-known vascular and renal abnormalities associated with SCD and the high prevalence of hypertension in African American adults [10-12]. Recently, in SCD children with no history of overt stroke or seizures, we have reported that higher systolic blood pressure (SBP) is associated with increased risk of an silent cerebral infarction (SCI) [13], a common form of neurological injury among children with SCD, occurring in at least 27% prior to six years of life and 37% by 14 years of life [14,15]. In addition, previous clinical studies have shown that the SCD may have a deleterious effect on myocardium, which contributes to abnormal rates of change in left ventricular cavity size and systolic/diastolic function in SCD patients [16-18]. Indeed, nearly one third of adults with SCD also develop an elevated tricuspid regurgitant velocity (TRV) that is associated with a much higher death rate in SCD patients compared to patients with SCD without elevated TRV. About 5-10% of these patients have true pulmonary hypertension [19]. Identifying genetic factors associated with systolic blood pressure (SBP) may help define both patho-physiological mechanisms, as well as identify patients at increased risk for SCI.
Studies of familial aggregation provide significant evidence that blood pressure is a highly heritable trait [20]; however, these estimates provide no information as to whether the same genetic variants influence blood pressure across human populations. In 2009, two genome-wide association studies (GWAS) and meta-analysis of inter-individual blood pressure variation in adults were conducted by the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium [21] and the Global Blood Pressure Genetics (Global BPgen) Consortium [22], leading to the identification of a number of genomic loci implicated with these traits, including 7 for SBP. Subsequently, these consortia were combined and expanded to form the International Consortium for Blood Pressure (ICBP), who reported many additional novel loci for these traits utilizing individuals of European (N=200,000), East Asian (N=30,000), South Asian (N=24,000) and African (N=20,000) descents [23]. In the present study, we sought to apply two complementary approaches for identifying SBP variants in individuals with SCD. First, we performed a genome-wide association study for SBP in SCD cohorts of African American ancestry. Second, we attempted to validate the ICBP identified SBP loci in these patients. Finally, we assessed whether these reported variants are also associated with SCI, exploring the use of SBP as an endo-phenotype for SCI.
Methods
Study and Population Samples
This study includes two unrelated admixed African American ancestry SCD cohorts. Study protocols of both cohorts were approved by the Institutional Review Board (IRB) of Johns Hopkins University and Boston Medical Center. Additionally, IRB approval was acquired from all of the participating sites for subject enrollment and conducted in accordance with institutional guidelines.
Silent Infarct Transfusion (SIT) Trial cohort
The Silent Infarct Transfusion (SIT) Trial is an international, multi-center clinical study funded by the National Institute of Neurological Disorders and Stroke (NINDS) (http://sitstudy.wustl.edu/) [24]. All the participants included in our study are of African American ancestry and written informed consent was obtained from parents of the SCD-affected individuals. For each patient, DNA was collected from Epstein-Barr virus (EBV) transformed lymphoblasts using Puregene Genomic DNA Purification kits (Gentra Systems, Inc). Demographic and phenotypic information were collected for each participant and the inclusion criteria for the recruitment were age (5-15 years) and hemoglobinopathy diagnosis (either Hb SS or Hb SB0-thalassemia). Details of the SIT Trial study design are given elsewhere [24].
Cooperative Study of Sickle Cell Disease
The Cooperative Study of Sickle Cell Disease (CSSCD) was a multi-institutional prospective longitudinal study of SCD funded by the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) [25]. In our study, we only included CSSCD participants who are of African American ancestry, and to match the samples with the SIT Trial cohort, age inclusion criteria of <15 years was used. Details of the CSSCD study design are given elsewhere [25].
Phenotype Assessment
Systolic Blood Pressure (SBP)
In the SIT Trial, a single measurement of blood pressure was obtained at well-visit for children with SCD. No guidelines were formulated to include uniform assessment. In the CSSCD cohort, longitudinal blood pressure measurements were collected during routine visits but not during episodes of acute illness. Blood pressure measurements were made by study nurses or physicians following the procedure described in the study manual of operations [25]. Subjects were asked not to smoke for at least 30 minutes prior to the examination and were allowed to rest quietly for at least 5 minutes before the measurement was made. A single measurement of pressure was made with the patient in the sitting position. Mercury sphygmomanometers were used for the measurement with a cuff size sufficient to cover two thirds of the upper arm. Systolic and diastolic pressures were reported as the first and fifth Korotkoff sounds, respectively.
Silent Cerebral Infarction (SCI)
A magnetic resonance imaging (MRI) was obtained from all participants and the presence or absence of SCI was adjudicated by a blinded panel of three expert neuro-radiologists.
Genotyping and Quality Control
Genotyping of the SIT Trial cohort was performed in two stages. For stage 1, a subset of 573 samples, along with 24 International HapMap Consortium [26] controls and 13 known duplicates, were genotyped at the Center for Inherited Disease Research (CIDR) at Johns Hopkins University using the Illumina HumanHap650Y SNP array (Illumina Inc., San Diego CA, USA). This array contains approximately 661,000 SNPs, of which ~100,000 were selected as tags for populations with African ancestry [27]. The Beadstudio software (Illumina Inc.) was used to cluster the data and samples with <96.5% call rate were re-genotyped. The reproducibility, calculated from duplicate pairs was 99.98% and genotype concordance with HapMap data was 99.76%. In stage 2, 509 samples were genotyped at the Center for Disease Control (CDC) at Washington University using the Illumina Infinium HumanOmni1-Quad SNP array (Illumina Inc.) and achieved the call rate of ≥96%.
For quality control (QC), we performed several rounds of data cleaning and a 96.5% cutoff was used for the sample call rate and SNP coverage in the combined SIT Trial data (n=1082), and resulted the exclusion of 6 individuals and 1,260 SNPs from the study. Cryptic relatedness was determined by examining pair-wise identity-by-descent (IBD), and 77 samples were identified as first-degree relatives and dropped from the study. Given the admixed nature of the study participants, we used principal component analysis (PCA) as implemented in EIGENSTRAT [28] to both identify genetic outliers (>6 standard deviations on any of the top ten principal components) and correct for any potential residual population substructure. In the SIT Trial, twenty-six individuals were identified as genetic outliers and further excluded from the analysis. Additionally, 48 samples, due to incomplete phenotype data, were also dropped from the study, leaving 925 samples (males: 51.9% & females: 48.1%) for the subsequent GWAS analysis. Among them, 89% of the samples (n=826/924) had a confirmed SCI status and 98 individuals (11%) were not classified. In total, 251 SCI positive and 575 SCI negative samples were assigned as cases and controls, respectively.
In the CSSCD cohort, DNA samples were genotyped at Boston University using Illumina Human610-Quad SNP arrays (Illumina, San Diego, CA, USA) with approximately ~600,000 SNPs. All samples were processed according the manufacturer’s protocol and the BeadStudio software was used to make genotype calls utilizing the Illumina pre-defined clusters. Samples with <95% call rate were removed and SNPs with a call rate <97.5% were re-clustered. After re-clustering, SNPs with call rates >97.5%, cluster separation score >0.25, and excess heterozygosity between -0.10 and 0.10 were retained in the analysis. The pair-wise IBD was used to identify cryptic relatedness and PCA was applied to detect genetic outliers. After excluding these samples and following the SIT Trial age inclusion criteria (<15 years), analysis was restricted to a dataset of 692 samples (males: 52% & females: 48%).
