Abstract
The storage of triacylglycerols (TAGs) is essential for non-replicating persistence relevant to survival and the re-growth of mycobacteria during their exit from non-replicating state stress conditions. However, the detailed structures of this lipid family in mycobacteria largely remain unexplored. In this contribution, we describe a multiple-stage linear ion-trap mass spectrometric approach with high resolution mass spectrometry toward direct structural analysis of the TAGs, including a novel lipid subclass previously defined as monomeromycolyl diacylglycerol (MMDAG) isolated from biofilm of Mycobacterium smegmatis, a rapidly growing, non-pathogenic mycobacterium that has been used as a tool for molecular analysis of mycobacteria. Our results demonstrate that the major isomer in each of the molecular species of TAGs and MMDAGs consists of the common structure in which Δ918:1- and 16:0-fatty acyl substituents are exclusively located at sn-1 and sn-2, respectively. Several isomers were found for most of the molecular species, and thus hundreds of structures are present in this lipid family. More importantly, this study revealed the structures of MMDAG, a novel subclass of TAG that has not been previously reported by direct mass spectrometric approaches.
Keywords: apolar lipid, triacylglycerol, mycobacteria smegmatis, meromycolyl chain, mass spectrometry
Introduction
Mycobacteria store triacylglycerols (TAGs) under various stress conditions, such as hypoxia, exposure to nitric oxide, and acidic environments [1] [2] [3]. These stress conditions are known to induce non-replicating persistence relevant to survival in mycobacteria and utilization of TAGs is essential for the re-growth of mycobacteria during their exit from non-replicating state. This notion was recently supported with the evidence that diacylglycerol acyltransferases involved in TAG biosynthesis are upregulated in stationary-phase [4]. Accumulation of TAG was also observed in the cells at late stages of growth [5]. However, the importance of TAG accumulation and utilization during re-growth is not clearly understood.
TAGs occurs as components of the cell envelope [6] and TAGs that accumulate within M. tuberculosis are generated mainly by the incorporation of fatty acids released from host TAG [4]. They are the predominant class of lipids, in the wild-type strain of M. smegmatis grown on the glycerol-rich Sauton’s medium, representing more than 50% of the surface-exposed lipids [7].
Previous investigations by Kremer et al. [8] described the presence of monomeromycolyl diacylglycerol (MMDAG), an unusual TAG subclass consisting of a novel meromycolate substituent and two fatty acyl moieties in mycobacteria. This minor lipid family was ubiquitously found in the TAG pool in all mycobacterial species, including pathogenic species and non-pathogenic M. smegmatis, emphasizing an important function in mycobacterial physiology, as it may play a potential role as a source of meromycolates during mycolic acid biosynthesis in growing cells.
Despite the many previous reports of TAGs in mycobacteria [2,9,8,10,11], their detailed structures have never been reported and the identification of the structure of MMDAG achieved by Kremer et al. was only established upon NMR spectroscopy and GC-MS analysis of the fatty acid methyl ester (FAME) derivatives of the alkaline deacylation product followed by esterification. The attempts to obtain the structural information of MMDAG by mass spectrometry, i.e., the molecular species of this lipid family have been unsuccessful [8] and thus, tandem mass spectrometric approaches leading to direct characterization of this unique lipid family have never been reported. Here, we applied linear ion-trap (LIT) multiple-stage (MSn) and high resolution mass spectrometry [12,13] toward complete characterization of TAG and MMDAG lipids isolated from biofilm of M. smegmatis.
Methods
Sample preparation
Biofilms of M. smegmatis strain mc2155 [14] obtained from ATCC were grown in polystyrene petri dishes at 30°C in modified Sauton’s medium without Tween 80. Sauton’s medium contained 0.5 g/L K2HPO4, 0.5 g/L MgSO4, 4.0 g/L L-asparagine, 0.05 g/L ferric ammonium citrate, 4.76 % glycerol, and 1.0 mg/L ZnSO4, to a final pH of 7.0. To obtain total lipids, cultures were harvested by centrifugation and resuspended in chloroform:methanol (2:1, v:v). Following extraction, total lipids were dried under N2 gas and the apolar lipids were harvested as described [15]. The total lipids were resuspended in 5 mL of methanol: 0.3% NaCl (10:1, v:v) and 2.5 mL petroleum ether. Samples were rocked for 30 min at room temperature. After centrifugation, the upper layer containing apolar lipids was retained and dried under N2 gas. The crude lipids in 300 uL chloroform:methanol (2:1, v:v) were loaded to a 3 mL/200 mg Macherey-Nagel amino Chromabond Sep-Pak column (Duren, Germany). The column was first washed with 2 mL EtOAc:Hexane (15:85, v:v), followed by 1.5 mL di-isopropyl ether:HOAc (98:2, v:v) (Fraction 2), and then eluted with 2 mL acetone:methanol (9:1.35, v:v) (Fraction 3) by gravity. The eluants containing MMDAG (fraction 2) and TAG (fraction 3) lipids were dried under a stream of nitrogen. The dried samples were re-dissolved in chloroform:methanol (1:2, v/v) containing 1 μM 7LiOH or NH4OAc before ESI-MS analysis.
Mass spectrometry and LC-MS analysis
Both high-resolution (R=100,000 at m/z 400) and low-energy CAD tandem mass spectrometry experiments were conducted on a Thermo Scientific (San Jose, CA) LTQ Orbitrap Velos mass spectrometer (MS) with Xcalibur operating system. Apolar lipids in chloroform:methanol (1/2) containing 1 μM 7LiOH or NH4OAc were infused (1.5 μL/min) to the ESI source to give rise to abundant [M + 7Li]+ or [M + NH4]+ ions. The skimmer of the ESI source was set at ground potential, the electrospray needle was set at 4.0 kV, and temperature of the heated capillary was 300°C. The automatic gain control of the ion trap was set to 5×104, with a maximum injection time of 100 ms. Helium was used as the buffer and collision gas at a pressure of 1×10−3 mbar (0.75 mTorr). The MSn experiments were carried out with an optimized relative collision energy ranging from 35–45% and with an activation q value at 0.25, and the activation time at 10 ms to leave a minimal residual abundance of precursor ion (around 20%). The mass selection window for the precursor ions was set at 1 Da wide to admit the monoisotopic ion to the ion-trap for collision-induced dissociation (CID) for unit resolution detection in the ion-trap or high resolution accurate mass detection in the Orbitrap mass analyzer. The instrument was tuned and calibrated according to the instructions in the users’ manual; and dimyristoylphosphatidylcholine (elemental composition: C36H72NO8P) was used as lock mass (calculated m/z for [M + H]+: 678.5068; for [M + Li]+: 684.5150). Mass spectra were accumulated in the profile mode, typically for 3–10 min for MSn spectra (n=2,3,4). LC-MS analysis of apolar lipid extract was conduct on Thermo Scientific (San Jose, CA) Vantage TSQ mass spectrometer with Thermo Accela UPLC operated by Xcalibur software. Separation of apolar lipid was achieved by a Supelco 100 × 2.1 mm (2.7 u particle size) AscentisC-8 column at a flow rate of 260μL/min. The mobile phase contained 10 mM ammonium formate (pH 5.0) in solvent A: acetonitrile:water (60:40, v:v); solvent B: 2-propanol:acetonitrile (90:10, v:v); and a gradient elution in the following manner was applied: 68% A, 0–1.5 min; 68-55% A, 1.5–4 min; 55-48% A, 4–5 min; 48-42% A, 5–8 min; 42-34% A, 8–11 min; 34-30% A, 11–14 min; 30-25% A, 14–18 min; 25-3% A, 18–23 min; 3-0% A, 25–30 min. The MMDAG fraction was eluted at 24.5–26 min, and TAG was eluted at 18–23.5 min (See Electronic Supplementary Material, Fig. S1). Fractions containing the above lipids were collected for further structure analysis.
