Abstract
A critical analysis of virtual screening results published between 2007 and 2011 was performed. The activity of reported hit compounds from over 400 studies was compared to their hit identification criteria. Hit rates and ligand efficiencies were calculated to assist in these analyses and the results were compared with factors such as the size of the virtual library and the number of compounds tested. A series of promiscuity, drug-like, and ADMET filters were applied to the reported hits to assess the quality of compounds reported and a careful analysis of a subset of the studies which presented hit optimization was performed. This data allowed us to make several practical recommendations with respect to selection of compounds for experimental testing, defining hit identification criteria, and general virtual screening hit criteria to allow for realistic hit optimization. A key recommendation is the use of size-targeted ligand efficiency values as hit identification criteria.
Keywords: Virtual screening, hit identification, hit criteria, ligand efficiency, hit optimization
INTRODUCTION
In recent years, the drug discovery arena has seen an exponential increase in the application of computer-based methodologies toward the identification of hit or lead compounds. Successful examples of virtual screening (VS) using ligand- or structure-based strategies have been frequently reported. Often, virtual screening techniques are employed in parallel with or in place of traditional high-throughput screening methods, particularly within academic laboratories. Although the hit identification, or ‘hit-calling’, criteria for traditional high-throughput screens (HTS) are well defined, there is less of a consensus in the literature as to how to define a hit compound identified from computational screening methods based upon the experimental activity of the compounds tested.1 With the expanding use of these techniques and numerous publications of their application, it is reasonable to ask: how should a virtual screening ‘hit’ be defined in terms of experimental activity? More to the point: What level of experimental activity can one realistically expect to obtain when employing VS methods? Is it different from that seen when the traditional HTS hit criteria are employed (i.e. based upon an activity cutoff or a specified number of standard deviations above a libraries mean activity)? Should it be?
To address these questions, we have performed an extensive analysis of the last five year’s literature reporting ‘active’ compounds identified by virtual screening methods and confirmed by experimental testing. The activity of the reported compounds from over 400 published studies has been compared with respect to the initial activity cut-off (if defined), lowest reported hit activity, activity range, or other hit identification criteria. Hit rates and ligand efficiencies were calculated to assist in these analyses and the results were compared with contributing factors such as the size of library virtually screened, and the number of compounds tested. Additionally, we have performed a careful analysis of a subset of the studies (approximately 80 reports), which presented the activities of next generation compounds derived from initial VS hits. This data, in particular the hit optimization results, have allowed us to perform a critical analysis of the virtual screening that has been performed over the last several years and to suggest several practical recommendations with respect to selection of virtually screened compounds for experimental testing, defining initial hit identification criteria, as well as general VS hit criteria that will allow for realistic hit optimization. An analysis of additional contributing factors such as the nature of the biological drug target (type of enzyme, active site classification, etc.), publication year, publishing journal, VS method, and research setting has been placed in Supporting Information, as these data have previously been quantified in an excellent literature survey.2
Extensive literature analyses of applied virtual screening have been previously published. Among these, a comprehensive literature survey of prospective VS approaches has been presented and provides a detailed picture of the applied VS field.2 This survey identified the scientific journals and target families where VS applications are most often published and the potency distribution of VS hits.2 The field has been further characterized by two following studies with in-depth analyses of available prospective ligand-based and structure-based VS applications and several scientific quality criteria for prospective VS have been formulated.3, 4 Additionally, several recent studies have critically examined method design and application in virtual screening, presented practical recommendations and discussed shortcomings.5–7 In this work, we instead focus on a critical analysis of the results of virtual screening, with particular attention given to identification of initial hits and hit optimization. Papers describing virtual screening results that were published from 2007 to 2011 were extracted from three databases, PubMed, Web of Science and Embase, using the MeSH terms and key word queries described in detail in the Supporting Information. The search queries were (‘virtual screen’ OR ‘in silico screen’ OR ‘ligand-based’ OR ‘structure-based’ OR ‘receptor-based’) AND (‘identified’ OR ‘identify’ OR ‘identification’ OR ‘discover’ OR ‘discovered’ OR ‘discovery’) AND [2007-2011]. There was no impact factor restriction on journal selections. The resulting 4029 publications were individually reviewed to confirm both that they were prospective VS studies and that experimental testing of compounds identified was performed and reported. This resulted in 421 publications that were identified for further analysis. In addition, a subset of 80 publications reporting both identification and optimization of initial hits were further analyzed, as discussed below.
SCREENING HIT CRITERIA IN DRUG DISCOVERY
Virtual screening, traditional high-throughput screening, and fragment-based screening are now established as three important hit identification paradigms in drug discovery. The goal of these screens is often to discover initial hit compounds for further medicinal chemistry optimization. Hit selection methods for high throughput screens typically include statistical analyses and/or a manually set threshold (i.e., a percentage inhibition at a given screening concentration).8 Hit selection criteria for fragment-based screens often employ a ligand efficiency (LE) metric, which normalizes the experimental activity to molecular weight, heavy atom count, or other molecular size quantifier (i.e., LE ≥ 0.3 kcal/mol/heavy atom).9, 10 The smaller number of experimentally tested compounds from most virtual screens, typically a fraction of the higher scoring compounds, can limit the use of statistical analyses for the determination of hit cutoffs. There was at least one report that employed statistical analyses for hit selection, but this study performed secondary HTS for biological validation of the virtually screened compounds, with hundreds of compounds experimentally tested.11 Percentage inhibition is widely used in virtual screening for hit selection while ligand efficiency has not been well employed as an activity cutoff method. From our analyses, only 121 (approximately 30%) of the studies reported a clear, predefined hit cutoff and no clear consensus on hit selection criteria was identified. Both concentration-response endpoints (IC50, EC50, Ki, or Kd) and single concentration percentage inhibition were used as biological metrics for hit cutoffs in the VS studies; 38 of the former and 85 of the latter (see Table 1). The activity cut-off for hit identification was often an arbitrary number (ranging from sub-micromolar to high micromolar activity) or, in some cases, determined from a positive control (known active).12, 13 Interestingly, ligand efficiency was not used as a hit selection metric in any of these reports.
Table 1.
Summary of hit identification and hit rate data collected.
| Hit Calling Metric | Screening Library Size | Compounds Tested | Calculated Hit Rate (%) | Validation Assays | |||||
|---|---|---|---|---|---|---|---|---|---|
| EC50 | 4 | <1,000 | 16 | 1 – 10 | 50 | < 1 | 8 | Binding a | 74 |
| IC50 | 30 | 1,000 – 10000 | 30 | 10 – 50 | 161 | 1 – 5 | 60 | Secondary assay b | 283 |
| % Inhibition | 85 | 10,001 –100,000 | 89 | 50 – 100 | 71 | 6 – 10 | 65 | Counter screen c | 116 |
| Ki/Kd | 4 | 100,001 –1,000,000 | 169 | 100 – 500 | 95 | 11 – 15 | 65 | ||
| Other | 8 | 1,000,001 –10,000,000 | 78 | 500 – 1000 | 13 | 16 – 20 | 25 | ||
| Not Reported | 290 | >10,000,001 | 13 | ≥ 1000 | 16 | 21 – 25 | 29 | ||
| Not Reported | 26 | Not Reported | 15 | ≥ 25% | 103 | ||||
| ND | 40 | ||||||||
Studies included evidence that hits were directly binding to the biological target, either via competition in an orthogonal assay, direct biophysical binding, or crystallography for structurally enabled targets.
Studies included secondary assays after primary assay to confirm the activity of hits.
Studies included counter screens to confirm the selectivity of hits.
To quantify and compare the hit identification criteria from the collected reports, we have analyzed the activity cutoff distributions in two different ways. First, from the studies with clearly defined activity cutoffs, the reported cutoffs were used for further analysis (Figure 1, blue). From the studies without clear hit cutoffs listed, the activity of the least active compound reported was used to estimate the hit cutoff, with the assumption that the least active compound’s activity was near to the hit cutoffs of these studies (Figure 1, red). For the activity cutoff distribution analysis, both concentration-response endpoints and percentage inhibition were considered. We note that when a concentration-response endpoint activity value was not used as the hit identification metric, the value was extrapolated for these studies from the specified percent inhibition cutoffs using the four-parameter logistic Hill equation recently discussed by Gubler and coworkers.14 The activity spectrum was divided into six categories: <1, 1–25, 25–50, 50–100, 100–500, and >500 μM.
Figure 1. Hit cutoff ranges.

