Abstract
Aims/hypothesis
Type 2 diabetes is a chronic, heterogeneous disease and a major risk factor for cardiovascular diseases. The underlying mechanisms leading to progression to type 2 diabetes are not fully understood and genetic tools may help to identify important pathways of glycaemic deterioration.
Methods
Using prospective data on American Indians from the Strong Heart Family Study, we identified 373 individuals defined as progressors (diabetes incident cases), 566 individuals with transitory impaired fasting glucose (IFG) and 1,011 controls (normal fasting glycaemia at all visits). We estimated the heritability (h2) of the traits and the evidence for association with 16 known variants identified in type 2 diabetes genome-wide association studies.
Results
We noted high h2 for diabetes progression (h2=0.65±0.16, p=2.7×10−6) but little contribution of genetic factors to transitory IFG (h2=0.09±0.10, p=0.19) for models adjusted for multiple risk factors. At least three variants (in WFS1, TSPAN8 and THADA) were nominally associated with diabetes progression in age- and sex-adjusted analyses with estimates showing the same direction of effects as reported in the discovery European ancestry studies.
Conclusions/interpretation
Our findings do not exclude these loci for diabetes susceptibility in American Indians and suggest phenotypic heterogeneity of the IFG trait, which may have implications for genetic studies when diagnosis is based on a single time-point measure.
Keywords: Heritability, Impaired fasting glucose, Single nucleotide polymorphisms, Type 2 diabetes
Introduction
Type 2 diabetes is a chronic, heterogeneous, complex disease and a major risk factor for cardiovascular and renal diseases. It is defined by abnormalities of fasting or postprandial glucose, which result from insulin resistance and pancreatic beta cell dysfunction [1]. Progressive deterioration in beta cell function is associated with beta cell loss due to multiple factors including apoptosis [2]. In insulin-resistant individuals, such as those with obesity, beta cell dysfunction precedes type 2 diabetes onset [3, 4]. Recent evidence suggests that defects in beta cell function occur early in the pathogenesis of type 2 diabetes, when individuals progress from normal fasting glucose (NFG) to prediabetes stages of impaired fasting glucose (IFG) and/or impaired glucose tolerance (IGT) measured using a 2 h OGTT [1, 5, 6]. American Indians have a higher rate of type 2 diabetes compared with other racial/ethnic populations [7–10]. Studies of Pima Indians have shown that beta cell dysfunction is a major determinant of progression from normoglycaemia to type 2 diabetes [6]; impairment in first-phase insulin secretion and a high basal hepatic endogenous glucose output were observed in prediabetic American Indians who progressed to type 2 diabetes [6].
IFG and IGT prevalence is approximately 15% and 26%, respectively, in the US adult population [11, 12], but higher in American Indians [13]. Prediabetes, a known risk factor for incident type 2 diabetes [8, 9], is phenotypically heterogeneous [14] with only 25–30% of individuals progressing to type 2 diabetes over 5 years [15]. In the Strong Heart Study (SHS), for example, 36.6% of American Indians with prediabetes developed type 2 diabetes at a median follow-up of 7.8 years [13]. Intense lifestyle and/or drug therapy have been shown to prevent or delay progression to type 2 diabetes in some individuals [16–20]. The underlying mechanisms leading to progression to type 2 diabetes are not fully elucidated [21], with a critical barrier being few prospective studies.
Studies suggest a strong genetic component to type 2 diabetes risk (familial aggregation, high concordance rates in monozygotic twins and increased risk in first-degree relatives of affected individuals) [22–24]. Over 60 genetic loci have been identified in candidate gene and genome-wide association (GWA) studies of type 2 diabetes [25–28]. Many of the identified loci are in or nearby genes affecting pancreatic beta cell development and function and insulin secretion [25, 29, 30], supporting a role for beta cell dysfunction and insulin resistance in type 2 diabetes. However, few large-scale genetic studies have addressed the genetic determinants of the progressive deterioration of glycaemic status and a limited number of candidate single nucleotide polymorphism (SNP) studies have evaluated progression to type 2 diabetes [31]. We hypothesise that progression to type 2 diabetes has a strong genetic component and that common variants in type 2 diabetes loci identified in European ancestry are also associated with progression to type 2 diabetes in American Indians. We also sought to identify the genetic contribution to IFG prediabetes subgroups to sort out some of the described phenotypic heterogeneity. Here we report the epidemiology and genetic characterisation of the diabetes progressor trait in American Indians, a population with high rates of progression to diabetes.
Methods
Study population
The NHLBI-funded Strong Heart Family Study (SHFS) is a collaborative project involving investigators at the Texas Biomedical Research Institute (formerly the Southwest Foundation for Biomedical Research), the University of North Carolina, Medstar Health Research Institute, the University of Oklahoma Health Sciences Center, Missouri Breaks Industries Research, and Cornell Medical Center. The study has the cooperation of the Indian Health Service and the tribes in three geographical areas: Arizona, Oklahoma and North and South Dakota. The SHFS is a large, family-based genetic study of metabolic and cardiovascular disease risk factors in American Indians and it is a component of the SHS, a population-based cohort study of diabetes and cardiovascular disease in American Indians. The SHFS began as a pilot study in 1998, when ∼900 members of extended families of the SHS parent study were examined. Additional family members were recruited and evaluated in a clinical visit in 2001– 2003. The total sample was 3,798 individuals from 94 multigenerational families (mean family size of 40 individuals, range 5–110), which were then re-examined in 2006–2009. Extensive and detailed measures of diabetes, insulin resistance, glycaemic traits and cardiovascular outcomes are available in the SHFS for one or two follow-up clinical visits.