Nomenclature
TAG molecules including MMDAGs that differ only with respect to the fatty acyl groups at sn-1 and sn-3, such as (A/B/C)-TAG vs. (C/B/A)-TAG (where A, B, and C are distinct fatty acid residues) are enantiomers and cannot be distinguished by the present mass spectrometric approach. In this premise for assignment of the fatty acid substituents on the glycerol backbone as described in the text, only the fatty acid substituent assigned to sn-2 is specific, and the fatty acid substituents assigned to sn-1 and at sn-3 are exchangeable (i.e., A/B/C)-TAG and (C/B/A)-TAG are not distinguishable). No effort was made to determine the chirality of TAGs in this study. Thus, 18:1/16:0/18:0-TAG, for example, signifies that oleoryl, palmitoyl, and stearoyl fatty acyl substituents are located at sn-1, sn-2, and sn-3 of the glycerol backbone, respectively. Similarly, monomeromycolyl-diacylglycerol (MMDAG) possessing oleoryl, palmitoyl, and meromycolylic 50:2- fatty acyl substituents at sn-1, sn-2, and sn-3, respectively, is designated as 18:1/16:0/m50:2-TAG.
Results and Discussion
Apolar TAG and MMDAG lipids are readily separable by HPLC (Fig. S1, Electronic Supplementary Material). The mass spectrum of the lipid fraction eluted at 18–23.5 min (Panel b) contained an array of homologous [M + NH4]+ ions of TAGs with an intermittence of 28 Da, mainly in the m/z ranged from 750 to 1000 Da (Fig. 1a); the lipids eluted at 24.5–26 min (Panel c) consisted of a series of the homologous [M + NH4]+ ions of MMDAGs with intermittence of 14 Da (CH2) in the m/z ranged from 1250 to 1430 Da (Fig. 1, Panel b). High-resolution mass measurements (R = 100,000 at m/z 400) on the [M + NH4]+ ions (Table S1, Electronic Supplementary Material) and the corresponding [M + Li]+ ions (Table 1) gave elemental compositions that are consistent with the presence of TAG. Elemental compositions deduced from high resolution mass measurements on the [M + Li]+ ions (Table 2) and the corresponding [M + NH4]+ ions of MMDAG (Table S2, Electronic Supplementary Material) suggested the presence of a new TAG family. The structural characterization, including the identities of the fatty acyl substituents and their position on the glycerol backbone are described below.
Fig. 1.
The ESI-LC/MS spectra of the [M + NH4]+ ions of TAG (a), and MMDAG (b) isolated from biofilm of M. Smegmatis.
Table 1.
High resolution mass measurements and LIT MSn analysis of TAG species from M. smegmatis biofilm
| measured m/z [M + Li]+ | Theo. Mass Da | Deviat. mDa | Elemental Composition | Rel. Int. (%) | Structuresa |
|---|---|---|---|---|---|
|
| |||||
| 753.6577 | 753.6579 | −0.15 | C47 H86 O6 Li | 1.7 | 18:1/16:1/10:0; 18:1/14:0/12:1; 16:1/16:0/12:1 |
| 755.6735 | 755.6735 | −0.09 | C47 H88 O6 Li | 2.31 | 18:1/16:0/10:0; 18:1/14:0/12:0; 16:1/16:0/12:0 |
| 781.6892 | 781.6892 | 0.01 | C49 H90 O6 Li | 10.46 | 18:1/16:0/12:1; 18:1/14:0/14:1; 16:1/16:0/14:1 |
| 783.7048 | 783.7048 | −0.01 | C49 H92 O6 Li | 11.32 | 18:1/16:0/12:0; 18:1/14:0/14:0; 16:1/16:0/14:0; 20:1/16:0/10:0 |
| 795.7049 | 795.7048 | 0.1 | C50 H92 O6 Li | 2.72 | b |
| 797.7206 | 797.7205 | 0.09 | C50 H94 O6 Li | 3.27 | b |
| 807.7050 | 807.7048 | 0.16 | C50 H94 O6 Li | 5.33 | b |
| 809.7206 | 809.7205 | 0.12 | C51 H94 O6 Li | 33.2 | 18:1/16:0/14:1; 18:1/16:1/14:0; 16:1/16:0/16:1 |
| 811.7360 | 811.7361 | −0.15 | C51 H96 O6 Li | 27.95 | 18:1/16:0/14:0 |
| 823.7363 | 823.7361 | 0.13 | C52 H96 O6 Li | 3.24 | b |
| 825.7517 | 825.7518 | −0.11 | C52 H98 O6 Li | 7.65 | 18:1/16:0/15:0; 18:1/14:0/17:0; 16:1/16:0/17:0; 16:1/14:0/19:0 |
| 835.7364 | 835.7361 | 0.24 | C53 H96 O6 Li | 10.33 | 18:1/16:1/16:1 |
| 837.7516 | 837.7518 | −0.24 | C53 H98 O6 Li | 65.32 | 18:1/16:0/16:1 |
| 839.7671 | 839.7674 | −0.34 | C53 H100 O6 Li | 20.77 | 18:1/16:0/16:0; 18:1/14:0/18:0 |
| 851.7674 | 851.7674 | −0.04 | C54 H100 O6 Li | 8.46 | 18:1/16:0/17:1; 16:1/16:0/19:1; 18:1/16:1/17:0; 16:1/16:1/19:0 |
| 853.7824 | 853.7831 | −0.71 | C54 H102 O6 Li | 23.98 | 18:1/16:0/17:0; 18:1/14:0/19:0 |
| 863.7674 | 863.7674 | −0.02 | C55 H100 O6 Li | 17.06 | 18:1/16:1/18:1 |