Values in blue were obtained from studies with clearly defined hit cutoffs; values in red are estimated from the lowest experimental activity reported for a hit.
In general, activity cutoffs at sub-micromolar levels were rarely used in the virtual screening studies published (Figure 1). Because the aim of virtual screening is usually to provide a novel chemical scaffold for further optimization, virtual hits with sub-micromolar activity, while desirable, are not typically necessary. The majority of the studies analyzed here used activity cutoffs in the low to mid-micromolar range: 136 studies used 1–25 μM, 54 studies used 25–50 μM and 51 studies used 50–100 μM as their cutoffs. Surprisingly, there were 56 studies that used 100–500 μM and 25 studies used >500 μM as the initial activity cutoff. In the last case, this high micromolar to low millimolar activity requirement would be considered borderline active to inactive in many cases, with the possible exception of fragment-based screens. In these cases, roughly one-third of these studies involved the screening of fragment compounds (MW <300). For virtual screening of lead- or drug-like compounds with high micromolar activity cutoffs, we found there were two common reasons that the authors discussed for using these low activity levels to define hits. The first was to improve the structural diversity of the hit compounds and the second was the use of lower activity cutoff requirements when screening against novel drug targets without a priori inhibitor knowledge, for example previously reported active compounds.5, 15
Initial hit identification activity cutoff criteria must be considered carefully. High activity cutoffs may identify a low number of potent compounds for easier optimization by medicinal chemistry; however, it will also limit the diversity and chemical space of the hits. Low activity cutoffs improve the diversity of hits but suffer from difficulties in further optimization.8 Studies by the Shoichet group compared the results from HTS and VS against the same library and found that many of the potent inhibitors generated from HTS were promiscuous, covalent, or possessed a known active scaffold, while two moderately active VS hits were able to be optimized to novel, non-covalent inhibitors.16 This suggests that prioritization of weak-but-novel chemotypes can be fruitful, in some cases, even more fruitful than focusing on the prioritization of just highly active hits. It is also true that identification of low-activity compounds against a novel target with very limited prior information, even if only borderline active, could be attractive. Nevertheless, the question of whether such hits can be optimized by medicinal chemists to an acceptable, drug-like level of activity (typically low nanomolar levels) remains. A retrospective study of hit optimization programs at Abbott showed that from a starting hit with MW at 350 Da, 0.5 μM activity or better was desirable in order to be obtain a 10 nM affinity lead compound with MW at 500 Da.17 The direct implication of these studies is that ligand efficiency should be considered when defining initial hit identification criteria, even when experimentally testing compounds obtained from virtual screening.
Although ligand efficiency was not employed as the hit identification metric in any of the VS studies we examined, we felt that it would be informative to analyze the data from the initial hits reported in order to obtain a better picture of their LE landscape. Ligand efficiency is simply the estimated binding free energy of a ligand normalized by its molecular size. Ligand efficiency has become an important and useful metric for assessing fragment-like hits, HTS hits, VS hits, and the resulting optimized compounds. There are few targets for which VS, HTS, and FBDD data are all available. We identified several such targets and performed a limited LE comparison of the results from each screening approach for each target. Overall, no significant trend was observed with respect to any method outperforming another in terms of resulting hit LE values, though we note the difficulty in observing a trend with the low number of targets we identified and the difficulty in comparing hit LE results across targets, where the target’s properties themselves influence the resulting hit activities. These results are summarized in Supporting Information (Table S2).
In this work we have chosen to follow the lead of the Kuntz lab and define the molecular size by the number of heavy (non-hydrogen) atoms.18 The ligand efficiency of the most active initial hits from these prospective VS studies was analyzed. Concentration-response endpoint activity values were used to estimate binding free energy, with the assumption that experimental testing was performed at room temperature (300 °K). Of the 365 compounds used in these calculations, the average LE was 0.3±0.2 kcal/mol/heavy atom. 76 (approximately 20%) of the reported hits possessed an LE greater than 0.4 kcal/mol/heavy atom, and 184 (approximately 50%) had calculated LE values lower than 0.3 kcal/mol/heavy atom (Figure 2A). Additionally, we plotted the LE against HAC to determine whether there was a non-linear correlation of initial LE with the molecular size, as Reynolds and co-workers have reported (Figure 2B).19 As can be seen in Figure 2B, the LE values of the reported hits were demonstrably higher on average for smaller molecules, and an exponential decrease in the estimated LE was observed as the molecular size (HAC) of the reported hits increased. This data is consistent with that reported by Reynolds, et al. and may be explained by a non-linear correlation between the surface area (for making favorable interactions) and the number of heavy atoms as well as a conformational entropy penalty for larger ligands.19
Figure 2. Ligand efficiency analysis of the most active reported VS hits.
(A) LE sorted by virtual screening study, (B) LE plotted against HAC for each compound reported.
Ligand efficiency values also varied considerably based on the molecular target. For some targets such as HSP90, it was not uncommon to find inhibitors possessing LE greater than 0.5 kcal/mol/heavy atom, while for more challenging targets, such as inhibitors of protein-protein interactions, LE could fall significantly below 0.3 kcal/mol/heavy atom. A possible reason behind this observation is the differences in ‘druggability’ between the different targets.20 A drug candidate with a Kd of 10 nM and a MW of 500 Da (~38 heavy atoms) would have an LE of 0.29 kcal/mol/heavy atom. Therefore, it may be tempting for researchers to look for hits with LE of 0.3 kcal/mol/heavy atom or better. The work by Hopkins, et al. previously mentioned suggests that LE should be maintained at this level (0.3 kcal/mol/heavy atom) for lead selection, however, it should be emphasized that this pertains to selection of optimized compounds for further advancement, rather than the initial hit identification.9 Practically speaking, the use of target LE values adjusted by the molecular size of the screened compounds and the nature of the screening target appears to be the optimal strategy for hit identification. This approach is discussed in more detail below.
VIRTUAL SCREENING HIT RATES
Calculated hit rates can provide a loose measure of the success of the method used to perform the virtual screen, although the direct comparison of hit rates between different studies, such as those surveyed here, must be interpreted with caution as the definition of a hit in these studies varied quite dramatically. Several studies from the Shoichet group have compared the number and quality of hits from both traditional and virtual screening against the same experimental library.16, 21, 22 Typical hit rates from experimental HTS can range between 0.01% and 0.14%, while hit rates for prospective virtual screen typically range between 1% and 40%.23 Hit rates are often higher in retrospective VS studies than those typically seen in prospective studies. Retrospective, or benchmark studies, are standard practice prior to VS for comparison of different methods and improvement of performance.24 However, benchmark performance is not considered a very good predictor of prospective VS performance, in term of hit rates, due in part to the different compositions of benchmark libraries when compared with prospective screening libraries.5 An additional consideration regarding hit rates is that higher hit rates in prospective studies, such as these, are not always preferable. Low hit rates from studies identifying interesting and novel scaffolds are preferable to high hit rates from studies identifying known active scaffolds.25 An alternate measure of VS success may be assessed by how many novel scaffolds are identified rather than the overall number of hits or general hit rates.
