Abstract
CRISPR RNA-guided endonucleases (RGENs) have rapidly emerged as a facile and efficient platform for genome editing. Here, we use a human cell-based reporter assay to characterize off-target cleavage of Cas9-based RGENs. We find that single and double mismatches are tolerated to varying degrees depending on their position along the guide RNA (gRNA)-DNA interface. We readily detected off-target alterations induced by four out of six RGENs targeted to endogenous loci in human cells by examination of partially mismatched sites. The off-target sites we identified harbor up to five mismatches and many are mutagenized with frequencies comparable to (or higher than) those observed at the intended on-target site. Our work demonstrates that RGENs are highly active even with imperfectly matched RNA-DNA interfaces in human cells, a finding that might confound their use in research and therapeutic applications.
Recent work has demonstrated that clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems1–3 can serve as the basis of a simple and highly efficient method for performing genome editing in bacteria, yeast and human cells, as well as in vivo in whole organisms such as fruit flies, zebrafish and mice4–13. The Cas9 nuclease from S. pyogenes (hereafter simply Cas9) can be guided via simple base pair complementarity between the first 20 nucleotides of an engineered gRNA and a target genomic DNA sequence of interest that lies next to a protospacer adjacent motif (PAM) matching the sequence NGG5–12, 14 (Supplementary Fig. 1). Previous studies performed in vitro14, in bacteria7 and in human cells10 have shown that Cas9-mediated cleavage can be abolished by single mismatches at the gRNA/target site interface, particularly in the last 10–12 nucleotides located in the 3’ end of the 20 nt gRNA targeting region. Although Maraffini and colleagues7 recently performed a systematic investigation of Cas9 RGEN specificity in bacteria, the specificities of RGENs in human cells have not been extensively defined and, to our knowledge, bona fide off-target mutations induced by Cas9 have not been identified in any eukaryotic cell or organism. Understanding the scope of RGEN-mediated off-target effects in human and other eukaryotic cells will be critically essential if these nucleases are to be used widely for research and therapeutic applications.
To begin to define the specificity determinants of RGENs in human cells, we sought to perform a large-scale test in which we assessed the effects of systematically mismatching various positions within multiple gRNA/target DNA interfaces. To do this, we used a quantitative human cell-based enhanced green fluorescent protein (EGFP) disruption assay previously described by our lab15 that enables rapid quantitation of targeted nuclease activities (Fig. 1a). In this assay, the activities of nucleases targeted to a single integrated EGFP reporter gene can be quantified by assessing loss of fluorescence signal in human U2OS.EGFP cells caused by inactivating frameshift insertion/deletion (indel) mutations introduced by error prone non-homologous end-joining (NHEJ) repair of nuclease-induced double-stranded breaks (DSBs) (Fig. 1a and Methods). For the studies described here, we used three ~100 nt single gRNAs (sgRNAs) targeted to different sequences within EGFP (Supplementary Fig. 2); each of these sgRNAs can efficiently direct Cas9-mediated disruption of EGFP expression (Supplementary Results).
Fig. 1. Activities of RGENs harboring variant mismatched sgRNAs in a human cell-based EGFP disruption assay.