Merging GWAS Data and Imputation
To infer un-genotyped SNPs and fill-in missing data in the SIT Trial and CSSCD cohort, HumanHap650Y, HumanOmni1-Quad and Human610-Quad SNP array datasets were merged and subsequently imputation was performed for autosomes using a Hidden Markov model, as implemented in the MaCH software [29] (version 1.16) (http://www.sph.umich.edu/csg/abecasis/MaCH/), with 50 rounds and 200 states. QC was performed both before and after imputation and poorly imputed SNPs (RSq <0.5, squared correlation between imputed and true genotypes) were excluded and total 1,019,297 SNPs were analyzed.
SNP Selection for Validation
Due to the lack of any published studies that report genetic determinants for SBP in children, and/or more specifically in African American children at genome-wide significance, we used the ICBP identified SBP SNPs for validation. To validate results from the ICBP study in SCD patients, we examined the 28 SNPs that were reported associated with SBP [23]. For SNPs that were not available on our genotyping array, a close proxy for the index SNP with criteria of r2 ≥ 0.6 from HapMap Phase III (release 2, ASW panel) [30] or 1000 Genomes project (Pilot 1, YRI panel) [31] was used.
GWAS Meta-analysis and Statistics
All quality control measures in both cohorts were performed using the PLINK software package [32], version 1.06 (http://pngu.mgh.harvard.edu/purcell/plink/). In the SIT Trial cohort, to account for the uncertainty of the imputed data, the estimated allele dosage was analyzed using ProABEL [33] under a multivariate linear regression framework. Association for each SNP was assessed after adjusting for age, sex, height and the 1st principal component, and assuming an additive effect of allele dosage on SBP. In the CSSCD cohort, SBP measurements were available for longitudinal time points and data was analyzed using a linear mixed effect model using the lme4 package (http://lme4.r-forge.r-project.org/) in R (http://www.r-project.org/) (version 2.14.1). In the mixed effect model, age, sex, height and the 1st principal component were used as fixed effect covariates, while multiple SBP measurements within each individual were treated as random effect. GWAS results from both the cohorts were meta-analyzed using inverse-variance weighted fixed-effect models as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/metal) [34]. The variance inflation factor for genomic control (λGC), as described by Devlin and Roeder [35], was evaluated in each cohort prior to meta-analysis and a total of 1,019,297 SNPs were meta-analyzed. To explore the previously published SBP associated SNPs [23], a one-sided test of significance was used. To estimate the effect of these SNPs on SCI (data available only from the SIT Trial cohort), multivariate logistic regression was used after adjustments for age, sex and 1st principal component. The genetic risk scores for SBP and SCI were constructed (based on previously published SBP variants and weighted according to their effect sizes) using an R package Genetics ToolboX (http://cran.r-project.org/web/packages/gtx/index.html). To construct the genetic risk scores, this R package uses the same underlying statistics which was used by the ICBP [23] and can be defined as follows: Assuming a set of m SNPs from a discovery panel, for the i-th SNP in the j-th individual denotes x ij as the coded genotype (for directly genotyped SNPs) or estimated allele dosage (in case of imputation). If the set of regression coefficients of the reported SNPs are w 1, w 2. . w m, then the risk score for individual j is defined as: s j = s o + w 1 x 1j + w 2 x 2j + … + w m x mj, where s o is the intercept. In our analyses, we specify the coefficient w 1, w 2. . w m, to be the effect sizes (in mmHg per coded allele). Further, to identify known functional regulatory variants within or in proximity to the loci of interest, the GTEx (Genotype-Tissue Expression) expression quantitative trait loci (eQTL) database was queried (http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi) [36].
Gene-based Association Testing
To increase power by combining independent associations within a gene into a single, stronger aggregated signal, gene-based association tests were performed using GWiS [37]. GWiS uses greedy Bayesian model selection (selecting a minimal subset of associated SNPs within a gene) to identify independent effects and estimates overall significance through permutation. For each test statistics, using meta-analysis summary data, the gene P-values were computed using 1,000,000 permutations and utilizing the 1000 Genomes Project ASW panel as a reference population to account for linkage disequilibrium (LD) between SNPs.
Results
Genome-wide single SNP association
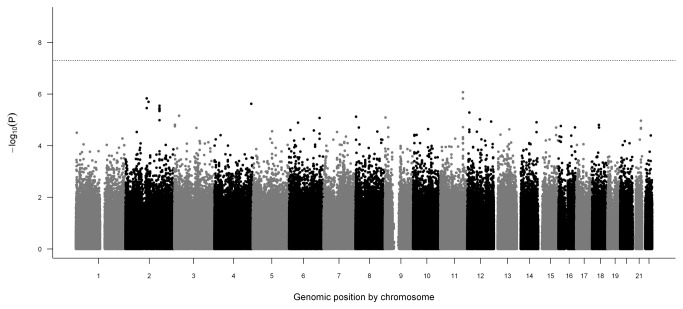
Genome-wide association and meta-analysis was performed for SBP in 1,617 subjects (843 males, 774 females) from the SIT Trial and CSSCD cohorts. The average ages of studied samples from the SIT Trial and CSSCD cohorts were 8.96 and 9.57 years, respectively. Detailed demographic and clinical characteristics for the study subjects are described in Table 1. The observed P-values show no early departure from the null (Figure S1), indicating minimal inflation (λGC=0.998) in test statistics due to potential population stratification and/or cryptic relatedness. None of the SNPs reached genome-wide significance (P-value <5.0x10-8) (Figure 1). However, a number of suggestive candidate loci (P-value <5.0x10-5) were identified that approached genome-wide significance (Table 2). The most significant signal was observed for rs7952106 (P-value: SIT Trial=6.40x10-3; CSSCD=3.94x10-5; Meta-analysis=8.57x10-7). This SNP is located ~78 kb 5’ upstream of the dopamine receptor D2 subtype (DRD2) gene on chromosome 11. rs7952106 is a common SNP with a minor allele frequency (MAF) of 23%, and directly genotyped in both the cohorts. The direction effect of the minor allele (G) is consistent across both GWAS (SIT Trial: Effect size=1.65 mmHg/allele; CSSCD: Effect size=1.50 mmHg/allele) and associated with increase in SBP. A second DRD2 intronic SNP (rs17529477; minor allele frequency [A] =12%) showed the same direction effect (Effect size=1.76 mmHg/minor allele; P-value=1.93x10-5) and was in low LD with rs7952106 (r2: SIT Trial=0.25 and CSSCD=0.24).