With these limitations in mind, we calculated the hit rate for each of the reports that were collected in this study for comparison with both typical HTS values and the previously reported VS hit rates described above. The results are summarized in Table 1. In our studies, hit rates were determined by dividing the number of initial hits by the number of experimentally tested compounds. In order to avoid any bias by very small number of experimental tested compounds, only studies with more than five compounds being biological tested were used for hit rate analysis. As expected, hit rates varied widely, with values below 1% to 100% of experimentally tested compounds described as hits. The median value of hit rate is 13%, which would be considered outstanding overall performance for prospective virtual screening, if the definition of a hit were realistically defined. There were 103 studies with hit rates calculated over 25%, which represented nearly one-third of the total number of studies for which we were able to calculate hit rates. For these studies, we performed a closer examination with respect to the size of the library virtually screened and the number of compounds biologically tested. We did not observe any trend in the size of the library screened to a higher hit rate, with sizes ranging from under 1,000 to over 10 million; however we did note that many of the high hit rate studies tested a low number of compounds, with 75% testing fewer than 20 compounds and nearly 50% tested fewer than 10.
HIT VALIDATION, PROBLEMATIC STRUCTURES, AND EARLY ADMET TESTING
In both experimental and virtual screening, it is good practice during the process of hit confirmation to confirm activity, target selectivity, and also to analyze the hit compounds for potentially problematic structures, including reactive or toxic pharmacophores and known promiscuous scaffolds. To assess the potential of the reported hits and next generation compounds for further development, we performed an analysis of the studies collected to determine whether direct binding assays, secondary assays, and counter screens were included in the studies. The results are summarized in Table 1. Out of 421 studies, 17% of the analyzed studies employed direct binding assays to confirm that hits were directly binding to the biological target, 67% of the analyzed studies employed secondary assays to confirm the activity of hits, and 27% of the analyzed studies employed counter screens to confirm the selectivity of hits. This data indicates that a large number of the hits may not have been well validated, as the hits were seldom assessed for direct binding or selectivity. The importance of hit confirmation, particularly relevant for VS studies, cannot be overemphasized. There are a variety of possible mechanisms for assay artifacts and it is important to understand the screening assay results, potential artifacts, and to confirm the activity of hits by secondary assays, such as direct biophysical binding assays, orthogonal assays, or structural biology studies.26 Pharmacological promiscuity of hits should also be investigated by counter screens to confirm the selectivity, especially for the targets such as kinases, cytochrome P450 and GPCRs.
It is important to evaluate hit compound’s potential for pharmacological promiscuity and for assay interference as early as possible. To identify such problematic compounds, there are a variety of filters available that may be applied either to the entire library prior to screening, or to potential hit compounds after they have been identified.27 Table 2 lists a small sample of useful tools that can be used to assess the quality of screening hits. In our laboratory, we typically apply a series of substructure filters to virtual libraries prior to screening to eliminate compounds from screening that possess known reactive and toxic functional groups. Potential hit compounds are then filtered after screening to identify potentially promiscuous compounds, which allows for a more judicious approach to the elimination of hits from further consideration. Additionally, early ADMET prediction using one or more of a variety of useful software (see Table 2), can prove quite useful in the selection of hit compounds for advancement to the optimization stage, as late-stage attrition of hit or lead compounds in drug development can be very costly. Recent studies have suggested that the perceived benefit of high in vitro activity may be negated by poor ADMET properties and have highlighted the employment of ADMET filters in hit identification and optimization.28–30 In order to gain some insight into the both the quality and potential for optimization of the virtual screening hits that are being reported, we applied a series of these filters to the data that was collected and also performed preliminary ADMET predictions using the QikProp program of Schrödinger, LLC.31
Table 2.
Tools available to assess problematic and/or ADMET properties. More tools are listed at http://www.click2drug.org/directory_ADMET.html
| Tools | Name | Features | Developer/Reference |
|---|---|---|---|
| Substructure Filters | PAINS | To identify frequent hitters (promiscuous compounds) in many HTS assays. | Cancer Therapeutics-CRC P/L 33 |
| REOS | A hybrid method that combines some simple counting schemes similar to rule-of-5 with a set of functional group filters to identify reactive, toxic and other problematic structures. | Vertex 32 | |
| Eli Lilly Rules | A hybrid method that combines physicochemical properties with a set of functional group filters to identify reactive, promiscuous structures. | Eli Lilly 34 | |
| Physicochemical Property Filters | Lipinski (Rule-of-5) | A rule of thumb to evaluate drug-likeness for orally active drugs. | Lipinski at Pfizer 57 |
| Oprea Lead-like and drug-like | A series properties rules for drug-likeness and lead-likeness based on extensive comparison studies of lead-drug pairs. | Oprea Group58 | |
| Veber | Molecular properties rules that influence the bioavailability of drug candidates. | Veber at GlaxoSmithKline 45 | |
| Software/Servers for ADMET prediction | QikProp | Provides rapid ADME predictions of drug candidates. Distributed by Schrödinger. | Jorgensen Group and Schrödinger31 |
| VolSurf | Calculate ADME properties and create predictive ADME Models. Distributed by Tripos. | Tripos59 | |
| admetSAR | A comprehensive source and free tool for assessment of chemical ADMET properties. | Yun Tang Group 60 |
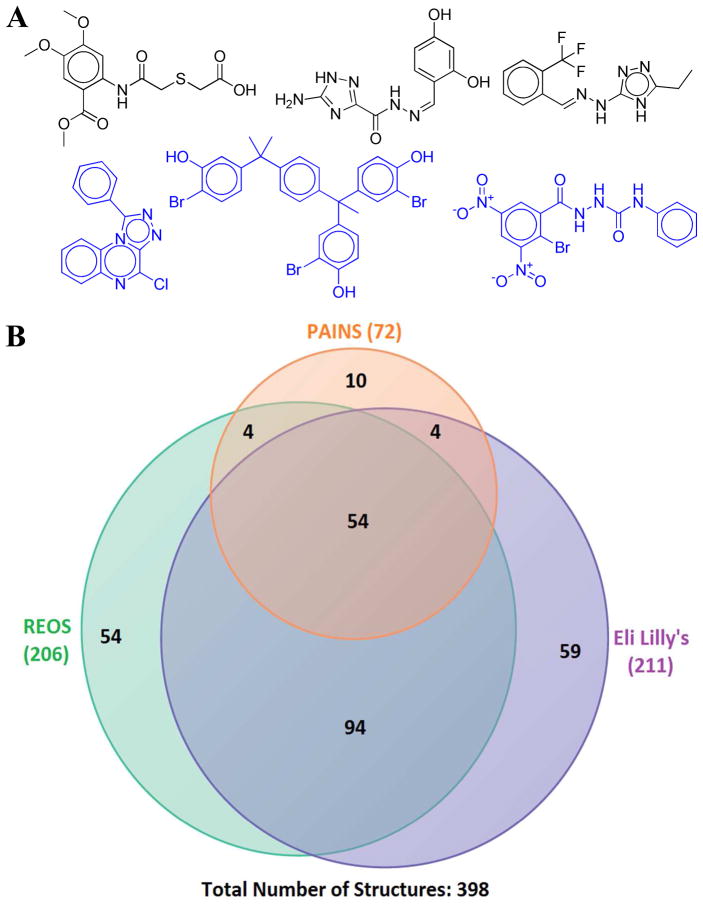
The structure of the initial hits (most active reported in each study) and the next generation compounds from the prospective VS studies were analyzed to assess potentially problematic structures. Three filters were used for this purpose: REOS (Rapid Elimination of Swill) published by the Walters and coworkers at Vertex Pharmaceuticals, the PAINS (Pan Assay Interference Compounds) filters and the recently published rules for identifying potentially reactive or promiscuous compounds from Eli Lilly.32–34 The REOS filters flag compounds containing functional groups that may lead to false positives due to reactivity, assay interference or poor ADMET properties. PAINS filters consist of a variety of functional groups extracted from “frequent hitters” in HTS assays, and the “rules for identifying potentially reactive or promiscuous compounds” from Eli Lilly are a series of demerit based rules that can be applied to filter out a variety of compounds known to be problematic.
Out of 398 structures extracted from the initial VS hits, 206 were flagged by REOS filters, 72 were flagged as PAINS and 211 were flagged as potentially reactive or promiscuous according to Eli Lilly’s filters (Figure 3A, black). The problematic structures flagged by these filters had a high degree of overlap (Figure 3B). This seems to indicate that there are a large number of molecules reported to be bioactive against their targets by virtual screening that are false positives or are unsuitable for optimization. While they may be acceptable as initial hits, they would likely have to be eliminated from consideration prior to optimization. Interestingly, of the 80 next generation compounds reported, 38 were flagged by REOS filters, none were flagged as PAINS, and 38 were flagged as potentially reactive or promiscuous according to Eli Lilly’s filters (Figure 3A, blue). Thus, although the fraction of potentially problematic compounds decreased during optimization compared to the initial VS hits, it is likely that a substantial number of these next generation compounds are also unsuitable for further development.
Figure 3. Flagged problematic compounds.
