Abstract
Epileptic encephalopathies (EE) are a devastating group of severe childhood epilepsy disorders for which the cause is often unknown. Here, we report a screen for de novo mutations in patients with two classical EE: infantile spasms (IS, n=149) and Lennox-Gastaut Syndrome (LGS, n=115). We sequenced the exomes of 264 probands, and their parents, and confirmed 329 de novo mutations. A likelihood analysis showed a significant excess of de novo mutations in the ~4,000 genes that are the most intolerant to functional genetic variation in the human population (p=2.9 × 10−3). Among these are GABRB3 with de novo mutations in four patients and ALG13 with the same de novo mutation in two patients; both genes show clear statistical evidence of association. Given the relevant site-specific mutation rates, the probabilities of these outcomes occurring by chance are p=4.1 × 10−10 and p=7.8 × 10−12, respectively. Other genes with de novo mutations in this cohort include: CACNA1A, CHD2, FLNA, GABRA1, GRIN1, GRIN2B, HDAC4, HNRNPU, IQSEC2, MTOR, and NEDD4L. Finally, we show that the de novo mutations observed are enriched in specific gene sets including genes regulated by the Fragile X protein (p<10−8), as was reported for autism spectrum disorders (ASD)1.
Genetics is believed to play an important role in many epilepsy syndromes; however, specific genes have been discovered in only a small proportion of cases. Genome-wide association studies for both focal and generalized epilepsies have revealed few significant associations, and rare copy number variants explain only a few percent of cases2–5. An emerging paradigm in neuropsychiatric disorders is the major impact of de novo mutations on disease risk6,7. We searched for de novo mutations associated with EE, a heterogeneous group of severe epilepsy disorders characterized by early onset of seizures with cognitive and behavioral features associated with ongoing epileptic activity. We focused on two “classic” forms of EE: IS and LGS, recognizing that some patients with IS evolve to LGS.
Exome sequencing of 264 trios (Additional Methods) identified 439 putative de novo mutations. Sanger sequencing confirmed 329 de novo mutations (Supplementary Table 2), and the remainder were either false positives, a result of B cell immortalization, or in regions where the Sanger assays did not work (Supplementary Table 3).
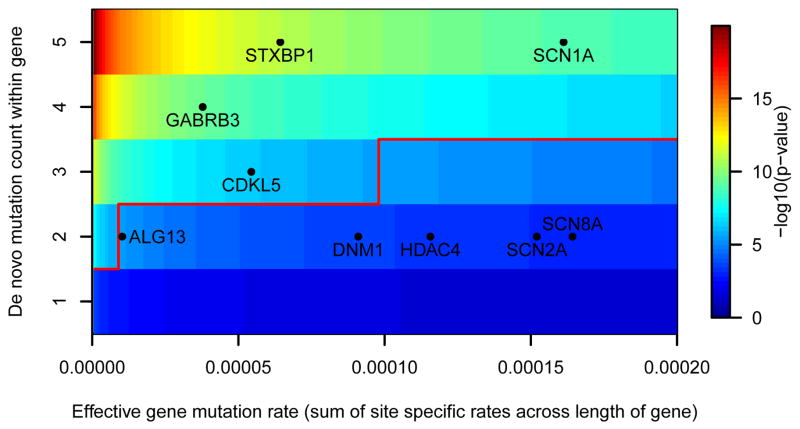
Across our 264 trios, we found nine genes with de novo SNV mutations in two or more probands (SCN1A n=7, STXBP1 n=5, GABRB3 n=4, CDKL5 n=3, SCN8A n=2, SCN2A n=2, ALG13 n=2, DNM1 n=2, and HDAC4 n =2). Of these, SCN1A, STXBP1, SCN8A, SCN2A, and CDKL5 are generally considered known EE genes.8–13 To assess whether the observations in the other genes implicate them as risk factors for EE, we determined the probability of seeing multiple mutations in the same gene given the sequence specific mutation rate, size of the gene, and the number and gender of patients evaluated in this study (Additional Methods). The number of observed de novo mutations in HDAC4 and DNM1 are not yet significantly greater than the null expectation. However, observing four unique de novo mutations in GABRB3 and two identical de novo mutations in ALG13 were found to be highly improbable (Table 1, Figure 1). We performed the same calculations on all the genes with multiple de novo mutations observed in 610 control exomes and found no genes with a significant excess of de novo mutations (Supplementary Table 4). While mutations in GABRB3 have previously been reported in association with another type of epilepsy14, and through in vivo studies in mice GABRB3 haploinsufficiency has been suggested to be one of the causes of epilepsy in Angelman’s syndrome15, our observations implicate it, for the first time, as a single gene cause of EE and provide the strongest evidence yet available for any epilepsy. Likewise, ALG13, an X-linked gene encoding a subunit of the uridine diphosphate-N-acetylglucosamine transferase, was previously shown to carry a novel de novo mutation in a male patient with a severe congenital glycosylation disorder with microcephaly, seizures, and early lethality16. Furthermore, the exact same ALG13 de novo mutation identified in this study was observed as a de novo mutation in an additional female patient with severe intellectual disability (ID) and seizures17.
Table 1.
Genes with greater than one de novo SNV mutation in this cohort of 264 trios, and the probabilities of getting greater than or equal observed de novo mutation tally by chance.
| Gene | Chr | Average effectively captured length (bp) | Weighted mutation rate | De novo mutation number | p-value† | |
|---|---|---|---|---|---|---|
|
| ||||||
| SCN1A | 2 | 6063.70 | 1.61×10−4 | 5* | 1.12×10−9 | *** |
| STXBP1 | 9 | 1917.51 | 6.44×10−5 | 5 | 1.16×10−11 | *** |
| GABRB3 | 15 | 1206.86 | 3.78×10−5 | 4 | 4.11×10−10 | *** |
| CDKL5 | X | 2798.38 | 5.44×10−5 | 3 | 4.90×10−7 | ** |
| ALG13# | X | 475.05 | 1.03×10−5 | 2 | 7.77×10−12 | *** |
| DNM1 | 9 | 2323.37 | 9.10×10−5 | 2 | 2.84×10−4 | |
| HDAC4 | 2 | 2649.82 | 1.16×10−4 | 2 | 4.57×10−4 | |
| SCN2A# | 2 | 5831.21 | 1.52×10−4 | 2 | 1.14×10−9 | *** |
| SCN8A | 12 | 5814.48 | 1.64×10−4 | 2 | 9.14×10−4 | |
Adjusted α is equivalent to 0.05/18,091 = 2.76×10 (*), 0.01/18,091 = 5.53×10 (**) and 0.001/18,091 = 5.53×10−8 (***).