Table 1. Demographic and clinical characteristics of the SIT Trial and CSSCD cohorts.
| Variables | SIT Trial | CSSCD | P-value* |
|---|---|---|---|
| Sex, n (%) | |||
| Men | 483 (52.2%) | 360 (52%) | 0.89 |
| Age (in years), mean ± SD | 8.96 ± 2.44 | 9.57 ± 2.91 | 0.002 |
| Baseline SBP† (mm Hg), mean ± SD | 108.2 ± 11.51 | 100.8 ± 10.99 | <0.001 |
| Baseline DBP‡ (mm Hg), mean ± SD | 60.58 ± 8.06 | 59.21 ± 10.27 | 0.04 |
| Height (cm), mean ± SD | 128.70 ± 14.37 | 131.5 ± 16.08 | 0.008 |
| Hemoglobin (g/dl), mean ± SD | 8.11 ± 1.08 | 8.13 ± 0.96 | 0.41 |
| Hematocrit (%), mean ± SD | 23.34 ± 3.42 | 23.57 ± 3.10 | 0.12 |
| White blood counts (Cu), mean ± SD | 12.58 ± 5.25 | 12.01 ± 2.56 | 0.13 |
| Fetal hemoglobin (%), mean ± SD | 8.93 ± 5.75 | 8.00 ± 7.45 | 0.014 |
The shown laboratory assessments for both the cohorts are at baseline
P-values for continuous and categorical variable comparisons were generated using wilcoxon rank sum and Kruskal-Wallis test, respectively
SBP indicates systolic blood pressure
DBP indicates diastolic blood pressure
Figure 1. Manhattan plot showing the association of SNPs with systolic blood pressure (SBP).
The genome-wide distribution of -log10 P-values are plotted against the physical position of each SNP on each chromosome. The threshold for genome-wide significance (P-value < 5.0x10-8) is indicated by the horizontal dashed line.
Table 2. Association summary of top scoring independent loci of systolic blood pressure GWAS.
| Chr | SNP | Position | Nearest Gene | Location | CA/OA | CAF | RSq* |
SIT Trial† (n=925, age <15 yrs)
|
CSSCD‡ (n=692, age <15 yrs)
|
Meta-analysis
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect† | SE | P-value | Effect‡ | SE | P-value | Effect§ | SE | P-value | Direction | ||||||||||
| 2 | rs1669539 | 105,981,706 | NCK2- C2orf40 | Intergenic | T/C | 0.74 | 0.92 | 1.31 | 0.62 | 0.036 | 1.6 | 0.37 | 1.29x10-5 | 1.52 | 0.32 | 1.47x10-6 | ++ | ||
| 2 | rs3105491 | 114,697,437 | ACTR3- DPP10 | Intergenic | T/G | 0.3 | 0.76 | 0.61 | 0.78 | 0.43 | 1.71 | 0.35 | 1.08x10-6 | 1.52 | 0.32 | 2.0x10-6 | ++ | ||
| 2 | rs11568377 | 169,561,466 | ABCB11 | Coding Synonymous | A/G | 0.17 | 1 | -0.81 | 0.68 | 0.23 | -1.95 | 0.41 | 2.41x10-6 | -1.64 | 0.35 | 3.54x10-6 | − | ||
| 3 | rs17586876 | 2,939,182 | CNTN4 | Intron | A/G | 0.92 | 0.92 | 0.58 | 1.13 | 0.61 | -2.95 | 0.58 | 3.02x10-7 | -2.22 | 0.51 | 1.56x10-5 | +- | ||
| 3 | rs826221 | 24,243,440 | THRB | Intron | A/G | 0.93 | 0.87 | -1.9 | 1.03 | 0.06 | -3.03 | 0.72 | 2.61x10-5 | -2.66 | 0.59 | 6.96x10-6 | − | ||
| 4 | rs10520528 | 183,497,595 | ODZ3 | Intron | T/G | 0.43 | 1 | 1.63 | 0.52 | 0.0018 | 1.17 | 0.32 | 0.0003 | 1.3 | 0.27 | 2.40x10-6 | ++ | ||
| 6 | rs7769148 | 44,412,354 | AARS2- SPATS1 | Intergenic | A/G | 0.23 | 0.63 | -2.43 | 0.68 | 0.0004 | -1.51 | 0.56 | 0.0066 | -1.88 | 0.43 | 1.30x10-5 | − | ||
| 6 | rs4869931 | 151,051,582 | PLEKHG1 | Intron | A/G | 0.47 | 0.97 | 0.12 | 0.53 | 0.81 | 1.61 | 0.32 | 3.90x10-7 | 1.21 | 0.27 | 8.48x10-6 | ++ | ||
| 8 | rs1442407 | 3,078,991 | CSMD1 | Intron | T/G | 0.1 | 1 | 1.45 | 0.87 | 0.1 | 2.16 | 0.51 | 2.33x10-5 | 1.98 | 0.44 | 7.58x10-6 | ++ | ||
| 9 | rs7045640 | 2,968,377 | KIAA0020- RFX3 | Intergenic | A/C | 0.76 | 0.76 | -2.18 | 0.6 | 0.0003 | -1.43 | 0.52 | 0.0056 | -1.75 | 0.39 | 8.13x10-6 | − | ||
| 11 | rs17529477 | 112,822,277 | DRD2 | Intron | A/G | 0.12 | 1 | 1.79 | 0.81 | 0.027 | 1.75 | 0.48 | 0.00024 | 1.76 | 0.41 | 1.93x10-5 | ++ | ||
| 11 | rs7952106 | 112,929,768 | DRD2- MIR4301 | 5'-Upstream | G/T | 0.23 | 1 | 1.65 | 0.61 | 0.0064 | 1.5 | 0.36 | 3.94x10-5 | 1.54 | 0.31 | 8.57x10-7 | ++ | ||
| 12 | rs11053548 | 10,061,994 | CLEC12B | Intron | A/G | 0.91 | 1 | 2.77 | 0.92 | 0.0022 | 1.88 | 0.54 | 0.00049 | 2.12 | 0.46 | 5.19x10-6 | ++ | ||
| 12 | rs1989584 | 62,619,479 | SRGAP1 | Intron | T/G | 0.36 | 1 | -1.14 | 0.53 | 0.034 | -1.27 | 0.33 | 9.68x10-5 | -1.23 | 0.28 | 9.64x10-6 | − | ||
| 12 | rs11064853 | 118,544,491 | LOC387890 | Intron | A/G | 0.07 | 0.67 | 2.37 | 1.14 | 0.038 | 3.54 | 0.9 | 7.75x10-5 | 3.09 | 0.71 | 1.17x10-5 | ++ | ||
| 14 | rs9743626 | 97,844,745 | C14orf64- C14orf177 | Intergenic | A/G | 0.1 | 1 | 2.37 | 0.86 | 0.0063 | 1.82 | 0.53 | 0.00056 | 1.97 | 0.45 | 1.24x10-5 | ++ | ||
| 16 | rs289673 | 12,338,371 | SNX29 | Intron | A/G | 0.67 | 0.68 | -1.63 | 0.61 | 0.0077 | -1.53 | 0.45 | 0.00073 | -1.57 | 0.36 | 1.75x10-5 | − | ||
| 16 | rs7499397 | 82,177,970 | CDH13 | Intron | T/C | 0.38 | 0.92 | -1.8 | 0.57 | 0.0017 | -1 | 0.32 | 0.0017 | -1.19 | 0.28 | 1.98x10-5 | − | ||
| 18 | rs1543073 | 34,149,356 | CELF4 | 5' Upstream | A/G | 0.41 | 0.93 | -0.52 | 0.56 | 0.36 | -1.44 | 0.32 | 7.97x10-6 | -1.21 | 0.28 | 1.57x10-5 | − | ||
| 21 | rs2210268 | 40,685,128 | DSCAM | Intron | T/G | 0.83 | 1 | -1.8 | 0.66 | 0.0062 | -1.43 | 0.41 | 0.00049 | -1.54 | 0.35 | 1.08x10-5 | − | ||
Chr: chromosome; CA: coded allele; OA: other allele; CAF: coded allele frequency; RSq: Imputation quality (squared correlation between imputed and true genotypes)
Genomic positions are in reference to NCBI build 36.3
Table includes SNPs with independent effects and whose significance in the meta-analysis is <5.0x10-5.