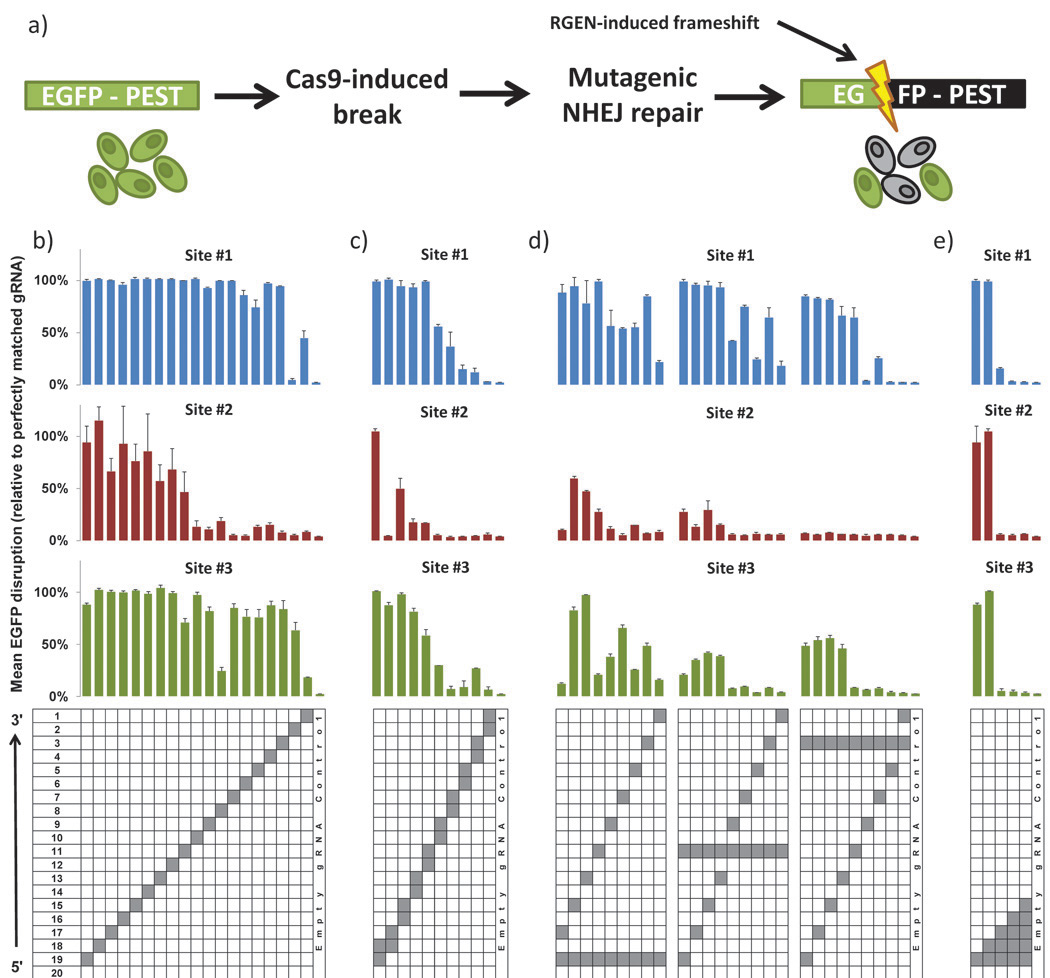
(a) Schematic overview of the EGFP disruption assay. Repair of targeted Cas9-mediated double-stranded breaks in a single integrated EGFP-PEST reporter gene by error-prone NHEJ-mediated repair leads to frameshift mutations that disrupt the coding sequence and to associated loss of fluorescence in cells. Activities of RGENs harboring sgRNAs bearing (b) single mismatches, (c) adjacent double mismatches, (d) variably spaced double mismatches, and (e) increasing numbers of adjacent mismatches assayed on three different target sites in the EGFP reporter gene sequence. Mean activities of replicates (see Online Methods) are shown, normalized to the activity of a perfectly matched sgRNA. Error bars indicate standard errors of the mean. Positions mismatched in each sgRNA are highlighted in grey in the grid below. Sequences of the three EGFP target sites are shown in Supplementary Figure 2a.
In initial experiments, we tested the effects of single nucleotide mismatches at 19 of 20 nucleotides in the complementary targeting region of our three EGFP-targeted sgRNAs. To do this, we generated variant sgRNAs for each of the three target sites harboring Watson-Crick transversion mismatches at positions 1 through 19 (numbered 1 to 20 in the 3’ to 5’ direction; see Supplementary Fig. 1) and tested the abilities of these various sgRNAs to direct Cas9-mediated EGFP disruption in human cells. (We did not generate variant sgRNAs bearing a substitution at position 20 because this nucleotide is part of the U6 promoter sequence and therefore must remain a guanine to avoid affecting expression.) For EGFP target site #2, single mismatches in positions 1 – 10 of the sgRNA have dramatic effects on associated Cas9 activity (Fig. 1b, middle panel), consistent with previous studies that suggest mismatches at the 5’ end of gRNAs are better tolerated than those at the 3’ end7, 10, 14. However, we found with EGFP target sites #1 and #3 that single mismatches at all but a few positions in the sgRNA appear to be well tolerated, even within the 3’ end of the sequence. Furthermore, the specific positions that are sensitive to mismatch differ for these two targets (Fig. 1b, compare top and bottom panels) – for example, target site #1 is particularly sensitive to a mismatch at position 2 whereas target site #3 is most sensitive to mismatches at positions 1 and 8.