The SHFS protocols were approved by the Indian Health Services (IHS) Institutional Review Board, by Institutional Review Boards of all Institutions and by the Indian tribes [32, 33]. All participants gave informed consent for genetic testing. The study was conducted according to the principles expressed in the Declaration of Helsinki. This study includes 2,011 American Indians free of diabetes at baseline visit and with follow-up data in 2006–2009.
Measurements
Baseline sociodemographic, obesity and glycaemic measures and cardiometabolic risk factors were obtained through interview, physical examination and laboratory measures. Physical examinations at baseline and follow-up were performed by centrally trained nurses and medical assistants following standardised protocols. Data collected included height, weight, body fat, waist and hip circumference and systolic and diastolic blood pressure. Twelve-hour fasting blood samples were also collected at each visit [32, 33] and stored at −70°C. Laboratory assays were performed for fasting glucose (measured by enzymatic methods), HbA1c (measured by HPLC, standardised to the DCCT assay, available at all visits in the SHFS among individuals without a diagnosis of type 2 diabetes), cholesterol, and creatinine. All samples were run blinded in a single laboratory and 5% blinded paired samples were included for quality control (QC). BMI (kg/m2) was calculated from weight and height. Additional available measures of obesity were total body fat (measured by bioelectrical impedance) and waist circumference. Hypertension was defined by a blood pressure of 140/90 mmHg or higher, or use of antihypertensive drugs. Demographic data (age, sex, education), lifestyle and behaviours (24 h dietary recall, smoking and alcohol intake), medical history and medications were obtained using standardised questionnaires [32, 33]. Physical activity was assessed using an Accusplit AE120 pedometer (Accusplit, San Jose, CA, USA), as previously described [34]. The average number of steps taken per day was calculated for any person who had data available for 3 or more days of the sampled week. We used the sex- and centre-specific 75% percentile of physical activity as an arbitrary cut-off point for the analyses, because 7,000–8,000 steps/day correspond to approximately the average daily steps of 30 min of moderate-intensity activity.
Trait definitions: progression to diabetes, non-progression (control) and transitory IFG
Type 2 diabetes, IFG and NFG were defined using the recent ADA criteria, which include fasting HbA1c values [1, 35]. Briefly, diabetes was defined as a fasting blood glucose (FBG) of ≥7.0 mmol/l (≥126 mg/dl), self-reported diabetes, use of diabetic medications or an HbA1c >6.5% (>48 mmol/mol). An FBG level of 6.1–6.9 mmol/l (100–125 mg/dl) or an HbA1c value of 5.7–6.4% (39–48 mmol/mol) was considered to indicate IFG and an FBG level of <6.0 mmol/l (<100 mg/dl) and an HbA1c value of <5.7% (<39 mmol/mol) was considered to indicate NFG. Our definition of type 2 diabetes progressors was adapted from the description given by Weyer et al for the natural history of insulin secretory dysfunction and insulin resistance in progression to diabetes in Pima Indians [6]. We defined diabetes progressors as individuals with new-onset diabetes at follow-up (n=373) and excluded prevalent diabetes at baseline visit. Individuals with persistent IFG over all visits (n=61) were included in a separate category. Controls were individuals aged 25 years or older at baseline visit with NFG at baseline and through all follow-up visits (up to three visits including baseline). Individuals presenting with new-onset IFG or prospectively changing from IFG to NFG over the observed period were defined as transitory IFG (as the progression to diabetes could not be established within available follow-up). Eighty-nine per cent of individuals with transitory IFG changed from NFG to IFG over 5–10 years of observation, and the remainder changed from IFG to NFG at follow-up. The median follow-up time for individuals classified as diabetes progressors, persistent IFG, transitory IFG and controls were 6.9, 5.2, 5.9 and 5.9 years, respectively, and the minimum follow-up was 3 years for all the groups.
Genotyping and existing markers
SNPs were selected from type 2 diabetes GWA publications of European and Asian ancestry (available at the genome catalogue up to January 2011) [36] and our analysis was thus restricted to these loci. SNPs were genotyped using the TaqMan genotyping assays (Life Technology, Carlsbad, CA, USA) or the multiplex VeraCode technology from Illumina (San Diego, CA, USA), according to the manufacturers’ protocols. Details of both technologies are reported elsewhere [37]. Replica samples were included as controls. Extensive standard QC was applied to genotyped data and included sample call rates (>95%), concordance of blinded replicates (>98%) and deviation from Hardy–Weinberg equilibrium among founders (p>0.01). Individuals with more than 10% of missing genotypes (n=64) were excluded. SNPs that failed genotyping were rs4430796 (HNF1B), rs2074196 (KCNQ1) and rs1326634 (SLC30A8).
