Abstract
The development of a safe, effective vaccine to prevent human immunodeficiency virus (HIV) infection is a key step for controlling the disease on a global scale. However, many aspects of HIV biology make vaccine design problematic, including the sequence diversity and structural variability of the surface envelope glycoproteins and the poor accessibility of neutralization-sensitive epitopes on the virus. In this review, we discuss recent progress in understanding HIV in a structural context using emerging tools in 3D electron microscopy, and outline how some of these advances could be important for a better understanding of mechanisms of viral entry and for vaccine design.
Keywords: Cryo-electron tomography, ion abrasion scanning electron microscopy (IA-SEM), focused ion beam scanning electron microscopy (FIB-SEM), vaccine design, virus–cell interaction
HIV infection
Human immunodeficiency virus (HIV) infection is a major health issue world-wide, with more than 34 million people currently living with HIV/AIDS (http://www.who.int). Despite decades of intensive research, a vaccine that can prevent transmission of the virus remains elusive. HIV presents unique challenges to the design of effective vaccines: the rapid mutation rate of the virus generates abundant variants in response to selective pressures, resulting in extensive sequence and structural variation. These characteristics enable the virus to escape the immune system in infected individuals. Further, they complicate the process of eliciting antibodies that can provide protection from the extraordinarily broad spectrum of antigenic variety displayed by the diverse strains of circulating HIV. Rational immunogen design in the face of this diversity is best served by a fundamental understanding of essential features of both envelope glycoprotein structure and function and mechanisms underlying the entry of HIV into target cells.
HIV is an enveloped virus. A lipid bilayer membrane envelope, acquired from the cellular membrane during the process of budding from an infected cell, surrounds the internal nucleocapsid, which contains two copies of the viral genome. Embedded in the envelope membrane and displayed on the surface of the virion are envelope glycoprotein (Env) spikes [1]. Each individual Env spike is composed of a trimer of heterodimers of the transmembrane envelope glycoprotein gp41 and the surface envelope glycoprotein gp120 [2], derived from a common gp160 precursor. This structure mediates infection through a multistep process in which gp120 