To test the effects of more than one mismatch at the sgRNA/DNA interface, we created a series of variant sgRNAs bearing double Watson-Crick transversion mismatches in adjacent (Fig. 1c) and separated (Fig. 1d) positions and tested the abilities of these to direct Cas9 nuclease activity in human cells using our EGFP disruption assay. All three target sites generally showed greater sensitivity to double alterations in which one or both mismatches occur within the 3’ half of the sgRNA targeting region (Figs. 1c and 1d). However, the magnitude of these effects exhibited site-specific variation, with target site #2 showing the greatest sensitivity to these double mismatches and target site #1 generally showing the least (Figs. 1c and 1d). To test the number of adjacent mismatches that can be tolerated, we constructed variant sgRNAs bearing increasing numbers of mismatched positions ranging from positions 19 to 15 in the 5’ end of the sgRNA targeting region (where single and double mismatches appear to be better tolerated) (Fig. 1e). Testing of these increasingly mismatched sgRNAs revealed that for all three target sites, the introduction of three or more adjacent mismatches results in significant loss of RGEN activity (Fig. 1e). Taken together, our results in human cells confirm that the activities of RGENs can be more sensitive to mismatches in the 3’ half of the sgRNA targeting sequence. However, our data also clearly reveal that the specificity of RGENs is complex and target site-dependent, with single and double mismatches often well tolerated even when one or more mismatches occur in the 3’ half of the sgRNA targeting region. Furthermore, our data also suggest that not all mismatches in the 5’ half of the sgRNA/DNA interface are necessarily well tolerated.
We next sought to determine whether we could identify off-target mutations for RGENs targeted to endogenous human genes. To accomplish this, we used six sgRNAs that target three different sites in the VEGFA gene, one in the EMX1 gene, one in the RNF2 gene, and one in the FANCF gene (Table 1 and Supplementary Table 1). These six sgRNAs efficiently directed Cas9-mediated indels at their respective endogenous loci in human U2OS.EGFP cells as detected by T7 Endonuclease I (T7EI) assay (Online Methods and Table 1). For each of these six RGENs, we then examined dozens of potential off-target sites (ranging in number from 46 to as many as 64) for evidence of nuclease-induced NHEJ-mediated indel mutations in U2OS.EGFP cells. The loci we assessed included all genomic sites that differ by one or two nucleotides as well as subsets of genomic sites that differ by three to six nucleotides and with a bias toward those that had one or more of these mismatches in the 5’ half of the sgRNA targeting sequence (Supplementary Table 2). Using the T7EI assay, we readily identified four off-target sites (out of 53 candidate sites examined) for VEGFA site 1, twelve (out of 46 examined) for VEGFA site 2, seven (out of 64 examined) for VEGFA site 3 and one (out of 46 examined) for the EMX1 site (Table 1 and Supplementary Table 2). No off-target mutations were detected among the 43 and 50 potential sites examined for the RNF2 or FANCF genes, respectively (Supplementary Table 2). The rates of mutation at verified off-target sites were very high, ranging from 5.6% to 125% (mean of 40%) of the rate observed at the intended target site (Table 1). These bona fide off-targets included sequences with mismatches in the 3’ end of the target site and with as many as a total of five mismatches, with most off-target sites occurring within protein coding genes (Table 1). DNA sequencing of a subset of off-target sites provided additional molecular confirmation that indel mutations occur at the expected RGEN cleavage site (Supplementary Fig. 3).
Table 1. On- and off-target mutations induced by RGENs designed to endogenous human genes.