Directly genotyped SNPs are marked as 1
Effect size is based on multivariate linear regression (adjusted for age, sex, height and 1st principal component)
Effect size is based on linear mixed model (age, sex, height, 1st principal component and the index SNP were treated as fixed effect covariates and longitudinal measurements of systolic blood pressure of each individual was used as a random effect)
Effect size is based on inverse-variance weighted fixed-effect meta-analysis
The effect size estimates corresponds to mmHg per coded allele for SBP
Gene-based association analysis
Given the suggestion of multiple independent signals in DRD2, we performed a gene-based test combining independent associations within a gene (using 20kb flanking region) and obtained a P-value for each gene using permutation. In total, 32,155 autosomal genes were tested, and results are shown in Table 3 . Among all the tested genes, none met the genome-wide significant criteria (P-value < 2.0x10-6). The most significant gene was MIR4301 (micro-RNA4301, Gene ID: 100422855) with a P-value=5.2x10-5 (Table 3 ). MIR4301 is a 65 base-pair long non-coding RNA at chromosome 11 and contained within the DRD2 intronic region, and shares the same set of associated SNPs as observed for the DRD2 gene. To examine micro-RNA target binding predictions, we used the software RNA22 (version 1.0) (http://cbcsrv.watson.ibm.com/rna22.html) [38]. MIR4301 showed a predicted target site in the 3’ UTR region of the DRD2 transcript (ENSEMBL: ENST00000355319). In our study, the observed suggestive genetic signals of DRD2 region SNPs and the prediction of MIR4301 binding to DRD2 as a potential target, suggests the plausible involvement of DRD2 region in the regulation of SBP in SCD cohorts. Examining the GTEx database, which queries lymphoblastoid, liver, brain cerebellum, frontal cortex, and temporal cortex tissue, we found no known eQTLs within or in close proximity of this locus.
Table 3. Gene-based association analysis of systolic blood pressure.
| Chr |
Gene
|
SNPs† | Tests‡ | K§ | P-value* | ||
|---|---|---|---|---|---|---|---|
| Name | Start | End | |||||
| 1 | LOC400750 | 39,154,602 | 39,196,561 | 13 | 6.31 | 1 | 0.0011 |
| 2 | ABCB11 | 169,759,449 | 169,907,833 | 172 | 22.99 | 1 | 3.9x10-4 |
| 4 | MRFAP1 | 6,622,445 | 6,664,449 | 14 | 7.59 | 1 | 7.4x10-4 |
| 5 | MIR583 | 95,394,842 | 95,434,916 | 14 | 4.77 | 1 | 1.5x10-4 |
| 6 | LOC134997 | 24,956,605 | 24,997,415 | 29 | 7.30 | 1 | 7.1x10-4 |
| 6 | SPATS1 | 44,290,397 | 44,364,904 | 38 | 11.54 | 1 | 4.1x10-4 |
| 6 | PLEKHG1 | 150,940,999 | 151,184,799 | 141 | 29.94 | 1 | 0.0011 |
| 8 | LOC100507422 | 48,484,706 | 48,525,170 | 3 | 2.95 | 1 | 9.0x10-4 |
| 9 | CARM1P1 | 2,923,562 | 3,073,404 | 89 | 17.04 | 1 | 3.1x10-4 |
| 10 | NUDT13 | 74,850,210 | 74,911,581 | 10 | 4.55 | 1 | 3.1x10-4 |
| 10 | USP54 | 75,237,296 | 75,355,433 | 10 | 3.59 | 1 | 2.6x10-4 |
| 11 | OR5D15P | 55,534,441 | 55,575,382 | 13 | 7.34 | 1 | 0.001 |
| 11 | OR5W1P | 55,650,819 | 55,691,621 | 31 | 8.51 | 2 | 3.3x10-4 |
| 11 | DRD2 | 113,260,317 | 113,366,001 | 61 | 14.46 | 2 | 0.0011 |
| 11 | MIR4301 | 113,300,745 | 113,340,810 | 17 | 7.13 | 2 | 5.2x10-5 |
| 12 | CLEC1B | 10,125,660 | 10,171,899 | 49 | 9.47 | 1 | 2.0x10-4 |
| 12 | PRKAG1 | 49,376,055 | 49,432,629 | 10 | 3.69 | 1 | 2.9x10-4 |
| 12 | SRGAP1 | 64,218,541 | 64,561,613 | 101 | 25.84 | 1 | 9.3x10-4 |
| 12 | TMEM233 | 120,011,264 | 120,099,363 | 24 | 8.01 | 2 | 1.4x10-4 |
| 13 | KCTD12 | 77,434,304 | 77,480,540 | 23 | 10.78 | 1 | 6.7x10-4 |
| 15 | BLM | 91,240,579 | 91,378,686 | 41 | 11.18 | 1 | 9.5x10-4 |
| 19 | MIR4746 | 4,425,975 | 4,466,045 | 13 | 2.70 | 1 | 0.0010 |
| 20 | CYB5AP4 | 22,846,128 | 22,886,676 | 13 | 6.16 | 1 | 6.9x10-4 |
| 21 | DSCAM-AS1 | 41,735,010 | 41,777,285 | 23 | 9.36 | 1 | 1.8x10-4 |
Gene start and end positions includes ±20 Kb of 5' and 3'-untranslated regions of the genes.
The threshold for genome-wide gene significance (2.0x10-6) was established using permutation.
Number of SNPs tested in the gene
Effective number of SNPs in the gene
Number of SNPs in the model
Significance for the gene model
Association of previously reported SBP loci
A total of 29 independent chromosomal loci associated with blood pressure have been reported from the ICBP meta-analysis, of which 28 show strong evidence for association with SBP. We attempted to validate these loci in the combined SCD cohorts. To ensure uniform comparison of the genetic effect and its direction, the SNPs were analyzed according to the reported coded alleles (under an additive genetic model). We were able to test 27 of these SNPs directly or with a proxy SNP (r2≥0.6), and 15/27 SNPs showed the same direction affect on SBP as reported in the ICBP study (Table 4 , Figure S2 ). Of the directly genotyped/imputed or proxy SNPs, 4 were nominally significant in the combined SCD cohorts, and 6 were significant at P-value <0.10 (Table 4 ), demonstrating a clear enrichment of signal (P-value =0.0045). However, none of the 27 tested SNPs was significant after Bonferroni correction (α=0.05/27=1.85x10-3). Given the limited power to detect significance for individual SNPs in the relatively smaller SCD samples (Figure S3 ), we constructed a genetic risk score for SBP incorporating the 27 previously reported SNPs; weighted according to effect sizes observed in the ICBP meta-analysis. The risk score derived from these 27 directly genotyped or proxy SNPs was nominally associated with SBP (P-value= 0.05), demonstrating the role of these SNPs in aggregate in the modulation of SBP in the SCD cohort.
