Abstract
Combined QM/MM molecular dynamics simulations reveal that the M20 loop conformational dynamics of dihydrofolate reductase (DHFR) is severely restricted at the transition state of the hydride transfer as a result of the M42W/G121V double mutation. Consequently, the double mutant enzyme has a reduced entropy of activation, i.e., increased entropic barrier, and altered temperature dependence of kinetic isotope effects in comparison with wild-type DHFR. Interestingly, both in the wild-type DHFR and the double mutant, the average donor-acceptor distances are essentially the same in the Michaelis complex state (about 3.5 Å) and the transition state (2.7 Å). It was found that an additional hydrogen bond is formed to stabilize the M20 loop in the closed conformation in the M42W/G121V double mutant. The computational results reflect a similar aim designed to knock out precisely the dynamic flexibility of the M20 loop in a different double mutant, N23PP/S148A.
I. Introduction
Protein dynamics are essential to the catalytic function of enzymes,(1-3) which range from femtosecond promoting vibrations and picosecond-nanosecond local fluctuations to millisecond and second conformational motions. The question, however, is the precise mechanism by which large-scale protein motions and fast, local dynamic fluctuations are connected to the chemical step to lower the free energy barrier.(4-5) Mutations distant from the active site, which do not show noticeable structural variations in enzyme-inhibitor complexes, provided indirect evidence, suggesting that altered protein motions may affect the reaction rate and transition state stabilization.(6-10) The effects are reflected in the observed kinetic data, including kinetic isotope effects (KIEs) and activation parameters.(11) Recently, the role of conformational dynamics on the chemical step of E. coli dihydrofolate reductase (DHFR) was directly probed experimentally by mutations designed to block dynamic fluctuations of the M20 loop,(12)Loveridge, 2012 #11221} which has previously been shown to be critical to the enzymatic function.(6) It was concluded that the decreased rate of hydride transfer in the “dynamic knockout” mutants is due to impaired flexibility on the millisecond time scale,(12) but a different conclusion was drawn on the same system from measurements of KIEs.(13)
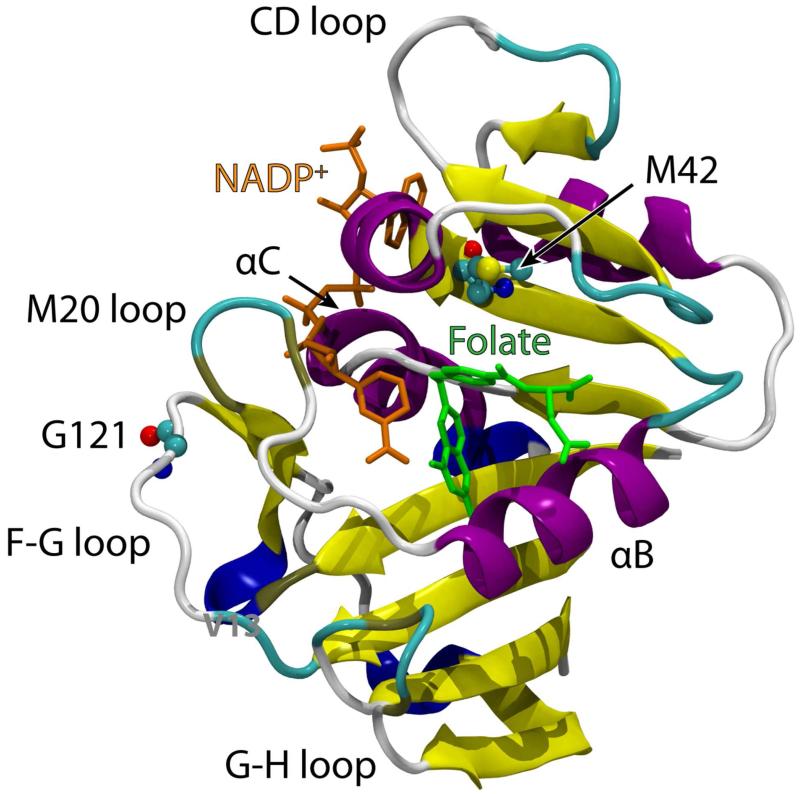
In this Current Topic article, we focus on a specific study of the hydride transfer in E. coli dihydrofolate reductase (DHFR) and the M42W/G121V double mutant (Figure 1) to illustrate the procedure and analyses used in computer simulations to understand the role of protein conformational dynamics in the catalyzed reaction. The methods used and the principles emerged from this specific investigation are likely to be generally applicable to other enzymes. DHFR is a relatively small, flexible protein that catalyzes the nicotinamide adenine dinucleotide phosphate (NADPH)-dependent reduction of 7,8-dihydrofolate to 5,6,7,8-tetrahydrofolate.(6, 14-15) The catalytic mechanism of DHFR has been extensively studied, although we cannot afford to provide a thorough review of the large body of literature. NMR relaxation experiments established that conformational changes of flexible loops spread out of the protein play a critical role in modulating ligand specificity and catalytic turnover.(6, 12, 15-17) Furthermore, single and double mutations at the M42 and G121 sites as well as other locations that are remote from the catalytic center can nevertheless produce profound, nonadditive effects on reaction rate, activation parameters and the temperature dependence of KIEs.(13, 18-26) Computational studies helped identify networks of interatomic distance variations along the reaction coordinate, and distal mutations can induce structural perturbations that alter these conformational change.(24-25, 27-30) Other studies have probed various aspects of DHFR catalysis.(31-37) On the other hand, it has been argued that all these effects can be attributed to changes in reorganization energy.(26) We show that restriction in conformational flexibility of the M20 loop at the transition state in the M42W/G121V double mutant is responsible for the impaired catalytic activity by increasing the entropic barrier.
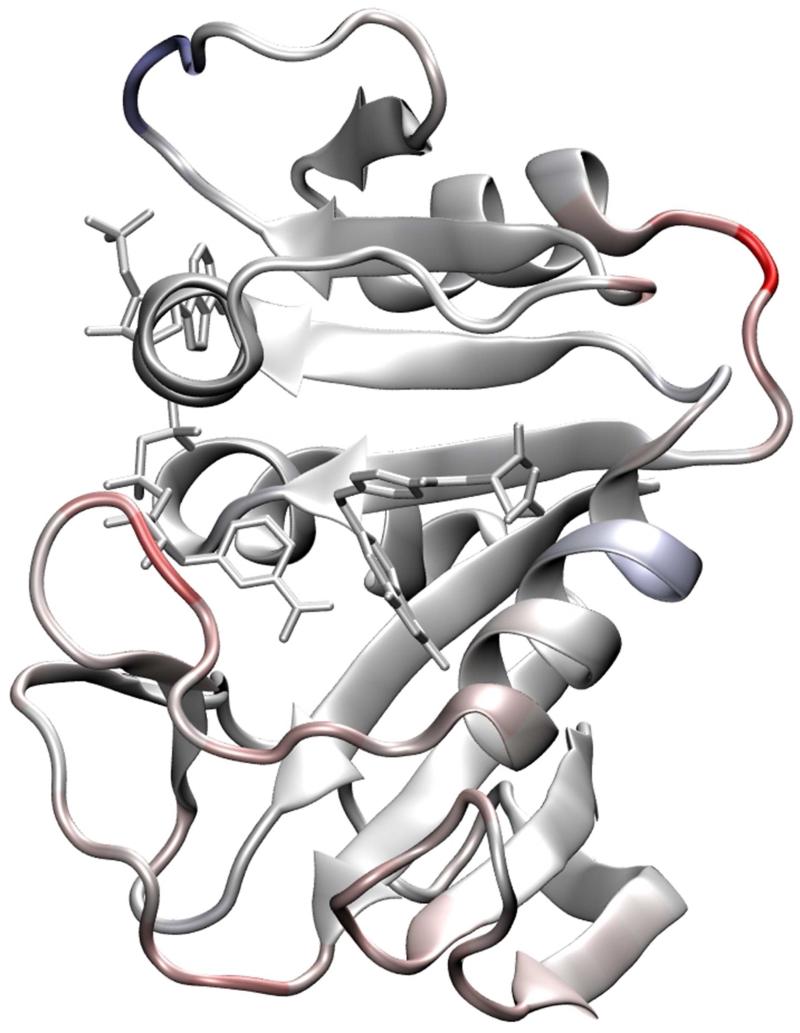
Figure 1.
X-ray structure of the ternary complex between E. coli dihydrofolate reductase, folate substrate and NADP+ cofactor (PDB code: 1RX2). Secondary structure and loop nomenclature are indicated.
Before we begin, it is necessary to make clear the meaning of the term dynamics used here since the language itself is not without controversy.(1) Roughly, interpretations of dynamics in enzyme catalysis may be grouped into three categories: (i) transmission coefficient in rate theory, (ii) coherent protein motions with transition state, and (iii) allosteric conformational change. One may argue that there is a fourth category – that there is no dynamic effect in enzyme catalysis – but it does not provide insight into enzyme mechanism and shall not be discuss further. In the first category, some researchers argue that the free energy of activation is an equilibrium quantity, which can be computed using either molecular dynamics or Monte Carlo simulations, and thereby any dynamic effects must be in the transmission coefficient in rate theory. However, the transmission coefficient and the free energy of activation are interrelated, both depending on the reaction coordinate, which can be chosen to yield a unity transmission coefficient.(1, 38) Thus, in this framework, “dynamics” lose meaning, except in nonequilibrium processes which are not known to play a significant role in enzyme catalyzed reactions.(39) For the second proposal, it has been pointed out that there is no theoretical or experimental evidence supporting coherent energy transfer from protein motions to the transition state.(40) In this article, we adopt the third definition, which is used by most enzymologists,(20, 41) to understand the specific role of protein conformational changes induced by the chemical transformation. These conformational changes, which may involve small shifts in the interatomic distances discussed by Benkovic and Hammes-Schiffer(24-25, 29) or open-closed conformational fluctuations of loops in or remote from the active site(6, 12, 16-17, 28, 42) or even without changes in the mean backbone conformation,(43) are allosteric transitions accompanying the chemical step,(10) and thus, are thermally averaged over the fast, local atomic fluctuations which may be important to barrier crossing at the transition state.(1)
In Section II, we first present computational results on activation parameters and temperature-dependent KIEs. In this section, we establish that the impaired catalytic function in the mutant M42W/G121V is due to reduced protein flexibility at the transition state relative to that in the wild-type DHFR. This accounts for the finding that the increased free energy barrier results predominantly from reduction in entropy of activation, i.e., increased entropic barrier in the double mutant.(13, 20) Then, in Section III, we identify a mechanism and show that the altered protein flexibility is related to restriction in loop conformational motions, mainly in the M20 and αE-F loop regions. The protein “dynamic knockout” in the M42W/G121V mutant can be attributed to increased steric congestion at the mutation sites, particularly in the M42W mutation, resulting in an extra hydrogen bond to stabilize the closed conformation of the M20 loop. In section IV, we make concluding remarks, and Section V provides a summary of a novel path integral free energy simulation method for computing kinetic isotope effects in enzymes along with computational details.