“OT” indicates off-target sites (with numbering of sites as in Supplementary Table 2). Mismatches from the on-target (within the 20 bp region to which the sgRNA hybridizes) are highlighted as bold, underlined text. Mean indel mutation frequencies in U2OS.EGFP, HEK293, and K562 cells were determined as described in Online Methods. Genes in which sites were located (if any) are shown. All sites listed failed to show any evidence of modification in cells transfected with Cas9 expression plasmid and a control U6 promoter plasmid that did not express a functional sgRNA.
| Target | Site name |
Sequence | Indel Mutation Frequency (%) ± SEM | Gene | ||
|---|---|---|---|---|---|---|
| U2OS.EGFP | HEK293 | K562 | ||||
|
Target 1 (VEGFA Site 1) |
T1 | GGGTGGGGGGAGTTTGCTCCTGG | 26.0 ± 2.9 | 10.5 ± 0.07 | 3.33 ± 0.42 | VEGFA |
| OT1-3 | GGATGGAGGGAGTTTGCTCCTGG | 25.7 ± 9.1 | 18.9 ± 0.77 | 2.93 ± 0.04 | IGDCC3 | |
| OT1-4 | GGGAGGGTGGAGTTTGCTCCTGG | 9.2 ± 0.8 | 8.32 ± 0.51 | N.D. | LOC116437 | |
| OT1-6 | CGGGGGAGGGAGTTTGCTCCTGG | 5.3 ± 0.2 | 3.67 ± 0.09 | N.D. | CACNA2D | |
| OT1-11 | GGGGAGGGGAAGTTTGCTCCTGG | 17.1 ± 4.7 | 8.54 ± 0.16 | N.D. | ||
|
Target 2 (VEGFA Site 2) |
T2 | GACCCCCTCCACCCCGCCTCCGG | 50.2 ± 4.9 | 38.6 ± 1.92 | 15.0 ± 0.25 | VEGFA |
| OT2-1 | GACCCCCCCCACCCCGCCCCCGG | 14.4 ± 3.4 | 33.6 ± 1.17 | 4.10 ± 0.05 | FMN1 | |
| OT2-2 | GGGCCCCTCCACCCCGCCTCTGG | 20.0 ± 6.2 | 15.6 ± 0.30 | 3.00 ± 0.06 | PAX6 | |
| OT2-6 | CTACCCCTCCACCCCGCCTCCGG | 8.2 ± 1.4 | 15.0 ± 0.64 | 5.24 ± 0.22 | PAPD7 | |
| OT2-9 | GCCCCCACCCACCCCGCCTCTGG | 50.7 ± 5.6 | 30.7 ± 1.44 | 7.05 ± 0.48 | LAMA3 | |
| OT2-15 | TACCCCCCACACCCCGCCTCTGG | 9.7 ± 4.5 | 6.97 ± 0.10 | 1.34 ± 0.15 | SPNS3 | |
| OT2-17 | ACACCCCCCCACCCCGCCTCAGG | 14.0 ± 2.8 | 12.3 ± 0.45 | 1.80 ± 0.03 | ||
| OT2-19 | ATTCCCCCCCACCCCGCCTCAGG | 17.0 ± 3.3 | 19.4 ± 1.35 | N.D. | HDLBP | |
| OT2-20 | CCCCACCCCCACCCCGCCTCAGG | 6.1 ± 1.3 | N.D. | N.D. | ABLIM1 | |
| OT2-23 | CGCCCTCCCCACCCCGCCTCCGG | 44.4 ± 6.7 | 28.7 ± 1.15 | 4.18 ± 0.37 | CALY | |
| OT2-24 | CTCCCCACCCACCCCGCCTCAGG | 62.8 ± 5.0 | 29.8 ± 1.08 | 21.1 ± 1.68 | ||
| OT2-29 | TGCCCCTCCCACCCCGCCTCTGG | 13.8 ± 5.2 | N.D. | N.D. | ACLY | |
| OT2-34 | AGGCCCCCACACCCCGCCTCAGG | 2.8 ± 1.5 | N.D. | N.D. | ||
|
Target 3 (VEGFA Site 3) |
T3 | GGTGAGTGAGTGTGTGCGTGTGG | 49.4 ± 3.8 | 35.7 ± 1.26 | 27.9 ± 0.52 | VEGFA |
| OT3-1 | GGTGAGTGAGTGTGTGTGTGAGG | 7.4 ± 3.4 | 8.97 ± 0.80 | N.D. | (abParts) | |
| OT3-2 | AGTGAGTGAGTGTGTGTGTGGGG | 24.3 ± 9.2 | 23.9 ± 0.08 | 8.9 ± 0.16 | MAX | |
| OT3-4 | GCTGAGTGAGTGTATGCGTGTGG | 20.9 ± 11.8 | 11.2 ± 0.23 | N.D. | ||
| OT3-9 | GGTGAGTGAGTGCGTGCGGGTGG | 3.2 ± 0.3 | 2.34 ± 0.21 | N.D. | TPCN2 | |
| OT3-17 | GTTGAGTGAATGTGTGCGTGAGG | 2.9 ± 0.2 | 1.27 ± 0.02 | N.D. | SLIT1 | |
| OT3-18 | TGTGGGTGAGTGTGTGCGTGAGG | 13.4 ± 4.2 | 12.1 ± 0.24 | 2.42 ± 0.07 | COMDA | |
| OT3-20 | AGAGAGTGAGTGTGTGCATGAGG | 16.7 ± 3.5 | 7.64 ± 0.05 | 1.18 ± 0.01 | ||
|
Target 4 (EMX1) |
T4 | GAGTCCGAGCAGAAGAAGAAGGG | 42.1 ± 0.4 | 26.0 ± 0.70 | 10.7 ± 0.50 | EMX1 |
| OT4-1 | GAGTTAGAGCAGAAGAAGAAAGG | 16.8 ± 0.2 | 8.43 ± 1.32 | 2.54 ± 0.02 | HCN1 | |
|
Target 5 (RNF2) |
T5 | GTCATCTTAGTCATTACCTGTGG | 26.6 ± 6.0 | --- | --- | RNF2 |
|
Target 6 (FANCF) |
T6 | GGAATCCCTTCTGCAGCACCAGG | 33.2 ± 6.5 | --- | --- | FANCF |
N.D. = none detected; --- = not tested.
