Summary
A naturally occurring p53 isoform that lacks 39 residues at the N-terminus (denoted ΔNp53), when expressed with wild-type p53 (WTp53), forms mixed ΔNp53:WTp53 tetramers and causes accelerated aging in mice. Cellular alterations specific to ΔNp53:WTp53 have been difficult to assess because ΔNp53 and WTp53 co-expression results in tetramer heterogeneity, including formation of contaminating WTp53 tetramers. Based upon the p53 tetramer structure, we expressed ΔNp53 and WTp53 as a single transcript that maintained tetramer architecture, ensuring a 2:2 ΔNp53:WTp53 stoichiometry. As expected, ΔNp53:WTp53 tetramers were stable and transcriptionally active in vitro and in cells, largely mimicking the function of WTp53 tetramers. Microarray analyses, however, revealed about 80 genes whose expression was altered 2-fold or more in ΔNp53:WTp53 cells. Moreover, global metabolomic profiling quantitated hundreds of biochemicals across different experiments (WTp53, ΔNp53:WTp53, plus controls). When evaluated collectively, these data suggested altered mTOR signaling and mitochondrial function—each canonical regulators of longevity—in cells expressing ΔNp53:WTp53 vs. WTp53. Increased levels of free amino acids, increased expression of IRS-1, and decreased expression of INPP5D/SHIP-1 suggested activated mTOR signaling in ΔNp53:WTp53 cells; this was confirmed upon comparative analyses of several mTOR pathway intermediates. We also observed changes in mitochondrial function in ΔNp53:WTp53 cells, which correlated with increased MARS2 expression and elevated levels of carnitine, acetyl CoA, ATP, and Krebs cycle intermediates. Finally, elevated levels of succinate and 2-hydroxyglutarate indicate potential epigenetic means to propagate ΔNp53:WTp53-induced gene expression changes to cell progeny. This may be especially important for aging, as biological effects manifest over time.
Keywords: DOT1L, urea cycle, polyamines, glutathione, 2HG, epigenetic
Introduction
The role of p53 in mammalian aging is best exemplified by mouse studies with p53 truncation mutants (Maier et al., 2004; Tyner et al., 2002). Both artificial and naturally occurring p53 isoforms that lack the N-terminal p53 activation domain, when expressed together with wild-type p53 (WTp53), cause a progeroid phenotype that includes early-onset osteoporosis, memory loss, and premature death (Maier et al., 2004; Pehar et al., 2010; Tyner et al., 2002). The molecular mechanisms by which ΔNp53 cause accelerated aging remain incompletely understood. It is clear, however, that ΔNp53 requires WTp53 to mediate its physiological effects; mice expressing only ΔNp53 do not age rapidly (Maier et al., 2004). Because p53 binds DNA as a tetramer, the formation of mixed ΔNp53:WTp53 hetero-tetramers is believed to mediate the accelerated aging phenotype (Courtois et al., 2002). Although ΔNp53 has been well studied using knock-in mouse models, ΔNp53 has not been thoroughly studied in human cells. Moreover, global gene expression and metabolic changes induced by ΔNp53:WTp53 have not been assessed in human cells or any model organism. One potential reason for this is that co-expression of ΔNp53 and WTp53 will confound analyses due to heterogeneity, including formation of “contaminating” WTp53 tetramers. As one consequence, it has been difficult to define cellular changes that are specifically dependent upon ΔNp53:WTp53, because a means to examine a single entity with fixed ΔNp53:WTp53 stoichiometry was not evident. For these reasons, the underlying mechanisms that link ΔNp53 and its progeroid phenotype remain incompletely understood, even in model organisms.
To further define the molecular pathways impacted by ΔNp53 expression in human cells, we expressed 2:2 ΔNp53:WTp53 tetramers (i.e. two copies of ΔNp53 and two copies of WTp53 per tetramer) in p53-null H1299 cells. Based upon the p53 tetramer structure (Okorokov et al., 2006), we implemented a strategy that expressed ΔNp53 and WTp53 as a single transcript that maintained the native tetramer architecture, ensuring a 2:2 ΔNp53:WTp53 stoichiometry. These ΔNp53:WTp53 tetramers were stable and transcriptionally active in vitro and in cells, largely mimicking the function of WTp53 tetramers. Microarray analyses identified 84 genes whose expression was up- or down-regulated 2-fold or more in ΔNp53:WTp53 cells compared with WTp53. This set of genes included many with established roles in longevity and metabolism. We next completed global metabolomic profiling, which identified and quantitated hundreds of biochemicals across different experiments (WTp53, ΔNp53:WTp53, plus controls). When evaluated collectively, the gene expression and metabolomics data provided clear predictions (validated via pathway analyses) about cellular processes disrupted by ΔNp53:WTp53 expression. Among the most striking were mitochondrial function and the mTOR pathway, each well-established regulators of longevity. Biochemical and cellular assays confirmed a hyper-active mTOR pathway and altered mitochondrial function in cells expressing ΔNp53:WTp53 (vs. WTp53). We also uncovered metabolites whose elevated levels could contribute to heritable epigenetic changes upon ΔNp53:WTp53 expression. Because ΔNp53 is a naturally occurring isoform whose levels can increase with cell stress, the ΔNp53-induced changes identified here could play a role in normal human aging.
Results
Expression and purification of 2:2 ΔNp53:WTp53 tetramers
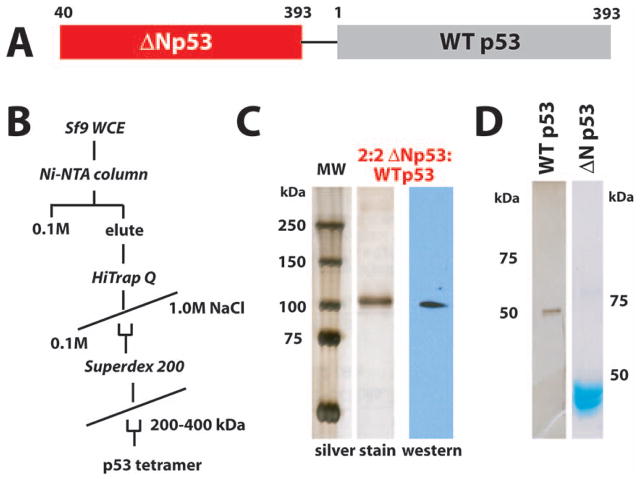
The p53 activation domain (p53AD) resides in the N-terminus of the protein. Two separate activation motifs have been identified, with residues 14–26 comprising p53AD1, and residues 41–57 comprising p53AD2. Thus, the ΔNp53 isoform lacks p53AD1 but retains p53AD2. The structure of the native p53 tetramer indicates that p53 assembles as a dimer of dimers, with the C-terminus of one p53 polypeptide interacting with the N-terminus of a second p53 protein (Okorokov et al., 2006). The juxtaposition of the N- and C-termini suggested that a flexible linker could tether two p53 monomers to yield a 1:1 ΔNp53:WTp53 dimer (Fig. S1). Implementing the strategy outlined in Figure 1A, we expressed a tethered ΔNp53:WTp53 protein as a single transcript in insect cells. Purification of p53 tetramers from these cells matched that of WTp53 tetramers; in particular, ΔNp53:WTp53 migrated similar to the native p53 tetramer on a size-exclusion column (Fig. 1B). Silver stain and western blot analysis (Fig. 1C) revealed a molecular weight of approximately 100 kDa, consistent with a tethered ΔNp53:WTp53 dimer. For functional comparisons, we purified WTp53 tetramers and tetramers consisting of only ΔNp53 proteins (Fig. 1D).
Fig. 1.