binding to the cellular receptor CD4, followed by binding to a chemokine receptor (typically CXCR4 or CCR5), results in conformational changes that expose a fusion domain in gp41, leading to fusion of the viral and target cell membranes and internalization of the viral nucleocapsid [3]. The primary target cells for HIV are CD4+ T cells; however, there are many other cell types that interact with and may become infected by HIV, including macrophages and dendritic cells [4]. In turn, these cells can harbor HIV, sometimes for very long periods of time, and transmit the virus to other cells.
In this review, we provide an overview of selected examples of recent progress using new approaches in 3D electron microscopy to better understand structural aspects of the biology of HIV infection. We address three separate topics, each with important implications for preventing the spread of HIV: mechanisms of antibody neutralization, transfer of virus at a virological synapse, and our current understanding of the interactions at the virus-cell contact zone.
Neutralizing antibodies and Env
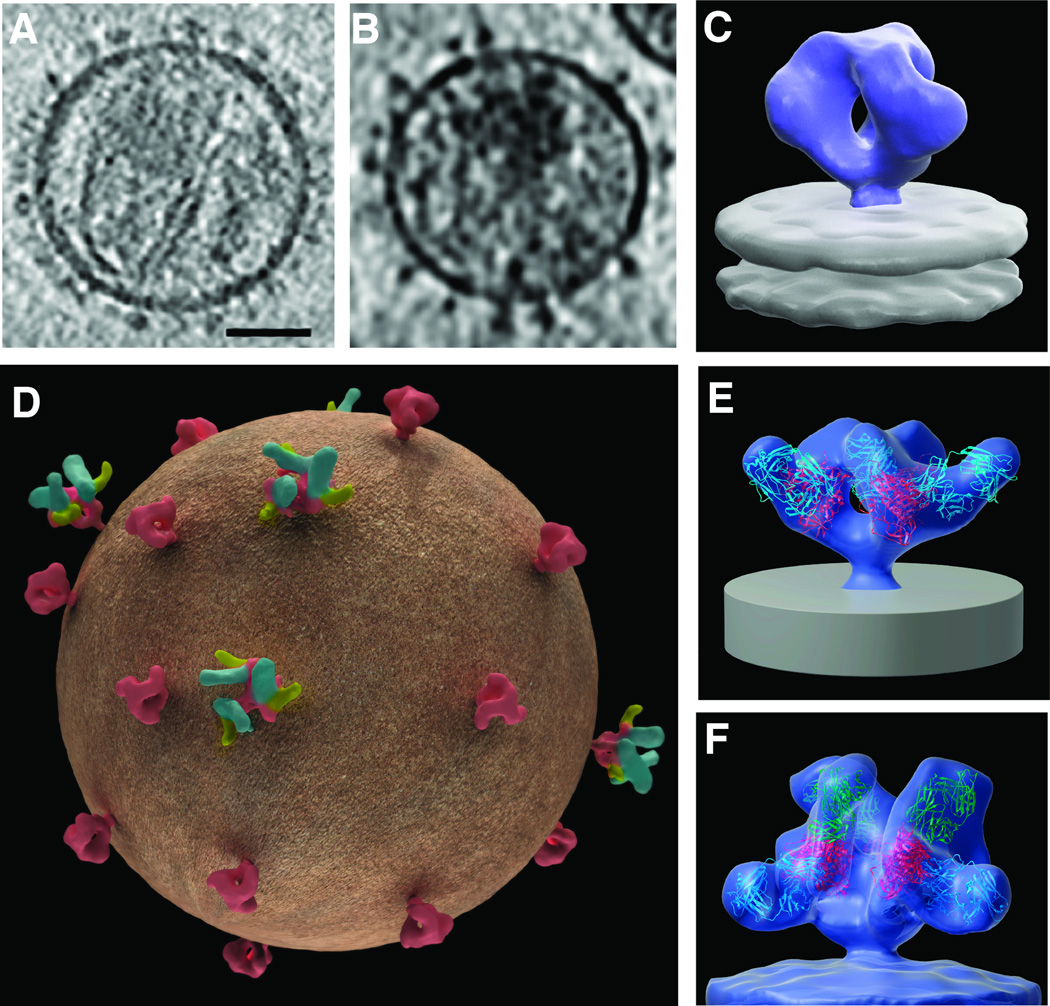
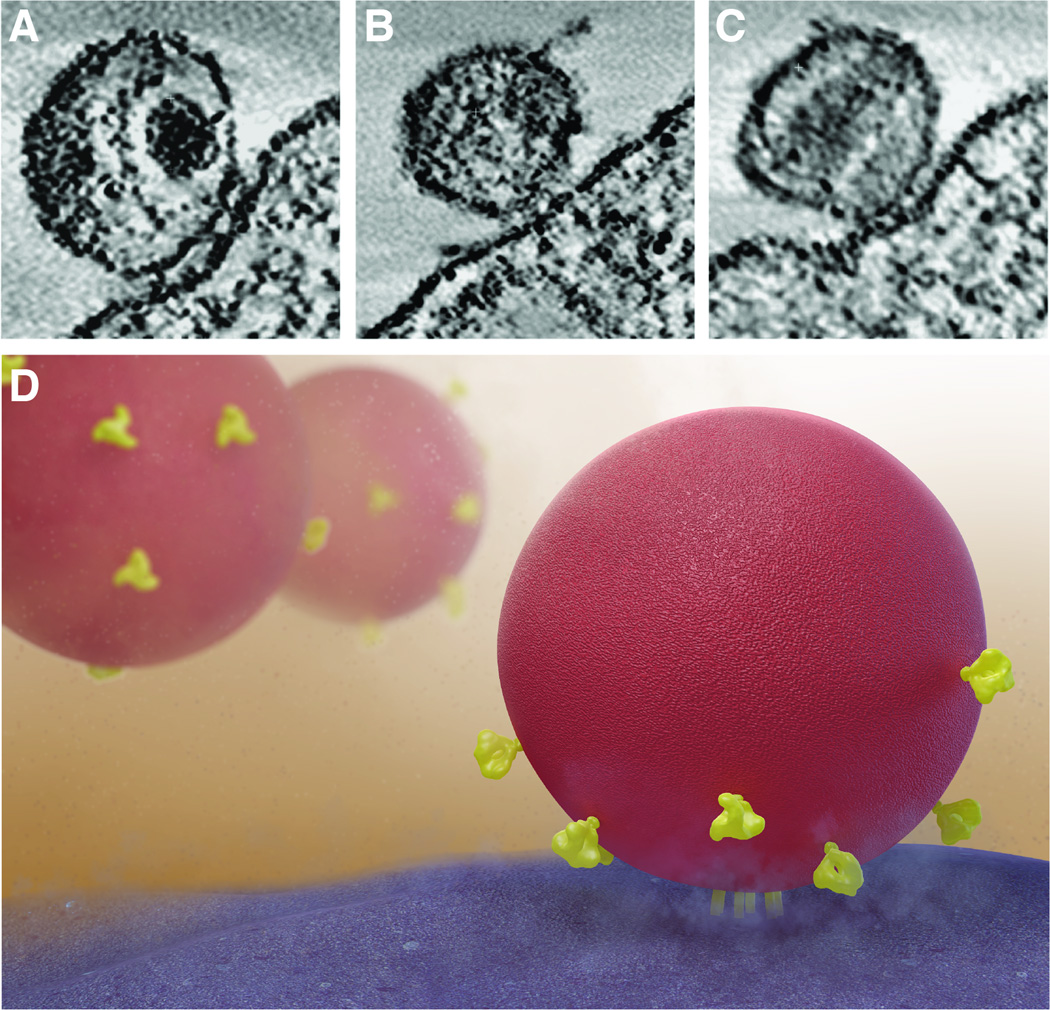
The identification of broadly neutralizing antibodies, i.e. antibodies that can bind and block entry of a large proportion of circulating HIV strains, has been an important objective in the quest for a vaccine [5]. The first such antibodies were discovered nearly two decades ago [6–8], but were thought to be extremely rare. In recent years however, new methods have determined that up to 25% of HIV-infected patients may develop such antibodies [9–13], and have allowed the rapid discovery of a number of highly potent broadly neutralizing antibodies [14–17]. While the structures of fragments of gp120 monomers bound to many of these neutralizing antibodies have been determined by X- ray crystallography [18–21], we do not yet have atomic-resolution structures for trimeric Env in either native or antibody-bound states [22]. However, significant insights into Env structure are beginning to emerge from studies of the Env complex using cryo-electron microscopy and cryo-electron tomography [1, 22–27]. Individual Env spikes can be visualized in tomograms (i.e. 3D volumes) of intact viruses and visualized either in slices through the tomogram (Figures 1A, 1B) or as three-dimensional density maps at resolutions of ~ 20 Å (Figure 1C). These density maps are acquired by averaging thousands of tomograms of individual spikes, each of which provide information on the structure of the