Having established that RGENs can induce off-target mutations with high frequencies in U2OS.EGFP cells, we next sought to determine whether these nucleases would also have these effects in other types of human cells. We had chosen U2OS.EGFP cells for our initial experiments because we previously used these cells to evaluate the activities of TALENs15 but human HEK293 and K562 cells have been more widely used to test the activities of targeted nucleases. Therefore, we also assessed the activities of the four RGENs targeted to VEGFA sites 1, 2, and 3 and the EMX1 site in HEK293 and K562 cells. We found that each of these four RGENs efficiently induced NHEJ-mediated indel mutations at their intended on-target site in these two additional human cell lines (as assessed by T7EI assay) (Table 1), albeit with somewhat lower mutation frequencies than those observed in U2OS.EGFP cells. Assessment of the 24 off-target sites for these four RGENs originally identified in U2OS.EGFP cells revealed that many were again mutated in HEK293 and K562 cells with frequencies similar to those at their corresponding on-target site (Table 1). As expected, DNA sequencing of a subset of these off-target sites from HEK293 cells provided additional molecular evidence that alterations are occurring at the expected genomic loci (Supplementary Figure 4). We do not know for certain why in HEK293 cells four and in K562 cells eleven of the off-target sites identified in U2OS.EGFP cells did not show detectable mutations. However, we note that many of these off-target sites also showed relatively lower mutation frequencies in U2OS.EGFP cells. Therefore, we speculate that mutation rates of these sites in HEK293 and K562 cells may be falling below the reliable detection limit of our T7EI assay (~2–5%) because RGENs generally appear to have lower activities in HEK293 and K562 cells compared with U2OS.EGFP cells in our experiments. Taken together, our results in HEK293 and K562 cells provide evidence that the high-frequency off-target mutations we observe with RGENs will be a general phenomenon seen in multiple human cell types.
Our results reveal that predicting the specificity profile of any given RGEN is neither simple nor straightforward. Our EGFP reporter assay experiments show that single and double mismatches can have variable effects on RGEN activity in human cells that do not strictly depend upon their position(s) within the target site. For example, consistent with previously published reports, alterations in the 3’ half of the sgRNA/DNA interface generally have greater effects than those in the 5’ half7, 10, 14; however, single and double mutations in the 3’ end sometimes also appear to be well tolerated whereas double mutations in the 5’ end can greatly diminish activities. In addition, the magnitude of these effects for mismatches at any given position(s) appears to be site-dependent. Comprehensive profiling of a large series of RGENs with testing of all possible nucleotide substitutions (beyond the Watson-Crick transversions used in our EGFP reporter experiments) may help provide additional insights into the range of potential off-targets. In this regard, the recently described bacterial cell-based method of Maraffini and colleagues7 or the in vitro, combinatorial library-based cleavage site-selection methodologies previously applied to ZFNs by Liu and colleagues16 might be useful for generating larger sets of RGEN specificity profiles.