Expression and purification of ΔNp53:WTp53 tetramers with 2:2 stoichiometry. (A) Schematic of ΔNp53:WTp53. See also Figure S1. To allow conformational flexibility, the linker was 14 residues in length (sequence: GGGGENLYFQGGGG) and contained a TEV cleavage site. (B) Purification protocol used for isolation of WTp53, ΔNp53:WTp53, and ΔNp53 tetramers. (C) Silver stain gel and p53 western blot for purified ΔNp53:WTp53. (D) Silver stained and coomassie stained gel for purified WTp53 and ΔNp53.
ΔNp53:WTp53 tetramers are functionally equivalent to WTp53 in vitro
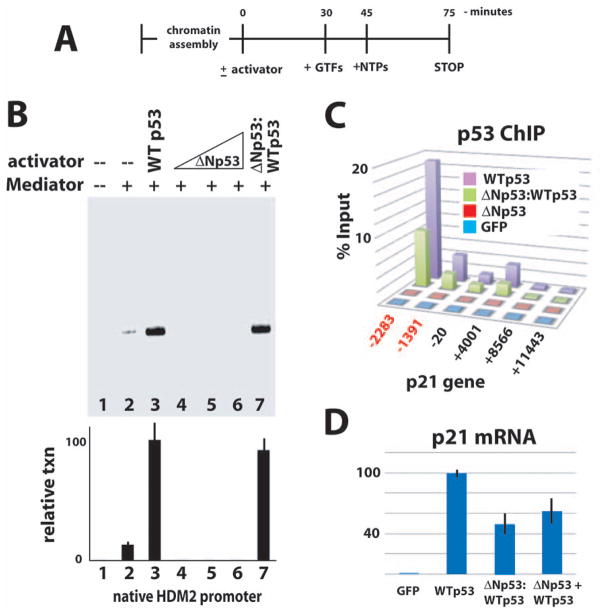
To compare the activity of the 2:2 ΔNp53:WTp53 tetramers (hereafter called ΔNp53:WTp53) with WTp53 tetramers, we used an in vitro transcription system reconstituted from purified human factors (Knuesel et al., 2009). An outline of the assay is shown in Figure 2A. Transcription activation at the native HDM2 promoter was assessed in the absence of p53, or in the presence of WTp53 tetramers, ΔNp53 tetramers, or ΔNp53:WTp53 tetramers. As expected, transcription activation required Mediator and WTp53 (Fig. 2B, compare lanes 1 – 3); ΔNp53 tetramers, which lack p53AD1 but retain p53AD2, were unable to activate transcription (Fig. 2B, lanes 4 – 6). By contrast, the ΔNp53:WTp53 tetramers could activate HDM2 transcription equally well compared with WTp53 (Fig. 2B, lane 7). This result demonstrated that ΔNp53:WTp53 had an inherent activity equal to WTp53, at least in this cell-free reconstituted transcription assay.
Fig. 2.
Like WTp53, ΔNp53:WTp53 tetramers bind p53 target genes and are transcriptionally active in vitro and in cells. (A) Schematic of in vitro transcription reaction. GTFs refers to purified TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, RNA polymerase II, and Mediator (Knuesel et al., 2009). (B) Reconstituted transcription. Activators and Mediator added as indicated. Representative data shown; plot summarizes results (mean and standard error) from multiple experiments (n = 3, 4, 6, 2, 2, 2, 6 for lanes 1 – 7). Note that ΔNp53 tetramers were titrated over a 200-fold range, with no evidence of transcription activation. (C) ChIP data (vs. p53) across the CDKN1A/p21 locus in H1299 cells. (The p53 antibody targeted residues 10–16; this epitope is absent in ΔNp53.) Binding sites for p53 are highlighted in red. The six probe locations are shown relative to the transcription start site. (D) Expression of the CDKN1A/p21 gene was probed by RT-qPCR in H1299 cells expressing WTp53, ΔNp53:WTp53, or ΔNp53 + WTp53. Cells expressing GFP vector only shown as a negative control. Note that ΔNp53 + WTp53 refers to separate expression of ΔNp53 and WTp53.
Evaluation of ΔNp53:WTp53 in cells
To establish whether ΔNp53:WTp53 tetramers would bind a p53-regulated promoter in cells, we expressed either WTp53 or ΔNp53:WTp53 in p53-null H1299 cells and performed chromatin immunoprecipitation assays (ChIP). As shown in Figure 2C, both WTp53 and ΔNp53:WTp53 tetramers bound the p21 (CDKN1A) promoter at the consensus p53 binding sites. Because WTp53 and ΔNp53:WTp53 tetramers each contain four intact DNA-binding domains, these data were not unexpected. Note that the p53 antibody (DO-2) used for ChIP targets residues 10–16, which are absent in ΔNp53. Consequently, the occupancy of ΔNp53:WTp53 appears reduced because 50% fewer epitopes are present. Next, we used RT-qPCR to measure p21 (CDKN1A) expression. The p21 gene represents one of only a few genes thus far identified as differentially expressed (reduced expression relative to WTp53) in cells expressing both ΔNp53 and WTp53 (Powell et al., 2008; Ungewitter & Scrable, 2010). In agreement, we observed a decrease in p21 expression, relative to WTp53, in cells expressing ΔNp53:WTp53 (Fig. 2D). We also examined p21 expression in H1299 cells expressing matched levels of both ΔNp53 and WTp53 (i.e. as separate transcripts). Whereas the stoichiometry of p53 tetramers could not be controlled with such experiments, p21 expression in these “ΔNp53 + WTp53” cells showed a decrease relative to WTp53 that was similar to ΔNp53:WTp53 cells (Fig. 2D).
ΔNp53:WTp53 induces a gene expression program slightly distinct from WTp53
Having confirmed that tetramers assembled from ΔNp53:WTp53 were stable, could bind p53-regulated promoters in cells, and were transcriptionally active in vitro and in cells, we next assessed their impact on global gene expression patterns. We used p53-null H1299 cells, which do not express any p53 isoform at detectable levels. Expression of p53 (WTp53 or ΔNp53:WTp53) was achieved by transfection using a vector that enabled bicistronic expression of Green Fluorescent Protein (GFP); this allowed p53-expressing cells to be sorted by GFP fluorescence. Prior to harvesting RNA for microarray analysis, we confirmed that p53 protein levels (WTp53 or ΔNp53:WTp53) were matched by quantitative western blot (Fig. S2A). Also, at the time of the analysis, each cell population (WTp53, ΔNp53:WTp53, or controls) displayed similar cell cycle distribution, as revealed by flow cytometry (Fig. S2B); relative levels of apoptosis and senescence were measured in each cell population, and only live cells were isolated (Fig. S3).
Microarray analysis was completed for four different samples: mock transfection (reagent only), empty vector (reagent plus GFP vector), WTp53, and ΔNp53:WTp53. The gene expression profile for cells expressing WTp53 (compared with mock or empty vector controls) yielded predictable results, and Gene Set Enrichment Analysis (GSEA) of the genes differentially expressed in WTp53 cells identified canonical p53-regulated processes, as expected (Fig. S4). The overall gene expression profiles of WTp53 vs. ΔNp53:WTp53 were similar when compared with control (GFP vector only) cells, as shown by the Venn diagrams in Figure S5 as well as the GSEA (Fig. S4). These similarities were not unexpected, as ΔNp53:WTp53 tetramers retain all four DNA-binding domains and only differ in two of four activation domains. About 80 genes, however, showed greater than 2-fold differences in expression in ΔNp53:WTp53 cells (up or down, compared with WTp53). These genes are shown in Table 1 and Table 2. Several hundred genes showed altered expression at levels 1.5-fold or greater in ΔNp53:WTp53 cells, compared with WTp53. These genes are shown in Table S1 and S2. The expression levels of numerous genes listed in Table 1 and 2 were probed independently using qRT-PCR. These experiments verified the microarray results (Fig. S6).
Table 1.