trimer. Tomograms of viruses complexed with neutralizing antibodies or other ligands provide insights into the location of the bound antibody on the surface of Env spikes in situ on virions (Figure 1D). By fitting three copies of the crystal structures of relevant gp120–Fab complexes into these density maps, molecular models for trimeric Env bound to one or more neutralizing antibodies can be obtained (Figures 1E, 1F).
Figure 1.
Quaternary structure of trimeric HIV-1 spikes on intact viruses in complex with neutralizing antibodies. (A, B) Slices through tomograms of plunge-frozen SIVmneE11S [23] and HIV-1 BaL variants [25], respectively. Scale bar in (A) is 35nm. (C) 3D tomographic map of unliganded native HIV-1 BaL [1]. (D) Schematic representation of a virus illustrating the possible heterogeneity of antibody and ligand binding. Env (red) is either unliganded or ligand (yellow) and antibody (green) bound. (E, F) 3D tomographic density maps allow the determination of the location and orientation of antibody binding on native Env spikes. Env is bound to VRC01 antibody alone (E) or to VRC01 antibody and 17b antibody (F) [33]. Crystal coordinates for gp120 (red), VRC01 antibody (blue) and 17b antibody (green) are fitted into the maps. Coordinates for VRC01 are from the gp120-VRC01 complex (PDB ID:3NGB) and are aligned to coordinates for gp120 and 17b from the gp120-CD4/17b complex (derived from PDB ID:1GC1).
Structural and biochemical studies have revealed that broadly neutralizing antibodies can bind to the Env spike at a variety of locations. Antibodies such as VRC01, b12, HJ16, NIH45-46, 12A12, and 3BNC117 bind to the CD4 binding site [6, 16, 28, 29]. Other broadly neutralizing antibodies such as PG9, PG16, PGT145 and CH04 are thought to bind primarily to the variable V1/V2 loops of gp120 [30–32]. Some others such as the glycan dependent antibodies 2G12 or PGT121 appear to mainly interact with V3 variable loop [31] while 17b or the llama antibody fragment m36 target the co-receptor binding site on gp120 [25, 33]. Others, such as 2F5 [34], 4E10 [35], 10E8 [20], and Z13e1 [36], bind to the highly conserved membrane-proximal external region (MPER) of gp41 [37, 38]. While the binding location and clonal specificities of many of these antibodies have been characterized in detail, many of the antibodies have unusual features, including high levels of somatic hypermutation. Development of an immunogen that can elicit these types of broadly neutralizing antibodies in the context of a vaccine remains an unsolved problem [17].