Despite these challenges in comprehensively predicting RGEN specificities, we were able to identify bona fide off-targets of RGENs with relative ease simply by examining a subset of genomic sites that differed from the on-target site by one to five mismatches. Notably, under conditions of our experiments, the frequencies of RGEN-induced mutations at many of these off-target sites were similar to (or higher than) those observed at the intended on-target site, enabling us to identify them using the simple and relatively insensitive T7EI assay (which, as performed in our laboratory, has a reliable detection limit of ~2 to 5% mutation frequency). Because these mutation rates were very high, we were able to avoid using deep sequencing methods previously required to detect much lower frequency ZFN- and TALEN-induced off-target alterations16–19. Our analysis of RGEN off-target mutagenesis in human cells also confirmed the difficulties of predicting RGEN specificities – not all single and double mismatched off-target sites show evidence of mutation whereas some sites with as many as five mismatches can also show alterations. Furthermore, the bona fide off-target sites we identified do not exhibit any obvious bias toward transition or transversion differences relative to the intended target sequence (Supplementary Table 2; grey highlighted rows).
Although we have unveiled off-target sites for a number of RGENs, we note that our identification of these sites was neither comprehensive nor genome-wide in scale. For the six RGENs we studied, we only examined a very small subset of the much larger total number of potential off-target sequences in the human genome (sites that differ by three to six nucleotides from the intended target site; compare Supplementary Tables 2 and 3). Although examining such large numbers of loci for off-target mutations by T7EI assay is neither a practical nor a cost-effective strategy, the use of high-throughput sequencing in future studies might enable the interrogation of larger numbers of candidate off-target sites and provide a more sensitive method for detecting bona fide off-target mutations. For example, such an approach might enable the unveiling of additional off-target sites for the two RGENs for which we failed to uncover any off-target mutations. In addition, an improved understanding both of RGEN specificities and of any epigenomic factors (e.g.--DNA methylation and chromatin status) that may influence RGEN activities in cells might also reduce the number of potential sites that need to be examined and thereby make genome-wide assessments of RGEN off-targets more practical and affordable.
It will be interesting to investigate whether the specific choice of RGEN target site can be used to minimize the frequencies of genomic off-target mutations. Given that off-target sites that differ at up to five positions from the intended target site can be efficiently mutated by RGENs, attempting to choose target sites with minimal numbers of off-target sites as judged by simple mismatch counting seems unlikely to be effective; thousands of potential off-target sites that differ by four or five positions within the 20 bp RNA:DNA complementarity region will typically exist for any given RGEN targeted to a sequence in the human genome (see, for example, Supplementary Table 3). It is also possible that the nucleotide content of the gRNA complementarity region might influence the range of potential off-target effects. For example, high GC-content has been shown to stabilize RNA:DNA hybrids20 and therefore might also be expected to make gRNA/genomic DNA hybridization more stable and more tolerant to mismatches. Additional experiments with larger numbers of gRNAs will be needed to assess if and how these two parameters (numbers of mismatched sites in the genome and stability of the RNA:DNA hybrid) influence the genome-wide specificities of RGENs. However, it is important to note that even if such predictive parameters can be defined, the effect of implementing such guidelines would be to further restrict the targeting range of RGENs.
Another potential general strategy for reducing RGEN-induced off-target effects might be to reduce the concentrations of gRNA and Cas9 nuclease expressed in the cell. We tested this idea using the RGENs for VEGFA target sites 2 and 3 in U2OS.EGFP cells but found that transfecting less sgRNA- and Cas9-expressing plasmid decreased the mutation rate at the on-target site but did not appreciably change the relative rates of off-target mutations (Table 2 and Supplementary Table 4). Consistent with this, we note that we also observe high-level off-target mutagenesis in two other human cell types (HEK293 and K562 cells) even though the absolute rates of on-target mutagenesis are lower than in U2OS.EGFP cells. Although additional work is clearly needed to further explore this strategy, these initial experiments suggest that reducing expression levels of gRNA and Cas9 in cells is not likely to provide a simple solution for reducing off-target effects. Furthermore, these results also suggest that the high rates of off-target mutagenesis we observe in human cells are not caused by overexpression of sgRNA and/or Cas9.