II. Reduced protein flexibility at the transition state is responsible for impaired catalytic effects in the M42W/G121 double mutant
The change in reaction rate due to amino acid mutations, particularly those remote from the active site, is often smaller than the enormous rate enhancement by the enzyme relative to the uncatalyzed process in water,(44) but an understanding of the precise role of the altered protein structure in catalysis can be much more informative than the rate change itself. Thus, comparative studies of the wild-type enzyme and its mutants can be very useful for helping identify a connection or absence of protein conformational dynamics with the catalyzed chemical step.(24-26, 28) The main challenge is to evaluate accurately the small free energy changes induced by distal mutations to allow for analyses of structural and dynamic effects. This requires an accurate description of the potential energy surface for the chemical reaction,(45-46) and an extensive sampling to yield the potential of mean force as well as nuclear quantum effects (NQEs).(38, 47) In the next section, we present detailed analyses of the dynamic trajectories, pointing to a role of reduced dynamic flexibility at the transition state in the M42W/G121V mutant relative to wild-type DHFR, and the restriction of a closed-open conformational transition of the M20 loop is responsible for the reduced dynamic fluctuation and increased entropy barrier in the chemical step.
A. Differences in catalytic effect
The starting point in computational study of enzyme catalysis is to determine the free energy of activation of the chemical step, and the relatively small changes in barrier height due to mutations.(39, 46) This requires an accurate potential energy surface to describe the chemical transformation, incorporating explicitly polarization effects in response to the dynamic fluctuations of the protein environment. We have developed a combined QM/MM approach, in which the electronic structure of the hydride transfer is explicitly modeled by electronic structure theory.(39, 45, 48-50) In the present case, the semiempirical AM1 Hamiltonian is used, along with a simple valence bond correction term specifically designed for the DHFR system,(30) and it has been used in a number of subsequently studies of DHFR.(39, 49, 51-52) We note that recently Major and coworkers reported a new parameterized AM1 model for the DHFR hydride transfer.(34-36)
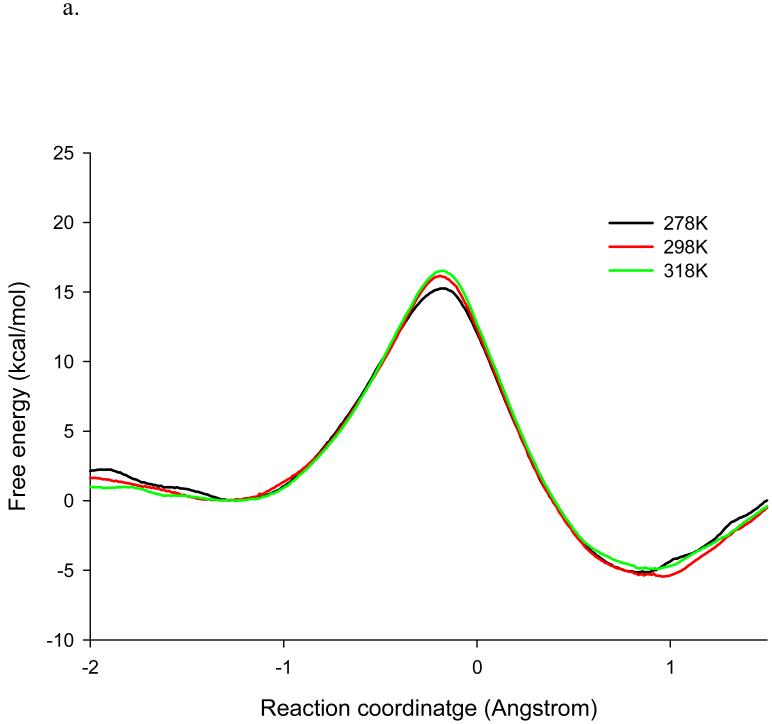
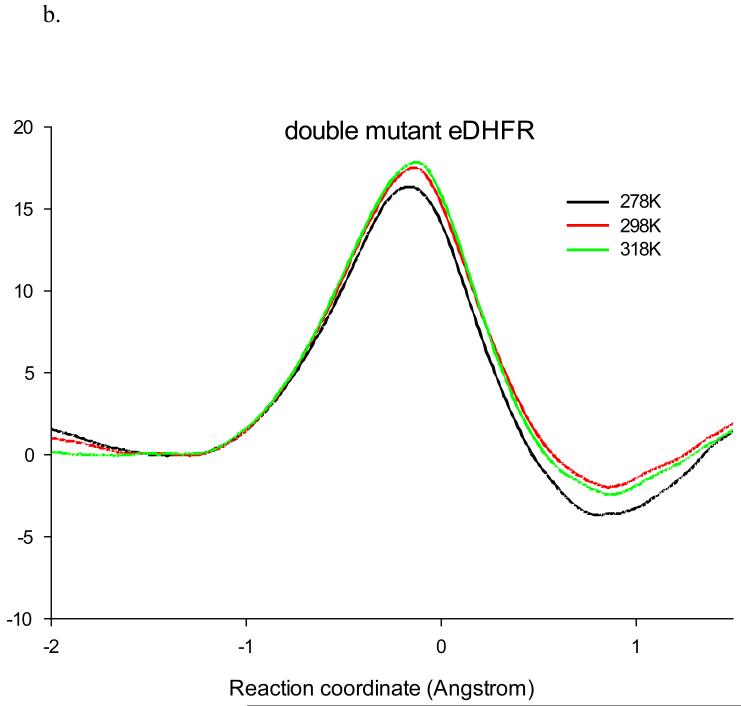
Using the same QM/MM methodology as in our earlier studies,(39, 49, 51-52) we obtained the potentials of mean force for the hydride transfer from NADPH to DHF catalyzed by wt-DHFR and the M42W/G121V dm-DHFR at 5 °C, 25 °C, and 45 °C (Figure 2). In both cases, the free energy of activation, ΔG≠, increases with temperature. At 25 °C, the computed ΔG≠ for the hydride transfer is 16.4 kcal/mol for the wt-DHFR, which may be compared with the experimental value of 16.7 kcal/mol determined at pH = 9, under which the hydride transfer step is rate limiting.(20) The activation free energy is increased by 1.2 kcal/mol in the M42W/G121V mutant, which is smaller than the corresponding experimental data (2.3 kcal/mol). Importantly, the experimental trends due to mutation are correctly reproduced. Inclusion of NQEs (see below) reduces the computed barrier by about 3 kcal/mol both for the wt- and dm-DHFR at 25 °C,(30, 52) and the trend is not changed. The estimated free energies of reaction, ΔGrxn, are about −5 and −2 kcal/mol for the wt- and dm-DHFR, respectively, reflecting that there is a small change in driving force for the hydride transfer due to the M42W/G121V mutations. This is in accord with experimental data on the M42W/G121S mutant.(18, 28) A similar effect of raising the driving force and a correlation between free energy barrier and ΔGrxn for a number of single and double mutant DHFR enzymes have been noted.(26)
Figure 2.
Computed classical mechanical potential of mean force for the hydride transfer in wild-type DHFR (a) and in the double mutant M42W/G121V at 5 °C, 25 °C, and 45 °C. The reaction coordinate is defined as the difference in the distance of the transferring hydrogen (H4) from the donor carbon (C4N) of the cofactor and the acceptor carbon (C6) of the substrate.
B. Enthalpy and Entropy of Activation
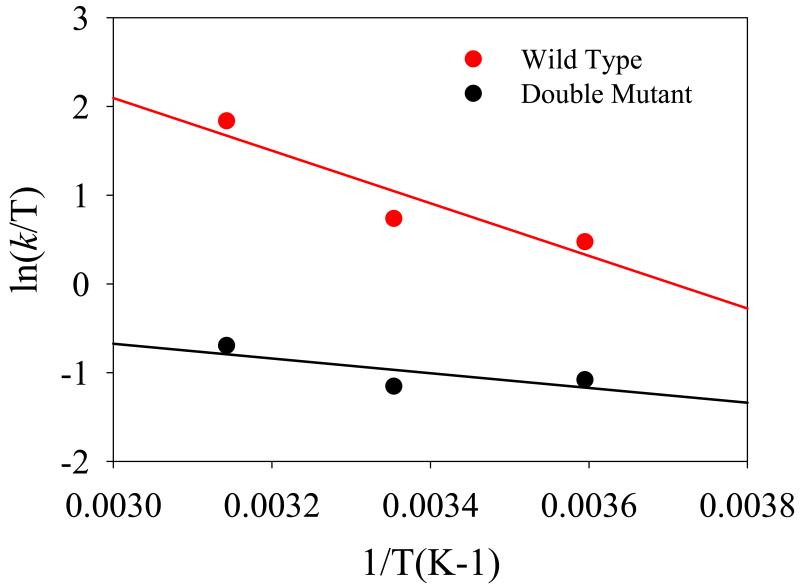
The computed free energies of activation ( ΔG≠) for the hydride transfer in wt- and in M42W/G121V dm-DHFR can be decomposed into enthalpy ( ΔH≠) and entropy ( ΔS≠) components using Eyring plot ( ln(k /T) vs. 1/T) and transition state theory (Figure 3).(20, 26, 53) The slope ( −ΔH≠ / R ) yields the enthalpy of activation and the intercept ( ln(kB / h) + ΔS≠ / R ) is converted to entropic contributions. This perhaps provides the most reliable estimate of entropy of activation for an enzymatic reaction since direct computation of entropies at the reactant state and transition state would have much larger uncertainties than the entropy change itself,(54) whereas free energy simulations converge much more rapidly. The activation parameters provide important insights into the thermodynamic stabilization of the transition state (via ΔH≠), and the contributions from dynamic fluctuations of the protein environment and the substrate that are transformed from the Michaelis complex to the transition state (through ΔS≠).
Figure 3.
Eyring plots for the hydride transfer in wild-type DHFR (red) and in M42W/G121V mutant (black). Rate constants are determined using quantum transition state theory in which nuclear quantum effects are included in the estimated free energy barrier.
The Eyring plots of the rate constants determined using the computed free energies of activation with the inclusion of NQEs are shown in Figure 3 for the wt-DHFR and dm-DHFR, and the activation parameters are listed in Table 1. The corresponding experimental results at pH 9 are also listed for comparison. The results in Table 1 and Figure 3 are in agreement with the experimental data,(20) from which several conclusions can be drawn immediately. First, not surprisingly, there is a major compensating effect between enthalpy and entropy components in the activation of the hydride transfer, both in wt-DHFR and in the M42W/G121V double mutant. Of the two systems, a larger enthalpic barrier (suggesting smaller stabilizing interactions at the transition state) is accompanied by a smaller negative entropy of activation, i.e., greater dynamic flexibility since reduction in negative ΔS≠ reflects enhanced flexibility at the transition state over the reactant state. Secondly, the enthalpy of activation is reduced by −4.3 kcal/mol in the double mutant relative to the wt-DHFR from computation, which may be compared with the experimental value of −1.7 kcal/mol.(20) Thus, counter-intuitively, the double mutant, in fact, has a greater enthalpic stabilization of the transition state (thus reducing ΔH≠) than that of the wild-type enzyme. Thirdly, the computed and experimental entropies of activation are negative for the hydride transfer both in the wild-type enzyme and in the M42W/G121V double mutant. Significantly, the entropy reduction becomes even more pronounced at the transition state as a result of the double mutation. In particular, the computed change in entropy of activation, ΔΔS≠, is −18.2 e.u. (cal mol−1 K−1), while the corresponding experimental value is −13.2 e.u.;(20) these translate to free energies of 5.4 and 3.9 kcal/mol at 25 °C, respectively. The implication of these results both from experiment and computation is that there is net reduction of disorder, or dynamic flexibility, in going from the Michaelis complex to the transition state in the hydride transfer, and that the reduction in dynamic flexibility is greater in the M42W/G121V double mutant than the change in the wild-type enzyme. A similar reduction in ΔΔS≠ has been noted in the G121V single mutant(19) and the N23PP/S148A double mutant.(12-13)
Table 1.