Statistical analysis
We estimated heritability (h2), using maximum likelihood variance decomposition methods [38] and a liability threshold model, in analyses adjusted for age and sex in the overall sample and within each recruiting centre, separately for diabetes progressors, persistent IFG and transitory IFG compared with controls. We also tested the effect of clinical and laboratory risk factors on diabetes progressors, persistent IFG and transitory IFG by further adjusting for the following covariates: education (12 years or more vs <12 years), smoking (ever vs never), BMI, HDL-cholesterol, LDL-cholesterol, systolic and diastolic blood pressures, hypertension treatment, physical activity (75% upper vs lower percentiles) and using α=0.05 for significance. Models were implemented in SOLAR, which accounts for family relatedness [38]. The number of relative-pairs overall and within each geographical region is shown in electronic supplementary materials (ESM) Table 1.
Association analyses were performed only for traits that showed significant heritability. We performed centre-stratified analyses and combined the evidence from each centre using fixed-effect meta-analyses. We used this strategy because of regional differences in allele frequencies of genetic markers. We assessed associations among diabetes progressors compared with controls using measured genotype (mixed models to account for family relatedness) and additive genetic models, adjusted for age and sex. We also provide results with further adjustments for BMI for main findings. Significance thresholds were Bonferroni adjusted for the number of independent SNPs (linkage disequilibrium r2<0.5) used in analyses and p<0.003 (16 SNPs) was considered significant. The between-centre variance was estimated and we considered evidence for heterogeneity as p<0.05. We also estimated the trait variability explained by significant variants by comparing liability models within and without using the SNPs as covariates and using a summary score of all SNPs based on reported coded allele (range 0–22, the highest having more at-risk alleles).
Results
Risk factor burden in diabetes progressors, transitory IFG and controls
After exclusions, 2,011 SHFS individuals were eligible for analysis of which 373 were diabetes progressors, 61 had persistent IFG, 566 had transitory IFG and 1,011 were controls. The cumulative incidence of diabetes progression was 28.5% for individuals recruited from Arizona, 13.9% for Oklahoma and 15.6% for the Dakotas (Table 1). The mean age for controls at the last follow-up visit was 47.3 years (SD=13.1), compatible with long-term normoglycaemia.
Table 1.
Descriptive characteristics at baseline of categories of progressive impairment of fasting glucose
| Characteristic/Category | Overall | T2D progressor | Persistent IFG | Transitory IFG | Control |
|---|---|---|---|---|---|
| n | 2,011 | 373 | 61 | 566 | 1,011 |
| Female sex, % | 61.0 | 60.1 | 47.5 | 55.3 | 65.1 |
| Mean age, years (SD) | 40.9 (14.5) | 40.7 (14.3) | 42.2 (14.2) | 38.5 (16.4) | 42.2 (13.2) |
| Education <12 years, No. (%) | 571 (28.4) | 115 (30.8) | 20 (32.8) | 225 (39.8) | 211 (21.0) |
| Current smoking, No. (%) | 721 (35.9) | 115 (30.8) | 19 (31.2) | 208 (36.8) | 379 (37.6) |
| Median physical activity, No. steps/day (percentiles 25–75) |
5,114 (3,281– 7,474) |
4,488 (2,686– 6,886) |
4,654 (3,032– 6,484) |
5,108 (3,118– 7,582) |
5,309 (3,535– 7,678) |
| Physical activity less than 75%, % | 74.9 | 68.1 | 73.8 | 73.4 | 74.0 |
| Mean BMI, kg/m2 (SD) | 32.5 (7.6) | 36.9 (8.4) | 35.4 (7.2) | 33.1 (7.6) | 30.4 (6.5) |
| Obese, % | 57.7 | 80.4 | 80.3 | 64.1 | 45.1 |
| Body fat, % | 37.6 (9.9) | 41.6 (9.1) | 39.5 (9.3) | 37.6 (10.1) | 36.1 (9.7) |
| Mean waist-to-hip-circumference ratio (SD) | 0.91 (0.08) | 0.94 (0.09) | 0.95 (0.07) | 0.92 (0.08) | 0.89 (0.07) |
| Mean SBP, mmHg (SD) | 122.5 (15.7) | 124.1 (15.8) | 123.8 (14.4) | 123.4 (15.4) | 121.4 (15.8) |
| Mean DBP, mmHg (SD) | 77.8 (10.8) | 78.8 (11.2) | 79.4 (10.4) | 78.2 (10.9) | 77.0 (10.7) |
| Hypertension, % | 27.0 | 33.8 | 34.4 | 27.4 | 24.1 |
| Mean HDL-cholesterol, mmol/l (SD) | 1.35 (0.39) | 1.21 (0.33) | 1.30 (0.36) | 1.31 (0.37) | 1.42 (0.41) |
| Mean LDL-cholesterol, mmol/l (SD) | 2.62 (0.77) | 2.58 (0.80) | 2.68 (0.69) | 2.61 (0.78) | 2.64 (0.76) |
| Mean fasting glucose, mmol/l (SD) | 5.5 (1.3) | 6.5 (2.4) | 6.4 (0.2) | 5.4 (0.6) | 5.1 (0.5) |
| Mean fasting insulin, pmol/l (SD) | 118.2 (119.0) | 170.1 (179.3) | 184.6 (163.3) | 127.2 (115.7) | 89.8 (71.6) |
| Centres, Arizona/Dakotas/Oklahoma, No. | 555/751/705 | 158/117/98 | 24/18/19 | 204/227/135 | 169/389/453 |
To convert HDL- and LDL-cholesterol to mg/dl, divide by 0.0259; to covert glucose to mg/dl, divide by 0.0555; to convert insulin to µIU/ml, divide by 6.945
DBP, diastolic blood pressure; SBP, systolic blood pressure; T2D, type 2 diabetes
Overall, diabetes progressors had higher BMI, percentage of body fat and measures of central obesity than individuals with transitory IFG and controls, although most individuals were overweight (BMI>25 kg/m2) or obese (BMI>30 kg/m2) (Table 1). These findings are consistent with those described in Pima Indians and other ethnic populations [6]. Compared with controls, individuals progressing to type 2 diabetes and those classified as having transitory IFG had more cardiometabolic risk factors including higher blood pressure levels and prevalent hypertension, and lower HDL-cholesterol. These individuals were also less likely to have undergone 12 years of education. However, controls were more often found to be current smokers. Transitory IFG risk factor burden was intermediate between diabetes progressors and controls. Individuals progressing to type 2 diabetes were also less active than those in the transitory IFG group and controls. Individuals with persistent IFG had cardiometabolic risk factors more closely resembling diabetes progressors than the other groups although the sample was small.