Counts exclude three additional patients with an indel or splice site mutation as these are not accounted for in the mutability calculation.
Two de novo mutations occur at the same position. The probability of these special cases obtain P = 7.77×10−12 and P = 1.14×10−9 for ALG13 and SCN2A, respectively (Additional Methods).
Figure 1. Heat map illustrating the probability of observing the number of de novo mutations in genes with an estimated gene mutation rate.
The number of de novo mutations required to achieve significance is indicated by the solid red line. The superimposed black dots reflect positions of all genes found to harbor multiple de novo mutations in our study. GABRB3, SCN1A, CDKL5, STXBP1 have significantly more de novo mutations than expected. The positions indicated for ALG13 and SCN2A reflect only the fact that there are two mutations observed, not that there are two mutations affecting the same site (Additional Methods).
Each trio harbored on average 1.25 confirmed de novo mutations, with 181 probands harboring at least one. Considering only de novo SNVs, each trio harbored on average 1.17 de novo mutations (Supplementary Figure 1). Seventy-two percent of the confirmed de novo SNV mutations were missense and 7.5% were loss-of-function (splice donor, splice acceptor, or stop-gain mutations). Compared to rates of these classes of mutations previously reported in controls (69.4% missense and 4.2% loss of function mutations)18–20, we observed a significant excess of loss-of-function mutations in patients with IS and LGS (Exact binomial p=0.01), consistent with data previously reported in ASD7,18–20.
Neale et al.7 recently established a framework for testing whether the distribution of de novo mutations in affected individuals differs from the general population. Here, we extend their simulation-based approach by developing a likelihood model that characterizes this effect and describes the distribution of de novo mutations among affected individuals in terms of the distribution in the general population, and a set of parameters describing the genetic architecture of the disease. These parameters include the proportion of the exome sequence that can carry disease-influencing mutations (η) and the relative risk (γ) of the mutations (Supplementary Methods). Consistent with what was reported in ASD7, we found no significant deviation in the overall distribution of mutations from expected (γ=1 and/or η=0). It is, however, now well-established that some genes tolerate protein-disrupting mutations without apparent adverse phenotypic consequences, while others do not. To take this into account, we employed a simple scoring system that uses polymorphism data in the human population to assign a tolerance score to every considered gene (Additional Methods). We then found that known EE genes rank amongst the most intolerant genes using this scheme (Supplementary Table 8). We therefore evaluated the distribution of de novo mutations within these 4,264 genes that are within the 25th percentile for intolerance and found a significant shift from the null distribution (p=2.9×10−3). The maximum likelihood estimates of η (percentage of intolerant genes involved in EE) was 0.021 and γ (relative risk) was 81, suggesting there are 90 genes amongst the intolerant genes that can confer risk of EE and that each mutation carries substantial risk. We also found that putatively damaging de novo variants in our cohort are significantly enriched in intolerant genes compared with control cohorts (Supplementary Methods).
We next evaluated whether the de novo mutations were drawn preferentially from six gene sets (Additional Methods, Supplementary Table 10), including ion channels21, genes known to cause monogenic disorders with seizures as a phenotypic feature22, genes carrying confirmed de novo mutations in patients with ASD7,18–20 and in patients with ID17,23, and FMRP-regulated genes. Taking into account the size of regions with adequate sequencing coverage to detect a de novo mutation (Additional Methods), we found significant over-representation for all gene lists in our data (Supplementary Table 10), and no over representation in controls 17–20,23.
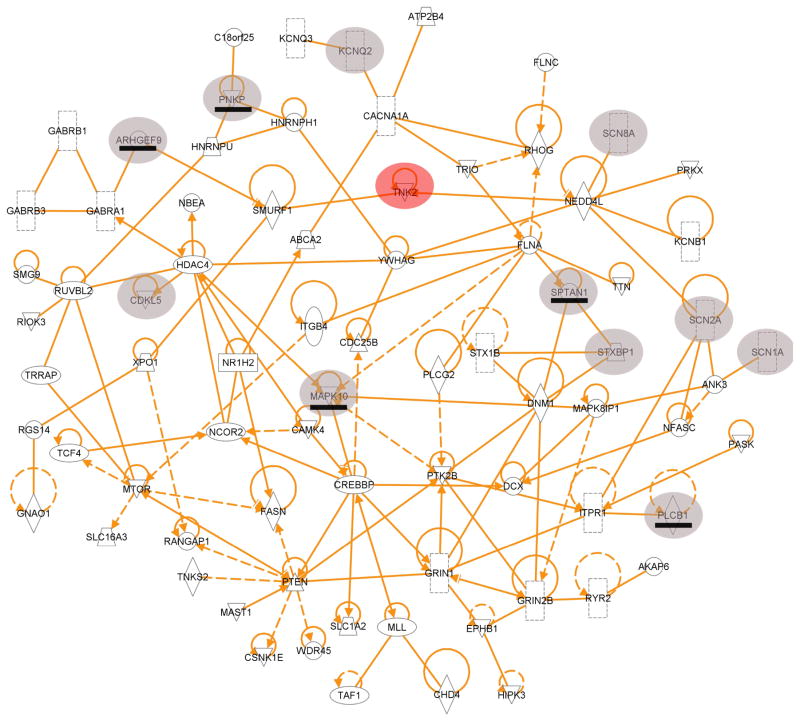
To determine possible interconnectivity among the genes carrying a de novo mutation, we performed a protein-protein interaction analysis and identified a single network of 71 connected proteins (Figure 2 and Supplementary Figure 7). These 71 proteins include six encoded by Mendelian Inheritance in Man (MIM, http://www.omim.org/) EE genes that have one or more de novo mutation in an EE patient in this study Genes in this protein-protein network were also found to far more likely overlap with the ASD77,18,20,24 and severe ID17,23 exome sequencing study genes, and with FMRP-associated genes, than the genes not in this network (Supplementary Table 11).
Figure 2. A protein-protein interaction network of genes with de novo mutations found in IS and LGS patients studied.
Six of the genes found to harbor de novo mutations in an IS or LGS patient are known MIM EE genes (shaded circles). Five additional known MIM EE genes that were not found to be mutated in the 264 EE patients, but are involved in this network, are also shown (shaded circles with the gene underlined). The previously identified severe infantile epilepsy gene TNK2 is superimposed into this network (red circle).