| 865.7828 | 865.7831 | −0.27 | C55 H102 O6 Li | 92.59 | 18:1/16:0/18:1 |
| 867.7985 | 867.7987 | −0.29 | C55 H104 O6 Li | 21.42 | 18:1/16:0/18:0; 16:1/16:0/20:0; 18:1/14:0/20:0 |
| 879.7990 | 879.7987 | 0.28 | C56 H104 O6 Li | 9.33 | b |
| 881.8139 | 881.8144 | −0.55 | C56 H106 O6 Li | 19.05 | 18:1/16:0/19:0; 18:1/18:0/17:0 |
| 891.7986 | 891.7987 | −0.2 | C57 H104 O6 Li | 3.45 | b |
| 893.8144 | 893.8144 | −0.04 | C57 H106 O6 Li | 21.82 | 18:1/16:1/20:0; 18:1/16:0/20:1; 16:1/16:0/22:1 |
| 895.8295 | 895.8300 | −0.53 | C57 H108 O6 Li | 37.95 | 18:1/16:0/20:0; 18:1/14:0/22:0; 16:1/16:0/22:0; 16:1/14:0/24:0; 18:1/12:0/24:0 |
| 907.8516 | 907.8512 | 0.44 | C55 H112 O8 Li | 1.47 | b |
| 909.8454 | 909.8457 | −0.29 | C58 H110 O6 Li | 2.24 | b |
| 919.8300 | 919.8300 | −0.05 | C59 H108 O6 Li | 3.22 | b |
| 921.8455 | 921.8457 | −0.16 | C59 H110 O6 Li | 27.76 | 18:1/16:1/22:0; 18:1/16:0/22:1; 18:1/14:0/24:1; 18:1/14:1/24:0;16:1/16:0/24:1; 16:1/16:1/24:0 |
| 923.8608 | 923.8613 | −0.52 | C59 H112 O6 Li | 56.1 | 18:1/16:0/22:0; 18:1/14:0/24:0; 16:1/16:0/24:0 |
| 935.8615 | 935.8613 | 0.13 | C60 H112 O6 Li | 2.21 | b |
| 937.8769 | 937.8770 | −0.13 | C60 H114 O6 Li | 2.45 | 18:1/15:0/24:0; 18:1/16:0/23:0; 18:1/17:0/22:0; 19:0/16:1/22:0; 17:1/16:0/24:0 |
| 939.8934 | 939.8926 | 0.72 | C60 H116 O6 Li | 7.96 | 19:0/16:0/22:0; 19:0/14:0/24:0 |
| 949.8763 | 949.8770 | −0.74 | C61 H114 O6 Li | 67.89 | 18:1/16:1/24:0; 18:1/16:0/24:1 |
| 951.8911 | 951.8926 | −1.55 | C61 H116 O6 Li | 100 | Δ918:1/16:0/24:0c |
| 963.8927 | 963.8926 | 0.02 | C62 H116 O6 Li | 3.47 | b |
| 965.9070 | 965.9083 | −1.25 | C62 H118 O6 Li | 2.36 | 19:0/16:1/24:0; 19:1/16:0/24:0; 18:1/17:0/24:0 |
| 967.9232 | 967.9239 | −0.71 | C62 H120 O6 Li | 3.54 | 19:0/16:0/24:0 |
| 977.9068 | 977.9083 | −1.48 | C63 H118 O6 Li | 4.69 | b |
| 979.9226 | 979.9239 | −1.39 | C63 H120 O6 Li | 3.03 | 18:1/18:0/24:0; 18:1/16:0/26:0 |
Only major isomers identified are listed;
structure not characterized;
double bond position of 18:1-FA (all) is at C-9
Table 2.
The high resolution mass measurements and LIT MSn analysis on the [M+Li]+ ions of mmDAG species from M. smegmatis biofilm
| measured m/z | Calcd. Mass | Deviat. | Elemental Composition | Rel. Intens. | Structure assignments | |
|---|---|---|---|---|---|---|
|
|
|
|||||
| [M + Li]+ | Da | mDa | (%) | major isomer | minor isomers | |
|
| ||||||
| 1130.0646 | 1130.0648 | −0.17 | C74 H138 O6 Li | 13.07 | 18:1/16:0/m37:2 | * |
| 1144.0806 | 1144.0804 | 0.13 | C75 H140 O6 Li | 28.5 | 18:1/16:0/m38:2 | * |
| 1158.0960 | 1158.0961 | −0.13 | C76 H142 O6 Li | 10.1 | 18:1/16:0/m39:2 | * |
| 1172.1116 | 1172.1117 | −0.12 | C77 H144 O6 Li | 16.16 | 18:1/16:0/m40:2 | * |
| 1186.1273 | 1186.1274 | −0.07 | C78 H146 O6 Li | 22.25 | 18:1/16:0/m41:2 | * |
| 1200.1429 | 1200.1430 | −0.11 | C79 H148 O6 Li | 24.55 | 18:1/16:0/m42:2 | * |
| 1214.1586 | 1214.1587 | −0.09 | C80 H150 O6 Li | 26.98 | 18:1/16:0/m43:2 | * |
| 1228.1746 | 1228.1743 | 0.29 | C81 H152 O6 Li | 30.46 | 18:1/16:0/m44:2 | * |
| 1240.1738 | 1240.1743 | −0.58 | C82 H152 O6 Li | 4.24 | 18:1/16:1/m45:2 | * |
| 1242.1906 | 1242.1900 | 0.58 | C82 H154 O6 Li | 32.22 | 18:1/16:0/m45:2 | 16:1/16:0/m47:2; 18:1/14:0/m47:2; 18:1/18:0/m43:2 |
| 1254.1894 | 1254.1900 | −0.63 | C83 H154 O6 Li | 4.97 | 18:1/16:1/m46:2 | * |
| 1256.2060 | 1256.2056 | 0.39 | C83 H156 O6 Li | 43.34 | 18:1/16:0/m46:2 | * |
| 1268.2056 | 1268.2056 | −0.08 | C84 H156 O6 Li | 6.78 | 18:1/16:1/m47:2 | * |
| 1270.2217 | 1270.2213 | 0.42 | C84 H158 O6 Li | 47.98 | 18:1/16:0/m47:2 | 18:1/18:0/m45:2; 18:1/14:0/m49:2; 16:1/16:0/m49:2 |
| 1282.2210 | 1282.2213 | −0.28 | C85 H158 O6 Li | 9.46 | 18:1/16:1/m48:2 | * |
| 1284.2373 | 1284.2369 | 0.34 | C85 H160 O6 Li | 59.59 | 18:1/16:0/m48:2 | 18:1/14:0/m50:2; 16:1/16:0/m50:2; 18:1/18:0/m46:2 |
| 1296.2370 | 1296.2369 | 0.06 | C86 H160 O6 Li | 9.75 | 18:1/16:1/m49:2 | * |
| 1298.2529 | 1298.2526 | 0.34 | C86 H162 O6 Li | 69.89 | 18:1/16:0/m49:2 | 18:1/18:0/m47:2; 18:1/14:0/m51:2; 16:1/16:0/m51:2 |
| 1310.2523 | 1310.2526 | −0.29 | C87 H162 O6 Li | 12.89 | 18:1/16:1/m50:2 | * |