Genes up-regulated 2-fold or more in H1299 cells expressing ΔNp53:WTp53, relative to WTp53.
| GENE NAME | FOLD CHG | p VALUE | DESCRIPTION |
|---|---|---|---|
| CDKL2 | 3.6808 | 0.0085 | Kinase; development |
| CHAC1 | 2.8481 | 0.0009 | cation transport; unfolded protein response |
| TMEM229A | 2.8415 | 0.0285 | transmembrane protein; putative TF |
| DUSP5P | 2.7321 | 0.0016 | pseudogene |
| ZNF443 | 2.7007 | 0.0208 | zinc-finger protein |
| RAB27B | 2.6512 | 0.0008 | GTPase; exosome secretion pathway |
| PDE5A | 2.6027 | 0.0211 | cGMP phosphodiesterase; purine metab. |
| MARS2 | 2.4967 | 0.0095 | Met tRNA synthetase, mitochondrial |
| ZBTB42 | 2.4794 | 0.03 | linked to metabolic syndrome |
| LOC643923 | 2.4794 | 0.0351 | |
| PLK3 | 2.406 | 0.0152 | Kinase; mitosis; Insulin/PI3K/mTOR pathway |
| DUSP10 | 2.3839 | 0.0072 | Phosphatase; MAPK signaling |
| ANKRD36B | 2.308 | 0.0075 | ankyrin repeat domain |
| IRF1 | 2.2921 | 0.0003 | TF; immune function |
| IFF02 | 2.2501 | 0.0028 | intermediate filament |
| C6orf114 | 2.2449 | 0.0067 | |
| HIST1H3H | 2.2397 | 0.0191 | histone H3 |
| IRS1 | 2.214 | 0.0013 | Insulin/PI3K/mTOR pathway |
| ZNF799 | 2.2089 | 0.0138 | zinc-finger protein |
| ZBTB1 | 2.2089 | 0.0004 | cell cycle |
| CNKSR3 | 2.1685 | 0.0002 | sodium transport; MAPK pathway |
| SNX33 | 2.1685 | 0.003 | phospholipid binding |
| DOT1L | 2.1585 | 0.0057 | H3K79 methyltransferase; cell cycle/diff. |
| C3orf59 | 2.1386 | 0.0037 | |
| ZNF354B | 2.1238 | 0.0138 | zinc-finger protein |
| JMY | 2.1092 | 0.0045 | p53 cofactor; cell motility |
| MAP3K1 | 2.1092 | 0.0047 | Kinase; Insulin signaling; differentiation |
| ANKZF1 | 2.0849 | 0.0032 | binds p97 AAA ATPase |
| DHRS13 | 2.0801 | 0.0296 | glucose dehydrogenase |
| VLDLR | 2.0801 | 0.0013 | FA metabolism |
| SNORD44 | 2.0801 | 0.0044 | snoRNA |
| SEPTIN6 | 2.0705 | 0.0011 | cytokinesis; GTPase |
| DENND3 | 2.0658 | 0.002 | GTP-GDP exchange; reg. MAPK signaling |
| P2RY1 | 2.061 | 0.0441 | ATP/ADP-gated receptor |
| WHAMML1 | 2.0562 | 0.0089 | pseudogene |
| LRCH1 | 2.0515 | 0.0094 | CHO transporter |
| CYBRD1 | 2.0467 | 0.0148 | heme binding/iron uptake; reductase |
| LNX2 | 2.042 | 0.0017 | ligand of numb protein X; Notch signaling |
| HIST1H2BF | 2.0373 | 0.0084 | histone H2B |
| ANGPTL4 | 2.0373 | 0 | glucose/FA metab; mitochondrial function |
| BHLHE41 | 2.0326 | 0.0039 | TF; circadian rhythm; differentiation |
| PLEKHF1 | 2.0326 | 0.0263 | phosphatidyl-inositol signaling |
| ANKRD36 | 2.0279 | 0.002 | ankyrin repeat domain |
| MGC16384 | 2.0232 | 0.0072 | |
| PTCH1 | 2.0232 | 0.0007 | sonic hedgehog receptor; Insulin signaling |
| FGF5 | 2.0232 | 0.0165 | MAPK/insulin signaling; differentiation |
| FAT4 | 2.0139 | 0.0016 | cell-cell adhesion |
| NFIL3 | 2.0046 | 0.02 | TF; circadian rhythm |
Table 2.
Genes down-regulated at least 2-fold in H1299 cells expressing ΔNp53:WTp53, relative to WTp53.
| GENE NAME | FOLD CHG | p VALUE | DESCRIPTION |
|---|---|---|---|
| GDF9 | 0.163 | 0.0001 | cytokine; growth/differentiation |
| UGT2B7 | 0.1826 | 0.0062 | UDP-glucuronyltransferase |
| CLCA2 | 0.2333 | 0.0066 | chloride channel; secretion |
| SERPINB5 | 0.2541 | 0.0003 | Maspin; cell motility |
| WDR69 | 0.2973 | 0.0045 | dynein assembly factor |
| AREG | 0.3064 | 0.006 | cytokine; EGF family |
| SLC2A2 | 0.3157 | 0.0014 | glucose xporter; CHO metabolism |
| LCE1C | 0.3231 | 0.0139 | late envelope protein; differentiation |
| DISP1 | 0.3479 | 0.0138 | hedgehog signaling; differentiation |
| PALMD | 0.361 | 0.0188 | palmdelphin |
| CRYAB | 0.3686 | 0.0009 | chaperone; ER-assoc. degradation |
| GPR56 | 0.3772 | 0.006 | G-protein receptor; differentiation |
| INPP5D | 0.3833 | 0.0001 | Insulin/PI3K/mTOR pathway |
| CKS2 | 0.3959 | 0.027 | CDK reg. protein; Insulin signaling |
| PLK1 | 0.3978 | 0.0456 | Kinase; reg. AURKA; mitosis |
| AURKA | 0.4043 | 0.024 | Kinase; reg. PLK1; reg. p53; mitosis |
| PTH2R | 0.4108 | 0.0099 | parathyroid hormone receptor |
| KLHL30 | 0.4156 | 0.0157 | kelch-like 30 |
| SATL1 | 0.4156 | 0.0015 | spermidine acetyltransferase |
| NCF2 | 0.4166 | 0.0102 | NADPH OX complex; differentiation |
| CENPA | 0.4166 | 0.039 | Centromere; reg. by AURKA; mitosis |
| GDF15 | 0.4253 | 0.0006 | cytokine; growth/differentiation |
| SLA | 0.4263 | 0.0021 | immune function |
| PLA2G10 | 0.4569 | 0.0159 | FA metabolism; arachidonic acid |
| LOC285679 | 0.4569 | 0.0366 | |
| KIF2C | 0.458 | 0.0439 | kinesin; centromere-associated |
| SLC46A1 | 0.459 | 0.0003 | folate/heme transporter |
| CLEC17A | 0.4633 | 0.0064 | CHO binding; cell-cell signaling |
| NEK2 | 0.4676 | 0.0248 | Kinase; mitosis; centrosome-assoc. |
| PADI3 | 0.473 | 0.011 | Arg de-iminase; urea cycle |
| BDKRB1 | 0.473 | 0.0041 | Bradykinin receptor; cell motility |
| CCDC99 | 0.4841 | 0.0367 | mitosis |
| KIF20A | 0.4863 | 0.0395 | kinesin |
| CEP55 | 0.4875 | 0.0427 | centrosome;cytokinesis;reg. by PLK1 |
| BEND2 | 0.4908 | 0.0014 | BEN domain |
| KIF4A | 0.4943 | 0.0324 | kinesin; chromosome dynamics |
We also used qRT-PCR to probe expression of numerous genes in Saos-2 cells. Like H1299 cells, Saos-2 cells are p53-null. Because p53-induced gene expression patterns are not identical in different cell types, we expected to observe some differences in transcription response (Espinosa, 2008). As shown in Figure S7, most, but not all, genes probed by qRT-PCR showed similar changes in expression levels in Saos-2 cells compared with H1299 cells.