In support of a hypothesis that individual rare mutations in different genes may converge on pathways, we draw attention to the six mutations that all affect subunits of the gamma-aminobutyric acid (GABA) ionotropic receptor (four in GABRB3, and one each in GABRA1 and GABRB1), and highlight two interactions: HNRNPU interacting with HNRNPH1 and NEDD4L (identified here) binding to TNK2, a gene previously implicated in EE25 (Figure 2). Although the HNRNPU mutation observed here is an indel in a splice acceptor site, and therefore likely functional, the HNRNPH1 de novo mutation is synonymous and thus of unknown functional significance (Supplementary Table 2). Importantly, a minigene experiment suggests that this synonymous mutation induces skipping of exon 12 (Supplementary Methods).
Evaluation of the clinical phenotypes among patients revealed significant genetic heterogeneity underlying IS and LGS, and begins to provide information about the range of phenotypes associated with mutations in specific genes (Supplementary Table 13). We identified four genes, SCN8A, STXBP1, DNM1, and GABRB3, with de novo mutations in both patients with IS and patients with LGS. Although IS may evolve to LGS, in three of these cases, the patients with LGS did not initially present with IS, suggesting phenotypic heterogeneity associated with mutations in these genes yet supporting the notion of shared genetic susceptibility. Interestingly, in multiple patients we identified de novo mutations in genes previously implicated in other neurodevelopmental conditions, and in some cases with very distinctive clinical presentations (Supplementary Table 12). Most notably, we found a de novo mutation in MTOR, a gene recently found to harbor a causal variant in mosaic form in a case with hemimegalencephaly26. Our patient however showed no detectable structural brain malformation. Similarly, we found one patient with a de novo mutation in DCX and another with a de novo mutation in FLNA, previously associated with lissencephaly and periventricular nodular heterotopia (PVNH), respectively27,28; neither patient had cortical malformations detected on magnetic resonance imaging.
In addition to de novo variants, we also screened for highly penetrant genotypes by identifying variants that create newly homozygous, compound heterozygous, or hemizygous genotypes in the probands that are not seen in parents or controls (Supplementary Methods). No inherited variants showed significant evidence of association. Additional studies evaluating a larger number of EE patients will be required to establish the role of inherited variants in the risk IS and LGS.
In summary, we have identified novel de novo mutations implicating at least two genes, and also describe a genetic architecture that strongly suggests we have identified additional causal mutations in genes intolerant to functional variation. Given that our sample size already shows many genes with recurrent mutations, it is clear that even modest increases in sample sizes will confirm many new genes now seen in only one of our trios. Our results also emphasize that it may be difficult to predict with confidence the responsible gene, even among known genes, based upon clinical presentation. This makes it clear that the future of genetic diagnostics in EE will focus on the genome as a whole as opposed to single genes or even gene panels. In particular, several of the genes with de novo mutations in our cohort have also been identified in patients with ID or ASD. Finally, and perhaps most importantly, this work suggests a clear direction for both drug development and treatment personalization in the epileptic encephalopathies, as many of these mutations appear to converge on specific biologic pathways.
Additional Methods
Study subjects
IS and LGS patients evaluated in this study were collected through the Epilepsy Phenome/Genome Project (EPGP, www.epgp.org). Patients were enrolled across 23 clinical sites. Informed consent was obtained for all patients in accordance with the site specific Institutional Review Boards. Phenotypic information has been centrally databased and DNA specimens stored at the Coriell Institute – NINDS Genetics Repository (Supplementary Table 1). IS patients were required to have hypsarrhythmia or a hypsarrhythmia on EEG. LGS patients were required to have EEG background slowing or disorganization for age and generalized spike and wave activity of any frequency or generalized paroxysmal fast activity (GPFA). Background slowing was defined as <8Hz posterior dominant rhythm in patients over 3 years of age, and <5Hz in patients over 2 years of age. EEGs with normal backgrounds were accepted if the generalized spike and wave activity was 2.5 hertz or less and/or if GPFA was present.
All patients were required to have no evidence of moderate-to-severe developmental impairment or diagnosis of autistic disorder or pervasive developmental disorder prior to the onset of seizures. Severe developmental delay was defined by 50% or more delay in any area: motor, social, language, cognition, or activities of living; or global delay. Mild delay was defined as delay of less than 50% of expected milestones in one area, or less than 30% of milestones across more than one area. All patients had no confirmed genetic or metabolic diagnosis, and no history of congenital TORCH infection, premature birth (before 32 weeks gestation), neonatal hypoxic-ischemic encephalopathy or neonatal seizures, meningitis/encephalitis, stroke, intra-cranial hemorrhage, significant head trauma, or evidence of acquired epilepsy. All IS and LGS patients had an MRI or CT scan interpreted as normal, mild diffuse atrophy or focal cortical dysplasia. (Our case with the mutation in HNRNPU had had a reportedly normal MRI but on review of past records, a second more detailed MRI was found showing small regions of PVNH.) In order to participate, both biological parents had to have no past medical history of seizures (except febrile or metabolic/toxic seizures).
A final diagnosis form was completed by the local site EPGP principal investigator based on all collected information. A subset of cases was reviewed independently by two members of the EPGP Data Review Core to ensure data quality and consistency. All EEGs were reviewed by a site investigator and an EEG core member to assess data quality and EEG inclusion criteria. EEGs accepted for inclusion were then reviewed and scored by two EEG core members for specific EEG phenotypic features. Disagreements were resolved by consensus conference among two or more EEG core members before the EEG data set was finalized. MRI scans were reviewed by local investigators and an MRI core member to exclude an acquired symptomatic lesion.
Exome sequenced unrelated controls (n=436) used to ascertain mutation frequencies were sequenced in the Center for Human Genome Variation as part of other genetic studies.
Exome sequencing, alignment and variant calling
Exome sequencing was carried out within the Genomic Analysis Facility in the Center for Human Genome Variation (Duke University). Sequencing libraries were prepared from primary DNA extracted from leukocytes of parents and probands using the Illumina TruSeq library preparation kit following the manufacturer’s protocol. Illumina TruSeq Exome Enrichment kit was used to selectively amplify the coding regions of the genome according to the manufacturer’s protocol. Six individual barcoded samples (two complete trios) were sequenced in parallel across two lanes of an Illumina HiSeq 2000 sequencer. Alignment of the sequenced DNA fragments to Human Reference Genome (NCBI Build 37) was performed using the Burrows-Wheeler Alignment Tool (BWA) (version 0.5.10). The reference sequence we use is identical to the 1000 Genomes Phase II reference and it consists of chromosomes 1–22, X,Y, MT, unplaced and unlocalized contigs, the human herpesvirus 4 type 1 (NC_007605), and decoy sequences (hs37d5) derived from HuRef, Human Bac and Fosmid clones and NA12878.