| 1312.2685 | 1312.2682 | 0.29 | C87 H164 O6 Li | 85.21 | 18:1/16:0/m50:2 | 18:1/14:0/m52:2; 16:1/16:0/m52:2; 18:1/18:0/m48:2 |
| 1324.2683 | 1324.2682 | 0.09 | C88 H164 O6 Li | 13.88 | 18:1/16:1/m51:2 | 18:1/18:1/m49:2; 18:1/14:1/m53:2; 16:1/16:1/m53:2 |
| 1326.2841 | 1326.2839 | 0.24 | C88 H166 O6 Li | 80.46 | 18:1/16:0/m51:2 | 18:1/18:0/m49:2; 18:1/14:0/m53:2; 16:1/16:0/m53:2 |
| 1338.2830 | 1338.2839 | −0.86 | C89 H166 O6 Li | 14.43 | 18:1/16:1/m52:2 | 16:1/16:1/m54:2; 18:1/14:1/m54:2; 18:1/18:1/m50:2 |
| 1340.2998 | 1340.2995 | 0.27 | C89 H168 O6 Li | 97.8 | 18:1/16:0/m52:2 | 18:1/14:0/m54:2; 16:1/16:0/m54:2; 18:1/18:0/m50:2 |
| 1352.2995 | 1352.2995 | −0.08 | C90 H168 O6 Li | 11.21 | 18:1/16:1/m53:2 | 16:1/16:1/m55:2; 18:1/14:1/m55:2; 18:1/18:1/m51:2 |
| 1354.3157 | 1354.3152 | 0.46 | C90 H170 O6 Li | 59.94 | 18:1/16:0/m53:2 | 18:1/18:0/m51:2; 18:1/14:0/m55:2; 16:1/16:0/m55:2 |
| 1366.3147 | 1366.3152 | −0.49 | C91 H170 O6 Li | 14.03 | 18:1/16:1/m54:2 | * |
| 1368.3312 | 1368.3308 | 0.32 | C91 H172 O6 Li | 71.85 | 18:1/16:0/m54:2 | 18:1/18:0/m52:2; 18:1/14:0/m56:2; 16:1/16:0/m56:2 |
| 1380.3309 | 1380.3308 | 0.09 | C92 H172 O6 Li | 6.92 | 18:1/16:1/m55:2 | * |
| 1382.3468 | 1382.3465 | 0.29 | C92 H174 O6 Li | 34.15 | 18:1/16:0/m55:2 | * |
| 1394.3455 | 1394.3465 | −0.97 | C93 H174 O6 Li | 4.3 | 18:1/16:1/m56:2 | * |
| 1396.3628 | 1396.3621 | 0.65 | C93 H176 O6 Li | 14.2 | 18:1/16:0/m56:2 | * |
| 1410.3793 | 1410.3778 | 1.54 | C94 H178 O6 Li | 6.77 | 18:1/16:0/m57:2 | * |
structure not assigned
Characterization of triacylglycerols
We previously described LIT MSn on the [M + Li]+ ions to characterize TAGs with the notion that a complete structure assignment can be achieved, while structural information from MSn on, for example, [M + Na]+ ions is incomplete [12,13]. The assignments of the fatty acid substituents on the glycerol backbone are based on the findings that the ions arising from losses of the outer fatty acid substituents (i.e., the fatty acids at sn-1 and sn-3) are more abundant than the ions arising from loss of the fatty acid loss at sn-2. As shown in Fig. 2a, the MS2 spectrum of the [M + Li]+ ion at m/z 951.9 contained abundant ions at m/z 695, 669, and 583 arising from losses of 16:0-, 18:1, and 24:0-fatty acid substituents, respectively, along with ions at m/z 689, 663, and 577, corresponding to losses of 16:0-, 18:1-, and 24:0-fatty acid substituents as lithium salt, respectively. The ions at m/z 669 and at 583 are more abundant than the ion of m/z 695, and the ions at m/z 663 and at m/z 577 are also more abundant than the ion at m/z 689, indicating that the 16:0-fatty acyl group is located at sn-2, and the [M + Li]+ ion of m/z 951 represents a 18:1/16:0/24:0-TAG. MS3 on the ion of m/z 583 (951 → 583; Fig. 2b) gave rise to the ion of m/z 329, arising from further elimination of the 16:0-fatty acid as an α,β-unsaturated fatty acid (loss as 16:1-FA), along with ions at m/z 289 (lithiated 18:1-FA ), 263 (lithiated 16:0-FA), 301 (loss of 18:1-FA) and 319 (loss of 18:1-fatty acyl ketene) further confirms that the 18:1-fatty acid is indeed located at sn-1 (or sn-3) [12,13]. The spectrum also contained ions at m/z 485, 429, 387, and 373, arising from β-cleavages with γ-H shift, indicating that the double bond of the 18:1-fatty acid substituent is located at C-9 [13,16] (See Electronic Supplementary Material, Scheme S1). This structural assignment is further confirmed by the MS3 spectrum of the ion of m/z 695 (951 → 695; Fig. 2c), which contained the ions at m/z 597, 541, 499, and 485, arising from the similar β-cleavages with γ-H shift. The spectrum also contained ions at m/z 415, 329 arising from losses of the 18:1- and 24:0-fatty acid substituents as α, β-unsaturated fatty acids (losses as 18:2 and 24:1-FA), along with ions at m/z 375 (lithiated 24:0-FA) and 289 (lithiated 18:1-FA), further support that the compound is Δ918:1/16:0/24:0-TAG. The above structural assignment of the fragment ions were confirmed by the elemental compositions obtained by high resolution mass measurements (Table S3, Electronic Supplementary Material).
Fig. 2.