(A) Shown are representative structures of potentially problematic compounds from initial hits (colored as black) and next generation compounds (colored as blue), identified by the application of the REOS,32 PAINS,33 and Eli Lilly filters.34 (B) Venn diagram showing the overlap of potentially problematic compounds from the different filters against initial hits.
From QikProp analysis, we are able to quickly compare the calculated ADMET properties (including LogP, LogS, Caco-2 and MDCK cell permeability, LogKhsa, for human serum albumin binding, and LogIC50 for HERG K+-channel blockage) of the initial hit and optimized compounds to known drugs using the ‘star’ penalty system of the program (where a star is given to a compound for each instance that the predicted property falls outside the range covered by 95% of known drugs). The recommended ‘acceptable’ range is 5 or less ‘stars’.31 Surprisingly, 53% of the initial hits were predicted to possess properties within the 95% range of similar values for known drugs (0 stars), and 26.7% of the initial hits were predicted to have only one property dissimilar to 95% of the known drugs (1 star), (Figure 4). 18% of the compounds were predicted to have two to five properties dissimilar to 95 % of the known drugs, and very few compounds (~2%) were predicted to have more than five properties dissimilar to 95% of the known drugs. As shown in Figure 4, the predictions for the next generation compounds (red) were very similar, or slightly better, than the initial hits and none of them were predicted to have more than five properties dissimilar to 95% of the known drugs. Considering the poor performance of the ‘hit’ compounds in the reactive and promiscuous filters discussed above, the ADMET prediction results were somewhat surprising.
Figure 4. Summary of ADMET property predictions of initial hits and next generation compounds by QikProp.
Number of stars: Number of properties or descriptors that fall outside the 95% range of similar values for known drugs. (Recommended acceptable range is 0–5.)
VIRTUAL SCREENING HIT OPTIMIZATION
Once a number of hits with confirmed activities have been obtained from virtual screening, HTS or fragment based screening, the hits will be prioritized and refined in order to improve potency, selectivity, toxicity, and in some cases, PK properties. Typically, this work consists of selection of interesting scaffolds for intensive SAR analysis by analog search, synthesis of a focused library, and structure-based (x-ray crystallography, NMR, or modeling) optimization. For drug-like VS hits, chemical modifications of the initial hits by scaffold/similarity search across commercially available databases, synthetic SAR exploration, and hit fragmentation are the most frequently used techniques. Hit fragmentation (scaffold decomposition or ‘scaffold pruning’) is the structural decomposition of initial hits to identify key binding fragments. Fragments identified from this approach can be subjected to fragment-based optimization strategies, including fragments linking, growing and merging.35 The application of ligand efficiency in hit optimization can offer useful information to assess the quality of drug-target interactions, as they are improved. Several recently described hit optimization campaigns provide illustrative examples of carefully monitoring key characteristics such as ligand efficiency to generate higher quality leads.36, 37
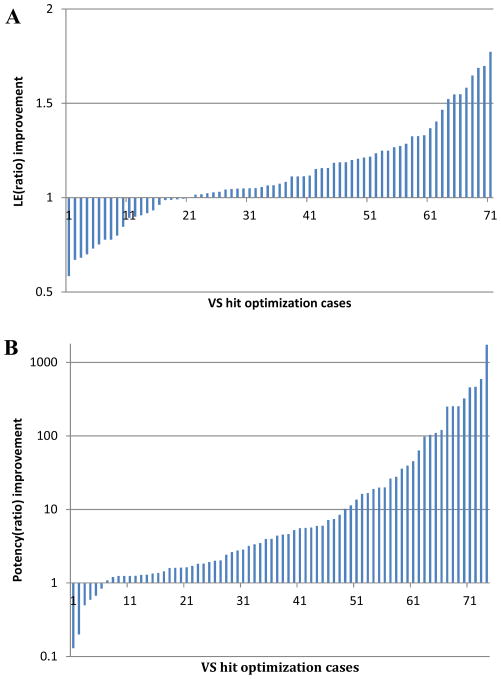
In these studies, we have analyzed 80 published VS hit optimization campaigns with respect to their optimization strategy, ligand efficiency and activity improvement, heavy atom count changes, and the chemical similarity between the initial hits and optimized compounds. The entire data set is included in Supporting Information. The ligand efficiency values were calculated as discussed above. LE and activity improvements during optimization were calculated by ratio changes as following: LE(ratio)= LE(after optimization)/LE(before optimization); Activity(ratio)= Activity(before optimization)/Activity(after optimization). The overall results are summarized in Table 3 and Figure 5. During VS hit-to-lead optimization, LE was increased in 72% of the cases, with an average LE ratio improvement of 1.12 ± 0.26; and activity was improved in 92% of cases, with an average activity ratio improvement of 71.1 ± 227.4. In most cases, LE remained relatively stable during hit optimization (LE ratio 1 ± 0.2), while activity improved significantly (10 – 100 fold, on average). This reflects, at least in part, the fact that the majority of the reports did not emphasize LE improvement during their hit optimization studies, instead focusing strictly on improvement in the absolute activity. The method of optimization, either analog search, synthetic SAR, or a combination thereof, did not significantly affect the LE ratio (Table 3); however the use of synthetic SAR methods in optimization resulted in approximately 10-fold better activity ratios when compared to the results from studies reporting analog searches or a combination strategy (although the results were highly variable). In 67% of the VS optimization cases, the optimized compounds had an equal or greater number of heavy atoms. In these studies the average LE ratio was 1.01 ± 0.19 (Table 3). The remaining studies decreased the number of heavy atoms during optimization which resulted in an average LE ratio of 1.35 ± 0.22. These results highlight the utility of hit fragmentation or scaffold decomposition approaches for hit optimization, particularly when the initial hits are lead-like or drug-like.
Table 3.
Summary of VS optimization process.
| Optimization methods | Same scaffold c | NHA decreased | |||||
|---|---|---|---|---|---|---|---|
| Analog search | Synthetic SAR | Analog search & synthetic SAR | Yes | No | Yes | No | |
| Number of cases | 31 | 44 | 5 | 25 | 55 | 27 | 53 |
| LE (ratio) a Mean | 1.14 ± 0.22 | 1.10 ± 0.28 | 1.21 ± 0.31 | 1.14 ± 0.22 | 1.10 ± 0.27 | 1.35 ± 0.22 | 1.01 ± 0.19 |
| Potency (ratio)b Mean | 12.30 ± 28.06 | 118.25 ± 297.82 | 14.20 ± 9.74 | 21.53 ± 68.55 | 92.11 ± 265.61 | 40.27 ± 110.04 | 86.67 ± 267.98 |
LE (ratio) = LE (after optimization)/LE (before optimization).
Potency (ratio) = Potency (before optimization)/Potency (after optimization).
Same scaffold at same level by scaffold decomposition.
Figure 5. Ligand efficiency and potency improvement during optimization.
(A) LE improvements during optimization was monitored by ratio changes as following: LE(ratio) = LE(after optimization)/LE(before optimization); (B) Potency improvements during optimization was monitored by Potency(ratio) = Potency(before optimization)/Potency(after optimization).
Interestingly, there were eight studies where LE was improved quite dramatically relative to the starting hit (LE ratio > 1.5); seven of these studies reduced the number of heavy atoms during the optimization (Table 4). There were several reasons for this reported by the authors. First, in four of the eight cases, hit fragmentation was employed during optimization. During the traditional hit optimization process the molecular weight of the hit series generally increases as functional groups are added or modified to achieve greater activity. In contrast to this, optimization studies that utilize a fragmentation approach can result in compounds with lower molecular weight, but with similar or improved activity. In these cases, the ligand efficiency of the next generation compounds can dramatically increase when compared to ligand efficiency improvement typically seen with the former. Second, some targets are very sensitive to ligand substituent patterns; therefore, compounds sharing the same scaffold can be optimized quite dramatically by simple chemical modifications. This concept of ‘activity cliffs’ is a recognized phenomenon that has been described in several recent publications.38–40 One exceptional example reported was the optimization of an essentially inactive VS hit (EC50 > 100 μM) to a potent inhibitor (EC50 = 310 nM) by computational substituent scanning and subsequent synthesis.41 This study introduces a computational strategy for promoting VS inactive or borderline active hits through chemical transformations against targets known to be very sensitive to the ligand’s substituent pattern.
Table 4.
Profiles of VS hit optimization where LE(ratio) is greater than 1.5. LE(ratio) = LE(after optimization)/LE(before optimization).
| Initial hit | Next generation compound | Optimization method | Same scaffold | NHA reduced | Ref | ||||
|---|---|---|---|---|---|---|---|---|---|
| Activity (nM) | Structure | LE (kcal/mol/atom) | Activity (nM) | Structure | LE (kcal/mol/atom) | ||||
| ~100,000 IC50 |