After alignments were produced for each individual separately using BWA, candidate de novo, recessive, and compound heterozygous genotypes were jointly called with the GATK Unified Genotyper for all family members in a trio. Loci bearing putative de novo mutations were extracted from the VCFs that met the following criteria: (1) the read depth in both parents should be greater than or equal to 10; (2) the depth of coverage in the child should be at least one tenth of the sum of the coverage in both parents; (3) for de novo variants, less than 5% of the reads in either parent should carry the alternate allele; (4) at least 25% of the reads in the child should carry the alternate allele; (5) the normalized, phred-scaled likelihood (PL) scores for the offspring genotypes AA, AB, and BB, where A is the reference allele and B is the alternate allele, should be >20, 0, and >0, respectively; (6) the PL scores for both parents should be 0, >20, and >20; (7) at least three variant alleles must be observed in the proband; and (8) the de novo variant had to be located in a CCDS exon targeted by the exome enrichment kit. PL scores are assigned such that the most likely genotype is given a score of 0, and the score for the other two genotypes represent the likelihood that they are not the true genotypes. SnpEff (version 3.0a) was used to annotate the variants according to Ensembl (version 69) and consensus coding sequencing (CCDS release 9, GRCh37.p5) and limited analyses to exonic or splice site (2 bp flanking an exon) mutations. All candidate de novo mutations that were absent from population controls, including a set of 436 internally sequenced controls and the ~6500 individuals whose single nucleotide variant data is reported in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) [date (August, 2012)] were also visually inspected using Integrative Genomics Viewer (IGV). All candidate de novo mutations were confirmed with Sanger sequencing of the relevant proband and parents. For comparison, we also called de novo variants from probands and parents individually for a subset of trios. Using this individual calling approach we identified and confirmed an additional 46 de novo mutations. These were included in all the downstream de novo mutation analyses.
Calculation of gene specific mutation rate
Point mutation rates were scaled to per base-pair, per generation, based on the human genome sequences matrix30 (kindly provided by Drs. Shamil Sunyaev and Paz Polak), and the known human average genome de novo mutation rate (1.2×10−8)31. The mutation rate (M) of each gene was calculated by adding up point mutation rates in effectively captured CCDS regions in the offspring of trios, and then dividing by the total trio number (S = 264). The p-value was calculated as [1 – Poisson cumulative distribution function (x-1, λ)], where x is the observed de novo mutation number for the gene, and λ is calculated as 2S*M for genes on autosome or (2f + m)*M for genes on chromosome X (f and m are the number of sequenced female and male probands, respectively). Genes on Y chromosome were not part of these analyses. Two de novo mutations in gene ALG13 are at the same position, likewise in gene SCN2A. We calculated the probability of this special case as [1 – Poisson cumulative distribution function (1, (2f + m)*r)], where r reflects the point mutation rate on that specific de novo mutation position. Further investigations indicated that it is unlikely for these de novo mutations, which occur at the same site across distinct probands, to have been caused by sequencing or mapping errors (Supplementary Methods).
Calculation of mutation tolerance for HGNC genes
To quantitatively assign a mutation tolerance score to genes in the human genome (HGNC genes), we calculated an empirical penalty based on the presence of common functional variation using the aggregate sequence data available from the 6,503 samples reported in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) [date (August, 2012) accessed]). We first filtered within the EVS database and eliminated from further consideration, genes where the number of 10-fold average covered bases was less than 70% of its total extent. In calculating a score, we focused on departures from the average common functional variant frequency spectrum, corrected for the total mutation burden in a gene. We construct this score as follows. Let Y be the total number of common, MAF>0.1%, missense and nonsense (including splice) variants and let X be total number of variants (including synonymous) observed within a gene. We regress Y on X and take the studentized residual as the score (S). Thus the raw residual is divided by an estimate of its standard deviation and thus account for differences in variability that comes with differing mutational burdens. S measures the departure from the average number of common functional mutations found in genes with a similar amount of mutational burden. Thus, when S = 0 the gene has the average number of common functional variants given its total mutational burden. Genes where S < 0 have less common functionals than average for their mutational burden and thus, would appear to be less tolerant of functional mutation, indicating the presence of weak purifying selection. We further investigated how different ‘intolerance’ thresholds of S captured known EE genes (Supplementary Table 8). Supplementary Figure 6 illustrates how different percentiles of S lead to the classification of different proportions of the known EE genes as ‘intolerant’. Note that ARX is not in these analyses as this gene did not meet a 70% of gene coverage threshold. The dashed vertical line in Supplementary Figure 6 illustrates the 25th percentile of S and shows that using this threshold results in 12 out of the 14 assessed known genes being considered ‘intolerant’. On the basis of this analysis, we used this 25th percentile threshold in classifying genes as intolerant in all subsequent analyses. Supplementary Table 9 lists the 25th percentile of most intolerant genes that had Sanger confirmed de novo mutations amongst the IS/LGS probands.
Defining the CCDS opportunity space for detecting de novo mutations
For each trio, we defined callable exonic bases, that had the opportunity for identification of a coding de novo mutation, by restricting to bases where each of the three family members had at least 10-fold coverage, obtained a multi-sampling (GATK) raw phred-scaled confidence score of ≥20 in presence or absence of a variant, and were within the consensus coding sequence (CCDS release 9, GRCh37.p5) or within the two base-pairs at each end of exons to allow for splice acceptor and donor variants. Using these three criteria, the average CCDS-defined de novo mutation opportunity space across 264 trios was found to be 28.84Mb ± 0.92Mb (range of 25.46Mb – 30.25Mb).
To explore at the gene level, we similarly assessed the de novo calling opportunity within any given trio for every gene with a CCDS transcript. For genes with instances of non-overlapping CCDS transcripts, we merged the corresponding regions into a consensus summary of all CCDS-defined bases for that gene. Using these criteria, over 85% of the CCDS-defined exonic regions were sequenced, to at least 10-fold coverage across the three family members, in over 90% of trios. All 264 trios covered at least 79% of the CCDS-defined regions under the CCDS opportunity space criteria. Calculations of CCDS opportunity space for calling a de novo mutation, aside from the Y chromosome, were used in both the gene-list enrichment and architecture calculations.
Supplementary Material
Supplementary Table 1. Coriell IDs for all exome sequenced 264 IS and LGS trios.
Supplementary Table 2. Comprehensive list of de novo mutations identified in 264 IS and LGS trios.
Supplementary Table 3. Summary of de novo mutations identified in 264 IS and LGS trios.
Supplementary Figure 1. Distribution of de novo mutations detected in 264 IS/LGS probands.