Epidemiology of diabetes progressor and IFG
Significant risk factors for diabetes progression in multivariate analyses were high BMI (p=4.8×10−6), lower HDL-cholesterol (p=5.5×10−9) and living in Arizona (p=5.9×10−3) compared with the Dakotas. For persistent IFG, the only significant predictor was male sex (p=0.003). For transitory IFG, male sex (p=7.0×10−3), lower level of education (p=3.5×10−7), higher BMI (p=2.5×10−4), higher systolic blood pressure (p=7.4×10−3), lower HDL-cholesterol (p=0.02) and belonging to the Oklahoma centre compared with the Dakotas (p=2.8×10−5) were significantly associated risk factors. These risk factors accounted for 15%, 11% and 8% of the phenotypic variability of diabetes progression, persistent IFG and transitory IFG, respectively, in models adjusted for age, sex, education, current smoking, BMI, percentage of body fat, systolic and diastolic blood pressure, hypertension treatment, HDL-cholesterol and LDL-cholesterol).
Genetic determinants of diabetes progression
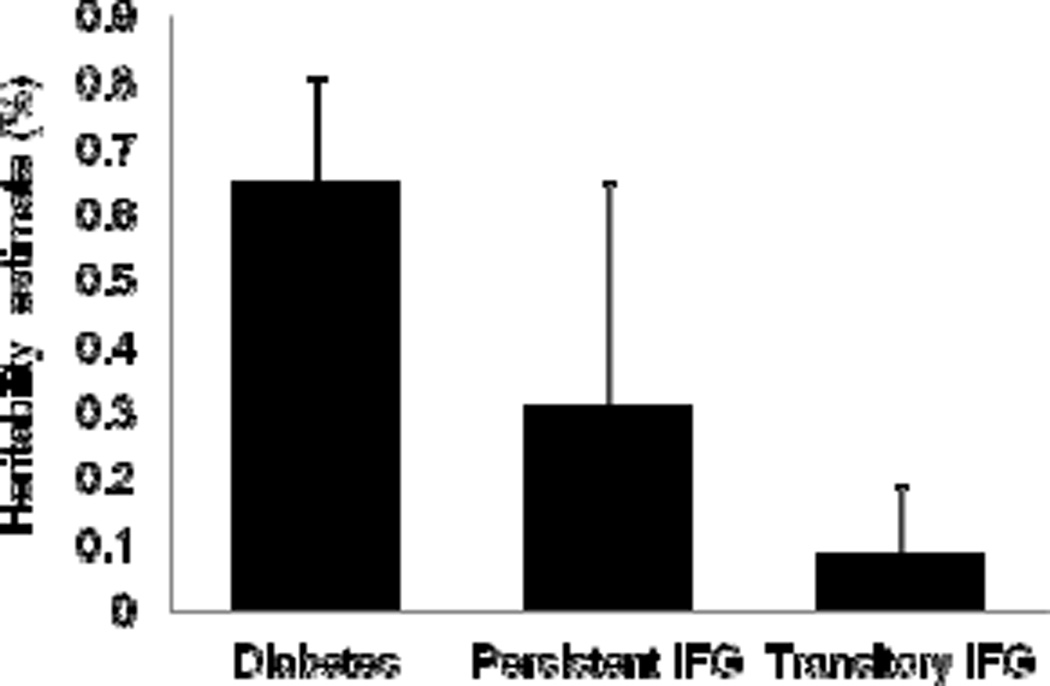
We first evaluated the proportion of the phenotypic variance due to genetic effects (heritability) among diabetes progressors and among individuals with transitory IFG compared with controls. Heritability is considered to be the single most useful measure of familial aggregation of disease [39]. In a fully adjusted model accounting for the risk factors described in Table 1, the overall heritability of diabetes progressor trait was 0.65 (SD=0.16, p=2.7×10−6) (Fig. 1). Interestingly, the heritability of persistent IFG was 31% and transitory IFG was 9%, neither of which was significantly greater than zero. We then examined the association of 16 known GWA type 2 diabetes SNPs in 16 loci with type 2 diabetes progression. SNPs that passed QC and their minor allele frequencies are shown in Table 2. All genotyped variants had allele frequencies higher than 1% in American Indians (Table 2). Association results for SNPs (and their loci) within each centre and in meta-analyses across all centres are shown in Table 2. Several SNPs were nominally associated with diabetes progression including SNPs nearby WFS1, TSPAN8, and THADA. There was no evidence for between-centre heterogeneity of effects for most of the associated variants (Table 2). The mean at-risk allele scores were 15.1 and 14.8 for individuals developing diabetes and for controls, respectively. The risk score was not associated with diabetes progression (p=0.20).