Table 4. Association of previously reported SBP-associated SNPs with SBP and SCI.
| Chr | Genes | SNP (Position) |
ICBP GWAS
|
Genotyped or Imputed data available in both cohorts (Proxy SNP) |
Systolic Blood Pressure (SIT Trial + CSSCD cohort) (n=1,617)
|
Silent Cerebral Infarction (SIT Trial cohort) (n=826)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA/OA | CAF | Reported effect of CA on SBP risk | ReportedP-value | CA/OA | CAF | Observed effect of CA on SBP risk† | Observed P-value* | OR‡ | 95% CI | P-value | |||||
| 1 | MTHFR-NPPB | rs17367504 (11,785,365) | G/A | 0.15 | Decreases | 8.72x10-22 | Yes | G/A | 0.1 | Decreases | 0.37 | 1.06 | (0.74-1.53) | 0.75 | |
| 1 | MOV10 | rs2932538 (113,018,066) | G/A | 0.75 | Increases | 1.17x10-9 | Yes | G/A | 0.83 | Decreases | NA | 1.33 | (0.87-2.02) | 0.18 | |
| 3 | SLC4A7 | rs13082711 (27,512,913) | T/C | 0.78 | Decreases | 1.51x10-6 | No (rs3755652) (27,447,940) (r2=0.71) | C/T | 0.93 | Increases | NA | 0.86 | (0.55-1.35) | 0.51 | |
| 3 | MECOM | rs419076 (170,583,580) | T/C | 0.47 | Increases | 1.78x10-13 | No (rs16853620) (170,582,811) (r2=0.97) | A/G | 0.41 | Increases | 0.42 | 0.85 | (0.69-1.05) | 0.13 | |
| 4 | FGF5 | rs1458038 (81,383,747) | T/C | 0.29 | Increases | 1.47x10-23 | Yes | T/C | 0.08 | Decreases | NA | 1.25 | (0.84-1.85) | 0.27 | |
| 4 | SLC39A8 | rs13107325 (103,407,732) | T/C | 0.05 | Decreases | 3.27x10-14 | Yes | T/C | 0.01 | Increases | NA | 0.62 | (0.27-1.39) | 0.24 | |
| 4 | GUCY1A3-GUCY1B3 | rs13139571 (156,864,963) | C/A | 0.76 | Increases | 1.16x10-6 | No Proxy Found | - | - | - | - | - | - | ||
| 5 | NPR3-C5orf23 | rs1173771 (32,850,785) | G/A | 0.6 | Increases | 1.79x10-16 | Yes | G/A | 0.78 | Decreases | NA | 0.91 | (0.70-1.18) | 0.48 | |
| 5 | EBF1 | rs11953630 (157,777,980) | T/C | 0.37 | Decreases | 3.02x10-11 | No (rs12187017) (157,781,873) (r2=0.92) | A/G | 0.19 | Decreases | 0.31 | 1.11 | (0.76-1.62) | 0.59 | |
| 6 | HFE | rs1799945 (26,199,158) | G/C | 0.14 | Increases | 7.69x10-12 | Yes | G/C | 0.02 | Decreases | NA | 1.27 | (0.38-4.22) | 0.7 | |
| 6 | BAT2-BAT5 | rs805303 (31,724,345) | G/A | 0.61 | Increases | 1.49x10-11 | Yes | G/A | 0.39 | Increases | 0.12 | 1.13 | (0.91-1.42) | 0.27 | |
| 10 | CACNB2(5') | rs4373814 (18,459,978) | G/C | 0.55 | Decreases | 4.81x10-11 | No (rs2357790) (18,463,386) (r2=0.93) | C/T | 0.55 | Decreases | 0.05 | 0.75 | (0.57-0.99) | 0.04 | |
| 10 | CACNB2(3') | rs1813353 (18,747,454) | T/C | 0.68 | Increases | 2.56x10-12 | No (rs11014171) (18,751,201) (r2=0.61) | C/T | 0.85 | Decreases | NA | 1.21 | (0.73-2.02) | 0.46 | |
| 10 | C10orf107 | rs4590817 (63,137,559) | G/C | 0.84 | Increases | 3.97x10-12 | No (rs12246717) (63,129,189) (r2=0.62) | T/G | 0.71 | Decreases | NA | 1.04 | (0.76-1.42) | 0.81 | |
| 10 | PLCE1 | rs932764 (95,885,930) | G/A | 0.44 | Increases | 7.10x10-16 | No (rs2901761) (95,885,117) (r2=0.60) | A/G | 0.17 | Increases | 0.01 | 1.01 | (0.76-1.35) | 0.94 | |
| 10 | CYP17A1-NT5C2 | rs11191548 (104,836,168) | T/C | 0.91 | Increases | 6.90x10-26 | Yes | T/C | 0.97 | Increases | 0.32 | 0.39 | (0.13-1.13) | 0.08 | |
| 11 | ADM | rs7129220 (10,307,114) | G/A | 0.89 | Decreases | 2.97x10-12 | No (rs7929332) (10,180,933) (r2=0.72) | T/C | 0.91 | Decreases | 0.28 | 1.57 | (0.90-2.76) | 0.11 | |
| 11 | PLEKHA7 | rs381815 (16,858,844) | T/C | 0.26 | Increases | 5.27x10-11 | Yes | T/C | 0.17 | Increases | 0.46 | 0.64 | (0.42-0.99) | 0.04 | |
| 11 | FLJ32810-TMEM133 | rs633185 (100,098,748) | G/C | 0.28 | Decreases | 1.21x10-17 | No (rs6590810) (100,083,885) (r2=0.63) | A/G | 0.27 | Decreases | 0.05 | 0.92 | (0.66-1.27) | 0.59 | |
| 12 | ATP2B1 | rs17249754 (88,584,717) | G/A | 0.84 | Increases | 1.82x10-18 | No (rs6538195) (88,586,507) (r2=1) | G/A | 0.89 | Decreases | NA | 0.92 | (0.61-1.41) | 0.72 | |
| 12 | SH2B3 | rs3184504 (110,368,991) | T/C | 0.48 | Increases | 3.83x10-18 | Yes | T/C | 0.07 | Increases | 0.43 | 0.7 | (0.46-1.06) | 0.1 | |
| 12 | TBX5-TBX3 | rs10850411 (113,872,179) | T/C | 0.7 | Increases | 5.38x10-8 | Yes | T/C | 0.63 | Increases | 0.38 | 0.79 | (0.54-1.15) | 0.22 | |
| 15 | CYP1A1-ULK3 | rs1378942 (72,864,420) | C/A | 0.35 | Increases | 5.69x10-23 | Yes | C/A | 0.9 | Decreases | NA | 1.36 | (0.80-2.30) | 0.26 | |
| 15 | FURIN-FÈS | rs2521501 (89,238,392) | T/A | 0.31 | Increases | 5.20x10-19 | No (rs1029420) (89,242,090) (r2=0.70) | C/T | 0.29 | Decreases | NA | 1.21 | (0.89-1.64) | 0.22 | |
| 17 | GOSR2 | rs17608766 (42,368,270) | T/C | 0.86 | Decreases | 1.13x10-10 | Yes | T/C | 0.98 | Increases | NA | 2.35 | (1.15-4.81) | 0.02 | |
| 17 | ZNF652 | rs12940887 (44,757,806) | T/C | 0.38 | Increases | 1.79x10-10 | No (rs17637472) (44,816,432) (r2=0.62) | A/G | 0.06 | Increases | 0.03 | 0.79 | (0.49-1.27) | 0.33 | |
| 20 | JAG1 | rs1327235 (10,917,030) | G/A | 0.46 | Increases | 1.87x10-8 | No (rs1887320) (10,913,998) (r2=1.0) | G/A | 0.48 | Increases | 0.08 | 0.98 | (0.79-1.20) | 0.82 | |
| 20 | GNAS-EDN3 | rs6015450 (57,184,512) | G/A | 0.12 | Increases | 3.87x10-23 | No(rs6026742) (57,174,000) (r2=0.85) | A/G | 0.22 | Increases | 0.06 | 1.22 | (0.89-1.69) | 0.22 | |
Genomic positions are in reference to NCBI build 36.3
Chr: chromosome; CA: coded allele; OA: other allele; CAF: coded allele frequency; OR: odds ratio; CI: confidence interval
In the absence of the genotype data for the reported index SNP, best proxy was selected using HapMap phase III (release 2, ASW panel) and 1000 Genomes project (Pilot 1, YRI panel ) with cutoff r2 ≥ 0.6
Observed effects are based on inverse variance weighted meta-analysis*P-values are based on one-tailed significanceNA indicates opposite direction of effect between ICBP and SCD cohorts
Odds ratio based on the multivariate logistic regression adjusted for age, sex and 1st principal component
Association of SBP reported loci with SCI
Given that higher SBP is associated with increased risk of an SCI in SCD patients, we determined whether any of the SNPs (or their proxies) associated with SBP in ICBP study was associated with SCI. Three SNPs were nominally associated with SCI (Table 4 ), with only the CACNB2 (5’) locus (rs2357790) consistent between the SBP (P-value=0.05) and SCI (P-value=0.04) analyses. None of these SNPs was significant after Bonferroni correction. Further, to estimate the aggregated effect of these SNPs on increased risk of SCI, we constructed the genetic risk score for SCI (weighted according to effect sizes of published SBP SNPs) and no significant association was observed (P-value= 0.95).