Supplementary Table 4. Estimating probability of multiple de novo events amongst control exomes.
Supplementary Methods: Investigating instances of multiple hits at the same site across distinct probands.
Supplementary Figure 2. The sequencing coverage of four de novo mutations.
Supplementary Methods: Genetic Architecture
Supplementary Table 5. Observed number of de novo coding mutations observed in autism spectral disorder trios as reported by Neale et al.
Supplementary Table 6. Observed number of de novo coding mutations observed in IS/LGS trio exomes.
Supplementary Table 7. Observed number of de novo coding mutations observed in intolerant genes among IS/LGS trios.
Supplementary Figure 3. Likelihood surface for autism spectrum data given in Neale et al. (Supplementary Table 5). Gray lines represent contours of the likelihood surface. Red lines represent 95% likelihood ratio confidence regions for η and “log”(γ). Maximum likelihood estimate given by black X (η=0.018,γ=15.6). Blue Xs represent 6 sets of parameter values for η and γ simulated by Neale et al.
Supplementary Figure 4. Likelihood surface for IS/LGS exome data (Supplementary Table 6). Gray lines represent contours of the likelihood surface. Red lines represent 95% likelihood ratio confidence regions for η and “log”(γ). Maximum likelihood estimate given by black X (η=0.007,γ=86).
Supplementary Figure 5. Likelihood surface for IS/LGS intolerant gene data (Supplementary Table 7). Gray lines represent contours of the likelihood surface. Red lines represent 95% likelihood ratio confidence regions for η and “log”(γ). Maximum likelihood estimate given by black X (η=0.021,γ=81).
Supplementary Table 8. Summary statistics of the opportunity space to call a de novo coding variant in the ‘known’ early epileptic encephalopathy MIM genes [last accessed OMIM® December 2012].
Supplementary Figure 6. Classification of known EE genes as intolerant.
Supplementary Table 9. List of “intolerant” CCDS-defined genes harboring a de novo mutation in an IS or LGS trio (n=109). Previously reported EE genes are highlighted in red.
Supplementary Methods: Assessment of mutation consequences in intolerant genes
Supplementary Table 10. Gene list enrichment results summary
Supplementary Figure 7. Full protein-protein network analysis of 267 genes.
Supplementary Table 11. Protein-Protein network enrichment.
Supplementary Methods: Functional work
Effects of a de novo synonymous HNRNPH1 mutation on splicing
Supplementary Figure 8. De novo HNRNPH1 synonymous mutation causes alternatively spliced transcript missing exon 12.
Supplementary Table 12. Likely functional (missense, nonsense, or splice site) de novo mutations identified in IS/LGS trios in genes that have been previously associated with a neuropsychiatric disorders† with or without seizures.
Supplementary Table 13. Clinical characteristics of patients harboring a likely-disease causing mutation summarized in Supplementary Table 12.
Supplementary Methods: Evaluation of disease causing recessive mutations
Supplementary Table 14. Newly hemizygous and homozygous* variants in genes associated with seizure phenotypes.
Supplementary Table 15. Compound heterozygous variants in genes associated seizure phenotypes.
Supplementary Note. Additional clinical details on EE patients harboring de novo mutations in HNRNPU, HNRNPH1, and NEDD4L.
Acknowledgments
We are deeply grateful to the patients, their families, clinical research coordinators and referring physicians for participating in the Epilepsy Phenome/Genome Project (EPGP) and providing the phenotype data and DNA samples used in this study. We thank the following professional and lay organizations for substantial assistance in publicizing EPGP and therefore enabling us to recruit participants effectively: AED Pregnancy Registry, American Epilepsy Society, Association of Child Neurology Nurses, California School Nurses Organization, Child Neurology Society, Citizens United for Research in Epilepsy, Dravet Syndrome Foundation, Epilepsy Alliance of Orange County, Epilepsy Foundation, Epilepsy Therapy Project, Finding a Cure for Epilepsy and Seizures, IDEA League, InfantileSpasms.com, Lennox-Gastaut Syndrome Foundation, PatientsLikeMe, People Against Childhood Epilepsy, PVNH Support & Awareness, and Seizures & Epilepsy Education.
We thank the EPGP Administrative Core (Catharine Freyer, Kristen Schardein, RN, MS, Robyn Fahlstrom, MPH, Sabrina Cristofaro, RN, BSN, Kathleen McGovern), EPGP Bioinformatics Core (Gerard Nesbitt, Kevin McKenna, Vickie Mays), staff at the Coriell Institute – NINDS Genetics Repository (Chi Tarn, PhD, Allison Scutti, MS), and members of the Duke Center for Human Genome Variation (Brian Krueger, PhD, Joshua Bridgers, MS, Jonathan Keebler, PhD, Hee Shin Kim, PhD, Erin Campbell, BS, Ken Cronin, BS, Linda Hong, BS, Melora McCall, BS) for their extraordinary dedication and commitment to this work. We also thank Shlomo Shinnar, MD, PhD (Albert Einstein College of Medicine) and Neil Risch, PhD (University of California, San Francisco) for valuable input into the creation of EPGP and Epi4K, and Randall Stewart, PhD, Katrina Gwinn, MD and Rod Corriveau, PhD from the National Institute of Neurological Disorders and Stroke for their careful oversight and guidance of both EPGP and Epi4K.
This work was supported by grants from the National Institute of Neurological Disorders and Stroke (The Epilepsy Phenome/Genome Project NS053998; Epi4K Project 1 – Epileptic Encephalopathies NS077364; Epi4K – Administrative Core NS077274; Epi4K Sequencing, Biostatistics and Bioinformatics Core NS077303 and Epi4K – Phenotyping and Clinical Informatics Core NS077276); Finding a Cure for Epilepsy and Seizures; and the Richard Thalheimer Philanthropic Fund.