Fig. 1.
Heritability of diabetes (n=373), persistent IFG (n=61) and transitory IFG (n=566). Estimates and SD are shown for polygenic models adjusted for age and sex using individuals with normal glycaemia as controls (n=1,011). Note that only diabetes had significant heritability and numbers were small for estimate heritability among individuals with persistent IFG
Table 2.
Results from associations of SNPs with type 2 diabetes progressors vs controls
| Nearby gene |
SNP | Effect allele |
Other allele |
Arizona |
Dakotas |
Oklahoma |
Meta-analyses |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF | OR (95% CI) | p | AF | OR (95% CI) | p | AF | OR (95% CI) | p | OR (95% CI) | p | n | phet | ||||
| WFS1 | rs1001013 1 |
G | A | 0.06 | 1.46 (1.00, 2.13) | 0.10 | 0.20 | 1.35 (1.06, 1.73) | 0.01 | 0.20 | 1.09 (0.87, 1.38) | 0.44 | 1.25 (1.07, 1.46) | 0.00 4 |
1,366 | 0.31 |
| TSPAN8 | rs7961581 | C | T | 0.03 | 1.19 (0.58, 2.41) | 0.63 | 0.10 | 1.54 (1.08, 2.19) | 0.01 | 0.11 | 1.30 (0.92, 1.83) | 0.12 | 1.39 (1.10, 1.75) | 0.00 54 |
1,366 | 0.71 |
| MTNR1B | rs1387153 | T | C | 0.13 | 0.85 (0.62, 1.16) | 0.98 | 0.29 | 0.96 (0.78, 1.18) | 0.69 | 0.25 | 0.80 (0.64, 1.00) | 0.05 | 0.88 (0.77, 1.00) | 0.06 | 1,357 | 0.47 |
| THADA | rs7578597 | T | C | 0.02 | 2.43 (0.72, 8.19) | 0.13 | 0.06 | 1.49 (0.88, 2.53) | 0.13 | 0.15 | 1.55 (0.91, 2.65) | 0.10 | 1.58 (1.10, 2.27) | 0.01 | 1,363 | 0.77 |
| FTO | rs8050136 | A | C | 0.10 | 0.95 (0.67, 1.34) | 0.74 | 0.16 | 1.53 (1.15, 2.04) | 0.00 2 |
0.15 | 1.02 (0.79, 1.32) | 0.88 | 1.15 (0.87, 1.52) | 0.09 | 1,365 | 0.05 |
| HHEX-IDE | rs1111875 | T | C | 0.52 | 0.96 (0.78, 1.17) | 0.66 | 0.44 | 0.88 (0.72, 1.08) | 0.20 | 0.38 | 1.24 (1.03, 1.50) | 0.03 | 1.03 (0.92, 1.15) | 0.65 | 1,358 | 0.04 |
|
CDKN2A/2 B |
rs1081166 1 |
C | T | 0.09 | 1.03 (0.69, 1.53) | 0.90 | 0.19 | 0.99 (0.77, 1.27) | 0.93 | 0.13 | 1.12 (0.81, 1.53) | 0.49 | 1.03 (0.97, 1.10) | 0.71 | 1,369 | 0.84 |
| NOTCH2 | rs1092393 1 |
T | G | 0.09 | 0.93 (0.65, 1.35) | 0.70 | 0.05 | 1.39 (0.90, 2.13) | 0.12 | 0.11 | 1.11 (0.95, 1.30) | 0.48 | 1.11 (0.96, 1.27) | 0.15 | 1,368 | 0.39 |
| CAMK1D | rs1277979 0 |
G | A | 0.12 | 1.27 (0.95, 1.70) | 0.11 | 0.16 | 0.99 (0.77, 1.27) | 0.95 | 0.15 | 0.98 (0.76, 1.27) | 0.90 | 1.06 (0.91, 1.23) | 0.46 | 1,362 | 0.35 |
| PPARG | rs1801282 | G | C | 0.10 | 0.72 (0.50, 1.05) | 0.09 | 0.10 | 0.84 (0.52, 1.37) | 0.30 | 0.21 | 1.01 (0.80, 1.28) | 0.92 | 0.91 (0.76, 1.09) | 0.32 | 1,365 | 0.31 |
| ADAMTS9 | rs4607103 | T | C | 0.42 | 0.93 (0.76, 1.15) | 0.52 | 0.30 | 0.97 (0.80, 1.17) | 0.74 | 0.35 | 1.05 (0.87, 1.28) | 0.59 | 0.99 (0.88, 1.11) | 0.82 | 1,366 | 0.68 |
| IGF2BP2 | rs4402960 | T | G | 0.21 | 1.00 (0.69, 1.44) | 0.99 | 0.19 | 1.32 (1.02, 1.70) | 0.03 | 0.18 | 0.81 (0.64, 1.03) | 0.08 | 1.01 (0.87, 1.18) | 0.88 | 1,359 | 0.02 |
| KCNJ11 | rs5219 | T | C | 0.39 | 0.97 (0.78, 1.20) | 0.76 | 0.33 | 0.89 (0.73, 1.09) | 0.24 | 0.37 | 1.00 (0.72, 1.40) | 0.99 | 0.94 (0.82, 1.07) | 0.33 | 1,344 | 0.78 |
| GCKR | rs780094 | T | C | 0.07 | 1.31 (0.82, 2.11) | 0.25 | 0.29 | 0.98 (0.81, 1.19) | 0.85 | 0.30 | 1.11 (0.90, 1.36) | 0.34 | 1.06 (0.92, 1.21) | 0.42 | 1,363 | 0.46 |
| TCF7L2 | rs7901695 | C | T | 0.09 | 1.56 (1.12, 2.15) | 0.00 6 |
0.11 | 0.95 (0.72, 1.25) | 0.69 | 0.16 | 0.77 (0.61, 0.96) | 0.03 | 0.96 (0.82, 1.11) | 0.56 | 1,355 | 0.00 2 |
| JAZF1 | rs864745 | C | T | 0.23 | 0.97 (0.77,1.22) | 0.77 | 0.40 | 1.00 (0.69, 1.44) | 1.00 | 0.33 | 1.07 (0.88, 1.31) | 0.49 | 1.02 (0.89, 1.18) | 0.75 | 1,363 | 0.79 |
Models adjusted for age and sex and accounted for family relatedness
AF, effect allele frequency among relative unrelated individuals (see text); phet, p for between-study heterogeneity
Discussion