The peptide linker itself does not alter biological function of p53
Collectively, the data in Figure 1 and 2 indicated that the flexible peptide linker inserted between ΔNp53 and WTp53, which was based upon the p53 tetramer structure (Okorokov et al., 2006), did not affect the biological activity of ΔNp53:WTp53 tetramers. To further address whether the linker itself might impact p53 function, we expressed tethered WTp53 tetramers (i.e. WTp53:WTp53) in p53-null H1299 cells. In other words, WTp53 dimers were expressed as a single transcript using the same linker sequence used for ΔNp53:WTp53. Microarray analyses were then completed in which we compared global gene expression changes induced by “standard” WTp53 vs. its tethered counterpart (i.e. WTp53:WTp53). These and other data, summarized in Figure S8A–J, indicate that the peptide linker itself does not appear to direct gene expression changes or otherwise alter WTp53 function, at least within the duration of these experiments.
In agreement with these findings, we performed qRT-PCR experiments in H1299 cells co-expressing equal levels of ΔNp53 and WTp53 (i.e. ΔNp53 + WTp53). As anticipated, and in agreement with data for p21 (Fig. 2D), gene expression from these “ΔNp53 + WTp53” cells was largely similar to ΔNp53:WTp53 cells (Fig. S9 and S10). Importantly, in cases where differences were observed, these could be attributed to tetramer heterogeneity (e.g. WTp53 tetramers) resulting from ΔNp53 and WTp53 co-expression (see legend, Fig. S8, S9 and S10).
Global metabolomics analyses
Whereas many genes up- or down-regulated in ΔNp53:WTp53 cells are directly or indirectly linked to aging, a significant fraction are involved in basic metabolic processes (Table 1 and Table 2). Given the major role that p53 plays in regulating cell metabolism (Feng & Levine, 2010; Vousden & Ryan, 2009), we compared the metabolomes of H1299 cells expressing WTp53 or ΔNp53:WTp53. The metabolomics analysis utilized an analytical platform that combines gas chromatography (GC) and liquid chromatography (LC) mass spectrometry (GC-MS and LC-MS/MS) techniques (Reitman et al., 2011).
Six biological replicate samples were prepared for metabolic profiling. In addition to WTp53 and ΔNp53:WTp53, we evaluated mock and GFP vector only transfected H1299 cells. Because cell cycle changes or changes in cell state (e.g. apoptosis or senescence) could potentially alter metabolite levels, we probed samples to ensure viability and that each cell population was similar in cell state and cell cycle distribution (Fig. S2B and S3). As with microarray analyses, we also confirmed that p53 levels were consistent and comparable in WTp53 vs. ΔNp53:WTp53 samples by quantitative western blot (Fig. S2A). A statistical summary of the biochemicals identified in each sample is shown in Table S3; a complete list of the metabolites identified, including statistical parameters and absolute or relative quantitation, is provided in Table S4.
Metabolic changes associated with WTp53 support roles in tumor suppression and metabolism
Relative to control cells (WTp53 vs. GFP vector; Table S3) 142 biochemicals were categorized as significantly altered in H1299 cells expressing WTp53 (p ≤ 0.05). As anticipated, the metabolic alterations induced by WTp53 support its known roles as a metabolic regulator and tumor suppressor (Vousden & Ryan, 2009). Although the primary focus of this study was how the ΔNp53 isoform altered WTp53 function, we highlight the metabolic changes induced by WTp53 in Table S5.
Metabolic changes associated with ΔNp53:WTp53 vs. WTp53
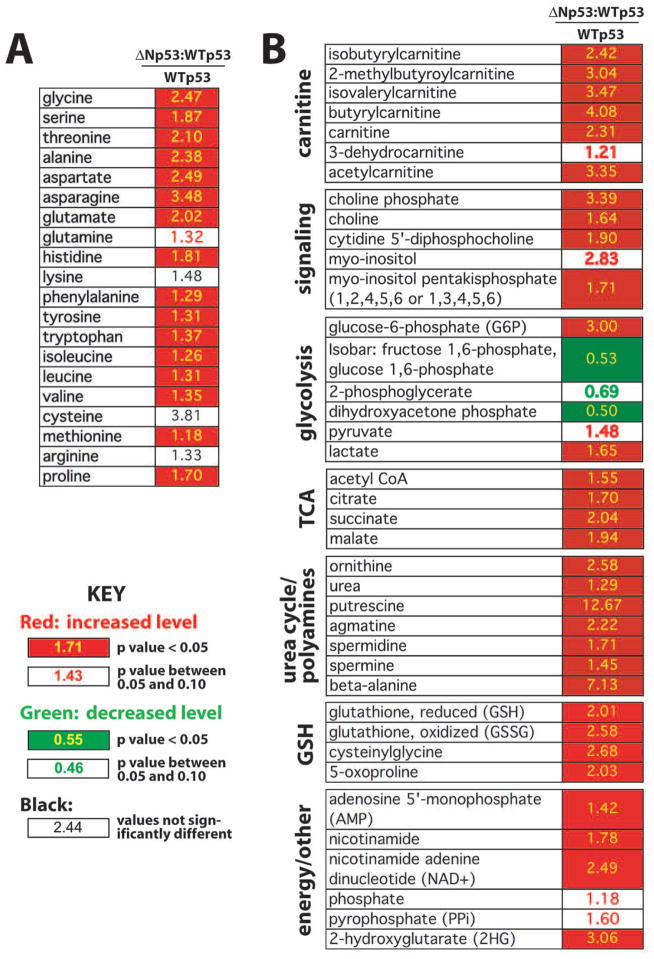
Compared with WTp53, the levels of 94 biochemicals were altered (p ≤ 0.05) in cells expressing ΔNp53:WTp53 (Table S3). Many intriguing results emerge from these data, and several key findings are highlighted in Figure 3. As shown in Figure 3A, amino acid levels were uniformly elevated in ΔNp53:WTp53 cells, compared with WTp53. Moreover, derivatives of these amino acids (e.g. N-acetylated forms) were more abundant in ΔNp53:WTp53 cells (Table S4). Figure 3B shows a subset of biochemicals whose levels were increased or decreased in ΔNp53:WTp53 cells, including carnitine and its derivatives, TCA/Krebs cycle intermediates, and glutathione metabolites.
Fig. 3.
Summary of metabolomics data. (A) Relative levels of amino acids in ΔNp53:WTp53 cells vs. WTp53. (B) Relative levels of select metabolites (ΔNp53:WTp53 vs. WTp53).
Elevated amino acids are known to activate the mTOR pathway, which acts as a nutrient sensor and regulates lifespan in yeast, worms, flies, and mammals (Laplante & Sabatini, 2012). Moreover, elevated levels of TCA intermediates and carnitine (Fig. 3B), which is critical for transporting fatty acids within mitochondria, suggested alterations in mitochondrial function in cells expressing ΔNp53:WTp53. As with the mTOR pathway, mitochondrial function is strongly linked to mammalian aging (Vendelbo & Nair, 2011).
Given the number of genes and metabolites altered in ΔNp53:WTp53 cells relative to cells expressing WTp53, pathway analyses were completed as an additional means to evaluate the data. Results are summarized in Table S6 – S9 and further implicate mTOR and mitochondrial function in ΔNp53:WTp53 cells. Therefore, we decided to probe ΔNp53:WTp53 cells for changes in mitochondrial function and mTOR signaling.