Computed (including nuclear quantum effects) and experimental activation parameters at 25 °C for the hydride transfer in wild-type (WT) dihydrofolate reductase and the M42W/G121V double mutant (DM).
| Computation | Experiment | |||||
|---|---|---|---|---|---|---|
| WT | DM | diff. | WT | DM | diff. | |
| ΔH‡ (kcal/mol) | 5.9 | 1.6 | −4.3 | 5.0±0.6 | 3.3±0.18 | −1.7 |
| ΔS‡ (cal/K·mol) | −25.4 | −43.6 | −18.2 | −39.6±1.0 | −52.8±1.3 | −13.2 |
| ΔG‡ (kcal/mol) | 13.5 | 14.7 | 1.2 | 16.7±0.8 | 19.0±0.4 | 2.3 |
The total entropy of activation for the hydride transfer in DHFR has several components (see discussions in Ref(55-56)), including the entropy change of the intrinsic reaction coordinate of the substrate and cofactor (), the change of protein () and solvent () fluctuations and the change in translational and rotational entropy () of the enzyme complex as the substrate is transformed from the reactant to the transition state:
| (1) |
The effect of single and double amino acid mutations on the overall translational and rotational entropy of the enzyme complex as well as on the surrounding solvent is expected to be negligible.(55-56) Consequently, the net change in ΔS≠ due to amino acid mutation has two major contributions:
| (2) |
where and are the (total) entropy of activation for the wild-type DHFR and the M42W/G121V double mutant, and and are variations of the intrinsic reactant and the protein environment.
Since the hydride transfer from NADPH to DHF brings two relatively large ligands together in covalent contact at the transition state, the first term in eq 1 is negative due to loss of translational motions and the asymmetric mode associated with the reaction coordinate; they are closely related to the hydride acceptor and donor length (ADL). For the hydride transfer in wt-DHFR and the M42W/G121V double mutant, we found that the average ADLs are 3.50 Å and 3.46 Å at the Michaelis complex, respectively, and they are changed to 2.71 Å and 2.70 Å at the transition state. As far as acceptor-donor distance is concerned, there is no real difference in preorganizing the substrate and cofactor in the active site and in reaching the transition state as a result of the M42W/G121V double mutation, indicating that the net change in is close to zero. Consequently, the large reduction in activation entropy estimated from molecular dynamics QM/MM simulations and determined experimentally (Table 1) may be attributed entirely to reduced protein flexibility at the transition state in the double mutant. Other studies have found wide-spread changes in the overall flexibility and coupled motions in mutant DHFR enzymes.(24-25, 28)
| (3) |
For comparison, an early study showed that the ADL is about 3.45 Å at the Michaelis complex in the wild-type DHFR,(26) nearly the same as the present result. However, it was found that this distance was stretched by half angstroms to about 4 Å in the M42W/G121V double mutant, and the distorted substrate-cofactor conformation in the Michaelis complex was found to increase the work term and thus the activation barrier by 1.8 kcal/mol relative to wt-DHFR.(26) Increased donor-acceptor distance was also proposed by Kohen and coworkers for the M42W/G121V mutant.(20) We do not observe this elongated ADL in the double mutation. If anything, there seems to be a slight compression both in the Michaelis complex and in the transition state since both mutations introduce steric congestions propagated to the active site,(21-22) especially in the M42W mutation site(28) (see below).
Analyses in the next section demonstrate that the reduced dynamic flexibility at the transition state of the hydride transfer in the M42W/G121V double mutant relative to wt-DHFR is directly connected to the altered M20 loop motions in the active site.
C. Temperature Dependence of Kinetic Isotope Effects
The altered protein motions due to amino acid mutations in the M42W/G121V enzyme are reflected in the observed KIEs. Wild-type E. coli DHFR exhibits a, now, general feature of temperature-independent KIEs for the hydride transfer step.(57) Intriguingly, the KIEs for the hydride transfer in M42W/G121V dm-DHFR become steeply temperature-dependent.(20) In fact, a large number of enzymes have been found to show temperature independence of KIEs in their optimal operating temperature range, but the KIEs are changed to temperature-dependent outside this temperature range, or as a result of mutations.(4, 11) The change in temperature dependence of KIEs provides a direct probe of variations in transition structure and its coupling to the enzyme environment.(13) Several studies have proposed that the wild-type enzyme has evolved to optimize the average donor-acceptor distance for tunneling at the transition state sampled by the mass-independent thermal activation, giving rise to temperature-independent KIEs (provided that tunneling is the dominant contribution to the observed KIEs).(4, 11, 20, 58-59) However, ADLs are altered in mutant enzymes. Consequently, thermally activated “gating fluctuations” are required for effective tunneling in mutant enzymes such as in the present M42W/G121V double mutant, leading to temperature dependence of KIEs.(20)
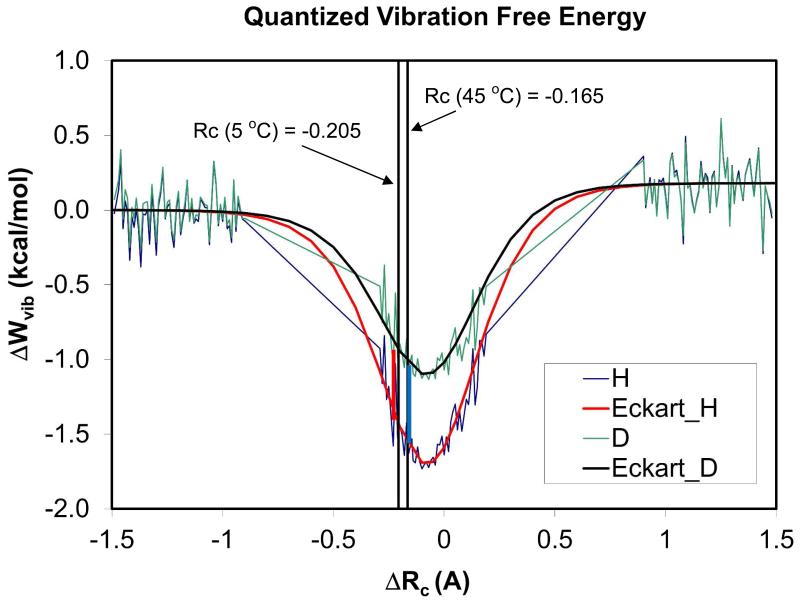
Conceptually, there are two major contributions to the overall NQEs, responsible for the observed KIEs: the change in quantum vibrational free energy, predominantly zero-point effects, and tunneling.(38, 49, 52, 60) For reactions where tunneling is dominant, such as the hydrogen atom transfer catalyzed by soybean lipoxygenase,(61) the tunneling mechanism above is quite reasonable. For the hydride transfer catalyzed by DHFR where the observed intrinsic primary KIEs are only about 3 both in the wt-DHFR and in mutants,(13, 20, 57) it would be important for a mechanism to also account for the contribution and change from vibrational free energy. Previously, our group performed microscopic simulations of the small temperature dependence of the KIEs for the hydride transfer in wt-DHFR; in our approach, the dynamic simulations always include ADL thermal fluctuations automatically.(52) Using ensemble averaged-transition state theory including semiclassical multidimensional tunneling (EA-VTST/MT),(60, 62) we identified two general features responsible for the observed temperature independence (or small temperature dependence) of KIEs in wt-DHFR; (a) variation of the transition-state position, and (b) temperature dependence of the effective potential for tunneling. In particular, we found that the location of the transition state for the hydride transfer coordinate is slightly shifted from −0.205 Å at 5 °C to −0.165 Å at 45 °C (Figure 4).(52) As a result, the difference in vibrational free energy between H-transfer and D-transfer is also increased at higher temperature, resulting in a nearly temperature invariant Boltzmann factor. In this sense, there are greater NQEs at higher temperatures. It is interesting to note that the difference in vibrational free energy is predominantly responsible for the computed H/D KIEs, with a net value of 2.83 and 2.81 (without tunneling) at 5 °C and 45 °C; inclusion of tunneling increases the overall KIEs to 3.22 and 3.01, which is a small factor of only 14%.(52) Subsequently, other studies have also investigated the temperature dependence of KIEs in DHFR and mutants.(63)
Figure 4.
Computed average vibrational free energies for H-transfer (blue) and D-transfer (green) in wild-type DHFR. The results are fitted to an inverse Eckart function, and the location of the transition state at 5 °C and 45 °C, determined after the vibrational free energies are included in the classical mechanical potential of mean force, are indicated by the vertical lines. The Boltzmann factor of between H- and D-transfers gives the vibrational free energy contribution (dominantly zero-point effects) to the overall KIE.
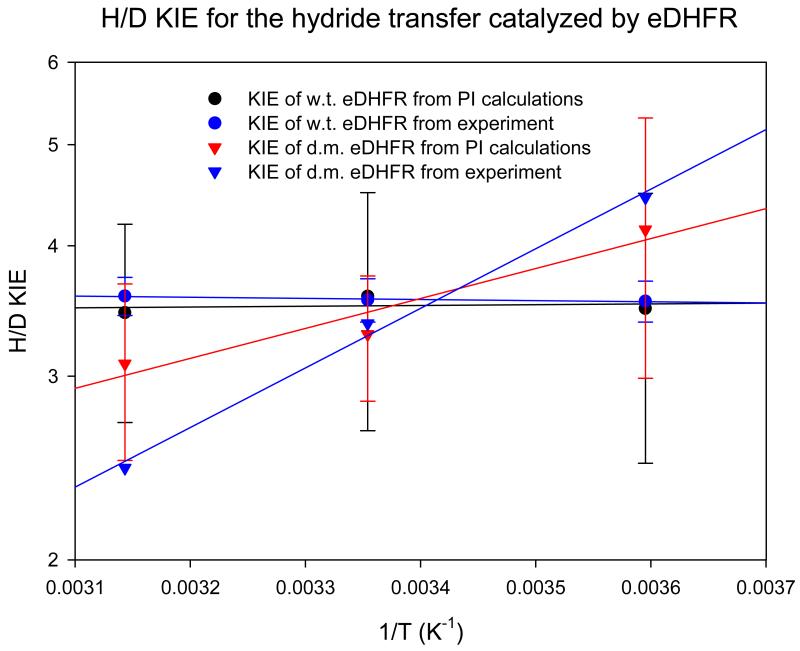
We further examined the temperature dependence of the KIEs for the hydride transfer both in wt-DHFR and the M42W/G121 dm-DHFR using Feynman path integral simulations along with free energy perturbation on isotopic masses (which yields the necessary computational accuracy) and umbrella sampling over the reaction coordinate (which incorporates protein dynamic fluctuations) – the method is called PI-FEP/UM.(64-65) Unlike the previous EA-VTST approach,(62) vibrational free energy and nuclear tunneling are not separable in path integral simulations, but the total NQEs as well as KIEs are obtained directly from statistical simulations. Nuclear quantum effects lower the free energy barrier from classical molecular dynamics simulations by about 2.8±0.5 kcal/mol both for the wt-DHFR and the dm-DHFR at 25 °C, in good accord with previous results (3.2 kcal/mol) computed using a completely different theoretical approach,(30, 52) namely the EA-VTST/MT method. The present computed NQEs are somewhat greater than that estimated by other methods (~2 kcal/mol) in which only the hydride atom was quantized.(66-67) Arrhenius plots of the calculated and experimental H/D KIEs are shown in Figure 5. Good accord was obtained for the wild-type DHFR both in the absolute value of KIEs, and in the temperature independent behavior. The agreement for the M42W/G121V double mutant is also good, although the slope of the temperature dependence plot is smaller than that measured experimentally. Since the average ADLs are not different in the Michaelis complex and in the transition state between wt-DHFR and the M42W/G121V mutant, suggesting similar zero-point effects, we attribute the change is temperature dependence of the KIEs in the double mutant to altered potential energy surface for tunneling. The results shown in Figure 5 demonstrate that the absolute values and the trends in kinetic parameters as illustrated by the KIEs for the hydride transfer, which directly report the structure and environment of the transition state, are correct reproduced by the theoretical model.