How do these antibodies function to block viral entry? Cryo-electron tomographic studies are beginning to provide clues to the mechanisms that may underlie the action of these different antibodies. Structural studies on native Env trimers show that a key step in the entry process, triggered by CD4 binding, is a dramatic opening of the quaternary conformation of Env, involving outward movement of the three gp120 protomers and exposure of buried regions of gp41. We now know that binding by 17b or m36 antibodies appears capable of triggering the open conformation [25, 33]. Despite this, these antibodies are neutralizing, presumably because binding of these antibodies at the apex of the spike blocks cell–virus contact. In contrast, the CD4 binding site-specific mAb VRC01 locks trimeric Env in a closed conformation that is very similar in structure to that of the native, unliganded Env trimer, preventing binding by co-receptor [33]. Antibodies that bind to the variable loop regions may block entry by a combination of these mechanisms, while MPER antibodies such as Z13e1 very likely recognize an intermediate structure that occurs after CD4 binding and spike opening, but before viral fusion is initiated [39], suggesting that these antibodies could block fusion by preventing gp41 rearrangements necessary for membrane fusion, if the kinetic challenges inherent in such a mode of action can be overcome. Although many broadly neutralizing antibodies can function to prevent cellular entry by HIV in vitro, and in fact have been shown to prevent infection by passive infusion in non-human primate studies [40–42], it is an open question whether such antibodies can be generated through vaccination, and be maintained at sufficient titer to block virus infection and spread, including from one cell to another [43].
Cell-to-cell transmission of HIV
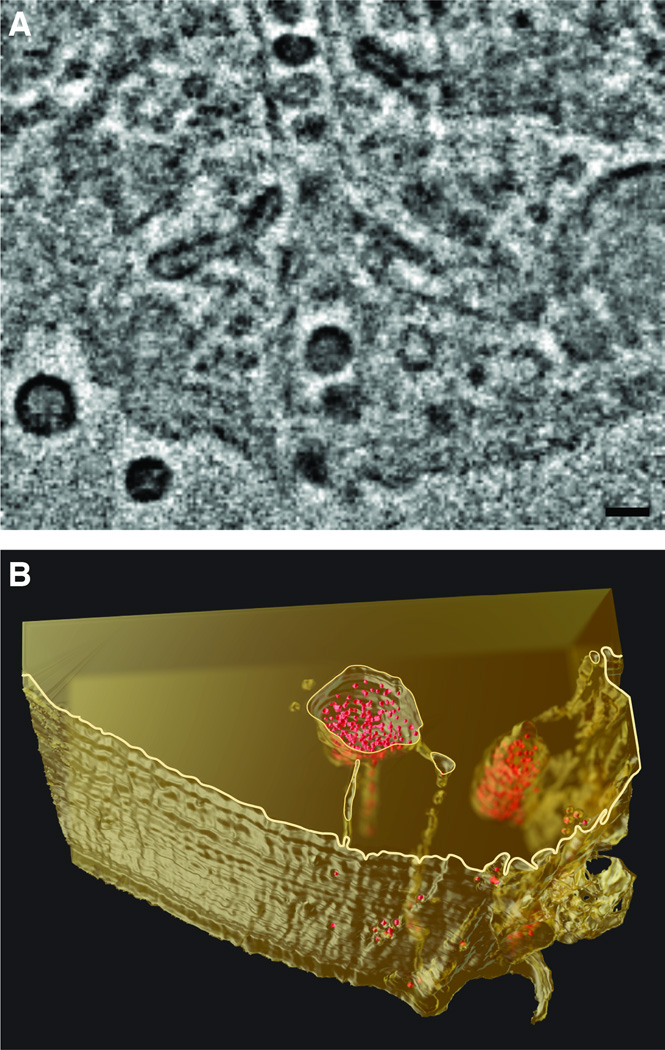
While CD4+ T cells are the primary targets of HIV infection, many other cell types, including long-lived cells like macrophages and dendritic cells, can either become productively infected by the virus or can take up and sequester infectious virus in a form that can be transmitted to other cells, despite not becoming productively infected themselves [44–48]. Viruses sequestered in these cells can evade detection by the immune system. Insights into the mechanism of sequestration of virus and the distribution of HIV in these infected cells has come from a 3D imaging technique called focused ion beam scanning electron microscopy. In this method, a scanning electron beam is used to image the surface of a macroscopic cell or tissue specimen block that is progressively abraded, layer by layer, by a focused ion beam that removes a thin layer of material (typically ~ 10 nm) at the surface. Electron microscopic images of the surface that are collected as the specimen is consumed by the progressive abrasion process can be combined to generate a 3D representation of the specimen, with resolution allowing visualization of ultrastructural features of a cell. 3D images of HIV-infected cells obtained using this approach led to the surprising discovery that HIV particles in infected macrophages can be present in virion channels (Figure 2) formed from invaginations of the plasma membrane. These channels are not closed internal compartments as was supposed from earlier 2D images of cross-sections through infected cells, but conduits, contiguous with the extracellular milieu that could allow rapid transport of sequestered HIV to and from the external medium [49].