|
0.24 | 13,500 IC50 |

|
0.39 | Analog search | Yes | Yes | 61 |
| 40,000 IC50 |

|
0.12 | 10,100 IC50 |

|
0.20 | Analog search and synthetic SAR | No | Yes | 54 |
| ~100,000 IC50 |

|
0.22 | 35,000 IC50 |

|
0.34 | Synthetic SAR | No | Yes | 55 |
| 2500 IC50 |

|
0.21 | 1240 IC50 |
|
0.37 | Synthetic SAR | No | Yes | 62 |
| 390 EC50 |

|
0.33 | 3.98 EC50 |

|
0.50 | Synthetic SAR | No | Yes | 63 |
| >100,000 EC50 |

|
<0.23 | 310 EC50 |

|
0.39 | Synthetic SAR | Yes | Yes | 41 |
| 16900 IC50 |

|
0.19 | 2000 IC50 |

|
0.29 | Analog search | No | Yes | 64 |
| 33000 IC50 |

|
0.18 | 19 IC50 |

|
0.29 | Synthetic SAR | No | No | 65 |
PRACTICAL RECOMMENDATIONS
Selection of compounds for biological testing
The number of compounds selected from a virtual screen for biological testing is often dependent upon the resources available. Typically only a small percentage of the top-ranked compounds are tested. Most commonly used selection methods focus on maximizing the diversity and drug-likeness of compounds to be experimentally tested, while minimizing false-positive rates. Clustering or scaffold decomposition based methods can be used to reduce costs by decreasing the number of compounds tested while maintaining structural diversity.42 The concept of drug-likeness is now widely utilized as a means to optimize PK properties early in drug discovery, given that PK issues are responsible for as high as 40% of the overall attrition rate in the drug development process.43 General physicochemical property filters are commonly used for drug-likeness evaluation of VS hits.44, 45 Additionally, certain physicochemical properties need to be individually evaluated based on the nature of the screening target. For example, increased weight and hydrophilicity are commonly observed for antimicrobials, and inhibitors of protein-protein interactions tend to have higher molecular weight and hydrophobicity. As discussed above, compounds possessing reactive functional groups and known toxic pharmacophores should be eliminated either during library preparation or after VS during compound selection (see Table 2). Promiscuous chemical scaffolds, and “frequent hitters” filters can also be used to reduce the number of false-positives. These filters are usually substructure-based and often must be modified based upon the assay conditions. For example, one should consider using a color substructure filter, such as has been incorporated in Eli Lilly’s filters, for VS follow-up using colorimetric assays and for biological assays using the classical malachite green assay, hydrophobic amines will have to be filtered out for potential assay interference.46 Other types of assay interference, including redox active compounds and intrinsically fluorescent compounds, may also be considered.
In addition, MW adjusted VS scores, particularly for structure-based VS, and scaffold novelty analyses can be also employed in the selection of VS compounds for experimental testing. It has been previously shown that structure-based VS approaches are biased toward the selection of high molecular weight compounds and normalization strategies based on the number of heavy atoms has been proposed for the chemical compounds being selected for screening.47, 48 This will be especially helpful if ligand efficiency is employed as a hit identification metric (see below). In order to avoid “rediscovering” known or very similar active compounds, scaffold novelty can be evaluated by comparing the similarity of ranked VS compounds with known actives by Tanimoto indices or other 2D metrics.49 This has the additional benefit of ensuring that compounds being screened are outside the current ‘patent space’ for the target being screened. Resources such as CAS SciFinder® and PubChem are excellent tools that can be employed for such evaluations.50
Defining initial hit identification criteria
Although the use of concentration-response values such as the IC50 or Ki are most desirable for use as hit cutoff metrics, they are less practical than percentage inhibition at a specific concentration for reasons including cost, manpower limitations, or limited compound supplies. For these reasons, percentage inhibition is most widely used as the initial hit identification criterion. However, it is possible to estimate the IC50 (assuming ideal conditions) using the four-parameter logistic Hill Equation (1).14, 51 It describes the ideal relationship of inhibition percentage y at given screening concentration x with IC50 and Hill coefficient n, and the asymptotic plateau A0 and Ainf values at low and high concentration.
| (1) |
In Figure 6A, we have plotted the percentage inhibition at several practical screening concentrations against the estimated Log (IC50). We note that a screening concentration of 100 μM is often a practical limit for non-fragment like compounds owing either to limited solubility of the compounds or general cell toxicity at higher concentrations.41 The estimated IC50 values can be taken one step further and used in the calculation of hit cutoff percent inhibition values for target Ligand Efficiency (LE), if the molecular weight or number of heavy atoms (HAC) for the compounds screened is known. For the screening of fragment-like compounds, a target ligand efficiency of greater than 0.3 kcal/mol/atom has been previously recommended as a practical hit identification metric.9 However, for lead- and drug-like compounds, the target LE value must be decreased for practical reasons, as 0.3 kcal/mol/atom for a drug-like compound (molecular weight roughly 500 Daltons, or 35 heavy atoms) corresponds to a roughly 10 nM activity, which is unrealistic for initial hit identification. As discussed above, it has been previously shown that ligand efficiency can be molecular size dependent and we have observed this trend in our current analysis of the initial VS hits.19 These authors recommended the use of molecular size adjusted LE values in the evaluation of HTS hit compounds.
Figure 6. Ligand efficiency as a hit identification metric.
(A) Expected relationship between Log (IC50) and percentage inhibition based on four-parameter logistic Hill equation at 10, 25, 50 and 100 μM screening concentrations. (B) Adjusted target LE values at different heavy atom count according to Equation (2). (C) Estimated percentage inhibition values at different HAC for hit selection cut-offs in order to maintain LE at target values shown in (B).
Additionally, we recommend the use of differing target LE values depending on the size of the compounds virtually screened. For fragment-like compounds (HAC ≤ 18) an LE of 0.32 is recommended, while for lead-like compounds (HAC 19 – 25) an LE of 0.25 is recommended and for drug-like compounds (HAC 26 – 35) an LE of 0.19 is recommended (the HAC/MW correlation is estimated by: MW = 14.248 x HAC + 2.5452; R2=0.9429 for all orally delivered drugs).52 Using the conversion calculations discussed above, these values roughly equate to IC50 values of 120 μM, 30 μM, and 15 μM, for compounds at 250, 350, and 500 Daltons, respectively. Another approach would be the application of a size-independent metric, such as the Fit Quality (FQ) metric, where LE values are normalized by a scaled value that takes molecular size into account.19 Alternatively, a weighting factor can be applied directly to the LE scale equation proposed by Reynolds and coworkers,19 that results in target LE values in the desired ranges for initial hit compounds. This approach is shown in Figure 6B, where the target LE is obtained using Equation (2) and is plotted against the heavy atom count.
| (2) |
This latter method is further demonstrated in Figure 6C using four typical screening concentrations and the corresponding percentage inhibition, where the calculated target LE was used to obtain the cutoff percent inhibition by combining Equations (1) and (2) to obtain Equation (3):
| (3) |