Author list
Epi4K
Andrew S. Allen 1, Samuel F. Berkovic 2, Patrick Cossette 3, Norman Delanty 4, Dennis Dlugos 5, Evan E. Eichler 6, Michael P. Epstein 7, Tracy Glauser 8, David B. Goldstein 9,Yujun Han 9, Erin L. Heinzen9, Yuki Hitomi9, Katherine B. Howell 10, Michael R. Johnson 11, Ruben Kuzniecky12, Daniel H. Lowenstein13, Yi-Fan Lu 9, Maura R.Z. Madou 13, Anthony G. Marson14, Heather C. Mefford 15, Sahar Esmaeeli Nieh16, Terence J. O’Brien 17, Ruth Ottman 18, Slavé Petrovski 2,9,17, Annapurna Poduri 19, Elizabeth K. Ruzzo9, Ingrid E. Scheffer 20, Elliott Sherr 21, Christopher J. Yuskaitis 22
EPGP
Bassel Abou-Khalil 23, Brian K. Alldredge 24, Jocelyn F. Bautista 25, Samuel F. Berkovic 2, Alex Boro 26, Gregory Cascino27, Damian Consalvo 28, Patricia Crumrine 29, Orrin Devinsky 30, Dennis Dlugos 5, Michael P. Epstein 7, Miguel Fiol 31, Nathan B. Fountain32, Jacqueline French 33, Daniel Friedman 34, Eric B. Geller 35, Tracy Glauser 8, Simon Glynn 36, Sheryl R. Haut37, Jean Hayward38, Sandra L. Helmers 39, Sucheta Joshi 40, Andres Kanner 41, Heidi E. Kirsch42, Robert C. Knowlton 43, Eric H. Kossoff 44, Rachel Kuperman45, Ruben Kuzniecky12, Daniel H. Lowenstein13, Shannon M. McGuire 46, Paul V. Motika47, Edward J. Novotny 48, Ruth Ottman 18, Juliann M. Paolicchi49, Jack Parent 50, Kristen Park51, Annapurna Poduri 19, Ingrid E. Scheffer 20, Renée A. Shellhaas 52, Elliott Sherr 21, Jerry J. Shih53, Rani Singh 54, Joseph Sirven55, Michael C. Smith41, Joe Sullivan13, K. Liu Lin Thio 56, Anu Venkat 57, Eileen P.G. Vining 58, Gretchen K. Von Allmen 59, Judith L. Weisenberg 60, Peter Widdess-Walsh 35, Melodie R. Winawer 61
Author contributions
Initial Design of EPGP: B.K.A., O.D., D.D., M.E., Ru.Ku., D.H.L., R.O., E.S., M.W. EPGP Patient Recruitment and Phenotyping: B.A., J.B., S.F.B., G.C., D.C., P.C., O.D., D.D., M.F., N.B.F., D.F., E.B.G., T.G,. S.G., S.H., J.H., S.L.H., H.K., R.Kn., E.K., Ra.Ku., Ru.Ku, D.H.L., S.M.M., P.V.M., E.J.N., J.M.P., J.P., K.P., A.P., I.E.S., J.S., R.S., J.S., M.S., L.L.T., A.V., E.V., G.K.V., J.W., P.W. Phenotype Data Analysis: B.A., B.K.A., A.B., G.C., O.D., D.D., J.F., T.G., S.J., A.K., R.Kn., Ru.Ku., D.H.L., R.O., J.M.P., A.P., I.E.S., R.S., E.S., J.S., J.S., P.W., M.W. Initial Design of Epi4K: S.F.B., P.C., N.D., D.D., E.E.E., M.E., T.G., D.B.G., E.L.H., M.R.J., R.K., D.H.L., A.G.M., H.C.M., T.J.O., R.O., A.P., I.E.S., E.S. Epileptic Encephalopathy Phenotyping Strategy: S.F.B., P.C., D.D., R.K., D.H.L., R.O., I.E.S., E.S. Encephalopathy Phenotyping: D.D., K.B.H., M.R.Z.M., H.C.M., A.P., I.E.S., E.S., C.J.Y. Sequence Data Analysis & Statistical Interpretation: A.S.A., D.B.G., Y.Ha., E.L.H., S.E.N., S.P., E.K.R., E.S. Functional Evaluation of Identified Mutations: D.B.G., E.L.H., Y.Hi., Y.L. Writing of Manuscript: A.S.A., S.F.B., D.D., D.B.G., Y.H., E.L.H., M.R.J., D.H.L., H.C.M., R.O., A.P., S.P., E.K.R., I.S., E.S.
Footnotes
Department of Biostatistics and Bioinformatics, Duke Clinical Research Institute, and Center for Human Genome Variation, Duke University Medical Center, Durham, North Carolina 27710, USA.
Epilepsy Research Centre, Department of Medicine, University of Melbourne (Austin Health), Heidelberg, Victoria 3084, Australia.
Centre of Excellence in Neuromics and CHUM Research Center, Université de Montréal, CHUM-Hôpital Notre-Dam Montréal, Quebec H2L 4M1e, Canada.
Department of Neurology, Beaumont Hospital and Royal College of Surgeons, Dublin 9 Ireland.
Department of Neurology and Pediatrics, The Children’s Hospital of Philadelphia, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, Pennsylvania 19104 USA.
Howard Hughes Medical Institute, Department of Genome Sciences, University of Washington School of Medicine, Seattle, Washington 98195 USA.
Department of Human Genetics, Emory University School of Medicine, Atlanta, Georgia 30322, USA.
Division of Neurology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio 45229 USA.
Center for Human Genome Variation, Duke University School of Medicine, Durham, North Carolina 27708 USA.
Neurology, Division of Medicine, Royal Children’s Hospital Melbourne, Parkville, Victoria 3052 Australia.
Centre for Clinical Translation Division of Brain Sciences, Imperial College London, London, SW7 2AZ United Kingdom
NYU School of Medicine, New York University, 10016 USA
Department of Neurology, University of California, San Francisco, San Francisco, California 94143 USA.
Department of Molecular and Clinical Pharmacology, University of Liverpool, Clinical Sciences Centre, Lower Lane, Liverpool, L9 7LJ, United Kingdom.
Department of Pediatrics, Division of Genetic Medicine, University of Washington, Seattle, WA 98115 USA.
University of California, San Francisco, California 94143 USA.
Departments of Medicine and Neurology, The Royal Melbourne Hospital, Parkville, Victoria, 3146 Australia.
Departments of Epidemiology and Neurology, and the G.H. Sergievsky Center, Columbia University; and Division of Epidemiology, New York State Psychiatric Institute, New York, New York 10032, USA.
Division of Epilepsy and Clinical Neurophysiology, Department of Neurology Boston Children’s Hospital, Boston, Massachusetts 02115 USA
Epilepsy Research Centre, Department of Medicine, University of Melbourne (Austin Health), Heidelberg, Victoria 3084, Australia, Florey Institute and Department of Pediatrics, Royal Children’ s Hospital, University of Melbourne, Victoria 3052, AUSTRALIA.
Departments of Neurology, Pediatrics and Institute of Human Genetics, University of California, San Francisco, San Francisco, California 94158 USA.
Department of Neurology, Boston Children’s Hospital Harvard Medical School, Boston, Massachusetts, 02115 USA.
Department of Neurology, Vanderbilt University Medical Center, Nashville, Tennessee 37232 USA.