Figure 2.
Deep, HIV-laden channels i n infected macrophages. Focused ion beam scanning electron microscopy (FIB-SEM) imaging established that these channels are connected to deep reservoirs that are contiguous with the extracellular medium, and not discrete intracellular compartments. (A) Transverse section of HIV-1 BaL-infected monocyte-derived macrophages imaged with FIB-SEM. Scale bar is approximately 100 nm. (B) 3D reconstruction of the HIV-1-infected macrophage demonstrating deep channels connecting a reservoir of HIV-1 virions (red) with the extracellular milieu [49].
Transmission of viruses from antigen presenting cells such as macrophages or dendritic cells to T cells can occur at cell–cell contact zones called virological synapses (reviewed in [50]) via a process that is more effective at infecting T cells than infection by cell free virus [51, 52]. Macrophages, for example, can become productively infected by HIV, and due to their longevity, can spread the infection to T cells through direct contact over extended periods [44, 45, 53, 54]. The virion channels described above may protect virions from antibody-mediated neutralization, while still allowing infection of T cells at virological synapses [49].
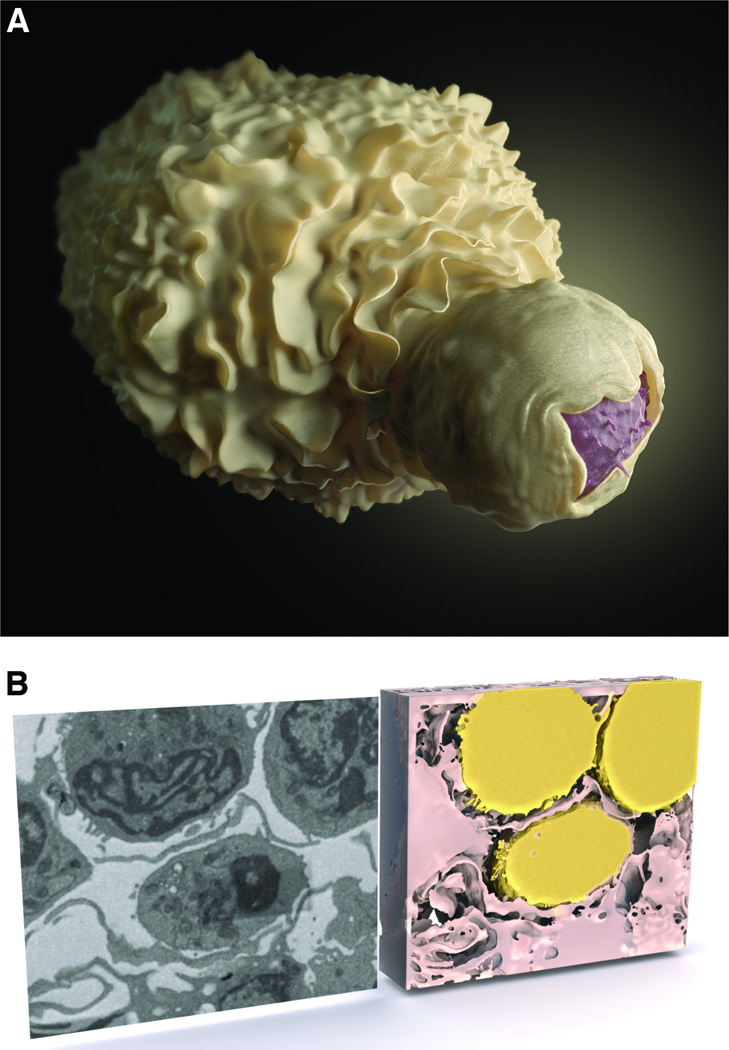
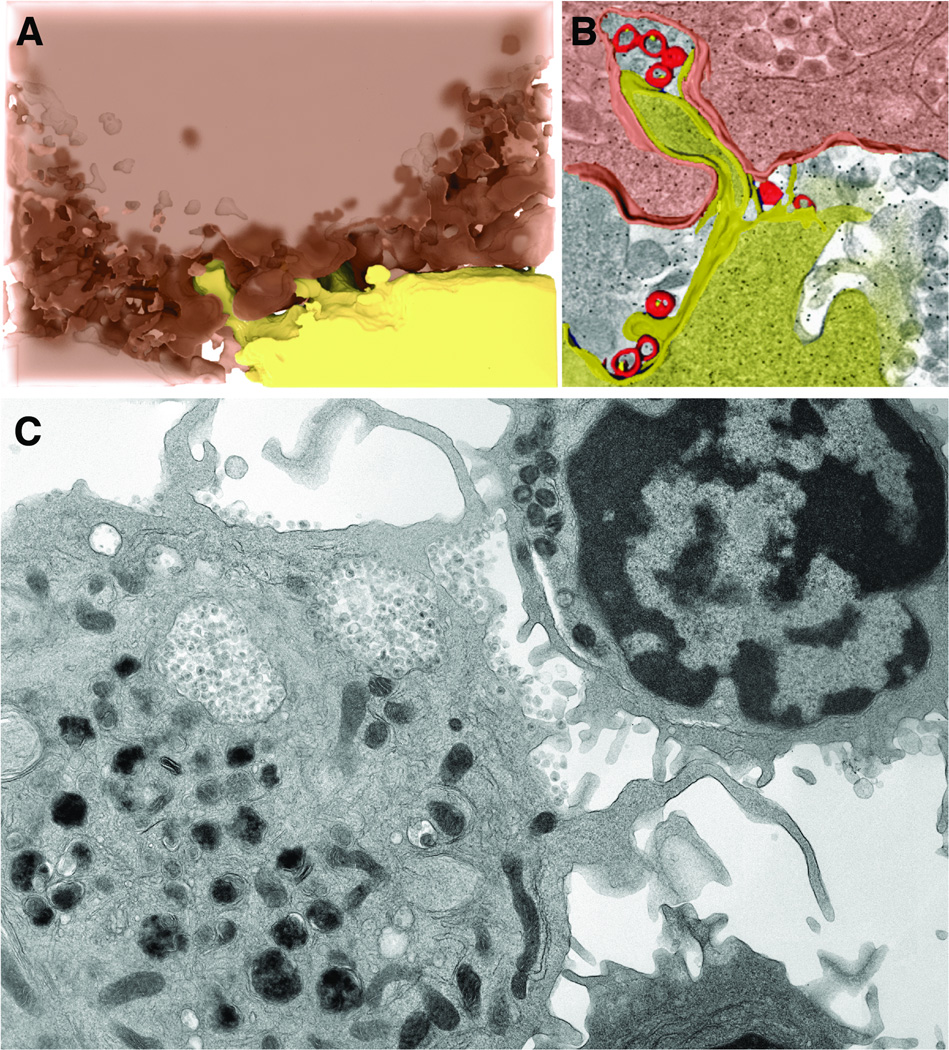
Dendritic cells, unlike macrophages, do not appear to become productively infected by HIV; nevertheless, the cells can also harbor infectious virus that can be transmitted to T cells across a virological synapse [4, 55, 56]. HIV can be taken up by dendritic cells by a number of different receptors, including the C-type lectin DC-SIGN. [57, 58]. Virus taken up by dendritic cells can be transferred to CD4+ T cells via virological synapses in a