Figure 5.
Comparison of the Arrhenius plots of experimental and computational intrinsic H/D kinetic isotope effects in wild-type and M42W/G121V mutant DHFR.
III. Reduced protein flexibility at the transition state in M42W/G121V mutant is due to changes in M20 loop conformational dynamics
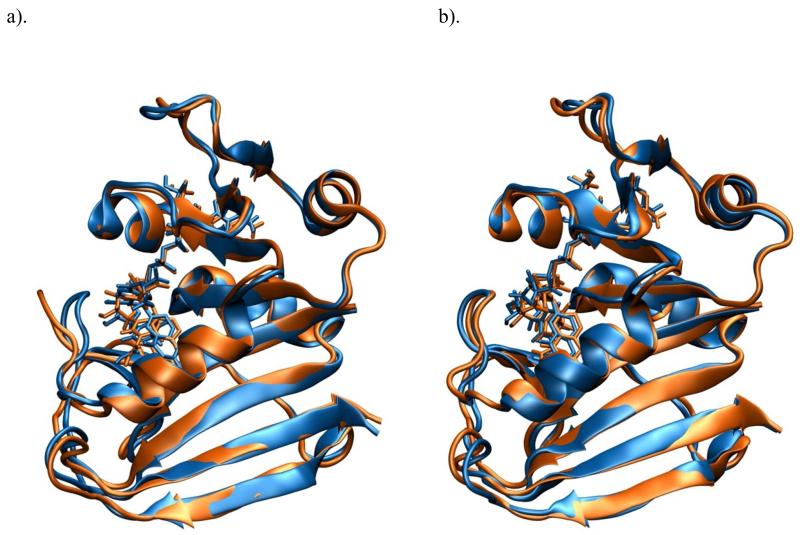
The qualitative structure difference between wild-type DHFR and the M42W/G121V double mutant in catalysis is immediately identified from inspection of the molecular dynamics trajectories along the hydride transfer pathway. In particular, at 25 °C, the M20 loop adopts a closed conformation in the Michaelis complex,(14) both in the wild-type and the double mutant enzymes (Figure 6, structures in blue). However, at the transition state of the hydride transfer, we found that the M20 loop undergoes a conformational transition in the wild-type enzyme, leading to a configuration resembling the open form (Figure 6a, brown) found in DHFR and folate complex (1RD7). Brooks and coworkers have noted that the closed conformation can be converted to the occluded state found in the product complex (1RX7) through the open conformation.(14, 28) The M20 loop remained in the closed conformation throughout the hydride transfer pathway in the M42W/G121V double mutant (Figure 6b).
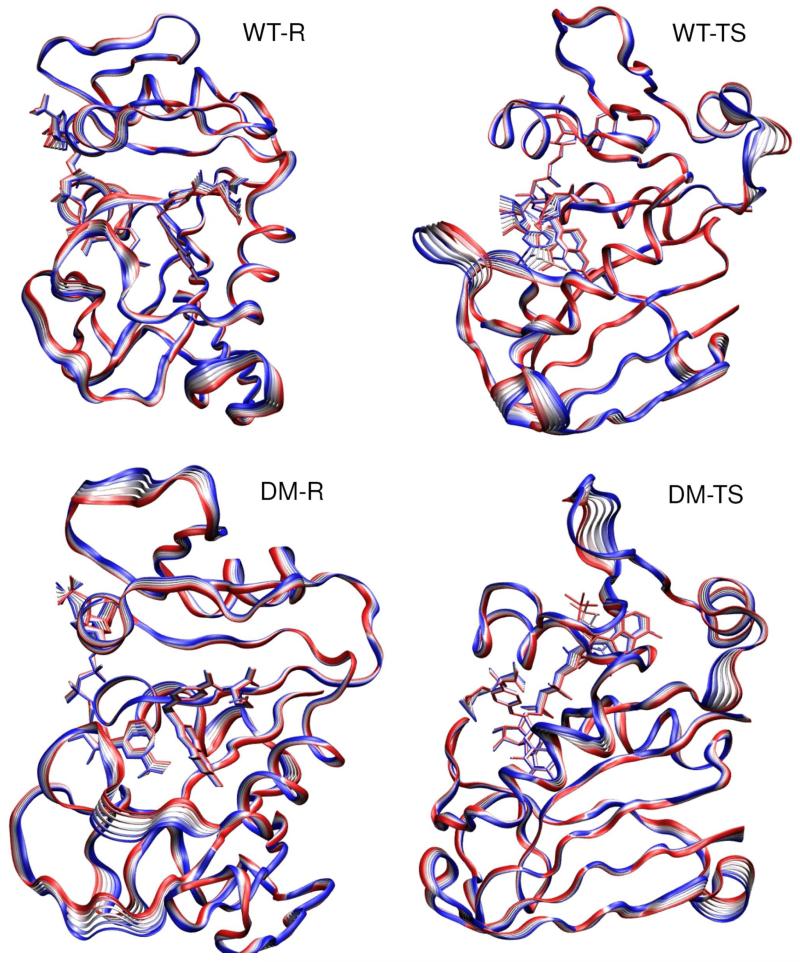
Figure 6.
Comparison of snap shot structures in the reactant state (Michaelis complex) in blue and in the transition state (brown) for the hydride transfer in wild-type DHFR (a) and in the M42W/G121V double mutant (b).
The M20 loop dynamics in the Michaelis complex and in the product complex have been studied by NMR relaxation experiments and it has been implicated to play a critical role in DHFR catalysis.(6, 12) To verify that the observed conformational transition in the transition state region was not due to artifacts of an incidental simulation during the umbrella-sampling free-energy calculations, we repeated these simulations twice in the regions of the reactant state (Michaelis complex) and the transition state, starting from different structures for 2.6 ns. In the Michaelis complex, the M20 loop fluctuates in the closed conformational substate in all cases both for wt-DHFR and dm-DHFR. On the other hand, in both repeats, closed-open conformational transitions of the M20 loop took place in less than100 ps in the wild-type enzyme, and it remained closed in the double mutant. We further compared the trajectories generated from simulations of the wt-DHFR at 5 °C and 45 °C, which showed a greater extent of M20 loop opening at 45 °C after the transition state, whereas the M20 loop was closed along the hydride transfer pathway at 5 °C. On the other hand, the M20 loop conformation was closed throughout the simulations of the M42W/G121V double mutant at all three temperatures. These findings suggest that, qualitatively, at physiologically relevant temperatures, the M20 loop undergoes conformational transitions as the hydride transfer reaction coordinate reaches the transition state region in wt-DHFR; however, this conformational change is quenched in the M42W/G121V double mutant enzyme. The differential M20-loop dynamics is likely to be responsible for the difference in temperature dependence of KIEs.
The coupling between M42 and G121 sites has been extensively studied and its impact on loop dynamic fluctuations of DHFR is well established through mutation, kinetics and NMR experiments.(18-19, 21-22, 68-70) Mauldin et al. showed that the M20 loop adopts the closed conformation in the ternary complex with NADPH and methotrexate both in the M42W and G121V mutants.(21-22) While the G121V mutantion results in a large response on the M42 dynamic fluctuations from NMR relaxation experiments,(21) the M42W mutation showed that the M42 site serves has a hob connecting the dynamic fluctuations throughout the protein.(22) The M20 conformational fluctuations in the wt-DHFR and single and double mutants at M42 and G121 as well as their coupled motions in the Michaelis complex have been examined by Brooks and coworkers.(27-28, 71-72) It was found that there are several pathways between the closed and the open conformations, with different mutants favoring some particular routes. For example, both the G121S and wt DHFRs went through a similar intermediate state to reach an open-like conformation, although they did not move all the way to the open conformation as in the crystal structure 1XD7.(28)
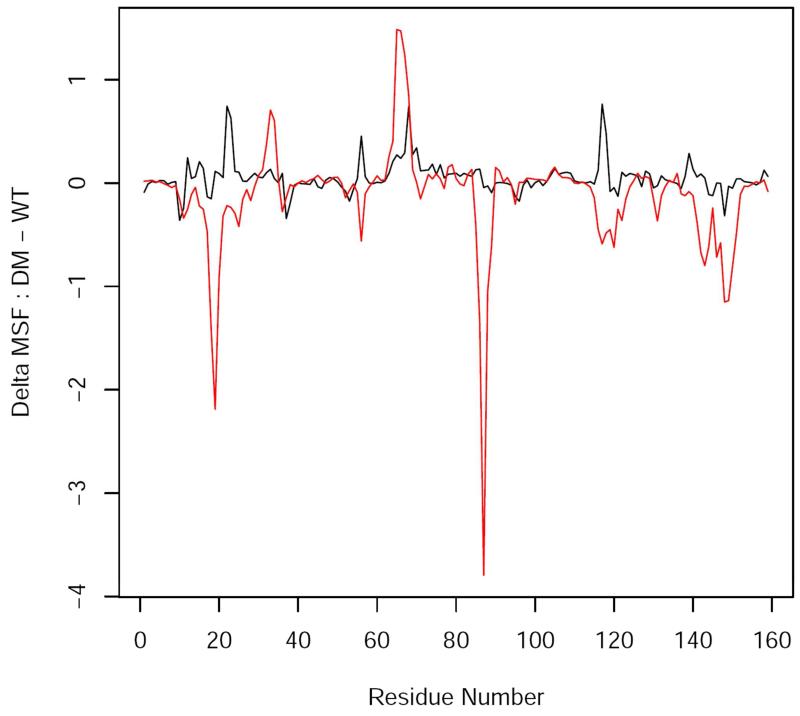
To provide a more quantitative assessment of the conformational fluctuations along the hydride transfer pathway, we computed the mean square fluctuations (MSF) of the backbone Cα atoms for the reactant state (RS) and transition state (TS) both for the wt-DHFR and dm-DHFR, making use of the 2.6 ns trajectories. The changes in MSF from wt-DHFR to the M42W/G121V dm-DHFR at the RS (black) and the TS (red) are displayed in Figure 7. In these plots, negative values correspond to reduced fluctuations in the double mutant (i.e., greater fluctuations in the wild-type enzyme), while positive values indicate enhanced dynamic fluctuations due to the double amino acid mutations. Overall, the dynamic fluctuations are very similar in the Michaelis complex between the wild-type and double mutant DHFR, although several regions exhibit somewhat increased fluctuations in the double mutant. However, significant differences between wt-DHFR and the M42W/G121V double mutant are found in several key regions in the DHFR structure at the transition state region of the hydride transfer (Figures 7 and 8). In particular, the M20 loop (around amino acid 18), and the αE-F loop (at amino acid 87) show significantly reduced fluctuations in the double mutant than in wt-DHFR, and dampened fluctuations are also found in the F-G loop (about residue 119) and the G-H loop in the region of residues 140 through 150. There are two regions in the double mutant that show increased motions relative to the wild-type enzyme at the transition state, although the magnitude is much smaller than reduced motions; they are located at the end of the αB helix and in the C-D loop. Figures 7 and 8 paint a picture that while the M42W/G121V double mutation has relatively small effects on the dynamic fluctuations in the Michaelis complex, it causes major changes in dynamic fluctuations at the transition state in key regions for the hydride transfer, predominantly reduction in nature relative to the wild-type DHFR. We attribute the restricted M20 and E-F loop-conformation dynamics at the transition state in M42W/G121V DHFR to be responsible for the computed decrease in activation entropy. (Note that we have not addressed the difference in loop flexibility of the apo enzymes and its influence on substrate and cofactor binding).