Discussion
In recent years, genome-wide scans demonstrated a successful means of identifying novel common genetic variants that contribute to susceptibility to complex diseases, including blood pressure [21-23,39,40]. Here, we present the results from a meta-analysis of SBP from two SCD cohorts comprised of 1,617 SCD patients, all with African American ancestry. No associations were genome-wide significant; however we observed suggestive association at rs7952106, a 5’ upstream SNP to DRD2 gene on chromosome 11, which showed consistent association evidence in both the studied cohorts. Further, in a gene-based test, a suggestive signal of non-coding RNA (MIR4301) and the prediction of MIR4301 binding to DRD2 as a potential target, suggests the plausible involvement of DRD2 region in the regulation of SBP in SCD cohorts.
Previously, several studies have shown that dopamine synthesis in the kidney has an important role in the regulation of fluid and electrolyte balance and systemic blood pressure [41-43]. Dopamine exerts its actions via 2 families of G-protein-coupled receptors D1-like receptors (DRD1 and DRD5) and D2-like receptors (DRD2, DRD3, and DRD4). Later, several lines of evidence also showed that an intact dopaminergic system is necessary to maintain normal blood pressure and that genetic hypertension is associated with alterations in dopamine production and receptor function [41-44]. Deletion of any dopamine receptor in mice results in increased blood pressure by mechanisms that are receptor dependent. In particular, mice lacking the DRD2 gene (DRD2-/-
) have reactive oxygen species (ROS)-dependent hypertension [44]. In addition, the DRD2 polymorphisms were also reported with decreased DRD2 expression [45] and shown to affect DRD2 mRNA stability and synthesis of the receptor [46]. These studies suggest that the DRD2 locus is plausibly involved in the regulation of SBP. Our results support these findings and the suggestive association from the DRD2 region SNPs may represents a true signal associated with SBP in SCD patients.
Recently, several large and well powered studies from European ancestry populations have identified 29 genomic loci associated with blood pressure [21-23]. We sought to validate these reported loci in the setting of SCD in populations of African American ancestry, and to further test whether any of these loci were involved in SCI. Our study reports that the derived genetic risk scores for SBP is significantly associated with SCD children of African American ancestry. The significant association of the aggregated genetic risk scores with SBP in our study highlights the importance of these loci in the modulation of SBP in the studied SCD cohort.
In SCD patients, neurovascular complications are common and largely due to tissue ischemia and infarctions [13,47]. SCI is a major cause of neurologic morbidity in SCD children with unclear genetic susceptibility [47,48]. Given that hypertension is a known risk factor for stroke and more recently, SBP has been reported associated with risk for SCI [13,49], identifying genetic variants associated with SBP in SCD patients may also lead to the identification of genes associated with SCI. Although we confirm the association of 4 loci with SBP, only the CACNB2 (5’) locus showed the consistent nominal significance for both SBP and SCI (Table 4 ).
A few limitations to the current study need to be acknowledged. First, although we observed a significant association of the aggregated genetic risk score with SBP, we failed to reproduce the significance of any individual SBP associated SNPs after multi-test correction and this may be due to the limited sample size of our study. As shown in Figure S3 , our study (n=1,617) was also under-powered to identify novel genome-wide significant variants. Secondly, biological differences that exist between ethnic groups and complex interaction of age with different genetic alteration may also have negatively impacted our ability to identify significant loci. Also, the admixed nature of the African ancestry population may lead to differences in local LD patterns. Since we are not likely to be genotyping the functional variant, changes in LD patterns between Europeans and the studied African American populations can change the nature of the observed associations. Thirdly, the ability to detect genetic determinants associated with any trait of interest largely depends on the quality and reliability of the data. In our study, it is noteworthy to highlight that we have used the longitudinal SBP measurements from the CSSCD cohort, as oppose to the single time point data available from the SIT Trial. In addition, CSSCD SBP measurements were taken after following the procedure described in the study manual of operations, and hence provides more certainty of less variability, whereas, no uniform guidelines were adopted in the SIT Trial. At last, it has also been known for over a decade that blood pressure is an age-dependent process [50,51]. The ICBP study was performed in adult individuals (38-72 yrs), whereas in our study, SCD patients are restricted to age < 15 years; therefore, it is possible that the lack of association for other loci may be due to an age-dependent genetic effect.
In summary, our results not only suggest the importance of DRD2 genetic variation in the modulation of SBP, but also extend the genetic significance of the 4 previously published loci in SBP in an African American population. Further, our study also identifies a significant association for the genetic risk score with SBP, suggesting the aggregated importance of previously reported SNPs in the modulation of SBP in the setting of SCD. This study provides new insight in SBP regulation in an admixed African American ancestry cohort, more specifically in children with SCD, and highlights the overlap in genetic signals between African American populations and European ancestry populations.
Supporting Information
A quantile-quantile (Q-Q) plot showing the distribution of observed χ2 statistics of analyzed SNPs in 1,617 SCD individuals (SIT Trial, n=925; CSSCD, n=692). The black diagonal line indicates expected results under the null hypothesis.