Among SHFS American Indians, a population with a high rate of obesity and insulin resistance, we identified subgroups that either progressed to type 2 diabetes or had persistent or transitory IFG. We confirmed the strong evidence for genetic susceptibility for diabetes progression in American Indians [40]. Individuals with persistent and transitory IFG had an intermediate risk factor burden compared with diabetes progressors and normoglycaemic controls, but the evidence for polygenic genetic effects was weak based on heritability estimates. The low contribution of genetic factors to transitory IFG may be related to phenotypic heterogeneity, previously described in longitudinal glycaemic studies [14]. Studies have shown that only one in four individuals with prediabetes will develop diabetes [15]. Obesity-related environmental factors, such as diet and physical activity, may contribute to early abnormalities of fasting glucose in a subset of individuals. Future work using longitudinal glycaemic measures may provide a better characterisation of prediabetes subgroups for which the contribution of genetic and environmental factors may vary. The sample size for persistent IFG was small and further studies are needed to confirm the findings for this subgroup. These findings may have implications in the genetic research of prediabetes when using a single time-point glycaemic measure and in the implementation of preventive measures for glycaemic deterioration in populations with a high burden of insulin resistance and cardiometabolic risk factors, given the global obesity epidemic.
We also explored the evidence for generalisation of associations of common genetic variants, identified in GWAs using type 2 diabetes case-control designs, with diabetes progression in American Indians. SNPs in the WFS1, TSPAN8 and THADA loci showed nominal associations in meta-analysis of all recruiting centres. Importantly, although there were regional differences in allele frequencies of genetic markers, we found no evidence for between-centre heterogeneity of genetic effects for these common variants. SNPs showed similar direction of effect as those described in individuals of European ancestry [41] but allele frequencies were lower in American Indians compared with European ancestry populations (ESM Table 2). Effects sizes were larger for associations in American Indians for diabetes progression compared with reported European population-based estimates in type 2 diabetes case-control studies. For example, the OR for each C allele copy of rs7961581 (TSPAN8) was 1.39 in American Indians compared with 1.09 in European ancestry meta-analysis [41]. Interestingly, TCF7L2 SNPs identified through a comprehensive genotyping of the locus were previously shown to not associate with diabetes in Pima Indians [42]. In our study, we found nominal associations with diabetes progression only in the Arizona centre (Table 2) but this SNP estimate also showed strong evidence for between-study heterogeneity (p for heterogeneity=0.002). We do not know the underlying genetic architecture of diabetes in American Indians and how well the available SNPs capture the functional variants in these loci.
The WFS1 variant has been prospectively associated with development of type 2 diabetes among individuals of European ancestry [43]. For remaining loci, differences in linkage disequilibrium of the genotyped SNP with the ‘causal’ variant(s) between American Indians and individuals of European ancestry, the population used in the discovery, may account for some of the negative findings. Because only the published SNP was available in these regions, we cannot rule out presence of additional variants in these and in other loci accounting for type 2 diabetes risk. It is possible that true associations may have been overlooked due to low power. We estimated that we only had 80% power for large effect sizes for analyses of diabetes progression, persistent IFG and transitory IFG compared with controls (ESM Table 3). The analyses were also limited to type 2 diabetes loci with validated GWAS variants published up until 2011, when the SNPs were selected.
Our findings do not exclude these loci as being important to diabetes in American Indians. Findings from other ethnic populations [44, 45] suggest that type 2 diabetes loci may be relevant to diverse ancestral populations. American Indians have a large burden of obesity, insulin resistance and early-onset type 2 diabetes, and studies of this population have the potential to uncover mechanisms related to progression to diabetes. However, a limited number of GWA studies have been performed in American Indians [46, 47].