process that usurps the normal physiological formation of dendritic cell–T cell conjugates to generate the immunological synapses involved in antigen presentation [59]. Structural studies of the close cell–cell contacts between HIV-carrying dendritic cells and uninfected T cells, using the focused ion beam scanning electron microscopy method described above, have shown that the dendritic cells encase the T cells in membranous sheets, creating extensive areas of membrane contact between the two cells (Figure 3). Moreover, within these areas of contact, a high local concentration of HIV becomes trapped between the dendritic cell and T cell [60]. T cells then appear to extend me mbrane protrusions towards pockets of virions (Figures 4A and 4B); these membrane extensions suggest HIV may trigger a reaction by uninfected T cells that could increase the likelihood of infection [61, 62]. Interestingly, CD4-blocking antibody appears to negate this effect, leaving the virions clustered next to the dendritic cell and away from the T cell, even in a close cell–cell contact (Figure 4C) [61].
Figure 3.
Visualization of dendritic cell–T cell contact in the context of cell–cell HIV transmission, and illustration of the sheets of dendritic cell membrane that encase the T cell. These sheets of dendritic cell membrane often appear in thin section images as dendrites. (A) Artistic representation of an HIV-1 virological synapse between a dendritic cell (white) and a T cell (pink). (B) 2D slice (left) and 3D reconstruction (right) of a virological synapse between a dendritic cell (pink) and three T cells (yellow) imaged by focused ion beam scanning electron microscopy. What appear as dendrites in 2D are shown to be membrane sheets in 3D [61].
Figure 4.