Altered mitochondrial function in ΔNp53:WTp53 cells
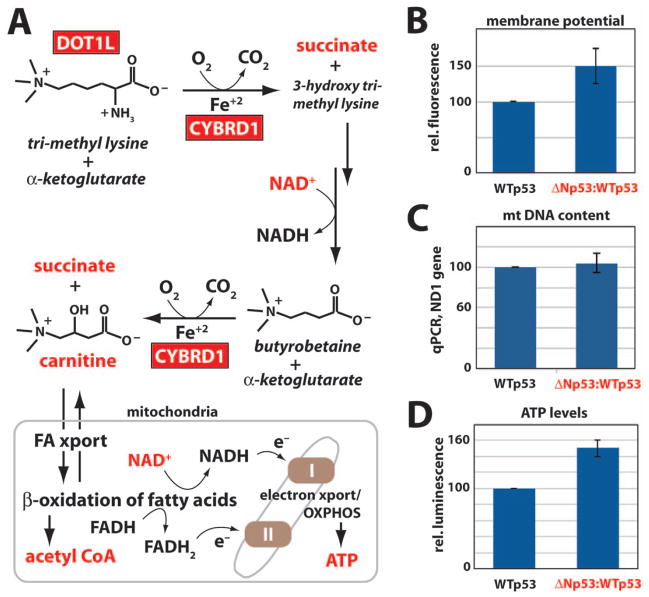
Gene expression (e.g. MARS2, CYBRD1) and metabolic changes (Fig. 3B) suggested altered mitochondrial function in ΔNp53:WTp53 cells (Bayat et al., 2012; Xu et al., 2010). Carnitine is critical for transporting fatty acids within mitochondria; subsequent β-oxidation of fatty acids generates acetyl CoA, NADH and FADH2. NADH and FADH2, in turn, provide electrons that drive mitochondrial electron transport and oxidative phosphorylation (Fig. 4A). The increase in carnitine and its intermediates suggested that mitochondrial function (in particular, ATP production) might be altered in cells expressing ΔNp53:WTp53. Gene expression changes may contribute to the increased carnitine levels in ΔNp53:WTp53 cells; for example, the lysine methyltransferase DOT1L and the iron import protein CYBRD1 were up-regulated over 2-fold (Fig. 4A).
Fig. 4.
Altered mitochondrial function in cells expressing ΔNp53:WTp53 relative to WTp53. (A) Simplified schematic of carnitine biosynthesis and β-oxidation of fatty acids in mitochondria. Red boxes indicate genes up-regulated in ΔNp53:WTp53 cells, and red font denotes biochemicals whose levels are increased in ΔNp53:WTp53 cells, relative to WTp53. (B) Mitochondrial membrane potential in H1299 cells expressing WTp53 or ΔNp53:WTp53. Plot shows mean and standard error of Mitotracker Red fluorescence intensities from three independent experiments. Data were normalized to WTp53. (C) Mitochondrial DNA copy number (as determined by real time PCR) in H1299 cells expressing WTp53 or ΔNp53:WTp53. Data were normalized to CDKN1A/p21 genomic DNA. (D) Relative ATP levels (normalized to WTp53) in H1299 cells. Plot shows mean and standard error from three independent experiments. See also Figure S11.
To determine whether mitochondrial function might be altered in H1299 cells expressing ΔNp53:WTp53, we measured mitochondrial membrane potential using Mitotracker Red. As shown in Figure 4B, cells expressing ΔNp53:WTp53 display increased Mitotracker staining, indicative of a greater potential difference across the inner mitochondrial membrane. The apparent increase in membrane potential did not result from an increased number of mitochondria in ΔNp53:WTp53 cells, because no significant changes in mitochondrial DNA content were observed in cells expressing WTp53 vs. ΔNp53:WTp53 (Fig. 4C). Together, these data confirm that an increase in membrane potential accompanies ΔNp53:WTp53 expression.
Mitochondrial membrane potential is generated via electron transport, which establishes a proton gradient that helps drive ATP synthesis. The increased membrane potential (Fig. 4B) suggested that ATP production might be enhanced in ΔNp53:WTp53 cells. To assess ATP levels, we implemented a quantitative, luciferase-based assay. This analysis revealed an ~50% increase in ATP production in cells expressing ΔNp53:WTp53 relative to WTp53 (Fig. 4D), consistent with the increased mitochondrial membrane potential shown in Figure 4B. Collectively, the data in Figure 4 (see also Fig. S11) indicate an activity for ΔNp53:WTp53 that alters mitochondrial function in a manner distinct from WTp53.
Upregulation of mTOR pathway in ΔNp53:WTp53 cells
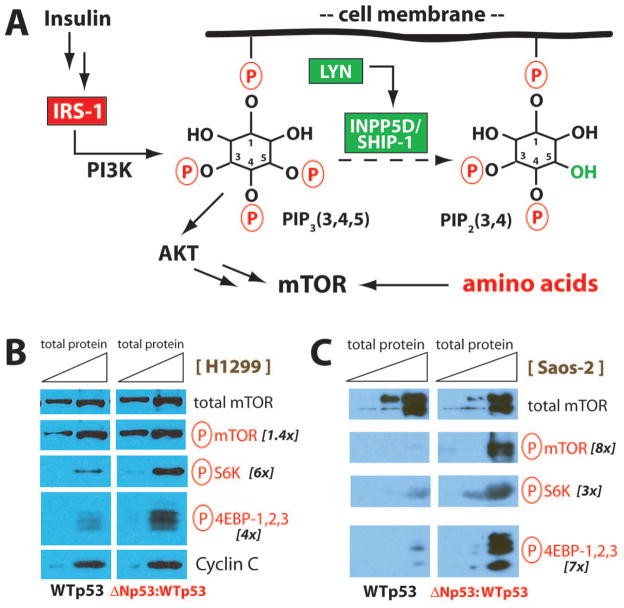
The across-the-board elevation in amino acids (Fig. 3A) suggested the mTOR pathway was activated in ΔNp53:WTp53 cells. Amino acids are known to activate mTOR via an elaborate mechanism involving lysosomes and a set of Rag GTPases (Laplante & Sabatini, 2012). Hyperactive mTOR signaling was further supported by the gene expression data, which indicated increased expression of IRS-1 and decreased expression of INPP5D/SHIP-1 in ΔNp53:WTp53 cells, compared with WTp53 (Table 1 and Table 2). As shown schematically in Figure 5A, IRS-1 activates PI3K to generate phosphoinositol (3,4,5) triphosphate; this serves to activate AKT kinase, an upstream activator of mTOR. Similarly, decreased expression of the 5-position inositol phosphatase INPP5D would promote phosphoinositol (3,4,5) triphosphate signaling by limiting its breakdown to phosphoinositol (3,4) diphosphate. In addition, reduced expression of LYN (which activates INPP5D) in ΔNp53:WTp53 cells (Table S2) could contribute to mTOR activation through a variety of mechanisms (Baran et al., 2003; Gardai et al., 2002).
Fig. 5.
The mTOR pathway is activated in cells expressing ΔNp53:WTp53. (A) Simplified schematic of signaling events that regulate the mTOR pathway. Red boxes indicate genes up-regulated in ΔNp53:WTp53 cells, and green boxes indicate genes down-regulated. Note the LYN kinase activates INPP5D (Baran et al., 2003; Gardai et al., 2002), and both gene products were down-regulated in ΔNp53:WTp53 cells. Amino acid levels were uniformly elevated in ΔNp53:WTp53 cells, relative to WTp53. (B) Western blot data from H1299 cells, showing increased levels of mTOR kinase targets (red font) in ΔNp53:WTp53 cells, compared with WTp53. mTOR substrates included phospho-mTOR (S2448), phospho-p70 S6 Kinase (S371), and phospho-4E-BP1/2/3 (T37/46). Each experiment (WTp53 or ΔNp53:WTp53) loaded the same amount of total protein (7.9 μg and 31.7 μg, respectively), with additional loading controls (total mTOR, CCNC) shown. Quantitation (fold change) is indicated at right. (C) Western experiments completed as described in B, except using p53-null Saos-2 cells. Total protein loaded in each lane was 0.9 μg, 3 μg, and 9 μg. Quantitation (fold change) shown at right. See also Figure S12.