where y is the percent inhibition cutoff, x is the screening concentration, and TargetLE is defined by Equation (2). The plots shown in Figure 6C allow an immediate judgment on the question that is frequently asked by drug discovery teams, that is, what percentage inhibition value at a specific screening concentration should be used for defining hits when we know the molecular size of compounds being screened. For example, 80% inhibition or greater at 100 μM is recommended for use as hit selection criteria when screening lead-like compounds with HAC ~25 in order to obtain the target LE discussed above. In order to demonstrate the potential for error propagation using this method, we have investigated the effect of 5%, 10% and 15% concentration jitter, for example the influence of typical liquid-handing uncertainties. Corresponding figures demonstrating this effect have been placed in Supporting Information (Figures S1–S4). We note the applicability of this approach to experimental HTS results as well as virtual screening and that the weighting factor used here can be adjusted based upon the nature of the screening target or the needs of the community.
Advancing initial hits to the optimization stage
Prior to advancement to the hit optimization stage, hits must be confirmed using secondary assays, dose-response assays (if not employed initially), and counter screens to determine selectivity and to confirm their mechanism of inhibition. In most cases, additional optimization aided by synthetic medicinal chemistry is necessary for hit to lead evolution. Initial hits will be prioritized for optimization based on multiple factors, including activity, novelty, ease of synthesis, toxicity and selectivity (if available). The use of cheminformatic techniques and computational filters can aid in this prioritization. Assessment of the hit compounds for problematic substructures and scaffold novelty is recommended at this stage if this has not been evaluated in the previous steps. Good practice includes reporting the similarity of newly identified hits compared to known actives by searching through chemical databases including CAS SciFinder® or PubChem using the similarity metrics described above. Attention in this phase has also turned to more detailed profiling of ADMET properties and these studies can be accelerated by computational tools.53
Once hit compounds are selected for optimization, the continued application of ligand efficiency can still offer useful information to assess the quality of drug-target interactions. Depending on the LE and the molecular size of the initial hits, the LE during optimization will need to be either maintained or improved (i.e. to achieve a target 0.3 kcal/mol/heavy atom for a 500 Dalton drug-like compound). To achieve this, hit fragmentation (scaffold pruning) can be very helpful in the identification of key substructures and optimization of the hit scaffolds. This strategy can reduce or maintain the molecular size of the hits while significantly improving their activity, which can be very useful if lead-like or drug-like compounds were used in the initial VS.54, 55 This can have the effect of increasing ligand efficiency and allowing for further synthetic optimization while minimizing the risk of exceeding lead-like or drug-like chemical space. Finally, for targets which show a strong sensitivity to substitution patterns, such as those seen with the ‘activity cliff’ examples described above, simple similarity search, affinity prediction, and analog synthesis (substitution scanning) can result in significant activity improvement while maintaining or slightly increasing the molecular size to achieve significant improvements in LE.56 Examples of the successful application of these approaches are shown in Table 4.
SUMMARY AND CONCLUSIONS
Our analysis of the data that was collected from virtual screening reports from a 5-year period (2007-2011), revealed a number of interesting trends. To summarize, we learned that hit identification criteria are typically not rigorously defined in prospective VS studies compared with experimental HTS or fragment-based screening. Only 30% of the studies that we pulled (121 of 421 reports) used a pre-defined hit cutoff metric of any sort. Understandably, percentage inhibition at a set screening concentration was the most widely employed, however the values were often arbitrarily determined and ranged from sub- to high-micromolar estimated activity. Ligand efficiency was not employed as a hit identification metric, although it was used in the prioritization of initial hits for advancement in some cases. With respect to ligand efficiency, our analyses showed that the LE of the reported initial hits was molecular size dependent; the trend seen being similar to previously reports. From the subset of studies reporting hit optimization, we noted that LE was also not carefully monitored during the optimization process, which was reflected in a decrease in LE in roughly 25% of the cases, though potency was increased in most cases (Figure 5). Importantly, it is likely that a substantial number of the initial hits and optimized compounds are problematic, and thus unsuitable for further development based on our analyses using several computational filters. The most likely reason for this is that computational filters were not regularly employed pre- or post-VS and the initial hits were not carefully validated by binding assays, secondary assays and counter screening as discussed previously. Finally, scaffold novelty was often poorly documented in the VS studies that were analyzed.
The questions of how to define a virtual screening hit with respect to experimental activity and what level of activity can be realistically expected when employing VS methods were posed in the introduction. From our analysis of the data, notwithstanding the poor hit definition and validation that was observed, we do not feel that there is any justification to the application of activity criteria that differ significantly from those commonly used in traditional HTS. The majority of the reports that were analyzed were able to identify lead- or drug-like compounds with low micromolar activity, which is on par with the level of activity and the typical hit identification criteria typically seen in traditional HTS. We do, however, strongly recommend the use of ligand efficiency as a hit identification metric, to assess the quality of initial hits, and also to monitor the hit optimization process. Considering our observations of the size-dependence of LE in the initial hits, as well as previous reports of the same, we recommend the use of target LE values (adjusted LE based on the molecular size of the screened compounds), as we have discussed above. We have shown how target LE can be calculated and used with a transformed percentage inhibition at a given screening concentration to estimate activity and determine an immediate hit identification cutoff. We note that these values are customizable based upon the nature of the screening target and the needs of the group. Additional factors that should be considered for hit confirmation and advancement were discussed and include the use of cheminformatics and computational substructure filters to remove potentially problematic compounds from consideration and analysis of the novelty of the compounds identified. The utility of early ADMET predictions was also highlighted. To conclude, virtual screening can add significant value to a drug discovery campaign when careful attention is paid to method design, validation and experimental confirmation of computational predictions.
Supplementary Material
Acknowledgments
Funding for this research was provided by National Institutes of Health grants AI077949 and AI089535. KEH was supported during a portion of this work by NIDCR DE018381, UIC College of Dentistry, MOST program. We thank ChemAxon, Ltd. for an academic research license of their cheminformatics suite including JChem and JChem for Excel for data analysis.
Abbreviations
- HTS
High throughput screening
- VS
Virtual screening
- LE
Ligand efficiency
- ADMET
Absorption, distribution, metabolism, excretion and toxicity
- REOS
Rapid elimination of swill
- PAINS
Pan-assay interference compounds
- HAC
Heavy atom count
- SAR
Structure-activity relationship
- QSPR
Quantitative structure-property relationship
- PK
Pharmacokinetics
- NMR
Nuclear magnetic resonance
- FQ
Fit quality
Biographies