The MS2 spectrum of the [M + Li]+ ion at m/z 951.9 (a), and its MS3 spectra of the ions at m/z 583 (951 →583) (b), at m/z 695 (951 → 695) (c); and the MS2 spectrum of the corresponding [M + NH4]+ ions at m/z 962.9 (d), and its MS3 spectrum of the ions at m/z 577 (962 → 577) (e). The ions marked with “●” in inset of Panels b and c locate the position of the double bond along the 18:1-FA chain.
By contrast, MS2 on the corresponding [M + NH4]+ ions at m/z 962.9 (Fig. 2d) contained ions at m/z 689, 663 and 577, arising from losses of 16:0-, 18:1-, and 24:0-fatty acid substituents as ammonium salt, respectively. The ions at m/z 663, and at 577 are more abundant than the ion of m/z 689, consistent with the notion that the 16:0-FA substituent is located at sn-2, and the 18:1-, and 24:0-fatty acids are located at sn-1 and sn-3 (or vise versa), respectively, similar to that deduced from the [M + Li]+ ions. The identity and the location of the fatty acids on the glycerol backbone were also recognized by the MS3 spectrum of the ion of m/z 577 (962 → 577; Fig. 2e), which possess ion pairs at m/z 265 (18:1-acylium ion), 247 (265 – H2O), that are respectively more abundant than the ion pairs at m/z 239 (16:0-acylium ion), 221 (239 – H2O), consistent with that 18:1- and 16:0-FA are located at sn-1 and sn-2, respectively [12,13].
Several isomeric structures were identified for most of the TAG species present in the biofilm (Table 1). For example, the MS2 spectrum of the [M + Li]+ ions of m/z 923.9 (Fig. 3a) contained ions at m/z 667, 641, and 583, arising from losses of 16:0-, 18:1-, and 22:0-FA substituents, respectively, and the ion at m/z 641 is the least prominent. The results indicate the presence of Δ918:1/16:0/22:0-TAG. The spectrum also contained ions at m/z 695 and 555 arising from losses of 14:0- and 24:0-FA substituents, respectively. These ions also paired with the ion at m/z 641 arising from loss of 18:1-FA moiety, led to the assignment of Δ918:1/14:0/24:0-TAG structure; while the ions observed at m/z 667, and 555 arising from losses of 16:0- and 24:0-FA substituent, respectively, combined with the ion at m/z 669 arising from loss of 16:1-FA substituent led to the identification of the Δ916:1/16:0/24:0-TAG isomer. The position of the double bond of the 16:1-FA at C-9 is determined by the observation of the ions at m/z 559 and 503, arising from β-cleavage with γ-H shift as seen earlier. The above structural assignments were further confirmed by the MS3 spectra of the ions of m/z 555 (923 → 555; Fig. 3b) and of m/z 583 (not shown). The former spectrum (Fig. 3b) contained ions at m/z 329 (loss of 14:0-FA as α,β-unsaturated FA), 289 (lithiated 18:1-FA), 235 (lithiated 14:0-FA), 291 (loss of 18:1-Fatty acyl ketene), 273 (loss of 18:1-FA), consistent with the assignment of Δ918:1/14:0/24:0-TAG; whereas ions at m/z 301 (loss of 16:1-FA), 261 (lithiated 16:1-FA), 263 (lithiated 16:0-FA), and 319 (loss of 16:1-fatty acyl ketene) support the presence of Δ916:1/16:0/24:1-TAG isomer, of which the double position of the Δ916:1 is recognized by the observation of the ions at m/z 485 and 429 (Fig. 3b, inset). The MS3 spectrum of the ion of m/z 583 is identical to that shown in Fig. 2d, indicating the presence of Δ918:1-, 16:0-FA at sn-1 and sn-2, respectively, consistent with the assignment of Δ918:1/16:0/22:0-TAG.
Fig. 3.
The MS2 spectrum of the [M + Li]+ ions of m/z 923.9 (a), its MS3 spectra of the ions of m/z 555 (923 → 555) (b); and the MS2 spectrum of the [M + Li]+ ion at m/z 965 (c), and its MS3 spectrum of the ion of m/z 597 (d). In inset of Panels b, the ions marked with “●” locate the position of the double bond of the 18:1-FA chain; while the ions marked with “○” locate the double bond of 16:1-FA substituent.
TAGs consisting of odd number carbon (branch) chain fatty acid substituents were also observed. For example, the MS2 spectrum of the [M + Li]+ ion at m/z 965 (Fig. 3c) contained the major ion set at m/z 711, 667, and 597 arising from losses of 16:1-, 19:0-, and 24:0-fatty acid substituents, respectively. The ions at m/z 667 and 597 are more abundant than the ion at 711, indicating that the 16:1-fatty acid is located at sn-2, while the 19:0- and 24:0-FA are located at sn-1 or sn-3. The structure assignment is further confirmed by the MS3 spectrum of the ion of m/z 597 (965 → 597; Fig. 3d), which is dominated by the ion of m/z 345 arising from loss of 16:1-fatty acid as α,β-unsaturated fatty acid (loss as 16:2-FA), supporting the notion that the 16:1-fatty acid is indeed located at sn-2, and the ions at m/z 305 (lithiated 19:0) and 261 (lithiated 16:1), consistent with the assignment of 19:0/16:1/24:0-TAG. The spectrum also contained the minor ion set at m/z 343 (loss of 16:0-FA as arising from loss of α,β-unsaturated fatty acid), 303 (lithiated 19:1) and 263 (lithiated 16:0); as well as the ion set at m/z 329 (loss of 17:0-FA as α,β-unsaturated fatty acid), 289 (lithiated 18:1) and 277 (lithiated 17:0). The former ion set indicates the presence of 19:1/16:0/24:0-TAG minor isomer, consistent with the observation of the minor ion set of the ions at m/z 709/703 (loss of 16:0-FA) and 669/663 (loss of 19:1-FA) in Fig. 3c. The latter set indicates the presence of 18:1/17:0/24:0-TAG, consistent with the observation of the ions at m/z 683/677 (loss of 18:1-FA), 695/689 (loss of 17:0-FA) in Fig. 3c.