To test whether the mTOR pathway was up-regulated in ΔNp53:WTp53 cells, we probed for well-established markers of mTOR activation, including phospho-S6 kinase, phospho-4EBP-1,2,3, and phospho-mTOR. Levels of the phosphorylated intermediates were compared with total mTOR and CCNC, as shown in Figure 5B. Phospho-S6 kinase, phospho-4EBP-1,2,3, and phospho-mTOR were uniformly increased in ΔNp53:WTp53 H1299 cells, compared with cells expressing WTp53 (Fig. 5B). We next evaluated levels of mTOR pathway intermediates in a different p53-null cell line, Saos-2. As shown in Figure 5C, phosphorylated mTOR, phospho-S6K, and phospho-4EBP-1,2,3 were each elevated in Saos-2 cells expressing ΔNp53:WTp53 compared with WTp53. Taken together, the data in Figure 5 (see also Fig. S12) demonstrate that ΔNp53:WTp53 expression activates the mTOR pathway in human cells. This outcome is opposed to WTp53, which suppresses mTOR signaling (Feng & Levine, 2010).
Discussion
The p53 transcription factor is expressed in most cell types, plays important roles throughout development, and helps sustain basic cellular functions, such as stress response. Whereas myriad factors influence aging, this study focused on the naturally occurring human ΔNp53 isoform, whose expression causes accelerated aging in mice (Maier et al., 2004; Tyner et al., 2002). To more thoroughly understand the distinct activities of human ΔNp53, we focused on differences relative to WTp53. Our goal was not to distinguish direct from indirect effects of ΔNp53, but rather to gain insight into the cellular pathways altered by its expression. A key feature of this study was analysis of ΔNp53:WTp53 tetramers with fixed 2:2 stoichiometry; this circumvented confounding issues associated with ΔNp53 and WTp53 co-expression and allowed clear delineation between WTp53 and ΔNp53:WTp53. Another key feature was the combination of gene expression and metabolomics data, which allowed better assessment of pathways altered by ΔNp53:WTp53. On their own, gene expression data correlate poorly with organismal aging. Age-associated changes in gene expression are heterogeneous and vary with cell or tissue type (Bahar et al., 2006; Somel et al., 2006). Also, age-associated gene expression changes are not consistent across model organisms (Zahn et al., 2007). Signaling and metabolic pathways, however, are more consistently linked to aging across different cell types and even model organisms (Vijg & Campisi, 2008). Collectively, our data suggest that in human cells, ΔNp53:WTp53 helps orchestrate changes in basic cellular processes that contribute to aging. In fact, mitochondrial function and mTOR signaling are linked to longevity in organisms as distinct as mammals and yeast (Laplante & Sabatini, 2012; Vendelbo & Nair, 2011).
We would like to point out some advantages and limitations of our study. Whereas the metabolomics analyses complemented the gene expression data and provide a wealth of information, they do not offer a fully comprehensive view of cellular alterations that accompany ΔNp53:WTp53 expression. The total number of chemically distinct metabolites in human cells exceeds a few thousand (Wishart et al., 2009), whereas we identified several hundred in this study. We also cannot delineate the cellular location of each metabolite, or its lifetime within each cell. Furthermore, non-coding RNAs can affect gene expression and cell physiology, and it remains unclear how ΔNp53:WTp53 affects expression of non-coding RNA genes. The effect of ΔNp53:WTp53 vs. WTp53 on H1299 cell growth, cell cycle, or senescence was essentially identical; however, a ~10% difference was observed in annexin V staining (early apoptosis). This could make some contribution to gene expression and metabolic differences between ΔNp53:WTp53 and WTp53 cells; however, our analyses focused on large-scale changes (typically 2-fold or greater) such that it is unlikely a major contributing factor. Consistent with this, phenotypic changes (e.g. mitochondrial membrane potential or mTOR signaling) were insensitive to removal of annexin V-positive cells and were consistent at 24 or 41 hours following ΔNp53:WTp53 or WTp53 expression. We acknowledge that the gene expression and metabolic changes induced by ΔNp53:WTp53 have similarities with aging studies in model organisms. Aging is controlled by a complex array of genetic, epigenetic, and environmental factors, however, and the data shown here focus only on ΔNp53:WTp53-dependent changes. Thus, our results will not correlate with all aging studies completed in model organisms. Moreover, the bulk of our analysis was completed in a single cell type. Although this has obvious advantages (isogenic; identical culture conditions) and avoids physical and physiological heterogeneity inherent in young vs. old comparisons, the ΔNp53:WTp53-induced changes identified here may not be identical in distinct cell types or contexts.
Analysis of ΔNp53:WTp53 tetramers as a single entity
The cryo-EM structure of the WTp53 tetramer has provided unprecedented insight about its structural organization (Okorokov et al., 2006) and revealed a straightforward means to link ΔNp53 and WTp53 as a single transcript while preserving p53 tetramer architecture. As expected, the flexible peptide linker—which was longer than necessary to enable conformational variability—did not notably affect the biological activity of p53, as tethered versions of WTp53 (i.e. WTp53:WTp53) mimicked phenotypic and gene expression changes induced by native WTp53 tetramers. Similarly, ΔNp53:WTp53 formed stable tetramers, bound p53 target sequences in cells, activated transcription similar to WTp53 in vitro, and displayed similar activity to co-expressed ΔNp53 + WTp53 in cells. It has been postulated that translation of p53 generates homo-dimers, followed by post-translational tetramer formation (Nicholls et al., 2002). Based upon this model, the structural organization of 2:2 ΔNp53:WTp53 tetramers would be slightly distinct, with one WT dimer merging with one ΔNp53 dimer. Whether this assembly mechanism predominates in cells remains unclear, in part because few studies have directly addressed this question, likely due to its enormous practical and technical challenges. The similar gene expression changes induced by ΔNp53:WTp53 compared with ΔNp53 + WTp53, however, suggest that either these potential architectural distinctions have little functional consequence, or that the linked ΔNp53:WTp53 tetramers evaluated here represent a predominant tetramer architecture.
Expression of linked ΔNp53:WTp53 dimers to ensure a 2:2 tetramer stoichiometry offered several advantages over co-expression of ΔNp53 and WTp53 (i.e. ΔNp53 + WTp53). Because ΔNp53:WTp53 stoichiometry cannot be controlled with separate co-expression (even when each protein is present at similar levels), up to five distinct entities could form, ranging from zero to four copies of ΔNp53 within any given tetramer. The strategy employed here avoided this heterogeneity, and ensured the presence of a single entity in which the ΔNp53:WTp53 stoichiometry was kept constant. Equally important, our strategy ensured that gene expression and metabolomics data would not be confounded by the presence of contaminating WTp53 tetramers that result from ΔNp53 and WTp53 co-expression. Thus, the gene expression and metabolomics comparison between ΔNp53:WTp53 and WTp53 completed here enabled a clear contrast that would simply not be possible with any series of co-expression studies.
Neomorphic function of human ΔNp53
Mutations can cause a gain- or loss-of-function in a transcription factor that modify its effect on global gene expression patterns; such “neomorphic” mutations are well documented for p53 (Freed-Pastor & Prives, 2012). In general, the effects of ΔNp53:WTp53 vs. WTp53 in H1299 cells were largely indistinguishable. Only a subset of genes were differentially expressed, and only minor changes, if any, were observed upon assessing cell growth, apoptosis, cell cycle, or senescence. Potentially, ΔNp53:WTp53 could gain functions that promote growth; however, this was not evident based upon our observations with cultured cells or from the gene expression and metabolomics data. Based upon our comparative analyses with WTp53, alterations in mTOR signaling and mitochondrial function appear to represent neomorphic outcomes of ΔNp53:WTp53 expression. Whereas WTp53 is a negative regulator of mTOR signaling (Feng & Levine, 2010), this function appears to be lost with ΔNp53:WTp53. Up-regulation of the mTOR pathway appears to result—at least in part—from altered gene expression (IRS-1, INPP5D) and metabolic changes (amino acids) in cells expressing ΔNp53:WTp53.