Department of Clinical Pharmacy, UCSF School of Pharmacy, Department of Neurology, UCSF School of Medicine 94143 USA.
Department of Neurology, Cleveland Clinic Lerner College of Medicine & Epilepsy Center of the Cleveland Clinic Neurological Institute, Cleveland, Ohio, 44195 USA.
Department of Neurology, Montefiore Medical Center, Albert Einstein College of Medicine, Bronx, New York, 10467 USA.
Divison of Epilepsy, Mayo Clinic, Rochester, Minnesota 55905 USA.
Epilepsy Center, Neurology Division, Ramos Mejía Hospital, Buenos Aires, 1221, Argentina.
Medical Epilepsy Program & EEG & Child Neurology, Children’s Hospital of Pittsburgh of UPMC, Pediatrics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15224 USA.
NYU and Saint Barnabas Epilepsy Centers, NYU School of Medicine, New York, New York 10016 USA.
Department of Neurology, Epilepsy Care Center, University of Minnesota Medical School, Minneapolis 55414 USA.
FE Dreifuss Comprehensive Epilepsy Program, University of Virginia, Charlottesville, Virginia 22908 USA.
NYU Comprehensive Epilepsy Center, New York, New York 10016 USA.
Department of Neurology, NYU School of Medicine, New York, New York, 10016 USA.
Division of Neurology, Saint Barnabas Medical Center, Livingston, New Jersey 07039 USA.
Department of Neurology, Comprehensive Epilepsy Program, University of Michigan Health System, Ann Arbor, Michigan 48109 USA.
Comprehensive Epilepsy Center, Montefiore Medical Center, Bronx, New York 10467 USA.
The Kaiser Permanente Group, Oakland, California 94618 USA.
Neurology and Pediatrics, Emory University School of Medicine, Atlanta, Georgia 30322 USA.
Pediatrics & Communicable Diseases, University of Michigan, Ann Arbor, Michigan 48109 USA.
Department of Neurological Sciences, Rush Epilepsy Center, Rush University Medical Center, Chicago, Illinois 60612 USA.
Departments of Neurology and Radiology, University of California, San Francisco, California 94143 USA.
Neurology, University of Texas Medical School, Houston, Texas 77030 USA.
Neurology and Pediatrics, Child Neurology, Pediatric Neurology Residency Program, Johns Hopkins Hospital, Baltimore, Maryland 21287 USA.
Epilepsy Program, Children’s Hospital & Research Center Oakland, Oakland, California 94609 USA.
Clinical Neurology, Children’s Hospital Epilepsy Center of New Orleans, New Orleans, Louisiana 70118 USA.
Comprehensive Epilepsy Center, Oregon Health and Science University, Portland, Oregon 97239 USA.
Departments of Neurology and Pediatrics, University of Washington School of Medicine, Seattle Children’s Hospital, Seattle, Washington 98105 USA.
Weill Cornell Medical Center, New York, New York 10065 USA.
Department of Neurology and Neuroscience Graduate Program, University of Michigan Medical Center, Ann Arbor, MI 49108 and Ann Arbor Veterans Administration Healthcare System, Ann Arbor MI 48105.
Department of Pediatrics, Pediatrics-Neurology, University of Colorado Hospital, Denver, Colorado 80218 USA.
University of Michigan, Pediatric Neurology, Ann Arbor, Michigan 48109 USA.
Department of Neurology, Mayo Clinic, Jacksonville, Florida 32224 USA.
Division of Pediatric Neurology, University of Michigan Health System, Ann Arbor, Michigan 48109 USA.
Department of Neurology, Mayo Clinic, Scottsdale, Arizona 85259 USA.
Department of Neurology, Washington University School of Medicine, St. Louis, Missouri 63110.
Neurology & Pediatrics, The Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania 19104 USA.
Department of Neurology, Johns Hopkins Hospital, Baltimore, Maryland 21287 USA.
Division of Child & Adolescent Neurology, Departments of Pediatrics, University of Texas Medical School, Houston, Texas 77030 USA.
Department of Neurology, Division of Pediatric Neurology, Washington University School of Medicine, St. Louis, Missouri 63110 USA.
Department of Neurology and the G.H. Sergievsky Center, Columbia University, New York, New York, 10032 USA.
The authors declare no competing financial interests.
References
- 1.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epicure Consortium et al. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum Mol Genet. 2012;21:5359–5372. doi: 10.1093/hmg/dds373. [DOI] [PubMed] [Google Scholar]
- 3.Heinzen EL, et al. Exome sequencing followed by large-scale genotyping fails to identify single rare variants of large effect in idiopathic generalized epilepsy. Am J Hum Genet. 2012;91:293–302. doi: 10.1016/j.ajhg.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulley JC, Mefford HC. Epilepsy and the new cytogenetics. Epilepsia. 2011;52:423–432. doi: 10.1111/j.1528-1167.2010.02932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasperaviciute D, et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain. 2010;133:2136–2147. doi: 10.1093/brain/awq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vissers LE, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 7.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalscheuer VM, et al. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet. 2003;72:1401–1411. doi: 10.1086/375538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claes L, et al. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitsu H, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008;40:782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 11.Otsuka M, et al. STXBP1 mutations cause not only Ohtahara syndrome but also West syndrome--result of Japanese cohort study. Epilepsia. 2010;51:2449–2452. doi: 10.1111/j.1528-1167.2010.02767.x. [DOI] [PubMed] [Google Scholar]
- 12.Veeramah KR, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya K, et al. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24:2690–2698. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M, DeLorey TM, Delgado-Escueta A, Olsen RW. In: Jasper’s Basic Mechanisms of the Epilepsies. Noebels JL, et al., editors. 2012. [PubMed] [Google Scholar]
- 15.Timal S, et al. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum Mol Genet. 2012;21:4151–4161. doi: 10.1093/hmg/dds123. [DOI] [PubMed] [Google Scholar]
- 16.de Ligt J, et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. N Engl J Med. 2012 doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 17.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klassen T, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145:1036–1048. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemke JR, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012;53:1387–1398. doi: 10.1111/j.1528-1167.2012.03516.x. [DOI] [PubMed] [Google Scholar]
- 22.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012 doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 23.Bhardwaj RD, et al. Neocortical neurogenesis in humans is restricted to development. Proc Natl Acad Sci U S A. 2006;103:12564–12568. doi: 10.1073/pnas.0605177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitomi Y, et al. Mutations in TNK2 in severe autosomal recessive infantile-onset epilepsy. Ann Neurol. 2013 doi: 10.1002/ana.23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gleeson JG, et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 27.Fox JW, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 28.The EPGP Collaborative. The Epilepsy Phenome/Genome Project. Clinical Trials: Journal of the Scoiety for Clinical Trials. 2013 doi: 10.1177/1740774513484392. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epicure Consortium et al. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum Mol Genet. 2012;21:5359–5372. doi: 10.1093/hmg/dds373. [DOI] [PubMed] [Google Scholar]