The number of isomeric structures increases as TAG species consists of two unsaturated fatty acid substituents (Table 1). For example, the MS2 spectrum of the ion of m/z 921.9 (Fig. 4a) contained ions at m/z 693, 667, 665, 639, 583, 581, 555, and 553, arising from losses of 14:0-, 16:1-, 16:0-, 18:1-, 22:1, 22:0-, 24:1, and 24:0-FA substituents, respectively. Further dissociation of the ion at m/z 639 (921 → 639; Fig. 4b) gave rise to ions at 385 (loss of α,β-unsaturated 16:1-FA), together with ions at m/z 345 (lithiated 22:1) and 263 (lithiated 16:0), indicating the presence of the major 18:1/16:0/22:1-TAG isomer. The spectrum also contained ions at m/z 387 (loss of α,β-unsaturated 16:2-FA), which paired with ions at m/z 347 (lithiated 22:0) and 261 (lithiated 16:1), indicating the presence of 18:1/16:1/22:0-TAG; while the ions at m/z 413 (loss of α,β-unsaturated 14:1-FA) m/z 373 (lithiated 24:1) and 235 (lithiated 14:0) led to assignment of minor 18:1/14:0/24:1-TAG isomer.
Fig. 4.
The MS2 spectrum of the ion of m/z 921.9 (a), its MS3 spectrum of the ion of m/z 639 (921 → 639) (b), of m/z 667 (921 → 667) (c), of m/z 665 (921 → 665) (d), of m/z 553 (921 → 553) (e), and of m/z 581 (921 → 581) (f).
Similarly, MS3 on the ion of m/z 667 (921 → 667; Fig. 4c) yielded abundant ions at m/z 415 (loss of α,β-unsaturated 16:2-FA), together with ions at m/z 375 (lithiated 24:0) and 261 (lithiated 16:1), indicating the presence of 16:1/16:1/24:0-TAG. The spectrum also contained ions at m/z 387 (loss of α,β-unsaturated 18:2-FA), 329 (loss of α,β-unsaturated 22:1-FA), 289 (lithiated 18:1), and 347 (lithiated 22:0), signifying the presence of 18:1-, and 22:0-FA at sn-1, and sn-3 respectively, consistent with the earlier structure assignment of 18:1/16:1/22:0-TAG isomer (Fig. 4b); while ions at m/z 413 (loss of α,β-unsaturated 16:1-FA) and 373 (lithiated 24:1) led to assignment of 16:1/16:0/24:1-TAG isomer. MS3 on the ion of m/z 665 (921 → 665; Fig. 4d) yielded ions at m/z 385 (loss of α,β-unsaturated 18:2-FA), 345 (lithiated 22:1), 289 (lithiated 18:1) and 329 (loss of α,β-unsaturated 22:2-FA), which gave assignment of 18:1/16:0/22:1-TAG; while the ions at m/z 413 (loss of α,β-unsaturated 16:2-FA), 373 (lithiated 24:1), 301 (loss of α,β-unsaturated 24:2-FA), and 261 (lithiated 16:1) led to the structure of 16:1/16:0/24:1-TAG. The assignments of these isomers, again, are consistent with the structures as seen earlier.
The MS3 spectra of the ions of m/z 583, and of 555 (not shown) are identical to those seen in Fig. 2b and 3b, leading to the assignments of the structures of 18:1/14:0/24:1-TAG, 16:1/16:0/24:1-TAG (from m/z 555) and 18:1/16:0/22:1-TAG (from m/z 583). The MS3 spectrum of the ions of m/z 553 (921 → 553; Fig. 4e) contained ions at m/z 301 (loss of α,β-unsaturated 16:2-FA), 261 (lithiated 16:1), 317 (loss of 16:1-ketene), indicating the presence of 16:1/16:1/24:0-TAG, along with ions at m/z 329 (loss of α,β-unsaturated 14:2-FA), 289 (lithiated 18:1), and 233 (lithiated 14:1), that identify the 18:1/14:1/24:0-TAG isomer. The MS3 spectrum of the ion of m/z 581 (921 → 581; Fig. 4f) are dominated by ions at m/z 329 ((loss of α,β-unsaturated 16:2-FA), 289 (lithiated 18:1), and 261 (lithiated 16:1), confirming the presence of 18:1/16:1/22:0-TAG isomer. The above results indicate that the ions at m/z 921 consist of the major 18:1/16:0/22:1-isomer along with 18:1/16:1/22:0-, 16:1/16:1/24:0-, 16:1/16:0/24:1-, 18:1/14:0/24:1-, 18:1/14:1/24:0-TAG isomers.
Structural Characterization of MMDAGs
The elemental compositions (C73H138(CH2)nO6; n=1,2, …, 25) of this lipid family deduced from high resolution mass measurements on the [M + NH4]+ (Table S2, Electronic Supplementary Material) and the corresponding [M + Li]+ (Table 2) adduct ions point to the notion that this lipid family possesses structures similar to TAG. The longer HPLC elution time of this lipid subclass (25–26 min) than that of TAG (18–23.5 min) in a C-8 reversed phase column (Fig. S1, Electronic Supplementary Material) is consistent with the presence of a very long meromycolyl fatty acyl chain at sn-3 (or sn-1) in the molecules. This structural feature permits its distinction from TAG lipids by mass spectrometry. The MSn spectra of the [M + Li]+ and the corresponding [M + NH4]+ ions as described below confirmed that this complex lipid also contains several isomers in each of the molecular species.
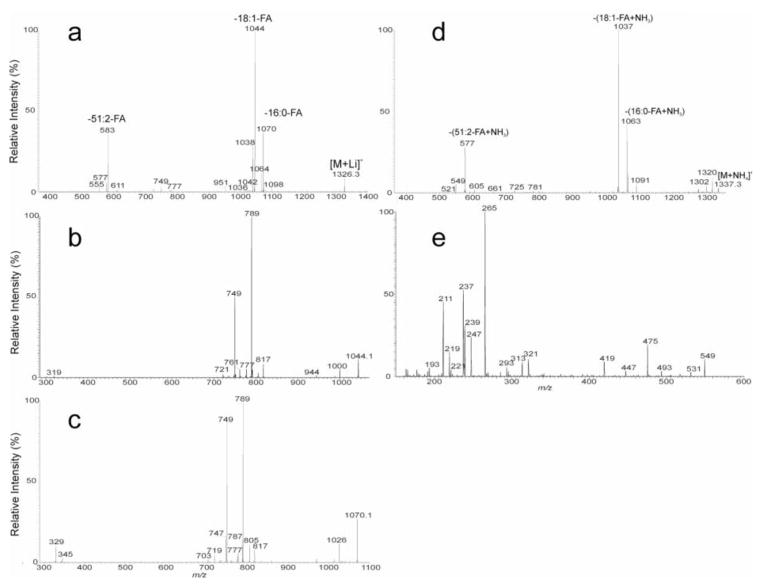
For example, the MS2 spectrum of the [M + Li]+ ion at m/z 1326 (Fig. 5a) contained prominent ions at m/z 1070, 1044 and 583 arising from losses of 16:0-, 18:1- and 51:2-FA substituents, respectively, and the ions at m/z 1064, 1038 and 577, arising from losses of the corresponding fatty acid substituents as lithium salt, respectively. The presence of 51:2-FA is consistent with the observation of the ion at m/z 749, representing a lithiated mycolyl 51:2-FA (m51:2-FA) ion. Further dissociation of the ion of m/z 1044 (1326 → 1044; Fig. 5b) also gave rise to the ion at m/z 749, corresponding to the lithiated m51:2-FA ion; together with the abundant ions at m/z 789 arising from loss of 16:0-FA moiety as an α,β-unsaturated fatty acid (loss as 16:1-FA), indicating that the 16:0-fatty acid substituent is located at sn-2. These results led to the assignment of the structure of 18:1/16:0/m51:2-TAG. The confirmation of 18:1- and 16:0-FA at sn-1 and sn-2, respectively, is further supported by the MS3 spectrum of the ion of m/z 583 (1326 → 583; data not shown), which is identical to the MS3 spectrum arising from 18:1/16:0/24:0-TAG (951 → 583) (Fig. 2b). The results also readily located the double bond of the 18:1-FA substituent at C-9. The combined structural information indicates the presence of the major Δ918:1/16:0/m51:2-TAG structure.