Table 2. Indel mutation frequencies at on- and off-target genomic sites induced by different amounts of Cas9- and sgRNA-expressing plasmids for the RGEN targeted to VEGFA Target Site 2.
Amounts of sgRNA- and Cas9-expressing plasmids transfected into U2OS.EGFP cells for these assays are shown at the top of each column. (Note that data for 250 ng sgRNA/750 ng Cas9 are the same as those presented in Table 1.) Mean indel frequencies were determined using the T7EI assay from replicate samples as described in Online Methods.
| Site | Sequence | 250ng sgRNA/750 ng Cas9 Mean indel frequency (%) ± SEM |
12.5ng sgRNA/50 ng Cas9 Mean indel frequency (%) ± SEM |
|---|---|---|---|
| T2 (On-target) | GACCCCCTCCACCCCGCCTCCGG | 50.2 ± 4.9 | 25.4 ± 4.8 |
| OT2-1 | GACCCCCCCCACCCCGCCCCCGG | 14.4 ±3.4 | 4.2 ± 0.2 |
| OT2-2 | GGGCCCCTCCACCCCGCCTCTGG | 20.0 ± 6.2 | 9.8 ± 1.1 |
| OT2-6 | CTACCCCTCCACCCCGCCTCCGG | 8.2 ± 1.4 | 6.0 ± 0.5 |
| OT2-9 | GCCCCCACCCACCCCGCCTCTGG | 50.7 ± 5.6 | 16.4 ± 2.1 |
| OT2-15 | TACCCCCCACACCCCGCCTCTGG | 9.7 ± 4.5 | 2.1 ± 0.0 |
| OT2-17 | ACACCCCCCCACCCCGCCTCAGG | 14.0 ± 2.8 | 7.1 ± 0.0 |
| OT2-19 | ATTCCCCCCCACCCCGCCTCAGG | 17.0 ± 3.3 | 9.2 ± 0.4 |
| OT2-20 | CCCCACCCCCACCCCGCCTCAGG | 6.1 ± 1.3 | N.D. |
| OT2-23 | CGCCCTCCCCACCCCGCCTCCGG | 44.4 ± 6.7 | 35.1 ± 1.8 |
| OT2-24 | CTCCCCACCCACCCCGCCTCAGG | 62.8 ± 5.0 | 44.1 ± 4.5 |
| OT2-29 | TGCCCCTCCCACCCCGCCTCTGG | 13.8 ± 5.2 | 5.0 ± 0.2 |
| OT2-34 | AGGCCCCCACACCCCGCCTCAGG | 2.8 ± 1.5 | N.D. |
OT = Off-target sites, numbered as in Table 1 and Supplementary Table 2. Mismatches from the on-target site (within the 20 bp region to which the gRNA hybridizes) are highlighted as bold, underlined text. N.D. = none detected
Our finding that significant off-target mutagenesis can be induced by RGENs in three different human cell types has important implications for broader use of this genome-editing platform. For research applications, the potentially confounding effects of high frequency off-target mutations will need to be considered, particularly for experiments involving either cultured cells or organisms with slow generation times for which the outcrossing of undesired alterations would be challenging. One way to control for such effects might be to utilize multiple RGENs targeted to different DNA sequences to induce the same genomic alteration as off-target effects are not random but instead related to the targeted site. However, for therapeutic applications, our findings clearly indicate that the specificities of RGENs will need to be carefully defined and/or improved if these nucleases are to be used safely in the longer term for treatment of human diseases.
Online Methods
Construction of guide RNAs
DNA oligonucleotides (Supplementary Table 1) harboring variable 20 nt sequences for Cas9 targeting were annealed to generate short double-strand DNA fragments with 4 bp overhangs compatible with ligation into BsmBI-digested plasmid pMLM3636. Cloning of these annealed oligonucleotides generates plasmids encoding a chimeric +103 single-chain guide RNA with 20 variable 5’ nucleotides under expression of a U6 promoter9, 11. pMLM3636 and the expression plasmid pJDS246 (encoding a codon optimized version of Cas9) used in this study are both available through the non-profit plasmid distribution service Addgene (http://www.addgene.org/crispr-cas).