Potential mechanisms that underlie the mitochondrial changes (e.g. increased membrane potential) can also be deduced from the metabolic and gene expression data. In addition to carnitine and its role in mitochondrial fatty acid transport, we observed an up-regulation of MARS2, the mitochondrial tRNAmet synthetase, which suggests enhanced translation of mitochondrial proteins in ΔNp53:WTp53 cells. Because translation initiates with the Met codon, it has been proposed that elevated levels of tRNAmet up-regulate translation (Marshall et al., 2008). Increased translation of mitochondrial proteins, in particular those involved in electron transport and oxidative phosphorylation, could result in the observed increase in membrane potential and ATP. Mitochondrial function is also sensitive to iron. In ΔNp53:WTp53 cells, we observed increased levels of glutathione and its derivatives, along with altered expression of the heme transporter SLC46A1 and the iron reductase/iron import protein CYBRD1. Glutathione is essential for regulating iron metabolism (Kumar et al., 2011), and increased levels of glutathione metabolites, coupled with changes in iron import-export factors, suggests that iron homeostasis might be disrupted in ΔNp53:WTp53 cells. Mitochondrial function is highly dependent upon iron, in part because many proteins and protein complexes important for electron transport contain iron-sulfur clusters. In fact, glutathione is required for proper biogenesis of iron-sulfur clusters (Lill, 2009).
Given the well-studied roles of the mTOR pathway and mitochondrial function in longevity, our results with ΔNp53:WTp53 are consistent with its accelerated aging phenotype in mice and implicate the naturally occurring ΔNp53 isoform as a potential regulator of human aging.
Potential links between ΔNp53 and normal physiological aging
A growing number of studies indicate that single metabolites are sufficient to trigger phenotypic changes in cells and within an organism. To cite a few examples, acetate can correct insulin signaling defects (Shin et al., 2011) and choline can promote atherosclerosis (Wang et al., 2011). The metabolic changes induced by ΔNp53:WTp53 expression are striking and encompass a limited set of pathways. We observed that amino acids, urea cycle intermediates, polyamines, and glutathione metabolites were significantly elevated in cells expressing ΔNp53:WTp53, relative to WTp53. Numerous studies have linked these same metabolites to aging and longevity in mammalian model organisms; however, far fewer studies have addressed the metabolic changes that accompany human aging. A metabolomic study of human plasma isolated from healthy old vs. young donors shows clear similarities to our metabolomic comparison of ΔNp53:WTp53 and WTp53 (Lawton et al., 2008). As seen in ΔNp53:WTp53 cells, amino acid levels were uniformly increased in old vs. young human plasma samples, as were polyamines, carnitine, TCA cycle, and urea cycle metabolites. Potentially, these shared features could reflect a distinct metabolic signature in aged human cells that is rapidly adopted upon ΔNp53 expression. Much additional work will be needed to rigorously test this hypothesis.
Among the various models for mammalian aging, a common theme includes stochastic, heritable changes that gradually alter gene expression patterns and metabolic pathways (Martin, 2009). Such mechanisms appear to represent a fundamental requirement for aging, as biological effects manifest over time. Because ΔNp53 is a naturally occurring isoform, its levels could change with age. Existing gene expression data have limited value, however, as ΔNp53 levels are controlled primarily by alternate translation (Candeias et al., 2006; Ray et al., 2006). In mice, relative levels of ΔNp53 change with developmental state (Ungewitter & Scrable, 2010), and several reports indicate that human ΔNp53 protein levels change during cell stress (Bourougaa et al., 2010; Candeias et al., 2006; Courtois et al., 2002). The links to cell stress imply that transient increases in ΔNp53 levels can occur throughout the human lifespan.
Could transient elevation of ΔNp53 levels have chronic, long-term consequences? We note two potential mechanisms by which transient ΔNp53 expression could initiate a self-reinforcing cascade, with possible implications for aging. First, it is established that ER stress increases relative levels of ΔNp53 vs. WTp53 in human cells by favoring alternate translation initiation from an IRES within the WTp53 mRNA (Bourougaa et al., 2010). Elevated levels of 2-hydroxyglutarate (2HG) can independently induce an ER stress response (Sasaki et al., 2012). Because 2HG is elevated in ΔNp53:WTp53 cells, ΔNp53 could promote its own translation by inducing ER stress. Second, we observed elevated levels of succinate and 2HG in cells expressing ΔNp53:WTp53, as well as increased expression of the histone H3K79 methyltransferase DOT1L. This suggests that DNA methylation and histone H3K79 methylation patterns may be globally altered by 3Np53 expression. Because 2HG or succinate can competitively inhibit demethylase enzymes that use α-ketoglutarate as a cofactor, elevated 2HG and succinate favors a general retention of methyl marks on both histones and DNA (Xiao et al., 2012; Xu et al., 2011). DNA methylation and histone H3K79 methylation are epigenetic regulators of transcription that may enable even transient 3Np53 expression to “lock in” gene expression patterns that propagate to cell progeny. Indeed, whereas both H3K79 methylation and DNA methylation marks are reversible, DNA methylation also appears to be heritable (Bonasio et al., 2010). Future studies will focus on whether ΔNp53:WTp53 alters patterns of DNA methylation and/or H3K79 tri-methylation genome-wide, and how this might contribute to the geriatric pathology of ΔNp53.
Accession Numbers
Microarray data were deposited in GEO (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE37271.
Experimental Procedures
In vitro transcription
In vitro transcription reactions and purification of factors for the reconstituted transcription system were completed as described (Knuesel et al., 2009).
Microarray analyses
Total RNA from FACS sorted cells was extracted with the RNeasy kit (Qiagen) and was analyzed at the Genomics and Microarray Shared Resource (University of Colorado Cancer Center) for transcriptional profiling. Per standard protocol, 100 to 150 ng of starting total RNA was converted to cDNA with the Ambion WT Expression kit, according to the manufacturer’s protocol. After standard labeling, each sample was hybridized to Affymetrix GeneChip Human Gene 1.0 ST array, followed by examination with an Affymetrix GeneChip Scanner 3000.
Global metabolic profiling
Metabolomic profiling analysis was performed by Metabolon (Durham, NC) as described (Reitman et al., 2011).
Real time quantitative polymerase chain reaction
Supplementary Material
Fig. S1 Schematic of ΔNp53:WTp53.
Fig. S2 Representative western blot and cell cycle data.
Fig. S3 Assessment of apoptosis, growth, and senescence
Fig. S4 GSEA of microarray data.
Fig. S5 Venn diagrams of gene expression data.
Fig. S6 qRT-PCR validation of microarray data.
Fig. S7 qRT-PCR data from Saos-2 cells.
Fig. S8 Evaluation of WTp53:WTp53 tetramers.
Fig. S9 qRT-PCR data comparing ΔNp53:WTp53 with ΔNp53 and WTp53 co-expressed at equal levels.
Fig. S10 Comparison of ΔNp53-induced gene expression changes in H1299 cells.
Fig. S11 Additional data for mitochondrial function.
Fig. S12 Additional data for mTOR pathway.
Table S1 Genes up-regulated 1.5-fold or greater in H1299 cells expressing ΔNp53:WTp53, compared with WTp53.
Table S2 Genes down-regulated 1.5-fold or greater in H1299 cells expressing ΔNp53:WTp53, compared with WTp53.
Table S3 Overview of metabolomics data.
Table S4 Metabolomics data.
Table S5 Summary of metabolic changes induced by WTp53.
Table S6 Pathway analysis of gene expression data: GO Processes and GO Metabolic Networks.
Table S7 Pathway analysis of gene expression data: GO Pathways.
Table S8 Pathway analysis of Metabolomics data.
Table S9 Merged pathway analysis of gene expression and metabolomics data.