Figure 7.
Difference is mean square fluctuations (MSF) of backbone Cα atoms between wild-type DHFR and the M42W/G121V double mutant, MSF[DM]-MSF[WT], in the Michaelis complex reactant state (black) and in the transition state (red). Negative values indicate regions with greater structural fluctuations in the WT enzyme, whereas positive regions reflect enhanced fluctuations in the DM enzyme.
Figure 8.
Illustration of regional variations between wild-type DHFR and the M42W/G121V double mutant in going from the reactant state (Michaelis complex) to the transition state. This corresponds to the difference of the two MSFs shown in Figure 7. Regions showing reduced fluctuations to reach the transition state in the double mutant relative to that in the WT enzyme are given in red, while enhanced fluctuations are in blue (almost nonexistent except the C-D loop region on the top).
Recently, Bhabha designed a different double mutant, N23PP/S138A, aimed to restrict the M20 loop conformational motions.(12) However, the N23PP/S138A mutant has a relatively minor effect on rate reduction,(12) and it was subsequently found that the temperature dependence of KIEs was not affected compared with the wild-type enzyme.(13) As a result, it was suggested that that double mutation did not achieve the original goal of restricting M20 loop dynamics.(13) Nevertheless, the entropy of activation is reduced by 5.3 e.u. in the N23PP/S138A enzyme,(13) indicating that protein fluctuations are reduced somewhere at the transition state. Here, we found that the impaired catalytic function in the DHFR mutant M42W/G121V is also a result of the M20 loop “dynamic knockout”.
The altered loop conformational dynamics between the wild-type and M42W/G121V double mutant DHFR enzymes can be characterized by low frequency quasiharmonic motions. To highlight this important property and the motions that are suppressed at the TS of the hydride transfer in the double mutant relative to wt-DHFR, we carried out principal component analyses (PCA)(54) by diagonalizing the mean covariant matrices of the mass-weighted interatomic distances for wt-DHFR and dm-DHFR in the RS and the TS. The PCA results show the directionality and frequency of protein dynamic motions, in which the lowest frequency modes are typically correlated with protein conformational changes and have been used to interpret conformational variations observed experimentally.(7, 73) The dynamic motions of the lowest frequency mode (stretched by variations up to 2σ from the mean) for the four states considered are illustrated in Figure 9. The fluctuations resulting from the lowest frequency mode contribute about 25% of the total dynamic motions of the enzyme in each case, whereas the contributions from the next mode are reduced to about 6%. Thus, Figure 9 provides key features of the dynamic motions in each system. Of the four different simulations (wt-RS, wt-TS, dm-RS, and dm-TS) in Figure 9, we immediately notice that the dominant motions of the wt-TS state is characterized by the closed-open conformational transition of the M20 loop. This is different from all other three states where the M20 loop remains in the closed form. The most significant motions at the TS in the M42W/G121V double mutant occur in the C-D loop and αE-F loop regions. The difference in quasiharmonic motions from a single, lowest-frequency mode is in remarkably good accord with the overall MSF in Figure 8, except in the αE-F loop region at the TS, where contributions from other modes are also important in the wild-type enzyme. Importantly, the PCA analyses provide a dynamic mechanism (not promoting modes or motions), connecting quasiharmonic motions of the protein – particularly in the closed-open conformation transition of the M20 loop – with average conformational changes along the hydride transfer coordinate in wt-DHFR.
Figure 9.
Illustration of the dynamic motions of the lowest frequency quasiharmonic mode for the wild-type (WT) and M42W/G121V double mutant (DM) in the reactant state (RS) and the transition state (TS). A sequence of six structures stretched up to 2σ from the mean are superimposed.
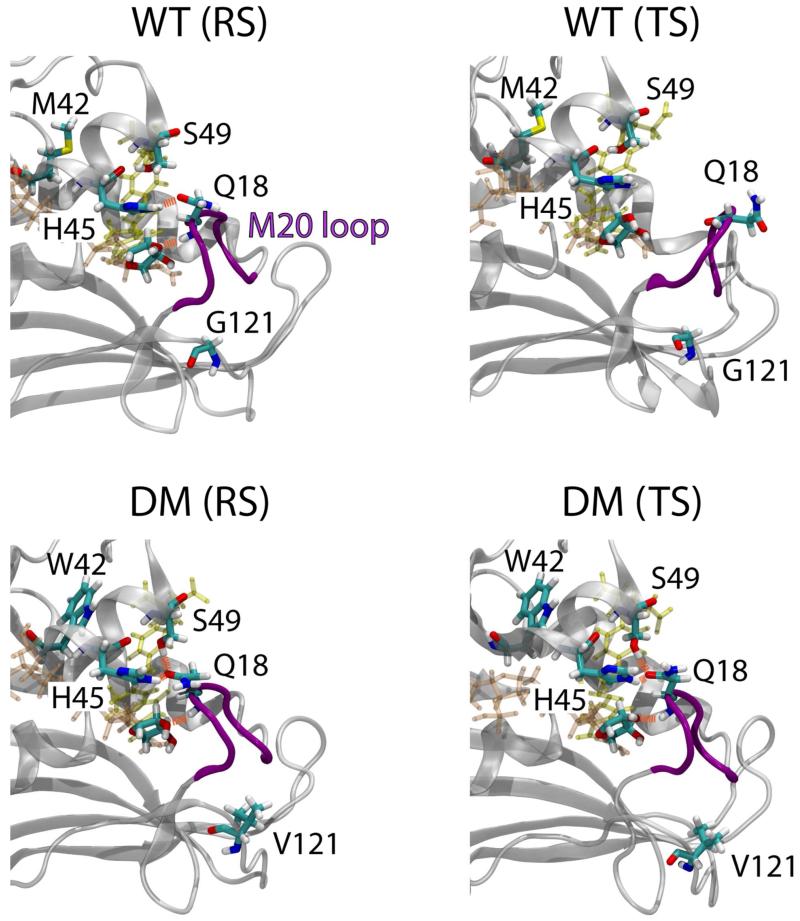
The structural origin responsible for the altered dynamic fluctuations of the enzyme at the transition state due to the M42W/G121V double mutation is illustrated in Figure 10, which depicts key hydrogen bonding interactions that maintain the M20 loop closure. In the reactant state (Michaelis complex) of the wild-type enzyme, Q18 forms two hydrogen bonds, one donating the backbone amide hydrogen to the 3′-hydroxyl oxygen, and another accepting from the side chain of H45 by the carbonyl group of Q18 side chain. Apparently, these interactions are not sufficient to maintain the M20 loop in the closed conformational substate, perhaps due to the altered electron density of the substrate and cofactor at the transition state, breaking away to adopt a more open form. In the double mutant, the M42W mutation imposes steric congestion due to the large size of Trp in comparison with Met, which pushes the αC helix towards the M20 loop. This shifts the side chain of S49 from donating a hydrogen bond to the backbone carbonyl group of H45 to form a hydrogen bond to the carbonyl oxygen of Q18 side chain. Thus, there is an additional hydrogen bond interacting with M20 loop, sufficient to keep its conformation in the closed form throughout the hydride transfer process. Note that W42 is sandwiched between the αC and αE helices, which in turn quench dynamic fluctuations of the αE-F loop.
Figure 10.
Structures highlighting hydrogen bonding interactions between Q18 of the M20 loop and the cofactor sugar and amino acids (H45 and S49) of the αC helix. M42 in the wild-type DHFR and W42 in the double mutant are shown as stick models in the back of the αC helix.
IV. Concluding Remarks
Combined QM/MM molecular dynamics simulations were carried out on the hydride transfer in wt-DHFR and the M42W/G121V double mutant at three different temperatures (5, 25, and 45 °C). The computed changes in activation parameters due to amino acid mutations are in reasonable accord with experiment. The dominant factor responsible for the impaired catalytic function in the mutant enzyme is due to reduced dynamic flexibility at the transition state, giving rise to increase free energy barrier. Although counterintuitive, the double mutation in fact enhances enthalpic stabilization of the transition state over the wild-type enzyme. It was found that the average acceptor-donor lengths for the hydride transfer are essentially the same in the Michaelis complex (about 3.5 Å) and in the transition state (2.7 Å) for wt-DHFR and the M42W/G121V double mutant, implicating that the change in the intrinsic entropy of activation of the cofactor and substrate due to mutations is negligible. Consequently, the overall reduction in ΔS≠ may be attributed to reduced protein flexibilities at the transition state in the double mutant relative to that of the wild-type enzyme.
Structural and principal component analyses of the dynamic trajectories reveal that loop conformational fluctuations are severely dampened in the transition state region as a result of mutation, whereas there is increased flexibility in the Michaelis complex. Significantly, the altered protein fluctuations are found to be associated with the allosteric conformational transition of the M20 loop between the closed (Michaelis complex) and open forms accompanying the chemical transformation. In particular, we found that the M20 loop, which is known to be critical to DHFR catalysis, can sample the open-like conformations near the transition state of the hydride transfer in wild-type DHFR. However, the M20 loop remains closed in the double mutant. We further identified that, due to the M42W/G121V double mutation, the hydrogen bond between S49 and H45 carbonyl oxygen in the αC helix is shifted to interact with the side chain of Q18 of the M20 loop. Thus, the extra hydrogen bonding interaction with the M20 loop in the M42W/G121V mutant stabilizes the closed M20 conformation, resulting in a net reduction in protein allosteric conformational flexibility at the transition state. The computational results appear to mirror the model of a different double mutant, N23PP/S148A,(12) which was designed precisely to knock out the dynamic flexibility of the M20 loop. In that case, the entropy of activation was also reduced, implicating reduced protein flexibility at the transition state relative to the wild-type enzyme; however, kinetic parameters, both in rate and in temperature dependence of KIEs, were not significantly affected.(13)
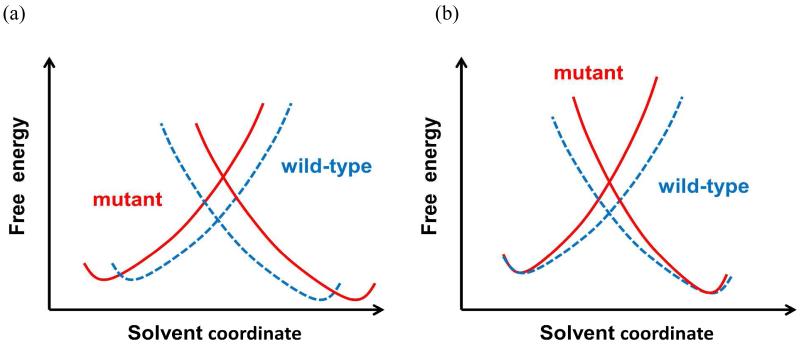
The present finding that restricted allosteric conformational flexibility of the M20 loop is a main factor responsible for the increased entropic barrier (more negative entropy of activation) in the M42W/G121V double mutant is not inconsistent with the reorganization model.(13, 20, 26, 63) It was pointed out that perturbations to the protein structure from distant mutations can propagate to the active site, resulting in elongated donor-acceptor distances(20) and increased reorganization energy relative to that in the wild-type enzyme.(26, 63) This hypothesis focuses on the “distance” of the energy gap coordinate, or preorganization of the substrate and cofactor in the enzyme environment. Implicitly, it emphasizes that the linear response force constant for protein reorganization is similar (but not required) in the wild-type enzyme and in the mutant enzyme. In addition, an elongated donor-acceptor distance would pay more reorganization energy, hence, increased free energy barrier (Scheme 2). This is very reasonable indeed. The present model can be put in the same context (Scheme 2), where the preorganization effects are similar in wild-type and mutant DHFR as far as the acceptor-donor distance for the hydride transfer is concerned. However, the force constants for the Marcus parabola are greater in the mutant enzyme than that in wild-type DHFR, thanks to reduced flexibility in loop conformational dynamics. Thus, the reduced entropy of activation is reported by an increased slope in the Marcus-type diabatic states (Scheme 2). Both mechanisms are possible depending on the specific enzymes.