In summary, using longitudinal data to define type 2 diabetes progression in American Indians who have previously been shown to have high burden of obesity, insulin resistance and type 2 diabetes, we showed evidence for a high genetic susceptibility to diabetes progression, and nominal replication of some SNPs in known type 2 diabetes loci to American Indians. Importantly, there was little evidence for contribution of genetic factors to transitory IFG, which could have public health implications when promoting healthy lifestyle [48, 49]. This study expands the knowledge on the genetics of progressive deterioration of glycaemic status in individuals at high risk of type 2 diabetes. Further work, including fine-mapping and/or sequencing of these regions, will be needed to better characterise the role of these genes in the susceptibility to diabetes in American Indians.
Supplementary Material
Acknowledgments
Funding
The SHS is supported by NHLBI grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654 and U01 HL65521. J. B. Meigs is supported in part by NIDDK K24 DK080140.
Abbreviations
- FBG
Fasting blood glucose
- GWA
Genome-wide association
- h2
Heritability
- IFG
Impaired fasting glucose
- IGT
Impaired glucose tolerance
- NFG
Normal fasting glucose
- OGTT
2 h oral glucose tolerance test
- QC
Quality control
- SHFS
Strong Heart Family Study
- SHS
Strong Heart Study
- SNP
Single nucleotide polymorphism
Footnotes
None of the authors have any disclosures to report.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
NF and SAC wrote the manuscript and researched data. NF, SAC, ETL, LGB, RRF and JWM contributed to the study design, and KH, HHHG, VSV, SL, LA, ETL, LGB, RRF, JWM, JBM, JSP and KEN contributed to acquisition of the data and reviewed/edited the manuscript. All authors approved the final version of the paper.
References
- 1.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2011;34(Suppl. 1):S62–S69. doi: 10.2337/dc11-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 3.Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest. 2006;116:1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin BC, Warram JH, Krolewski AS, Bergman RN, Soeldner JS, Kahn CR. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet. 1992;340:925–929. doi: 10.1016/0140-6736(92)92814-v. [DOI] [PubMed] [Google Scholar]
- 5.Godsland IF, Jeffs JA, Johnston DG. Loss of beta cell function as fasting glucose increases in the non-diabetic range. Diabetologia. 2004;47:1157–1166. doi: 10.1007/s00125-004-1454-z. [DOI] [PubMed] [Google Scholar]
- 6.Weyer C, Bogardus C, Mott DM, Pratley RE. The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J Clin Invest. 1999;104:787–794. doi: 10.1172/JCI7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabir MM, Hanson RL, Dabelea D, et al. The 1997 American Diabetes Association and 1999 World Health Organization criteria for hyperglycemia in the diagnosis and prediction of diabetes. Diabetes Care. 2000;23:1108–1112. doi: 10.2337/diacare.23.8.1108. [DOI] [PubMed] [Google Scholar]
- 8.Edelstein SL, Knowler WC, Bain RP, et al. Predictors of progression from impaired glucose tolerance to NIDDM: an analysis of six prospective studies. Diabetes. 1997;46:701–710. doi: 10.2337/diab.46.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vegt F, Dekker JM, Jager A, et al. Relation of impaired fasting and postload glucose with incident type 2 diabetes in a Dutch population: The Hoorn Study. Jama. 2001;285:2109–2113. doi: 10.1001/jama.285.16.2109. [DOI] [PubMed] [Google Scholar]
- 10.Vaccaro O, Ruffa G, Imperatore G, Iovino V, Rivellese AA, Riccardi G. Risk of diabetes in the new diagnostic category of impaired fasting glucose: a prospective analysis. Diabetes Care. 1999;22:1490–1493. doi: 10.2337/diacare.22.9.1490. [DOI] [PubMed] [Google Scholar]
- 11.Cowie CC, Rust KF, Byrd-Holt DD, et al. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health And Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29:1263–1268. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- 12.Harris MI, Flegal KM, Cowie CC, et al. Prevalence of diabetes, impaired fasting glucose, and impaired glucose tolerance in U.S. adults. The Third National Health and Nutrition Examination Survey, 1988–1994. Diabetes Care. 1998;21:518–524. doi: 10.2337/diacare.21.4.518. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Shara NM, Calhoun D, Umans JG, Lee ET, Howard BV. Incidence rates and predictors of diabetes in those with prediabetes: the Strong Heart Study. Diabetes Metab Res Rev. 2010;26:378–385. doi: 10.1002/dmrr.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meigs JB, Muller DC, Nathan DM, Blake DR, Andres R. The natural history of progression from normal glucose tolerance to type 2 diabetes in the Baltimore Longitudinal Study of Aging. Diabetes. 2003;52:1475–1484. doi: 10.2337/diabetes.52.6.1475. [DOI] [PubMed] [Google Scholar]
- 15.Haffner SM. Epidemiology of type 2 diabetes: risk factors. Diabetes Care. 1998;21(Suppl. 3):C3–C6. doi: 10.2337/diacare.21.3.c3. [DOI] [PubMed] [Google Scholar]
- 16.Pan XR, Li GW, Hu YH, et al. Effects of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care. 1997;20:537–544. doi: 10.2337/diacare.20.4.537. [DOI] [PubMed] [Google Scholar]
- 17.Walker KZ, O’Dea K, Gomez M, Girgis S, Colagiuri R. Diet and exercise in the prevention of diabetes. J Hum Nutr Diet. 2010;23:344–352. doi: 10.1111/j.1365-277X.2010.01061.x. [DOI] [PubMed] [Google Scholar]