3D imaging shows interdigitation of dendritic cell and T cell membranes at the virological synapse. (A, B) A virological synapse between an HIV-1-pulsed dendritic cell (pink) and an uninfected T cell (yellow) was imaged by focused ion beam scanning electron microscopy and reconstructed in 3D. (B) The reconstruction shows that T cell membrane protrusions extend towards HIV-1 virions (red) on the surface of the dendritic cell. (C) 2D section through a virological synapse between a dendritic cell pulsed with HIV-1 and an uninfected T cell treated with anti-CD4 antibody [61]. In the presence CD4 blocking antibodies, virions remain at the surface of the dendritic cells and prevented from touching the T cell, indicating that cellular receptors are required for virion transfer within the virological synapse.
The significant differences between HIV infection by cell-free virus and at the virological synapse have clear implications for vaccine design. Not only are viruses within synapses likely shielded from potentially neutralizing antibodies, but the local concentrations of viruses may be much higher in virological synapses than in the context of free diffusion (reviewed in [63]). If transmission of virus across virological synapses is involved in the process of viral infection and dissemination of infection in a new host, including this mode into our paradigm of rational vaccine design and development may be necessary.
Viral entry
Viral entry into cells is a multistep process (reviewed in [64]). As noted above, the initial stages of viral entry involve gp120 on Env trimers binding to the cellular receptor CD4, an opening of the Env spike, followed by binding of gp120 to a chemokine co-receptor, such as CXCR4 or CCR5. However, a number of studies have indicated that binding of a single Env spike is not sufficient to allow viral entry: multiple Env spikes are necessary for initiating viral fusion with the cell membrane [64]. Consistent with such a process, clustering of Env trimer spikes on the surface of virions can be observed directly by electron tomography (Figures 5A–C). This finding is particularly striking in light of the limited number of spikes pre sent on most HIV-1 virions [1]. Studies of the contact zone between HIV and T cells have shown that there are an average of 5–6 Env spikes at the interface between a bound virion and a target cell. This structure has been designated as an ‘entry claw’, and is speculated to be involved in mediating membrane fusion and viral entry [65] (Figure 5D). The presence of the entry claw suggests a mechanism by which neutralizing antibodies that fail to saturate the surface of an HIV virion may still be able to inhibit virus-cell fusion: if one of the spikes in the entry claw is blocked by a neutralizing antibody, it is possible that the claw will fail to initiate fusion [66]. Thus understanding the local concentration of antibodies at the virus–cell interface in vivo, and how this concentration corresponds to saturation of Env spikes on the surface of live virus, is key to optimizing the function of an antibody-based preventive or therapeutic drug.
Figure 5.
Visualization of entry claw formed at the contact zone between HIV and CD4+ T cells. The entry claw is thought to represent the formation of a macromolecular complex between HIV envelope glycoproteins and cellular receptors prior to fusion between viral and cellular membranes. (A–C) Transmission electron microscopic images of HIV-1 virions in contact with a CD4+ T cell. The entry claw is shown in a cluster of lines of density between the virus and cell. (D) Artistic visualization of the entry claw, illustrating the nature of the virus–cell contact and clustering of Env spikes on the surface of the virion.
SIV and SHIV versus HIV
Since HIV does not productively infect non-human primate species used in experimental models, preclinical evaluation of vaccine concepts and candidates in animal models typically entails two phases, and involves the use of a simian immunodeficiency virus (SIV), one of a family of HIV-related primate lentiviruses that infect and can cause AIDS in macaques [67]. In the initial proof of concept phase, the ability of a SIV vaccine that embodies the desired vaccine approach to protect against a pathogenic SIV challenge is evaluated. If protection can be confirmed, then the immunogenicity of the corresponding HIV-based vaccine is typically evaluated prior to a commitment to clinical testing. Alternatively, for Env based vaccines, HIV immunogens can be used in macaques, and protection assessed usi ng chimeric viruses, designated simian human immunodeficiency viruses (SHIV), comprised of HIV envelope encoding sequences grafted onto a SIV backbone. However, many of the SHIV that have been used to date for such studies have proven to be rather easier to protect against than typical SIV isolates, and by inference, HIV, and may not mediate authentic pathogenesis. Such viruses have been somewhat useful in studies where the endpoint is prevention of acquisition of infection, and substantially less useful in studies involving modulation of pathogenesis post-acquisition as an endpoint (reviewed in [67]). Consequently, SIV virions have been characterized, using some of the cryo-electron microscopy and tomography approaches described above, to study SIV Env spikes on virions in situ [23]. As is the case for many other aspects of AIDS virus biology, SIV largely recapitulates essential features of HIV infection, but there are subtle differences, especially in quaternary conformation changes in the entry spike [23], that must be borne in mind when using SIV as a model for HIV.