Tian Zhu received her B.S. in Bioengineering in 2006 under National Bioscience and Technology Base program and M.S. in Pharmacology in 2008 at the National Drug Screen Center from China Pharmaceutical University. She is a Ph.D. candidate in Medicinal Chemistry & Pharmacognosy at the University of Illinois at Chicago in the Center for Pharmaceutical Biotechnology under Dr. Michael Johnson, where she has been for four years. Her research interests are antimicrobial rational drug design, in silico ADMET prediction, cheminformatics, and HTS post analysis.
Kirk E. Hevener earned his B.S. in Chemistry from Tennessee State University in Nashville. He then moved to Memphis, TN where he completed his Pharm.D. in 2005 and Ph.D. in Pharmaceutical Sciences in 2008 at the University of Tennessee Health Science Center under Professor Richard Lee. This was followed by a 1 year postdoc at St. Jude Children’s Research Hospital in Chemical Biology and Therapeutics and his current postdoc at the University of Illinois at Chicago in the Center for Pharmaceutical Biotechnology under Dr. Michael Johnson, where he has been for three years. Dr. Hevener’s research focuses on the rational design of antimicrobial agents and involves the use of computational chemistry, molecular biochemistry, and structural biology techniques.
Michael E. Johnson received his B.S. from the University of Wyoming and his PhD at Northwestern University under Dr. Tai Te Wu. After a short period as an NIH postdoctoral fellow at the University of Pittsburgh, he joined the then University of Illinois Medical Center as an Assistant Professor, and is now Emeritus Professor at the University of Illinois at Chicago. He has had visiting appointments at The Scripps Research Institute; University of California, San Diego; Argonne National Laboratory; and Integrated Genomics. He is the author of over 100 scientific papers on structural biology and drug discovery, an editor of a textbook on Biotechnology, holder of several patents, and has been Principal Investigator of multiple funded projects focusing on discovery of antiviral and antimicrobial therapeutics.
Footnotes
The detailed data collection methods, factors influencing hit cut-offs, LE comparison of different screening approaches, the data set from the 80 optimization campaigns, propagated errors on Equation 1 and LE as a hit identification metric are included in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Shun TY, Lazo JS, Sharlow ER, Johnston PA. Identifying actives from HTS data sets: practical approaches for the selection of an appropriate HTS data-processing method and quality control review. J Biomol Screen. 2011;16:1–14. doi: 10.1177/1087057110389039. [DOI] [PubMed] [Google Scholar]
- 2.Ripphausen P, Nisius B, Peltason L, Bajorath J. Quo vadis, virtual screening? A comprehensive survey of prospective applications. J Med Chem. 2010;53:8461–8467. doi: 10.1021/jm101020z. [DOI] [PubMed] [Google Scholar]
- 3.Stumpfe D, Ripphausen P, Bajorath J. Virtual compound screening in drug discovery. Future Med Chem. 2012;4:593–602. doi: 10.4155/fmc.12.19. [DOI] [PubMed] [Google Scholar]
- 4.Ripphausen P, Nisius B, Bajorath J. State-of-the-art in ligand-based virtual screening. Drug Discovery Today. 2011;16:372–376. doi: 10.1016/j.drudis.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Scior T, Bender A, Tresadern G, Medina-Franco JL, Martinez-Mayorga K, Langer T, Cuanalo-Contreras K, Agrafiotis DK. Recognizing Pitfalls in Virtual Screening: A Critical Review. J Chem Inf Model. 2012;52:867–881. doi: 10.1021/ci200528d. [DOI] [PubMed] [Google Scholar]
- 6.Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: applications to targets and beyond. Br J Pharmacol. 2007;152:21–37. doi: 10.1038/sj.bjp.0707306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guido RV, Oliva G, Andricopulo AD. Virtual screening and its integration with modern drug design technologies. Curr Med Chem. 2008;15:37–46. doi: 10.2174/092986708783330683. [DOI] [PubMed] [Google Scholar]
- 8.Walters WP, Namchuk M. Designing screens: how to make your hits a hit. Nat Rev Drug Discovery. 2003;2:259–266. doi: 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- 9.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discovery Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 10.Abad-Zapatero C, Perisic O, Wass J, Bento AP, Overington J, Al-Lazikani B, Johnson ME. Ligand efficiency indices for an effective mapping of chemico-biological space: the concept of an atlas-like representation. Drug Discovery Today. 2010;15:804–811. doi: 10.1016/j.drudis.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Kalid O, Mense M, Fischman S, Shitrit A, Bihler H, Ben-Zeev E, Schutz N, Pedemonte N, Thomas PJ, Bridges RJ, Wetmore DR, Marantz Y, Senderowitz H. Small molecule correctors of F508del-CFTR discovered by structure-based virtual screening. J Comput-Aided Mol Des. 2010;24:971–991. doi: 10.1007/s10822-010-9390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carosati E, Budriesi R, Ioan P, Ugenti MP, Frosini M, Fusi F, Corda G, Cosimelli B, Spinelli D, Chiarini A, Cruciani G. Discovery of novel and cardioselective diltiazem-like calcium channel blockers via virtual screening. J Med Chem. 2008;51:5552–5565. doi: 10.1021/jm800151n. [DOI] [PubMed] [Google Scholar]
- 13.Drwal MN, Agama K, Wakelin LP, Pommier Y, Griffith R. Exploring DNA topoisomerase I ligand space in search of novel anticancer agents. PLoS One. 2011;6:e25150. doi: 10.1371/journal.pone.0025150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gubler H, Schopfer U, Jacoby E. Theoretical and Experimental Relationships between Percent Inhibition and IC50 Data Observed in High-Throughput Screening. J Biomol Screen. 2012;18:1–13. doi: 10.1177/1087057112455219. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Z, Qian L, Cao L, Tan H, Huang Y, Xue X, Shen Y, Zhou S. Virtual screening for novel quorum sensing inhibitors to eradicate biofilm formation of Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2008;79:119–126. doi: 10.1007/s00253-008-1406-5. [DOI] [PubMed] [Google Scholar]
- 16.Babaoglu K, Simeonov A, Irwin JJ, Nelson ME, Feng B, Thomas CJ, Cancian L, Costi MP, Maltby DA, Jadhav A, Inglese J, Austin CP, Shoichet BK. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase. J Med Chem. 2008;51:2502–2511. doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajduk PJ. Fragment-based drug design: how big is too big? J Med Chem. 2006;49:6972–6976. doi: 10.1021/jm060511h. [DOI] [PubMed] [Google Scholar]
- 18.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc Natl Acad Sci U S A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds CH, Tounge BA, Bembenek SD. Ligand binding efficiency: trends, physical basis, and implications. J Med Chem. 2008;51:2432–2438. doi: 10.1021/jm701255b. [DOI] [PubMed] [Google Scholar]
- 20.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discovery. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 21.Doman TN, McGovern SL, Witherbee BJ, Kasten TP, Kurumbail R, Stallings WC, Connolly DT, Shoichet BK. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem. 2002;45:2213–2221. doi: 10.1021/jm010548w. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira RS, Simeonov A, Jadhav A, Eidam O, Mott BT, Keiser MJ, McKerrow JH, Maloney DJ, Irwin JJ, Shoichet BK. Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors. J Med Chem. 2010;53:4891–4905. doi: 10.1021/jm100488w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truchon JF, Bayly CI. Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J Chem Inf Model. 2007;47:488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 24.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 25.Mannhold R, Kubinyi H, Folkers G. Preface. In: Sotriffer C, editor. Virtual Screening Principles, Challenges, and Practical Guidelines. Wiley-VCH Verlag & Co. KGaA; Weinheim, Germany: 2011. pp. XXIII–XXIV. [Google Scholar]
- 26.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 27.Oprea TI, Bologa CG, Olah M. Compound selection for virtual screening. In: Alvarez J, Shoichet B, editors. Virtual Screening in Drug Discovery. Taylor & Francis Group; Boca Raton, FL: 2005. pp. 89–106. [Google Scholar]