(TIFF)
Comparison of the effect size of variants from the reported regions associated with SBP in the ICBP meta-analysis and SCD cohort.
(TIFF)
Power calculation to detect genome-wide significant SNPs (minor allele frequency > 0.05) for systolic blood pressure in the combined SIT Trial and CSSCD cohort, utilizing 1617 SCD samples. The authors thank the staff, clinicians and patients for their participation in the Silent Infarct Transfusion (SIT) Trial study. Genotyping services for the SIT Trial were provided by the Center for Inherited Disease Research (CIDR) at Johns Hopkins University and the Division of Blood Disorders at the Centers for Disease Control and Prevention (CDC). The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
(TIFF)
Funding Statement
This study was supported by the National Heart, Lung and Blood Institute (NHLBI) (Award Number: U54HL090515, 5R01HL091759) and the National Institute of Neurological Disorders and Stroke (NINDS) (NIH-NINDS 5U01-NS042804-03). This work was also supported by 5T32 HL007501 (J.N.M.) and R01 HL 87681, R01 068970 and RC2 HL 101212 (M.H.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Steinberg MH (1999) Management of sickle cell disease. N Engl J Med 340: 1021–1030. doi:10.1056/NEJM199904013401307. PubMed: 10099145. [DOI] [PubMed] [Google Scholar]
- 2. Ballas SK, Lieff S, Benjamin LJ, Dampier CD, Heeney MM et al. (2010) Definitions of the phenotypic manifestations of sickle cell disease. Am J Hematol 85: 6-13. PubMed: 19902523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Driss A, Asare KO, Hibbert JM, Gee BE, Adamkiewicz TV et al. (2009) Sickle Cell Disease in the Post Genomic Era: A Monogenic Disease with a Polygenic Phenotype. Genomics Insights 2009: 23-48 PubMed: 20401335. [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson CS, Giorgio AJ (1981) Arterial blood pressure in adults with sickle cell disease. Arch Intern Med 141: 891-893. doi:10.1001/archinte.141.7.891. PubMed: 7235808. [PubMed] [Google Scholar]
- 5. de Jong PE, Landman H, van Eps LW (1982) Blood pressure in sickle cell disease. Arch Intern Med 142: 1239-1240. doi:10.1001/archinte.142.6.1239. PubMed: 7092436. [DOI] [PubMed] [Google Scholar]
- 6. Grell GA, Alleyne GA, Serjeant GR (1981) Blood pressure in adults with homozygous sickle cell disease. Lancet 2: 1166 PubMed: 6118599. [DOI] [PubMed] [Google Scholar]
- 7. Hatch FE, Crowe LR, Miles DE, Young JP, Portner ME (1989) Altered vascular reactivity in sickle hemoglobinopathy. A possible protective factor from hypertension. Am J Hypertens 2: 2-8. doi:10.1093/ajh/2.2.2S. PubMed: 2643968. [DOI] [PubMed] [Google Scholar]
- 8. Rodgers GP, Walker EC, Podgor MJ (1993) Is "relative" hypertension a risk factor for vaso-occlusive complications in sickle cell disease? Am J Med Sci 305: 150-156. doi:10.1097/00000441-199303000-00004. PubMed: 8447334. [DOI] [PubMed] [Google Scholar]
- 9. Homi J, Homi-Levee L, Gentles S, Thomas P, Serjeant G (1993) Adolescent blood pressure in a cohort study of sickle cell disease. Arch Intern Med 153: 1233-1236. doi:10.1001/archinte.1993.00410100063009. PubMed: 8494475. [PubMed] [Google Scholar]
- 10. JAMA (1977) Blood pressure studies in 14 communities. A two-stage screen for hypertension. JAMA 237: 2385-2391. doi:10.1001/jama.237.22.2385. PubMed: 576938. [DOI] [PubMed] [Google Scholar]
- 11. Stamler J, Stamler R, Riedlinger WF, Algera G, Roberts RH (1976) Hypertension screening of 1 million Americans. Community Hypertension Evaluation Clinic; (CHEC; ) program, 1973 through 1975. AMA 235: 2299-2306. [DOI] [PubMed] [Google Scholar]
- 12. Cornoni-Huntley J, LaCroix AZ, Havlik RJ (1989) Race and sex differentials in the impact of hypertension in the United States. The National Health and Nutrition Examination Survey I Epidemiologic Follow-up Study. Arch Intern Med 149: 780-788. doi:10.1001/archinte.149.4.780. PubMed: 2784957. [PubMed] [Google Scholar]
- 13. DeBaun MR, Sarnaik SA, Rodeghier MJ, Minniti CP, Howard TH et al. (2012) Associated risk factors for silent cerebral infarcts in sickle cell anemia: low baseline hemoglobin, sex, and relative high systolic blood pressure. Blood 119: 3684-3690. doi:10.1182/blood-2011-05-349621. PubMed: 22096242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kwiatkowski JL, Zimmerman RA, Pollock AN, Seto W, Smith-Whitley K et al. (2009) Silent infarcts in young children with sickle cell disease. Br J Haematol 146: 300-305. doi:10.1111/j.1365-2141.2009.07753.x. PubMed: 19500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bernaudin F, Verlhac S, Arnaud C, Kamdem A, Chevret S et al. (2011) Impact of early transcranial Doppler screening and intensive therapy on cerebral vasculopathy outcome in a newborn sickle cell anemia cohort. Blood 117: 1130-1140. doi:10.1182/blood-2010-06-293514. PubMed: 21068435. [DOI] [PubMed] [Google Scholar]
- 16. San M, Demirta scedil M, Burgut R, Birand A, Bascedillami scedilli F (1998) Left Ventricular systolic and diastolic functions in patients with sickle cell anemia. Int J Angiol 7: 185-187. doi:10.1007/BF01617389. PubMed: 9585446. [DOI] [PubMed] [Google Scholar]
- 17. Troy BL, Pombo J, Rackley JE (1945) Measurements of left ventricular wall thickness and mass by echocardiography. Circulation 45: 602. [DOI] [PubMed] [Google Scholar]
- 18. Balfour IC, Covitz W, Arensman FW, Eubig C, Garrido M et al. (1988) Left ventricular filling in sickle cell anemia. Am J Cardiol 61: 395-399. doi:10.1016/0002-9149(88)90952-6. PubMed: 3341219. [DOI] [PubMed] [Google Scholar]
- 19. Parent F, Bachir D, Inamo J, Lionnet F, Driss F et al. (2011) A hemodynamic study of pulmonary hypertension in sickle cell disease. N Engl J Med 365: 44-53. doi:10.1056/NEJMoa1005565. PubMed: 21732836. [DOI] [PubMed] [Google Scholar]
- 20. Havlik RJ, Garrison RJ, Feinleib M, Kannel WB, Castelli WP et al. (1979) Blood pressure aggregation in families. Am J Epidemiol 110: 304-312. PubMed: 474567. [DOI] [PubMed] [Google Scholar]
- 21. Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M et al. (2009) Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 41: 666-676. doi:10.1038/ng.361. PubMed: 19430483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ et al. (2009) Genome-wide association study of blood pressure and hypertension. Nat Genet 41: 677-687. doi:10.1038/ng.384. PubMed: 19430479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. International Consortium for Blood Pressure Genome-Wide Association Studies. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103-109. doi:10.1038/nature10405. PubMed: 21909115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Casella JF, King AA, Barton B, White DS, Noetzel MJ et al. (2010) Design of the silent cerebral infarct transfusion (SIT) trial. Pediatr Hematol Oncol 27: 69-89. doi:10.3109/08880010903360367. PubMed: 20201689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaston M, Rosse WF (1982) The cooperative study of sickle cell disease: review of study design and objectives. Am J Pediatr Hematol/Oncol 4: 197-201. PubMed: 7114401. [PubMed] [Google Scholar]