Scheme 2.
A schematic description of extreme models using protein reorganization energy. In model (a), the preorganization effect is disrupted due to distant mutantions along with increased donor-acceptor distance (not shown in the energy gap coordinate). This results in a greater reorganization energy, i.e., free energy barrier, to reach the transition state at the diabatic crossing point. In model (b), the donor and acceptor species have the same distance and preorganization effect in the Michaelis complex both in the wild-type and double mutant enzymes, but remote mutations affect protein conformational flexibility towards the transition state, which is reported by increased force constant for the protein response in the diabatic picture.
That the impaired catalytic activity by the M42W/G121V mutation is due to increased entropic barrier relative to the wild-type enzyme was known long ago;(18, 20) however, its mechanistic implication has not been emphasized. Benkovic and coworkers pointed out that while the coupling between the distal residues M42 and G121 and their roles on catalysis may be inferred from kinetic data, “the physical nature of the process is not known”.(18) The present study favors a mechanism in which regional changes of the conformational heterogeneity of the enzyme, accompanying the chemical transformation from the Michaelis complex to the transition state, are important to catalysis, and the effects are expressed primarily in entropy of activation, ΔS≠. Consequently, increased dynamic fluctuations of the protein in the transition state over the Michaelis complex are entropically favored to the catalyzed reaction, although stabilization of the transition state can be a dominant factor to lower the free energy barrier in catalysis.
This mechanism may be considered as an extension of the “ensemble” view of protein allostery that has emerged in recent years.(10, 43, 74-75) In this picture, rather than an induced transition from a well-defined inactive state to an active configuration as in the classical allosteric models,(76-77) the binding at the allosteric site shifts the population of a conformation ensemble towards the active state. In fact, this shift may not necessarily require a change in the mean backbone conformation of the protein.(43) Here, the connection between protein dynamics and enzyme catalysis is not in the precise time-dependent transition between the inactive and active states, e.g., from the Michaelis complex to the transition state. Instead, the conformational ensemble of the enzyme-substrate complex encompasses all these substates (the existence of such conformational ensembles has been demonstrated for the entire DHFR catalytic cycle).(6, 28) and it is the population of the protein corresponding to the Michaelis complex that is shifted towards a population in the transition state as a result of the thermal activation of the chemical reaction. One may prefer that this is purely an equilibrium, thermodynamic phenomenon, not a dynamic effect. However, there are differences in protein dynamic fluctuation and in conformational sampling between the Michaelis complex and the transition state in an enzymatic process, and an understanding of their contributions to catalysis and their changes as a result of amino acid mutation is useful. In DHFR, the wild-type enzyme is capable of sampling a greater range of conformational space, especially involving the M20 loop motions, whereas in the M42W/G121V mutant, the conformational heterogeneity is restricted at the transition state in several loop regions.
V. Methods
We provide a brief summary of the integrated path integral and free energy simulation method used to determine kinetic isotope effects for enzymatic reactions,(65) and a summary of the key computational details. The key novel contribution is to carry out free energy perturbation of different isotopic masses in a single dynamics simulations.(65) Consequently, both primary and secondary, as well as heavy atom, KIEs can be accurately determined from statistical simulations.(78-79)
A. Quantum Transition State Theory and Path Integral Simulations of KIEs
The theoretical framework in our discussion is path integral quantum transition state theory (QTST),(80-82) which is derived by writing the rate expression analogous to classical transition state theory.
| (4) |
where w(z) is the potential of mean force (PMF) as a function of the centroid reaction coordinate is the value of at the maximum of the PMF, and is a dynamical frequency factor approximated by the velocity for a free particle of effective mass Meff along the reaction coordinate direction. The centroid coordinate of particle (n) in path integral simulation is defined by , where P is the number of discrete particles with the corresponding coordinates . The exact rate constant is obtained by multiplying the QTST rate constant by a correction factor or transmission coefficient γq:(81)
| (5) |
Eqs 4 and 5 have identical forms to that of the classical rate constant, but unlike transition state theory, there is no variational upper bound in the QTST rate constant because the quantum transmission coefficient γq may be either greater or less than one. There is no practical procedure to compute the quantum transmission coefficient γq. For a model reaction with a parabolic barrier along the reaction coordinate coupled to a bath of harmonic oscillators, the quantum transmission coefficient is the Grote-Hynes (GH) classical transmission coefficient κGH.(83) Often, the classical γq is used to approximate the quantum transmission coefficient; however, there is no correspondence between classical and quantum dynamic trajectories and the effects of tunneling may greatly affect reaction dynamics near the barrier top.
As in classical transition state theory, the PMF including nuclear quantum effects, w(z) , can be computed from the equilibrium averages of a double-average procedure:(84-85)
| (6) |
where and wcm (z) are the centroid quantum mechanical and the classical mechanical (CM) PMF, is the difference in potential energy between the QM and CM system,(84, 86-87) the ensemble average <…>U is obtained from classical molecular dynamics simulations using the potential , and the inner average is carried out through path integral free-particle sampling over configurations generated from the classical trajectories. The double averaging approach was used by Sprik et al. in Monte Carlos simulations, called the hybrid classical and path integral,(86) and used in enzyme calculations by Hwang and Warshel, called quantized classical path,(87) and by our group in coupled free energy perturbation and umbrella sampling (PI-FEP/UM) simulations.(65)
The centroid path integral method enable us to conveniently determine KIEs by directly computing the ratio of the quantum partition functions for two different isotopes through free energy perturbation (FEP) theory.
| (7) |
with
| (8) |
where the subscripts L specifies that the ensemble averages are done using the light isotope, is the free energy of the free particle reference state for the quantized particles,(84) and represents the difference in “classical” potential energy at the heavy and light bead positions ri, H and ri,L.
In essence, only one simulation of a given isotopic reaction, e.g., the light isotope, is performed, while the ratio of the partition function, i.e., the KIE, to a different isotopic reaction, is obtained through FEP by perturbing the mass from the light isotope to the heavy isotope.(65) A bisection sampling technique was extended to centroid path integral simulations to obtain converged results.(64) This is in contrast to other approaches that have been reported in the literature employing centroid path integral simulations,(87-89) in which two separate simulations are performed. The use of “mass” perturbation in free-particle bisection sampling scheme results in a major improvement in computation accuracy for KIE calculations such that secondary kinetic isotope effects and heavy atom isotope effects can be reliably obtained.(65, 78, 90) To our knowledge, the coupled path-integral free energy perturbation and umbrella sampling (PI-FEP/UM) method is the only practical approach to yield computed secondary KIEs sufficiently accurate to be compared with experiments.(79)
B. Combined QM/MM Potential Energy Surface
The accuracy of the potential energy function used to carry out molecular dynamics simulations directly affects the reliability of the computed. We employ combined quantum mechanical and molecular mechanical (QM/MM) methods,(46, 91) in which the hydride transfer catalyzed by DHFR is modeled by a reaction specific parameterized AM1 model(92) with a simple valence bond correction described in ref. (30), and the rest of the system is represented by the CHARMM22 force field.(45, 50) The QM/MM potential is given by(91)
| (9) |
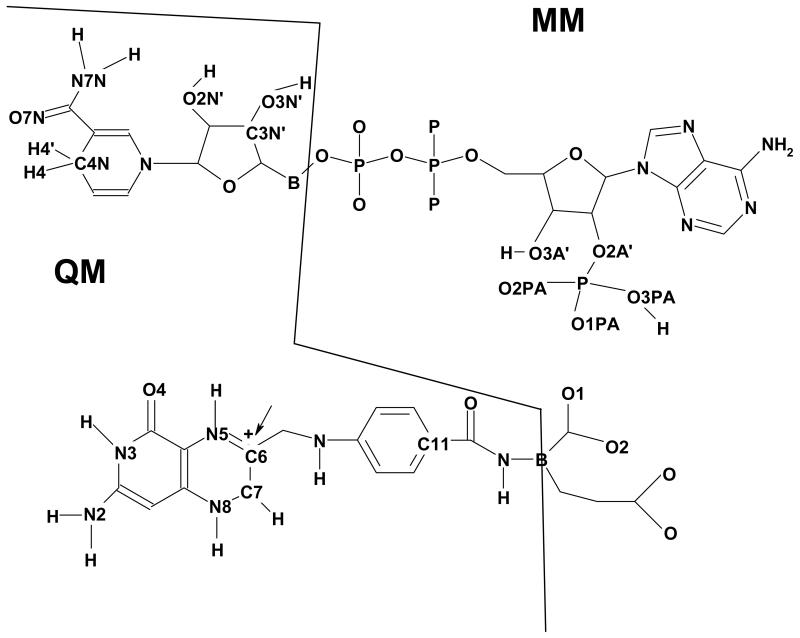
where is the Hamiltonian of the QM-subsystem, Umm is the classical (MM) potential energy for the MM region, and Hqm / mm(S) is the interaction Hamiltonian between the two regions. The wild-type DHFR system(14) was partitioned into NQM=69 quantum mechanical atoms and NMM= 21399 classical mechanical atoms. The QM system (Scheme 1) includes 39 atoms of the DHF substrate (the pteridine ring, the pABA moiety and the -NH-Cα group of the glutamate moiety), and 30 atoms of the NADPH cofactor (the dihydronicotinamide and ribose rings). This QM subsystem contains 2 boundary atoms: the Cα at the glutamate moiety and the C5 ribose atom, which are treated partly by MM and partly with the generalized hybrid orbital (GHO) method.
Scheme 1.
Schematic representation of atoms treated explicitly by electronic structure theory and by molecular mechanics for the NADPH cofactor (top) and 7,8-dihydrofolate substrate (bottom). The symbol B indicates a boundary atom which is treated both as a QM and an MM atom in the generalized hybrid orbital (GHO) method. Adapted from ref. (30) with permission from the American Chemical Society.