- 28.Gleeson MP, Hersey A, Montanari D, Overington J. Probing the links between in vitro potency, ADMET and physicochemical parameters. Nat Rev Drug Discovery. 2011;10:197–208. doi: 10.1038/nrd3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huggins DJ, Venkitaraman AR, Spring DR. Rational methods for the selection of diverse screening compounds. ACS Chem Biol. 2011;6:208–217. doi: 10.1021/cb100420r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J. Comprehensive assessment of ADMET risks in drug discovery. Curr Pharm Des. 2009;15:2195–2219. doi: 10.2174/138161209788682514. [DOI] [PubMed] [Google Scholar]
- 31.Suite 2012: QikProp, version 35. Schrödinger, LLC; New York, NY: 2012. [Google Scholar]
- 32.Walters WP, Murcko MA. Prediction of ‘drug-likeness’. Adv Drug Del Rev. 2002;54:255–271. doi: 10.1016/s0169-409x(02)00003-0. [DOI] [PubMed] [Google Scholar]
- 33.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 34.Bruns RF, Watson IA. Rules for identifying potentially reactive or promiscuous compounds. J Med Chem. 2012;55:9763–9772. doi: 10.1021/jm301008n. [DOI] [PubMed] [Google Scholar]
- 35.Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Sosa AT, Sild S, Takkis K, Maran U. Combined approach using ligand efficiency, cross-docking, and antitarget hits for wild-type and drug-resistant Y181C HIV-1 reverse transcriptase. J Chem Inf Model. 2011;51:2595–2611. doi: 10.1021/ci200203h. [DOI] [PubMed] [Google Scholar]
- 37.Efremov IV, Vajdos FF, Borzilleri KA, Capetta S, Chen H, Dorff PH, Dutra JK, Goldstein SW, Mansour M, McColl A, Noell S, Oborski CE, O’Connell TN, O’Sullivan TJ, Pandit J, Wang H, Wei B, Withka JM. Discovery and Optimization of a Novel Spiropyrrolidine Inhibitor of beta-Secretase (BACE1) through Fragment-Based Drug Design. J Med Chem. 2012;55:9069–9088. doi: 10.1021/jm201715d. [DOI] [PubMed] [Google Scholar]
- 38.Maggiora GM. On outliers and activity cliffs--why QSAR often disappoints. J Chem Inf Model. 2006;46:1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 39.Stumpfe D, Bajorath J. Exploring activity cliffs in medicinal chemistry. J Med Chem. 2012;55:2932–2942. doi: 10.1021/jm201706b. [DOI] [PubMed] [Google Scholar]
- 40.Hu Y, Bajorath J. Extending the Activity Cliff Concept: Structural Categorization of Activity Cliffs and Systematic Identification of Different Types of Cliffs in the ChEMBL Database. J Chem Inf Model. 2012;52:1806–1811. doi: 10.1021/ci300274c. [DOI] [PubMed] [Google Scholar]
- 41.Barreiro G, Guimaraes CR, Tubert-Brohman I, Lyons TM, Tirado-Rives J, Jorgensen WL. Search for non-nucleoside inhibitors of HIV-1 reverse transcriptase using chemical similarity, molecular docking, and MM-GB/SA scoring. J Chem Inf Model. 2007;47:2416–2428. doi: 10.1021/ci700271z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuffenhauer A, Ertl P, Roggo S, Wetzel S, Koch MA, Waldmann H. The scaffold tree--visualization of the scaffold universe by hierarchical scaffold classification. J Chem Inf Model. 2007;47:47–58. doi: 10.1021/ci600338x. [DOI] [PubMed] [Google Scholar]
- 43.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discovery. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 44.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 45.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 46.Feng J, Chen Y, Pu J, Yang X, Zhang C, Zhu S, Zhao Y, Yuan Y, Yuan H, Liao F. An improved malachite green assay of phosphate: mechanism and application. Anal Biochem. 2011;409:144–149. doi: 10.1016/j.ab.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 47.Pan Y, Huang N, Cho S, MacKerell AD., Jr Consideration of molecular weight during compound selection in virtual target-based database screening. J Chem Inf Comput Sci. 2003;43:267–272. doi: 10.1021/ci020055f. [DOI] [PubMed] [Google Scholar]
- 48.Carta G, Knox AJ, Lloyd DG. Unbiasing scoring functions: a new normalization and rescoring strategy. J Chem Inf Model. 2007;47:1564–1571. doi: 10.1021/ci600471m. [DOI] [PubMed] [Google Scholar]
- 49.Willett P, Barnard JM, Downs GM. Chemical similarity searching. J Chem Inf Comput Sci. 1998;38:983–996. [Google Scholar]
- 50.Haldeman M, Vieira B, Winer F, Knutsen LJ. Exploration tools for drug discovery and beyond: applying SciFinder to interdisciplinary research. Curr Drug Disc Technol. 2005;2:69–74. doi: 10.2174/1570163054064693. [DOI] [PubMed] [Google Scholar]
- 51.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curve. J Physiol (Lond) 1910;40(Suppl):iv–vii. [Google Scholar]
- 52.Albert JS, Edwards PD. Identification of high-affinity beta-secretase inhibitors using fragment-based lead generation. In: Zartler ER, Shapiro MJ, editors. Fragment-Based Drug Discovery A Practical Approach. John Wiley & Sons, Ltd; West Sussex, UK: 2008. pp. 261–279. [Google Scholar]
- 53.Jorgensen WL. Progress and issues for computationally guided lead discovery and optimization. In: Merz KM Jr, Ringe D, Reynolds CH, editors. Drug Design: Structure- and Ligand-Based Approaches. Cambridge University Press; New York, NY: 2010. pp. 1–10. [Google Scholar]
- 54.Chen CS, Chiou CT, Chen GS, Chen SC, Hu CY, Chi WK, Chu YD, Hwang LH, Chen PJ, Chen DS, Liaw SH, Chern JW. Structure-based discovery of triphenylmethane derivatives as inhibitors of hepatitis C virus helicase. J Med Chem. 2009;52:2716–2723. doi: 10.1021/jm8011905. [DOI] [PubMed] [Google Scholar]
- 55.De Luca L, Barreca ML, Ferro S, Christ F, Iraci N, Gitto R, Monforte AM, Debyser Z, Chimirri A. Pharmacophore-based discovery of small-molecule inhibitors of protein-protein interactions between HIV-1 integrase and cellular cofactor LEDGF/p75. ChemMedChem. 2009;4:1311–1316. doi: 10.1002/cmdc.200900070. [DOI] [PubMed] [Google Scholar]
- 56.Jorgensen WL. Efficient drug lead discovery and optimization. Acc Chem Res. 2009;42:724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Del Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 58.Oprea TI, Davis AM, Teague SJ, Leeson PD. Is there a difference between leads and drugs? A historical perspective. J Chem Inf Comput Sci. 2001;41:1308–1315. doi: 10.1021/ci010366a. [DOI] [PubMed] [Google Scholar]
- 59.Cruciani G, Pastor M, Guba W. VolSurf: a new tool for the pharmacokinetic optimization of lead compounds. Eur J Pharm Sci. 2000;11(Suppl 2):S29–39. doi: 10.1016/s0928-0987(00)00162-7. [DOI] [PubMed] [Google Scholar]
- 60.Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y. admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J Chem Inf Model. 2012;52:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 61.Cai H, Yan G, Zhang X, Gorbenko O, Wang H, Zhu W. Discovery of highly selective inhibitors of human fatty acid binding protein 4 (FABP4) by virtual screening. Bioorg Med Chem Lett. 2010;20:3675–3679. doi: 10.1016/j.bmcl.2010.04.095. [DOI] [PubMed] [Google Scholar]
- 62.Deng J, Feng E, Ma S, Zhang Y, Liu X, Li H, Huang H, Zhu J, Zhu W, Shen X, Miao L, Liu H, Jiang H, Li J. Design and synthesis of small molecule RhoA inhibitors: a new promising therapy for cardiovascular diseases? J Med Chem. 2011;54:4508–4522. doi: 10.1021/jm200161c. [DOI] [PubMed] [Google Scholar]
- 63.Brian Budzik VG, Shi Dongchuan, Walker Graham, Woolley-Roberts Marie, Pardoe Joanne, Lucas Adam, Tehan Ben, Rivero Ralph A, Langmead Christopher J, Watson Jeannette, Wu Zining, Forbes Ian T, Jian Jin. Novel N-Substituted Benzimidazolones as Potent Selective CNS-Penetrant, and Orally Active M1 mAChR Agonists. ACS Med Chem Lett. 2010;1:244–248. doi: 10.1021/ml100105x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartzoulakis B, Rossiter S, Gill H, O’Hara B, Steinke E, Gane PJ, Hurtado-Guerrero R, Leiper JM, Vallance P, Rust JM, Selwood DL. Discovery of inhibitors of the pentein superfamily protein dimethylarginine dimethylaminohydrolase (DDAH), by virtual screening and hit analysis. Bioorg Med Chem Lett. 2007;17:3953–3956. doi: 10.1016/j.bmcl.2007.04.095. [DOI] [PubMed] [Google Scholar]
- 65.Mollard A, Warner SL, Call LT, Wade ML, Bearss JJ, Verma A, Sharma S, Vankayalapati H, Bearss DJ. Design, Synthesis and Biological Evaluation of a Series of Novel Axl Kinase Inhibitors. ACS Med Chem Lett. 2011;2:907–912. doi: 10.1021/ml200198x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.