- 18.Tuomilehto J, Lindstrom J, Eriksson JG, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 19.Knowler WC, Barrett-Connor E, Fowler SE, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramachandran A, Snehalatha C, Mary S, Mukesh B, Bhaskar AD, Vijay V. The Indian Diabetes Prevention Programme shows that lifestyle modification and metformin prevent type 2 diabetes in Asian Indian subjects with impaired glucose tolerance (IDPP-1) Diabetologia. 2006;49:289–297. doi: 10.1007/s00125-005-0097-z. [DOI] [PubMed] [Google Scholar]
- 21.Hivert MF, Jablonski KA, Perreault L, et al. Updated genetic score based on 34 confirmed type 2 diabetes Loci is associated with diabetes incidence and regression to normoglycemia in the diabetes prevention program. Diabetes. 2011;60:1340–1348. doi: 10.2337/db10-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce M, Keen H, Bradley C. Risk of diabetes in offspring of parents with non-insulin-dependent diabetes. Diabet Med. 1995;12:6–13. doi: 10.1111/j.1464-5491.1995.tb02054.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaprio J, Tuomilehto J, Koskenvuo M, et al. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992;35:1060–1067. doi: 10.1007/BF02221682. [DOI] [PubMed] [Google Scholar]
- 24.Hemminki K, Li X, Sundquist K, Sundquist J. Familial risks for type 2 diabetes in Sweden. Diabetes Care. 2010;33:293–297. doi: 10.2337/dc09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 26.Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nature Genetics. 2011;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2012;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 29.Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24:613–621. doi: 10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfs MG, Hofker MH, Wijmenga C, van Haeften TW. Type 2 diabetes mellitus: new genetic insights will lead to new therapeutics. Curr Genomics. 2009;10:110–118. doi: 10.2174/138920209787847023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walford GA, Green T, Neale B, et al. Common genetic variants differentially influence the transition from clinically defined states of fasting glucose metabolism. Diabetologia. 2012;55:331–339. doi: 10.1007/s00125-011-2353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.North KE, Williams JT, Welty TK, et al. Evidence for joint action of genes on diabetes status and CVD risk factors in American Indians: the strong heart family study. Int J Obes Relat Metab Disord. 2003;27:491–497. doi: 10.1038/sj.ijo.0802261. [DOI] [PubMed] [Google Scholar]
- 33.Lee ET, Welty TK, Fabsitz R, et al. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 34.Storti KL, Arena VC, Barmada MM, et al. Physical activity levels in American-Indian adults: the Strong Heart Family Study. Am J Prev Med. 2009;37:481–487. doi: 10.1016/j.amepre.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 36.Haiman CA, Fesinmeyer MD, Spencer KL, et al. Consistent directions of effect for established type 2 diabetes risk variants across populations: the population architecture using Genomics and Epidemiology (PAGE) Consortium. Diabetes. 2012;61:1642–1647. doi: 10.2337/db11-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buyske S, Wu Y, Carty CL, et al. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS One. 2012;7:e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenesa A, Haley CS. The heritability of human disease: estimation, uses and abuses. Nat Rev Genet. 2013;14:139–149. doi: 10.1038/nrg3377. [DOI] [PubMed] [Google Scholar]
- 40.Hanson RL, Elston RC, Pettitt DJ, Bennett PH, Knowler WC. Segregation analysis of non-insulin-dependent diabetes mellitus in Pima Indians: evidence for a major-gene effect. Am J Hum Genet. 1995;57:160–170. [PMC free article] [PubMed] [Google Scholar]
- 41.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature genetics. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo T, Hanson RL, Traurig M, et al. TCF7L2 is not a major susceptibility gene for type 2 diabetes in Pima Indians: analysis of 3,501 individuals. Diabetes. 2007;56:3082–3088. doi: 10.2337/db07-0621. [DOI] [PubMed] [Google Scholar]
- 43.Cheurfa N, Brenner GM, Reis AF, et al. Decreased insulin secretion and increased risk of type 2 diabetes associated with allelic variations of the WFS1 gene: the Data from Epidemiological Study on the Insulin Resistance Syndrome (DESIR) prospective study. Diabetologia. 2011;54:554–562. doi: 10.1007/s00125-010-1989-0. [DOI] [PubMed] [Google Scholar]
- 44.Rong R, Hanson RL, Ortiz D, et al. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes. 2009;58:478–488. doi: 10.2337/db08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malhotra A, Kobes S, Knowler WC, Baier LJ, Bogardus C, Hanson RL. A genome-wide association study of BMI in American Indians. Obesity (Silver Spring) 2011;19:2102–2106. doi: 10.1038/oby.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanson RL, Bogardus C, Duggan D, et al. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes. 2007;56:3045–3052. doi: 10.2337/db07-0462. [DOI] [PubMed] [Google Scholar]
- 48.Lloyd-Jones DM, Hong Y, Labarthe D, et al. Defining and setting national goals for cardiovascular health promotion and disease reduction: the American Heart Association’s strategic Impact Goal through 2020 and beyond. Circulation. 2010;121:586–613. doi: 10.1161/CIRCULATIONAHA.109.192703. [DOI] [PubMed] [Google Scholar]
- 49.American Diabetes Association. Executive summary: Standards of medical care in diabetes--2013. Diabetes Care. 2013;36(Suppl. 1):S4–S10. doi: 10.2337/dc13-S004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.