C. Computational Details
The starting structures for the simulations of the wild-type eDHFR at the three temperatures were taken from the simulations carried previously.(30, 52) These structures were re-equilibrated at their respective temperatures with a constant pressure of 1 atm for 100 ps. The M42W-G121V double mutant structures were built by performing in silico mutation on the wild-type enzyme structures, and were subject to 200 ps equilibration using the NPT ensemble. All equilibrations were carried out using the QM/MM potential detailed in (30).
The periodic boundary conditions are employed to solvate the system. A cubic water box is used to solvate the entire Michaelis complex, with a dimension of about 60 Å × 60 Å × 60 Å. The simulation was performed corresponding to pH 7 by adjusting the protonation state of histidine residues, and all polar amino acid residue sidechains are protonated and deprotonated accordingly. The resulting Michaelis complex bears 15 units of negative charge, and it is neutralized by randomly placing 15 sodium ions in the water box. A spherical cutoff distance of 12 Å was used for the nonbonded interactions, with a switch function in the region between 11 and 12 Å. The pressure (at 1 atm) and the temperature of the systems were controlled with the Nosé-Hoover extended system method. A total of 16 simulation windows were used to encompass the reaction coordinate from −2.0 to 1.5 Å, which is defined as the difference between the transferring hydride from the donor carbon on NADPH and the acceptor carbon of DHF. Each simulation window was subjected to at least 100 ps of further equilibration followed by 170 to 220 ps averaging. Thus, each potential of mean force was averaged over 4 to 5 ns simulations. The kinetic isotope effects were averaged over at least 24000 configurations at each temperature.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM46367).
Abbreviations
- ADL
acceptor-donor length
- DHF
7,8-dihydrofolate
- DHFR
dihydrofolate reductase
- dm
double mutant
- EA-VTST/MT
ensemble averaged transition state theory with multidimensional tunneling
- KIE
kinetic isotope effects
- NADPH
nicotinamide adenine dinucleotide phosphate
- NQE
nuclear quantum effects
- PCA
principal component
- PI-FEP/UM
path integral free energy perturbation and umbrella smapling
- PMF
potential of mean force
- QM/MM
quantum mechanics and molecular mechanics
- QTST
quantum transition state theory
- RS
reactant state
- TS
transition state
- wt
wild-type
Footnotes
One table of computed data including free energies of activation at different temperatures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Antoniou D, Schwartz SD. Protein Dynamics and Enzymatic Chemical Barrier Passage. J. Phys. Chem. B. 2011;115:15147–15158. doi: 10.1021/jp207876k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem. Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 3.Hay S, Scrutton NS. Good vibrations in enzyme-catalysed reactions. Nat Chem. 2012;4:161–168. doi: 10.1038/nchem.1223. [DOI] [PubMed] [Google Scholar]
- 4.Nagel ZD, Klinman JP. A 21(st) century revisionist’s view at a turning point in enzymology. Nature Chem. Biol. 2009;5:543–550. doi: 10.1038/nchembio.204. [DOI] [PubMed] [Google Scholar]
- 5.Glowacki DR, Harvey JN, Mulholland AJ. Taking Ockham’s razor to enzyme dynamics and catalysis. Nat Chem. 2012;4:169–176. doi: 10.1038/nchem.1244. [DOI] [PubMed] [Google Scholar]
- 6.Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 7.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–U813. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 8.Loria JP, Berlow RB, Watt ED. Characterization of enzyme motions by solution NMR relaxation dispersion. Acc. Chem. Res. 2008;41:214–221. doi: 10.1021/ar700132n. [DOI] [PubMed] [Google Scholar]
- 9.Ghanem M, Li L, Wing C, Schramm VL. Altered thermodynamics from remote mutations altering human toward bovine purine nucleoside phosphorylase. Biochemistry. 2008;47:2559–2564. doi: 10.1021/bi702132e. [DOI] [PubMed] [Google Scholar]
- 10.Hilser VJ. An Ensemble View of Allostery. Science. 2010;327:653–654. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagel ZD, Klinman JP. Tunneling and dynamics in enzymatic hydride transfer. Chem. Rev. 2006;106:3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 12.Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE. A Dynamic Knockout Reveals That Conformational Fluctuations Influence the Chemical Step of Enzyme Catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loveridge EJ, Behiry EM, Guo JN, Allemann RK. Evidence that a ‘dynamic knockout’ in Escherichia coli dihydrofolate reductase does not affect the chemical step of catalysis. Nature Chem. 2012;4:292–297. doi: 10.1038/nchem.1296. [DOI] [PubMed] [Google Scholar]
- 14.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 15.Boehr DD, McElheny D, Dyson HJ, Wright PE. Millisecond timescale fluctuations in dihydrofolate reductase are exquisitely sensitive to the bound ligands. Proc. Nat. Acad. Sci. 2010;107:1373–1378. doi: 10.1073/pnas.0914163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oyeyemi OA, Sours KM, Lee T, Resing KA, Ahn NG, Klinman JP. Temperature dependence of protein motions in a thermophilic dihydrofolate reductase and its relationship to catalytic efficiency. Proc. Nat. Acad. Sci. 2010;107:10074–10079. doi: 10.1073/pnas.1003678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oyeyemi OA, Sours KM, Lee T, Kohen A, Resing KA, Ahn NG, Klinman JP. Comparative Hydrogen-Deuterium Exchange for a Mesophilic vs Thermophilic Dihydrofolate Reductase at 25 degrees C: Identification of a Single Active Site Region with Enhanced Flexibility in the Mesophilic Protein. Biochemistry. 2011;50:8251–8260. doi: 10.1021/bi200640s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopalan PT, Lutz S, Benkovic SJ. Coupling interactions of distal residues enhance dihydrofolate reductase catalysis: mutational effects on hydride transfer rates. Biochemistry. 2002;41:12618–12628. doi: 10.1021/bi026369d. [DOI] [PubMed] [Google Scholar]
- 19.Swanwick RS, Shrimpton PJ, Allemann RK. Pivotal Role of Gly 121 in Dihydrofolate Reductase from Escherichia coli: The Altered Structure of a Mutant Enzyme May Form the Basis of Its Diminished Catalytic Performance. Biochemistry. 2004;43:4119–4127. doi: 10.1021/bi036164k. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Goodey NM, Benkovic SJ, Kohen A. Coordinated effects of distal mutations on environmentally coupled tunneling in dihydrofolate reductase. Proc. Nat. Acad. Sc. U.S.A. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mauldin RV, Lee AL. Nuclear Magnetic Resonance Study of the Role of M42 in the Solution Dynamics of Escherichia coli Dihydrofolate Reductase. Biochemistry. 2010;49:1606–1615. doi: 10.1021/bi901798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauldin RV, Sapienza PJ, Petit CM, Lee AL. Structure and Dynamics of the G121V Dihydrofolate Reductase Mutant: Lessons from a Transition-State Inhibitor Complex. Plos One. 2012;7 doi: 10.1371/journal.pone.0033252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll MJ, Mauldin RV, Gromova AV, Singleton SF, Collins EJ, Lee AL. Evidence for dynamics in proteins as a mechanism for ligand dissociation. Nature Chemical Biology. 2012;8:246–252. doi: 10.1038/nchembio.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watney JB, Agarwal PK, Hammes-Schiffer S. Effect of Mutation on Enzyme Motion in Dihydrofolate Reductase. J. Am. Chem. Soc. 2003;125:3745–3750. doi: 10.1021/ja028487u. [DOI] [PubMed] [Google Scholar]
- 25.Wong KF, Selzer T, Benkovic SJ, Hammes-Schiffer S. Impact of distal mutations on the network of coupled motions correlated to hydride transfer in dihydrofolate reductase. Proc. Nat. Acad. Sci. 2005;102:6807–6812. doi: 10.1073/pnas.0408343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Warshel A. The Catalytic Effect of Dihydrofolate Reductase and Its Mutants Is Determined by Reorganization Energies. Biochemistry. 2007;46:6011–6025. doi: 10.1021/bi700201w. [DOI] [PubMed] [Google Scholar]
- 27.Radkiewicz JL, Brooks CL., III Protein Dynamics in Enzymatic Catalysis: Exploration of Dihydrofolate Reductase. J. Am. Chem. Soc. 2000;122:225–231. [Google Scholar]
- 28.Rod TH, Radkiewicz JL, Brooks CL., III Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc. Nat. Acad. Sci. 2003;100:6980–6985. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal PK, Billeter SR, Rajagopalan PTR, Benkovic SJ, Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc. Natl. Acad. Sci. 2002;99:2794–2799. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Viloca M, Truhlar DG, Gao J. Reaction-Path Energetics and Kinetics of the Hydride Transfer Reaction Catalyzed by Dihydrofolate Reductase. Biochemistry. 2003;42:13558–13575. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 31.Cui Q, Karplus M. Promoting Modes and Demoting Modes in Enzyme-Catalyzed Proton Transfer Reactions: A Study of Models and Realistic Systems. J. Phys. Chem. B. 2002;106:7927–7947. [Google Scholar]
- 32.Kamath G, Howell EE, Agarwal PK. The tail wagging the dog: insights into catalysis in R67 dihydrofolate reductase. Biochemistry. 2010;49:9078–9088. doi: 10.1021/bi1007222. [DOI] [PubMed] [Google Scholar]
- 33.Boekelheide N, Salomon-Ferrer R, Miller TF. Dynamics and dissipation in enzyme catalysis. Proc. Nat. Acad. Sci. 2011;108:16159–16163. doi: 10.1073/pnas.1106397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doron D, Major DT, Kohen A, Thiel W, Wu X. Hybrid Quantum and Classical Simulations of the Dihydrofolate Reductase Catalyzed Hydride Transfer Reaction on an Accurate Semi-Empirical Potential Energy Surface. J. Chem. Theory Comput. 2011;7:3420–3437. doi: 10.1021/ct2004808. [DOI] [PubMed] [Google Scholar]
- 35.Doron D, Kohen A, Major DT. Collective Reaction Coordinate for Hybrid Quantum and Molecular Mechanics Simulations: A Case Study of the Hydride Transfer in Dihydrofolate Reductase. J. Chem. Theory Comput. 2012;8:2484–2496. doi: 10.1021/ct300235k. [DOI] [PubMed] [Google Scholar]
- 36.Engel H, Doron D, Kohen A, Major DT. Momentum Distribution as a Fingerprint of Quantum Delocalization in Enzymatic Reactions: Open-Chain Path-Integral Simulations of Model Systems and the Hydride Transfer in Dihydrofolate Reductase. J. Chem. Theory Comput. 2012;8:1223–1234. doi: 10.1021/ct200874q. [DOI] [PubMed] [Google Scholar]