Table S10 Primer sequences used for cloning and qRT-PCR.
Acknowledgments
We thank Joaquin Espinosa and his lab for generous donation of the H1299 and Saos-2 cell lines, as well as use of RT-PCR and FACS instrumentation and valuable advice. Theresa Nahreini assisted with cell culture and cell sorting experiments. We thank Jim Goodrich and Joaquin Espinosa for comments on the manuscript. Microarray analyses were completed at the Genomics Shared Resource at the University of Colorado Cancer Center (UCCC); WTp53, ΔNp53:WTp53, and ΔNp53 proteins were expressed at the Tissue Culture Shared Resource at the UCCC. This work was supported by grants from the NCI (CA127364), the American Cancer Society, the Ellison Medical Foundation, and the NIH (R03 AG035228) to DJT.
Footnotes
Author contributions
Conceived and designed the experiments: SL and DT. Performed the experiments: SL, EK, and DT. Analyzed the data: SL, EK, DT. Wrote the paper: SL and DT.
The authors declare no conflict of interest.
References
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Baran CP, Tridandapani S, Helgason CD, Humphries RK, Krystal G, Marsh CB. The inositol 5′-phosphatase SHIP-1 and the Src kinase Lyn negatively regulate macrophage colony-stimulating factor-induced Akt activity. J Biol Chem. 2003;278:38628–38636. doi: 10.1074/jbc.M305021200. [DOI] [PubMed] [Google Scholar]
- Bayat V, Thiffault I, Jaiswal M, Tetreault M, Donti T, Sasarman F, Bernard G, Demers-Lamarche J, Dicaire MJ, Mathieu J, Vanasse M, Bouchard JP, Rioux MF, Lourenco CM, Li Z, Haueter C, Shoubridge EA, Graham BH, Brais B, Bellen HJ. Mutations in the Mitochondrial Methionyl-tRNA Synthetase Cause a Neurodegenerative Phenotype in Flies and a Recessive Ataxia (ARSAL) in Humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourougaa K, Naski N, Boularan C, Mlynarczyk C, Candeias MM, Marullo S, Fahraeus R. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol Cell. 2010;38:78–88. doi: 10.1016/j.molcel.2010.01.041. [DOI] [PubMed] [Google Scholar]
- Candeias MM, Powell DJ, Roubalova E, Apcher S, Bourougaa K, Vojtesek B, Bruzzoni-Giovanelli H, Fahraeus R. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene. 2006;25:6936–6947. doi: 10.1038/sj.onc.1209996. [DOI] [PubMed] [Google Scholar]
- Courtois S, Verhaegh G, North S, Luciani MG, Lassus P, Hibner U, Oren M, Hainaut P. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene. 2002;21:6722–6728. doi: 10.1038/sj.onc.1205874. [DOI] [PubMed] [Google Scholar]
- Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008;27:4013–4023. doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardai S, Whitlock BB, Helgason C, Ambruso D, Fadok V, Bratton D, Henson PM. Activation of SHIP by NADPH oxidase-stimulated Lyn leads to enhanced apoptosis in neutrophils. J Biol Chem. 2002;277:5236–5246. doi: 10.1074/jbc.M110005200. [DOI] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator co-activator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C, Igbaria A, D’Autreaux B, Planson AG, Junot C, Godat E, Bachhawat AK, Delaunay-Moisan A, Toledano MB. Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 2011;30:2044–2056. doi: 10.1038/emboj.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA, Milburn MV. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Martin GM. Epigenetic gambling and epigenetic drift as an antagonistic pleiotropic mechanism of aging. Aging Cell. 2009;8:761–764. doi: 10.1111/j.1474-9726.2009.00515.x. [DOI] [PubMed] [Google Scholar]
- Nicholls CD, McLure KG, Shields MA, Lee PW. Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J Biol Chem. 2002;277:12937–12945. doi: 10.1074/jbc.M108815200. [DOI] [PubMed] [Google Scholar]
- Okorokov AL, Sherman MB, Plisson C, Grinkevich V, Sigmundsson K, Selivanova G, Milner J, Orlova EV. The structure of p53 tumor suppressor protein reveals the basis for its functional plasticity. EMBO J. 2006;25:5191–5200. doi: 10.1038/sj.emboj.7601382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehar M, O’Riordan KJ, Burns-Cusato M, Andrzejewski ME, del Alcazar CG, Burger C, Scrable H, Puglielli L. Altered longevity-assurance activity of p53:p44 in the mouse causes memory loss, neurodegeneration and premature death. Aging Cell. 2010;9:174–190. doi: 10.1111/j.1474-9726.2010.00547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DJ, Hrstka R, Candeias M, Bourougaa K, Vojtesek B, Fahraeus R. Stress-dependent changes in the properties of p53 complexes by the alternative translation product p53/47. Cell Cycle. 2008;7:950–959. doi: 10.4161/cc.7.7.5626. [DOI] [PubMed] [Google Scholar]
- Ray PS, Grover R, Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006;7:404–410. doi: 10.1038/sj.embor.7400623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio, Cairns RA, McCracken S, Wakeham A, Haight J, Ten AY, Snow B, Ueda T, Inoue S, Yamamoto K, Ko M, Rao A, Yen KE, Su SM, Mak TW. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26:2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- Somel M, Khaitovich P, Bahn S, Paabo S, Lachmann M. Gene expression becomes heterogeneous with age. Curr Biol. 2006;16:R359–360. doi: 10.1016/j.cub.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Park SH, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010;24:2408–2419. doi: 10.1101/gad.1987810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendelbo MH, Nair KS. Mitochondrial longevity pathways. Biochim Biophys Acta. 2011;1813:634–644. doi: 10.1016/j.bbamcr.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Campisi J. Puzzles, promises and a cure for ageing. Nature. 2008;454:1065–1071. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S, Ye D, Xiong Y, Guan KL. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Marzetti E, Seo AY, Kim JS, Prolla TA, Leeuwenburgh C. The emerging role of iron dyshomeostasis in the mitochondrial decay of aging. Mech Ageing Dev. 2010;131:487–493. doi: 10.1016/j.mad.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, Lakatta EG, Boheler KR, Xu X, Mattson MP, Falco G, Ko MS, Schlessinger D, Firman J, Kummerfeld SK, Wood WH, 3rd, Zonderman AB, Kim SK, Becker KG. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Schematic of ΔNp53:WTp53.
Fig. S2 Representative western blot and cell cycle data.
Fig. S3 Assessment of apoptosis, growth, and senescence
Fig. S4 GSEA of microarray data.
Fig. S5 Venn diagrams of gene expression data.
Fig. S6 qRT-PCR validation of microarray data.
Fig. S7 qRT-PCR data from Saos-2 cells.
Fig. S8 Evaluation of WTp53:WTp53 tetramers.
Fig. S9 qRT-PCR data comparing ΔNp53:WTp53 with ΔNp53 and WTp53 co-expressed at equal levels.
Fig. S10 Comparison of ΔNp53-induced gene expression changes in H1299 cells.
Fig. S11 Additional data for mitochondrial function.
Fig. S12 Additional data for mTOR pathway.
Table S1 Genes up-regulated 1.5-fold or greater in H1299 cells expressing ΔNp53:WTp53, compared with WTp53.
Table S2 Genes down-regulated 1.5-fold or greater in H1299 cells expressing ΔNp53:WTp53, compared with WTp53.
Table S3 Overview of metabolomics data.
Table S4 Metabolomics data.
Table S5 Summary of metabolic changes induced by WTp53.
Table S6 Pathway analysis of gene expression data: GO Processes and GO Metabolic Networks.
Table S7 Pathway analysis of gene expression data: GO Pathways.
Table S8 Pathway analysis of Metabolomics data.
Table S9 Merged pathway analysis of gene expression and metabolomics data.
Table S10 Primer sequences used for cloning and qRT-PCR.