- 3.Heinzen EL, et al. Exome sequencing followed by large-scale genotyping fails to identify single rare variants of large effect in idiopathic generalized epilepsy. Am J Hum Genet. 2012;91:293–302. doi: 10.1016/j.ajhg.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulley JC, Mefford HC. Epilepsy and the new cytogenetics. Epilepsia. 2011;52:423–432. doi: 10.1111/j.1528-1167.2010.02932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasperaviciute D, et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain. 2010;133:2136–2147. doi: 10.1093/brain/awq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vissers LE, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 7.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalscheuer VM, et al. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet. 2003;72:1401–1411. doi: 10.1086/375538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claes L, et al. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saitsu H, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008;40:782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 11.Otsuka M, et al. STXBP1 mutations cause not only Ohtahara syndrome but also West syndrome--result of Japanese cohort study. Epilepsia. 2010;51:2449–2452. doi: 10.1111/j.1528-1167.2010.02767.x. [DOI] [PubMed] [Google Scholar]
- 12.Veeramah KR, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya K, et al. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24:2690–2698. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M, et al. Hyperglycosylation and reduced GABA currents of mutated GABRB3 polypeptide in remitting childhood absence epilepsy. Am J Hum Genet. 2008;82:1249–1261. doi: 10.1016/j.ajhg.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang YH, et al. Altered ultrasonic vocalization and impaired learning and memory in Angelman syndrome mouse model with a large maternal deletion from Ube3a to Gabrb3. PLoS One. 2010;5:e12278. doi: 10.1371/journal.pone.0012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timal S, et al. Gene identification in the congenital disorders of glycosylation type I by whole-exome sequencing. Hum Mol Genet. 2012;21:4151–4161. doi: 10.1093/hmg/dds123. [DOI] [PubMed] [Google Scholar]
- 17.de Ligt J, et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. N Engl J Med. 2012 doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 18.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klassen T, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145:1036–1048. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemke JR, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012;53:1387–1398. doi: 10.1111/j.1528-1167.2012.03516.x. [DOI] [PubMed] [Google Scholar]
- 23.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012 doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 24.Bhardwaj RD, et al. Neocortical neurogenesis in humans is restricted to development. Proc Natl Acad Sci U S A. 2006;103:12564–12568. doi: 10.1073/pnas.0605177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitomi Y, et al. Mutations in TNK2 in severe autosomal recessive infantile-onset epilepsy. Ann Neurol. 2013 doi: 10.1002/ana.23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gleeson JG, et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 28.Fox JW, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 29.The EPGP Collaborative. The Epilepsy Phenome/Genome Project. Clinical Trials: Journal of the Scoiety for Clinical Trials. 2013 doi: 10.1177/1740774513484392. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Coriell IDs for all exome sequenced 264 IS and LGS trios.
Supplementary Table 2. Comprehensive list of de novo mutations identified in 264 IS and LGS trios.
Supplementary Table 3. Summary of de novo mutations identified in 264 IS and LGS trios.
Supplementary Figure 1. Distribution of de novo mutations detected in 264 IS/LGS probands.
Supplementary Table 4. Estimating probability of multiple de novo events amongst control exomes.
Supplementary Methods: Investigating instances of multiple hits at the same site across distinct probands.
Supplementary Figure 2. The sequencing coverage of four de novo mutations.
Supplementary Methods: Genetic Architecture
Supplementary Table 5. Observed number of de novo coding mutations observed in autism spectral disorder trios as reported by Neale et al.
Supplementary Table 6. Observed number of de novo coding mutations observed in IS/LGS trio exomes.
Supplementary Table 7. Observed number of de novo coding mutations observed in intolerant genes among IS/LGS trios.
Supplementary Figure 3. Likelihood surface for autism spectrum data given in Neale et al. (Supplementary Table 5). Gray lines represent contours of the likelihood surface. Red lines represent 95% likelihood ratio confidence regions for η and “log”(γ). Maximum likelihood estimate given by black X (η=0.018,γ=15.6). Blue Xs represent 6 sets of parameter values for η and γ simulated by Neale et al.
Supplementary Figure 4. Likelihood surface for IS/LGS exome data (Supplementary Table 6). Gray lines represent contours of the likelihood surface. Red lines represent 95% likelihood ratio confidence regions for η and “log”(γ). Maximum likelihood estimate given by black X (η=0.007,γ=86).
Supplementary Figure 5. Likelihood surface for IS/LGS intolerant gene data (Supplementary Table 7). Gray lines represent contours of the likelihood surface. Red lines represent 95% likelihood ratio confidence regions for η and “log”(γ). Maximum likelihood estimate given by black X (η=0.021,γ=81).
Supplementary Table 8. Summary statistics of the opportunity space to call a de novo coding variant in the ‘known’ early epileptic encephalopathy MIM genes [last accessed OMIM® December 2012].
Supplementary Figure 6. Classification of known EE genes as intolerant.
Supplementary Table 9. List of “intolerant” CCDS-defined genes harboring a de novo mutation in an IS or LGS trio (n=109). Previously reported EE genes are highlighted in red.
Supplementary Methods: Assessment of mutation consequences in intolerant genes
Supplementary Table 10. Gene list enrichment results summary
Supplementary Figure 7. Full protein-protein network analysis of 267 genes.
Supplementary Table 11. Protein-Protein network enrichment.
Supplementary Methods: Functional work
Effects of a de novo synonymous HNRNPH1 mutation on splicing
Supplementary Figure 8. De novo HNRNPH1 synonymous mutation causes alternatively spliced transcript missing exon 12.
Supplementary Table 12. Likely functional (missense, nonsense, or splice site) de novo mutations identified in IS/LGS trios in genes that have been previously associated with a neuropsychiatric disorders† with or without seizures.
Supplementary Table 13. Clinical characteristics of patients harboring a likely-disease causing mutation summarized in Supplementary Table 12.
Supplementary Methods: Evaluation of disease causing recessive mutations
Supplementary Table 14. Newly hemizygous and homozygous* variants in genes associated with seizure phenotypes.
Supplementary Table 15. Compound heterozygous variants in genes associated seizure phenotypes.
Supplementary Note. Additional clinical details on EE patients harboring de novo mutations in HNRNPU, HNRNPH1, and NEDD4L.