- 37.Dametto M, Antoniou D, Schwartz SD. Barrier crossing in dihydrofolate reductase does not involve a rate-promoting vibration. Mol. Phys. 2012;110:531–536. doi: 10.1080/00268976.2012.655337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pu J, Gao J, Truhlar DG. Multidimensional tunneling, recrossing, and the transmission coefficient for enzymatic reactions. Chem. Rev. 2006;106:3140–3169. doi: 10.1021/cr050308e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. How enzymes work: Analysis by modern rate theory and computer simulations. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 40.Olsson MHM, Parson WW, Warshel A. Dynamical contributions to enzyme catalysis: Critical tests of a popular hypothesis. Chem. Rev. 2006;106:1737–1756. doi: 10.1021/cr040427e. [DOI] [PubMed] [Google Scholar]
- 41.Miller GP, Benkovic SJ. Chem. Biol. 1998;5:R105–R113. doi: 10.1016/s1074-5521(98)90616-0. [DOI] [PubMed] [Google Scholar]
- 42.Rozovsky S, McDermott AE. The time scale of the catalytic loop motion in triosephosphate isomerase. J. Mol. Biol. 2001;310:259–270. doi: 10.1006/jmbi.2001.4672. [DOI] [PubMed] [Google Scholar]
- 43.Tsai CJ, del Sol A, Nussinov R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J. Mol. Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc. Chem. Res. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 45.Gao J, Xia X. A prior evaluation of aqueous polarization effects through Monte Carlo QM-MM simulations. Science. 1992;258:631–635. doi: 10.1126/science.1411573. [DOI] [PubMed] [Google Scholar]
- 46.Gao J, Ma S, Major DT, Nam K, Pu J, Truhlar DG. Mechanisms and free energies of enzymatic reactions. Chem. Rev. 2006;106:3188–3209. doi: 10.1021/cr050293k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Wong K-Y, Major DT, Cembran A, Song L, Lin Y-L, Fan Y, Ma S. Kinetic isotope effects from hybrid classical and quantum path integral computations. In: Allemann RK, editor. Quantum Tunnelling in Enzyme-Catalysed Reactions. 2009. pp. 105–131. [Google Scholar]
- 48.Gao J. Hybrid Quantum Mechanical/Molecular Mechanical Simulations: An Alternative Avenue to Solvent Effects in Organic Chemistry. Acc. Chem. Res. 1996;29:298–305. [Google Scholar]
- 49.Truhlar DG, Gao J, Alhambra C, Garcia-Viloca M, Corchado J, Sanchez ML, Villa J. The Incorporation of Quantum Effects in Enzyme Kinetics Modeling. Acc. Chem. Res. 2002;35:341–349. doi: 10.1021/ar0100226. [DOI] [PubMed] [Google Scholar]
- 50.Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew. Chem., Int. Ed. 2009;48:1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Viloca M, Truhlar DG, Gao J. Importance of Substrate and Cofactor Polarization in the Active Site of Dihydrofolate Reductase. J. Mol. Biol. 2003;327:549–560. doi: 10.1016/s0022-2836(03)00123-2. [DOI] [PubMed] [Google Scholar]
- 52.Pu J, Ma S, Gao J, Truhlar DG. Small Temperature Dependence of the Kinetic Isotope Effect for the Hydride Transfer Reaction Catalyzed by Escherichia coli Dihydrofolate Reductase. Journal of Physical Chemistry B. 2005;109:8551–8556. doi: 10.1021/jp051184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagel ZD, Dong M, Bahnson BJ, Klinman JP. Impaired protein conformational landscapes as revealed in anomalous Arrhenius prefactors. Proc. Nat. Acad. Sci. 2011;108:10520–10525. doi: 10.1073/pnas.1104989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karplus M, Kushick JN. Method for estimating the configurational entropy of macromolecules. Macromolecules. 1981;14:325–332. [Google Scholar]
- 55.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wand AJ. The dark energy of proteins comes to light: conformational entropy and its role in protein function revealed by NMR relaxation. Curr Opin Struct Biol. 2012 doi: 10.1016/j.sbi.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sikorski RS, Wang L, Markham KA, Rajagopalan PTR, Benkovic SJ, Kohen A. Tunneling and Coupled Motion in the Escherichia coli Dihydrofolate Reductase Catalysis. J. Am. Chem. Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- 58.Stojkovic V, Perissinotti LL, Willmer D, Benkovic SJ, Kohen A. Effects of the Donor-Acceptor Distance and Dynamics on Hydride Tunneling in the Dihydrofolate Reductase Catalyzed Reaction. J. Am. Chem. Soc. 2012;134:1738–1745. doi: 10.1021/ja209425w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roston D, Cheatum CM, Kohen A. Hydrogen Donor-Acceptor Fluctuations from Kinetic Isotope Effects: A Phenomenological Model. Biochemistry. 2012;51:6860–6870. doi: 10.1021/bi300613e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, Truhlar DG. Quantum mechanical methods for enzyme kinetics. Ann. Rev. Phys. Chem. 2002;53:467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- 61.Knapp MJ, Rickert K, Klinman JP. Temperature-dependent isotope effects in soybean lipoxygenase-1: Correlating hydrogen tunneling with protein dynamics. J. Am. Chem. Soc. 2002;124:3865–3874. doi: 10.1021/ja012205t. [DOI] [PubMed] [Google Scholar]
- 62.Alhambra C, Corchado J, Sanchez ML, Garcia-Viloca M, Gao J, Truhlar DG. Canonical Variational Theory for Enzyme Kinetics with the Protein Mean Force and Multidimensional Quantum Mechanical Tunneling Dynamics. Theory and Application to Liver Alcohol Dehydrogenase. J. Phys. Chem. B. 2001;105:11326–11340. [Google Scholar]
- 63.Liu H, Warshel A. Origin of the Temperature Dependence of Isotope Effects in Enzymatic Reactions: the Case of Dihydrofolate Reductase. J. Phys. Chem. B. 2007 doi: 10.1021/jp070938f. ACS ASAP. [DOI] [PubMed] [Google Scholar]
- 64.Major DT, Gao J. Implementation of the bisection sampling method in path integral simulations. J. Mol. Graph. Model. 2005;24:121–127. doi: 10.1016/j.jmgm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 65.Major DT, Gao J. An Integrated Path Integral and Free-Energy Perturbation-Umbrella Sampling Method for Computing Kinetic Isotope Effects of Chemical Reactions in Solution and in Enzymes. J. Chem.Theory Comput. 2007;3:949–960. doi: 10.1021/ct600371k. [DOI] [PubMed] [Google Scholar]
- 66.Castillo R, Andres J, Moliner V. Catalytic Mechanism of Dihydrofolate Reductase Enzyme. A Combined Quantum-Mechanical/Molecular-Mechanical Characterization of Transition State Structure for the Hydride Transfer Step. J. Am. Chem. Soc. 1999;121:12140–12147. [Google Scholar]
- 67.Agarwal PK, Billeter SR, Hammes-Schiffer S. Nuclear Quantum Effects and Enzyme Dynamics in Dihydrofolate Reductase Catalysis. J. Phys. Chem. B. 2002;106:3283–3293. [Google Scholar]
- 68.Cameron CE, Benkovic SJ. Evidence for a Functional Role of the Dynamics of Glycine-121 of Escherichia coli Dihydrofolate Reductase Obtained from Kinetic Analysis of a Site-Directed Mutant. Biochemistry. 1997;36:15792. doi: 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- 69.Venkitakrishnan RP, Zaborowski E, McElheny D, Benkovic SJ, Dyson HJ, Wright PE. Conformational changes in the active site loops of dihydrofolate reductase during the catalytic cycle. Biochemistry. 2004;43:16046–16055. doi: 10.1021/bi048119y. [DOI] [PubMed] [Google Scholar]
- 70.Ohmae E, Fukumizu Y, Iwakura M, Gekko K. Effects of mutation at methionine-42 of Escherichia coli dihydrofolate reductase on stability and function: Implication of hydrophobic interactions. J.Biochem. 2005;137:643–652. doi: 10.1093/jb/mvi079. [DOI] [PubMed] [Google Scholar]
- 71.Thorpe IF, Brooks CL., III Conformational Substates Modulate Hydride Transfer in Dihydrofolate Reductase. J. Am. Chem. Soc. 2005;127:12997–13006. doi: 10.1021/ja053558l. [DOI] [PubMed] [Google Scholar]
- 72.Khavrutskii IV, Price DJ, Lee J, Brooks CL., III Conformational change of the methionine 20 loop of Escherichia coli dihydrofolate reductase modulates pKa of the bound dihydrofolate. Protein Science. 2007;16:1087–1100. doi: 10.1110/ps.062724307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc. Nat. Acad. Sci. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.del Sol A, Tsai CJ, Ma BY, Nussinov R. The Origin of Allosteric Functional Modulation: Multiple Pre-existing Pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc. Natl. Acad. Sci. 2011;108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 77.Koshland DE, Jr., Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 78.Gao J, Wong K-Y, Major DT. Combined QM/MM and path integral simulations of kinetic isotope effects in the proton transfer reaction between nitroethane and acetate ion in water. J. Comput. Chem. 2008;29:514–522. doi: 10.1002/jcc.20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Major DT, Heroux A, Orville AM, Valley MP, Fitzpatrick PF, Gao J. Differential quantum tunneling contributions in nitroalkane oxidase catalyzed and the uncatalyzed proton transfer reaction. Proc. Natl. Acad. Sci. 2009;106:20736–20739. doi: 10.1073/pnas.0911416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Voth GA, Chandler D, Miller WH. Rigorous formulation of quantum transition state theory and its dynamical corrections. J. Chem. Phys. 1989;91:7749–7760. [Google Scholar]
- 81.Voth GA. Feynman path integral formulation of quantum mechanical transition-state theory. J. Phys. Chem. 1993;97:8365–8377. [Google Scholar]
- 82.Messina M, Schenter GK, Garrett BC. Centroid-density, quantum rate theory: variational optimization of the dividing surface. J. Chem. Phys. 1993;98:8525–8536. [Google Scholar]
- 83.Grote RF, Hynes JT. The stable states picture of chemical reactions. II. Rate constants for condensed and gas phase reaction models. J. Chem. Phys. 1980;73:2715–2732. [Google Scholar]
- 84.Feynman RP, Hibbs AR. Quantum Mechanics and Path Integrals. McGraw-Hill; New York: 1965. [Google Scholar]
- 85.Feynman RP, Kleinert H. Effective classical partition functions. Phys. Rev. A. 1986;34:5080. doi: 10.1103/physreva.34.5080. [DOI] [PubMed] [Google Scholar]
- 86.Sprik M, Klein ML, Chandler D. Staging: a sampling technique for the Monte Carlo evaluation of path integrals. Phys. Rev. B. 1985;31:4234–4244. doi: 10.1103/physrevb.31.4234. [DOI] [PubMed] [Google Scholar]
- 87.Hwang JK, Warshel A. A quantized classical path approach for calculations of quantum mechanical rate constants. J. Phys. Chem. 1993;97:10053–10058. [Google Scholar]
- 88.Hwang J-K, Warshel A. How Important Are Quantum Mechanical Nuclear Motions in Enzyme Catalysis? J. Am. Chem. Soc. 1996;118:11745–11751. [Google Scholar]
- 89.Hinsen K, Roux B. Potential of mean force and reaction rates for proton transfer in acetylacetone. J. Chem. Phys. 1997;106:3567–3577. [Google Scholar]
- 90.Lin Y.-l., Gao J. Kinetic Isotope Effects of L-Dopa Decarboxylase. J. Am. Chem. Soc. 2011 doi: 10.1021/ja108209w. ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao J. Methods and applications of combined quantum mechanical and molecular mechanical potentials. In: Lipkowitz KB, Boyd DB, editors. Rev. Comput. Chem. VCH; New York: 1995. pp. 119–185. [Google Scholar]
- 92.Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. Development and use of quantum mechanical molecular models. 76. AM1: a new general purpose quantum mechanical molecular model. J. Am. Chem. Soc. 1985;107:3902–3909. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.