EGFP Activity Assays
U2OS.EGFP cells harboring a single integrated copy of an EGFP-PEST fusion gene were cultured as previously described15. For transfections, 200,000 cells were Nucleofected with the indicated amounts of sgRNA expression plasmid and pJDS246 together with 30 ng of a Td-tomato-encoding plasmid using the SE Cell Line 4D-Nucleofector™ X Kit (Lonza) according to the manufacturer’s protocol. Cells were analyzed 2 days post-transfection using a BD LSRII flow cytometer. Transfections for optimizing gRNA/Cas9 plasmid concentration were performed in triplicate and all other transfections were performed in duplicate.
PCR amplification and sequence verification of endogenous human genomic sites
PCR reactions were performed using Phusion Hot Start II high-fidelity DNA polymerase (NEB) with PCR primers and conditions listed in Supplementary Table 2. Most loci amplified successfully using touchdown PCR (98 °C, 10 s; 72–62 °C, −1 °C/cycle, 15 s; 72 °C, 30 s]10 cycles, [98 °C, 10 s; 62 °C, 15 s; 72 °C, 30 s]25 cycles). PCR for the remaining targets were performed with 35 cycles at a constant annealing temperature of 68 °C or 72 °C and 3% DMSO or 1M betaine, if necessary. PCR products were analyzed on a QIAXCEL capillary electrophoresis system to verify both size and purity. Validated products were treated with ExoSap-IT (Affymetrix) and sequenced by the Sanger method (MGH DNA Sequencing Core) to verify each target site.
Determination of RGEN-induced on- and off-target mutation frequencies in human cells
For U2OS.EGFP and K562 cells, 2 × 105 cells were transfected with 250 ng of sgRNA expression plasmid or an empty U6 promoter plasmid (for negative controls), 750 ng of Cas9 expression plasmid, and 30 ng of td-Tomato expression plasmid using the 4D Nucleofector System according to the manufacturer’s instructions (Lonza). For HEK293 cells, 1.65 × 105 cells were transfected with 125 ng of sgRNA expression plasmid or an empty U6 promoter plasmid (for the negative control), 375 ng of Cas9 expression plasmid, and 30 ng of a td-Tomato expression plasmid using Lipofectamine LTX reagent according to the manufacturer’s instructions (Life Technologies). Genomic DNA was harvested from transfected U2OS.EGFP, HEK293, or K562 cells using the QIAamp DNA Blood Mini Kit (QIAGEN), according to the manufacturer’s instructions. To generate enough genomic DNA to amplify the off-target candidate sites, DNA from three Nucleofections (for U2OS.EGFP cells), two Nucleofections (for K562 cells), or two Lipofectamine LTX transfections was pooled together before performing T7EI. This was done twice for each condition tested, thereby generating duplicate pools of genomic DNA representing a total of four or six individual transfections. PCR was then performed using these genomic DNAs as templates as described above and purified using Ampure XP beads (Agencourt) according to the manufacturer’s instructions. T7EI assays were performed as previously described15.
DNA sequencing of NHEJ-mediated indel mutations
Purified PCR products used for the T7EI assay were cloned into Zero Blunt TOPO vector (Life Technologies) and plasmid DNAs were isolated using an alkaline lysis miniprep method by the MGH DNA Automation Core. Plasmids were sequenced using an M13 forward primer (5’ – GTAAAACGACGGCCAG – 3’) by the Sanger method (MGH DNA Sequencing Core).
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health (NIH) Director’s Pioneer Award DP1 GM105378, NIH R01 GM088040, NIH P50 HG005550, Defense Advanced Research Projects Agency (DARPA) W911NF-11-2-0056, and the Jim and Ann Orr Massachusetts General Hospital Research Scholar Award. We thank Shengdar Q. Tsai for helpful discussions and encouragement.
Footnotes
Author Contributions
Y.F., J.D.S., and J.K.J. designed experiments; Y.F., J.A.F., C.K., M.L.M, D.R., and J.D.S. performed experiments; Y.F., M.L.M., D.R., J.D.S., and J.K.J. wrote the manuscript.
Competing Financial Interests
J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 3.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, et al. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen B, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013 doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dicarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 13.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotech. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 19.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto N, et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