Fig. 5.
The MS2 spectrum of the [M + Li]+ ion of MMDAG at m/z 1326.2 (a), its MS3 spectrum of the ions at m/z 1044 (1326 → 1044) (b), at m/z 1070 (1326 → 1070) (c); and the MS2 spectrum of the ion at m/z 1337.3 (d), and its MS3 spectrum of the ions at m/z 549 (1337 → 549) (e).
In Fig. 5b, minor ions at m/z 761 arising from loss of 18:0-FA as α,β-unsaturated fatty acid (loss as 18:1-FA) together with the ion at m/z 721 representing a lithiated m49:2-FA cation were also observed, indicating the presence of Δ918:1/18:0/m49:2-TAG isomer. This structural assignment is further supported by the observation of the minor ions at m/z 1042 and 1036 arising from losses of 18:0-FA as acid and as lithium salt respectively, and of the ion of m/z 611 arising from loss of m49:2-FA moiety seen in Fig. 5a.
The ions at m/z 1098 arising from loss of 14:0-FA substituent, and at m/z 555 arising from loss of 53:2-FA substituent were also observed in Fig. 5a, indicating that a minor 18:1/14:0/m53:2-TAG isomer is also present. This structural assignment is also in agreement with the observation of the ions at m/z 777, representing a lithiated m53:2-FA; and at m/z 817, arising from loss of 14:0-FA as α,β-unsaturated fatty acid (loss as 14:1-FA), and supporting the presence of 14:0-FA substituent at sn-2 (Fig. 5b). Further dissociation of the ion at m/z 1070 (1326 → 1070; Fig. 5c) gave rise to ions at m/z 789 and 329 arising from losses of 18:1- (loss as 18:2-FA) and 51:2-fatty acids (loss as 51:3-FA) as α,β-unsaturated fatty acids respectively, together with ions at m/z 805 and 345 arising from losses of the 18:1- and m51:2-FA as ketenes, respectively. These results located the 18:1- and m51:2-fatty acid at sn-1 and sn-3 (or vise verse), respectively. The structural information combined with the prominent ion seen at m/z 749, representing a lithiated m51:2-FA, gave the assignment of the major 18:1/16:0/m51:2-TAG isomer, consistent with the structure assignment as described earlier. The spectrum (Fig. 5c) also contained the minor ions at m/z 817 (loss of 16:1-FA as α,β-unsaturated fatty acid), and at m/z 777 (lithiated 53:2-FA), suggesting the presence of 16:1/16:0/m53:2-TAG isomer. The assignment is also consistent with the observation of the ion of 1072 (loss of 16:1-FA) in Fig. 5a. The above structural assignments of the fragment ions were confirmed by high resolution mass measurements (Table S4, Electronic Supplementary Material). The combined results revealed that the [M + Li]+ ion of m/z 1326 consists of the major Δ918:1/16:0/m51:2-TAG structure together with minor isomers of Δ918:1/14:0/m53:2-TAG, 16:1/16:0/m53:2-TAG, and Δ918:1/18:0/m49:2-TAG.
The above structure assignments were confirmed by MSn on the corresponding [M + NH4]+ ions at m/z 1337. As shown in Fig. 5d, the MS2 spectrum of the ion at m/z 1337 yielded prominent ions at m/z 1063, 1037, and 577, arising from losses of the 16:0-, 18:1-, and m51:2-FA substituents as ammonium salt, respectively, indicating the presence of 18:1-, 16:0-, and m51:2-fatty acyl substituents in the molecule. The MS3 spectrum of the ion of m/z 577 (1337 → 577; data not shown) is identical to that shown in Fig. 2e, consistent with the notion that the 18:1- and 16:0-FA substituents are located at sn-1 and sn-2 of the glycerol backbone, respectively; while the m51:2-FA moiety is located at sn-3. This gave assignment of the major 18:1/16:0/m51:2-TAG isomer. The spectrum (Fig. 5d) also contained minor ions at m/z 549 and 1091, arising from losses of m53:2- and 14:0-FA substituent as NH4+ salt, respectively, indicating the presence of 18:1/14:0/m53:2-TAG minor isomer. This structural assignment is further supported by the MS3 spectrum of the ion of m/z 549 (1337 → 549; Fig. 5e), in which the ions at m/z 265 (18:1-acylium ion) and 247 (265 – H2O) are respectively more prominent than the ions at m/z 211 (14:0-acylium ion) and 193 (211 – H2O), suggesting the presence of 18:1- and 14:0- fatty acid substituents at sn-1 and sn-2, respectively, and an m53:2-FA substituent is located at sn-3. This information led to assignment of 18:1/16:0/m53:2-TAG structure. The spectrum (Fig. 5e) also contained the ions at m/z 237 (16:1-acylium ion) and 219 (237 – H2O), which are respectively more abundant than the ions at m/z 239 (16:0-acylium cation) and 221 (239 – H2O), indicating an isomer with 16:1- and 16:0-Fatty acyl groups at sn-1 and sn-2, respectively, is also present, and consistent with the assignment of a minor 16:1/16:0/m53:2-TAG isomer, similar to the structural assignment deduced from the [M + Li]+ ions as seen earlier.