- 26. International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437: 1299-1320. doi:10.1038/nature04226. PubMed: 16255080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steemers FJ, Gunderson KL (2007) Whole genome genotyping technologies on the BeadArray platform. Biotechnol J 2: 41-49. doi:10.1002/biot.200600213. PubMed: 17225249. [DOI] [PubMed] [Google Scholar]
- 28. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904-909. doi:10.1038/ng1847. PubMed: 16862161. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34: 816-834. doi:10.1002/gepi.20533. PubMed: 21058334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. International HapMap3 Consortium, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature. 467: 52-58. doi:10.1038/nature09298. PubMed: 20811451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061-1073. doi:10.1038/nature09534. PubMed: 20981092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559-575. doi:10.1086/519795. PubMed: 17701901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aulchenko YS, Struchalin MV, van Duijn CM (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11: 134. doi:10.1186/1471-2105-11-134. PubMed: 20233392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190-2191. doi:10.1093/bioinformatics/btq340. PubMed: 20616382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55: 997-1004. doi:10.1111/j.0006-341X.1999.00997.x. PubMed: 11315092. [DOI] [PubMed] [Google Scholar]
- 36. Hardy J, Singleton A (2009) Genome-wide association studies and human disease. N Engl J Med 360: 1759-1768. doi:10.1056/NEJMra0808700. PubMed: 19369657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang H, Chanda P, Alonso A, Bader JS, Arking DE (2011) Gene-based tests of association. PLOS Genet 7: e1002177 PubMed: 21829371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL et al. (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203-1217. doi:10.1016/j.cell.2006.07.031. PubMed: 16990141. [DOI] [PubMed] [Google Scholar]
- 39. Sotoodehnia N, Isaacs A, de Bakker PI, Dörr M, Newton-Cheh C et al. (2010) Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet 42: 1068-1076. doi:10.1038/ng.716. PubMed: 21076409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weiss LA, Arking DE, Gene Discovery Project of Johns Hopkins & the Autism Consortium, Daly MJ, Chakravarti A (2009) A genome-wide linkage and association scan reveals novel loci for autism. Nature 461: 802-808 doi:10.1038/nature08490. PubMed: 19812673. [Google Scholar]
- 41. Jose PA, Soares-da-Silva P, Eisner GM, Felder RA (2010) Dopamine and G protein-coupled receptor kinase 4 in the kidney: role in blood pressure regulation. Biochim Biophys Acta 1802: 1259-1267. doi:10.1016/j.bbadis.2010.02.004. PubMed: 20153824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jose PA, Eisner GM, Felder RA (2003) Regulation of blood pressure by dopamine receptors. Nephron Physiol 95: 19-27. doi:10.1159/000073676. [DOI] [PubMed] [Google Scholar]
- 43. Hussain T, Lokhandwala MF (2003) Renal dopamine receptors and hypertension. Exp Biol Med 228: 134-142. [DOI] [PubMed] [Google Scholar]
- 44. Armando I, Wang X, Villar VA, Jones JE, Asico LD et al. (2007) Reactive oxygen species-dependent hypertension in dopamine D2 receptor-deficient mice. Hypertension 49: 672-678. doi:10.1161/01.HYP.0000254486.00883.3d. PubMed: 17190875. [DOI] [PubMed] [Google Scholar]
- 45. Laakso A, Pohjalainen T, Bergman J, Kajander J, Haaparanta M et al. (2005) The A1 allele of the human D2 dopamine receptor gene is associated with increased activity of striatal L-amino acid decarboxylase in healthy subjects. Pharmacogenet Genomics 15: 387-391. doi:10.1097/01213011-200506000-00003. PubMed: 15900211. [DOI] [PubMed] [Google Scholar]
- 46. Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR et al. (2003) Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet 12: 205-216. doi:10.1093/hmg/ddg055. PubMed: 12554675. [DOI] [PubMed] [Google Scholar]
- 47. Moser FG, Miller ST, Bello JA, Pegelow CH, Zimmerman RA et al. (1996) The spectrum of brain MR abnormalities in sickle-cell disease: a report from the cooperative study of sickle cell disease. AJNR Am J Neuroradiol 17: 965–972. PubMed: 8733975. [PMC free article] [PubMed] [Google Scholar]
- 48. DeBaun MR, Armstrong FD, McKinstry RC, Ware RE, Vichinsky E et al. (2012) Silent cerebral infarcts: a review on a prevalent and progressive cause of neurologic injury in sickle cell anemia. Blood 119: 4587-4596. doi:10.1182/blood-2011-02-272682. PubMed: 22354000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zia E, Hedblad B, Pessah-Rasmussen H, Berglund G, Janzon L et al. (2007) Blood pressure in relation to the incidence of cerebral infarction and intracerebral hemorrhage. Hypertensive hemorrhage: debated nomenclature is still relevant. Stroke 38: 2681-2685. doi:10.1161/STROKEAHA.106.479725. PubMed: 17761929. [DOI] [PubMed] [Google Scholar]
- 50. Franklin SS, Gustin W 4th, Wong ND, Larson MG, Weber MA et al. (1997) Hemodynamic patterns of age-related changes in blood pressure. The Framingham Heart Study. Circulation 96: 308-315. doi:10.1161/01.CIR.96.1.308. PubMed: 9236450. [DOI] [PubMed] [Google Scholar]
- 51. Safar ME, Lange C, Tichet J, Blacher J, Eschwège E et al. (2008) The Data from an Epidemiologic Study on the Insulin Resistance Syndrome Study: the change and the rate of change of the age-blood pressure relationship. J Hypertens 26: 1903-1911. doi:10.1097/HJH.0b013e32830b8937. PubMed: 18806613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A quantile-quantile (Q-Q) plot showing the distribution of observed χ2 statistics of analyzed SNPs in 1,617 SCD individuals (SIT Trial, n=925; CSSCD, n=692). The black diagonal line indicates expected results under the null hypothesis.
(TIFF)
Comparison of the effect size of variants from the reported regions associated with SBP in the ICBP meta-analysis and SCD cohort.
(TIFF)
Power calculation to detect genome-wide significant SNPs (minor allele frequency > 0.05) for systolic blood pressure in the combined SIT Trial and CSSCD cohort, utilizing 1617 SCD samples. The authors thank the staff, clinicians and patients for their participation in the Silent Infarct Transfusion (SIT) Trial study. Genotyping services for the SIT Trial were provided by the Center for Inherited Disease Research (CIDR) at Johns Hopkins University and the Division of Blood Disorders at the Centers for Disease Control and Prevention (CDC). The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
(TIFF)