Similarly, the MS2 spectrum of the [M + Li]+ ion of m/z 1340.3 (Fig. 6a) is dominated by the ion of m/z 1058 arising from loss of 18:1-FA substituent, along with ions at m/z 1084, and 583 arising from losses of 16:0-, and m52:2-FA substituents, respectively. The profile of the spectrum is nearly identical to that shown in Fig. 5a, indicating the presence of the major Δ918:1/16:0/m52:2-TAG structure, along with minor isomers of Δ918:1/14:0/m54:2-TAG, 16:1/16:0/m54:2-TAG, and Δ918:1/18:0/m50:2-TAG. The assignments of these minor isomers were also confirmed by the MS3 spectra of the ions at m/z 1058 (1340 → 1058; Fig. 6b), 1084 (1340 → 1084; Fig. 6c) and at 583 (data not shown). The profiles of the former spectrum (Fig. 6b) is similar to Fig. 5b, and the MS3 spectrum of the ion of m/z 1084 is also similar to Fig. 5c, indicating that the ion of m/z 1340 consist of the similar isomeric structures in which the mycolyl FA substituent at sn-3 is one carbon longer than those seen for the ion of m/z 1326. These structures with the common fatty acyl substituents located at sn-1 and sn-2 were observed for the entire lipid family (Table 2).
Fig. 6.
The MS2 spectrum of the [M + Li]+ ion of MMDAG at m/z 1340.3 (a), its MS3 spectra of the ions at m/z 1058 (1340 → 1058) (b), and at m/z 1084 (1340 → 1084) (c).
Conclusions
The structures of TAG (Table 1) and MMDAG (Table 2) found in the biofilm of M. smegmatis are complex. Interestingly, the predominate isomer in each of the molecular species all contains 1-oleory 2-palmitoyl residue. These results are in agreement with the previous findings by Walker et al. [17]. The presence of the very long fatty acyl chain in MMDAG altered the appearance of the MS2 spectrum, in which the abundance of the ions at m/z 583 arising from loss of the meromycolic acid residue (Fig. 5a and 6a) declines significantly as compared to that arising from the loss of fatty acid with shorter chain length (e.g., Fig. 3a). Nevertheless, spectra from MS3 are readily applicable for assignments of the position of the fatty acid substituents on the glycerol backbone, underscoring the utility of LIT MSn in the structural elucidation of complex lipid. Less is clear, however, its utility in the location of the position of the unsaturated bond or side chain along the meromycolic chain, due to the presence of many isomers of the meromycolic acid [8] that are not separable by mass spectrometry. The LIT MSn spectrometric approach as reported here is simple and affords the complex structures of TAGs and MMDAGs in the biofilm of M. smegmatis to be unfold in detail, without employment of the laborious steps that were required using other analytical approaches.
Supplementary Material
Acknowledgments
This research is supported by US Public Health Service Grants P41-RR-00954, P60-DK-20579, and P30-DK56341(mass spectrometry facility) and AI087840 (GEP). We acknowledge the technical assistance from Meei-Hua Lin and Alan Bohrer.
Abbreviations
- ESI-MS
electrospray ionization-MS
- HRMS
high resolution mass spectrometry
- LIT
linear ion-trap
- TAG
triacylglycerol
- MMDAG
monomeromycolyl diacylglycerol
- FA
Fatty acid
References
- 1.Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, Kolattukudy PE. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology. 2006;152 (9):2717–2725. doi: 10.1099/mic.0.28993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Low KL, Rao PSS, Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. Triacylglycerol utilization is required for regrowth of in vitro hypoxic nonreplicating Mycobacterium bovis Bacillus Calmette-Guerin. J Bacterio. 2009;191 (16):5037–5043. doi: 10.1128/jb.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garton NJ, Christensen H, Minnikin DE, Adegbola RA, Barer MR. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology. 2002;148 (10):2951–2958. doi: 10.1099/00221287-148-10-2951. [DOI] [PubMed] [Google Scholar]
- 4.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011;7 (6):e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa H, Kashiwabara Y, Matsuki G. Metabolism of Triacylglycerol in Mycobacterium smegmatis. Journal of Biochemistry. 1976;80 (5):923–928. doi: 10.1093/oxfordjournals.jbchem.a131378. [DOI] [PubMed] [Google Scholar]
- 6.Ortalo-Magné A, Lemassu A, Lanéelle MA, Bardou F, Silve G, Gounon P, Marchal G, Daffé M. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacterio. 1996;178 (2):456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etienne G, Laval F, Villeneuve C, Dinadayala P, Abouwarda A, Zerbib D, Galamba A, Daffé M. The cell envelope structure and properties of Mycobacterium smegmatis mc2155: is there a clue for the unique transformability of the strain? Microbiology. 2005;151 (6):2075–2086. doi: 10.1099/mic.0.27869-0. [DOI] [PubMed] [Google Scholar]
- 8.Kremer L, De Chastellier C, Dobson G, Gibson KJC, Bifani P, Balor S, Gorvel J-P, Locht C, Minnikin DE, Besra GS. Identification and structural characterization of an unusual mycobacterial monomeromycolyl-diacylglycerol. Mol Microbiol. 2005;57 (4):1113–1126. doi: 10.1111/j.1365-2958.2005.04717.x. [DOI] [PubMed] [Google Scholar]
- 9.Kim M-J, Wainwright HC, Locketz M, Bekker L-G, Walther GB, Dittrich C, Visser A, Wang W, Hsu F-F, Wiehart U, Tsenova L, Kaplan G, Russell DG. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Molecular Medicine. 2010;2 (7):258–274. doi: 10.1002/emmm.201000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS pathogens. 2011;7 (6):e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed MB, Gagneux S, DeRiemer K, Small PM, Barry CE. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J Bacterio. 2007;189 (7):2583–2589. doi: 10.1128/jb.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu F-F, Turk J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom. 1999;10 (7):587–599. doi: 10.1016/s1044-0305(99)00035-5. [DOI] [PubMed] [Google Scholar]
- 13.Hsu F-F, Turk J. Electrospray ionization multiple-stage linear ion-trap mass spectrometry for structural elucidation of triacylglycerols: Assignment of fatty acyl groups on the glycerol backbone and location of double bonds. J Am Soc Mass Spectrom. 2010;21 (4):657–669. doi: 10.1016/j.jasms.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4 (11):1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 15.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR, Hatfull GF. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008;69 (1):164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu FF, Turk J. Elucidation of the double-bond position of long-chain unsaturated fatty acids by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J Am Soc Mass Spectrom. 2008;19 (11):1673–1680. doi: 10.1016/j.jasms.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker RW, Barakat H, Hung JG. The positional distribution of fatty acids in the phospholipids and triglycerides of Mycobacterium smegmatis and M. bovis BCG. Lipids. 1970;5 (8):684–691. doi: 10.1007/BF02531435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.