Concluding remarks: perspectives for HIV vaccines
Numerous HIV vaccine candidates have been developed and evaluated over the past two decades, some of them rationally designed from our structural knowledge of HIV [68–71]. Nevertheless, no vaccine formulation has yet proven sufficiently safe and effective at preventing HIV infection to warrant licensure. The phase III vaccine trial that has reported the greatest efficacy to date has been the RV144 ‘Thai Trial’, based on pox virus vector priming and recombinant protein boosting, with an efficacy of 31% [70]. The limited protection observed has been correlated with non-neutralizing IgG antibody responses, directed a gainst the gp120 V1/V2 domain [72], and while additional studies are in progress to attempt to identify other potential correlates of protection, there is broad agreement that more efficacious vaccines will be required.
Dramatic progress in the number of broadly neutralizing monoclonal antibodies available and in the structural description of the way these antibodies interact with HIV envelope glycoproteins has increased our understanding of their function and mechanism of action. However, we have yet to bridge the gap between antigenicity and immunogenicity. We understand in detail the antigenic binding determinants on HIV envelope glycoproteins for broadly neutralizing antibodies that are developed through the course of HIV infection, but we do not yet understand how to induce formation of such antibodies by immunization of uninfected persons. Moreover, understanding how these antibodies act mechanistically to block infection by cell free virions and inhibit transmission of virus across virological synapses may be key to maximizing the efficacy of potential vaccines. In both these areas, future studies using 3D electron microscopy techniques hold promise for providing informative insights. For blockade of infection by cell free virus, a better understanding of the structural basis of interactions of antibodies with different functional states of the envelope spike, achieved through the methods described here, may help guide design of improved vaccine immunogens. Similarly, studies of the spatial architecture of virological synapses, in combination with functional studies, may help better define the challenges for antibody-mediated blockade of cell-to-cell virus transmission.
Highlights.
The native HIV Env structure can be determined by cryo-electron tomography.
Focused ion beam scanning electron microscopy (FIB-SEM) reveals 3D ultrastructure of HIV virological synapses.
Cryo-electron microscopy structures provide key information for vaccine design.
ACKNOWLEDGEMENTS
We thank Donald B liss for assistance with illustrations. This work was supported by funds from the NIH Intramural AIDS Targeted Antiviral Program and Center for Cancer Research at the National Cancer Institute (to S.S.) and National Cancer Institute, National Institutes of Health Contract HHSN261200800001E (to J.D.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Liu J, et al. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Dalgleish AG, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 4.Piguet V, Steinman RM. The interaction of HIV with dendritic cells: outcomes and pathways. Trends Immunol. 2007;28:503–510. doi: 10.1016/j.it.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nabel GJ, et al. Progress in the rational design of an AIDS vaccine. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2011;366:2759–2765. doi: 10.1098/rstb.2011.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 7.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. Journal of virology. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trkola A, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. Journal of virology. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhillon AK, et al. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. Journal of virology. 2007;81:6548–6562. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nature medicine. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. Journal of virology. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doria-Rose NA, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. Journal of virology. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamatatos L, et al. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nature medicine. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 14.Simek MD, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. Journal of virology. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 16.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pejchal R, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proceedings the National Academy of Sciences of the United States of America. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merk A, Subramaniam S. HIV-1 envelope glycoprotein structure. Current opinion in structural biology. 2013;23:268–276. doi: 10.1016/j.sbi.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS pathogens. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris A, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyerson JR, et al. Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:513–518. doi: 10.1073/pnas.1214810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank GA, et al. Computational separation of conformational heterogeneity using cryo-electron tomography and 3D sub-volume averaging. Journal of structural biology. 2012;178:165–176. doi: 10.1016/j.jsb.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu P, et al. Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. PLoS pathogens. 2008;4:e1000203. doi: 10.1371/journal.ppat.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corti D, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PloS one. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. Journal of virology. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran EE, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS pathogens. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. Journal of virology. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardoso RM, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Pejchal R, et al. A conformational switch in human immunodeficiency virus gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. Journal of virology. 2009;83:8451–8462. doi: 10.1128/JVI.00685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weissenhorn W, et al. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 38.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS pathogens. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris AK, et al. HIV-1 envelope glycoprotein trimers display open quaternary conformation when bound to the gp41 membrane-proximal external-region-directed broadly neutralizing antibody Z13e1. Journal of virology. 2013;87:7191–7196. doi: 10.1128/JVI.03284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nature medicine. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 41.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nature medicine. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 42.Hessell AJ, et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS pathogens. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen BK. T cell virological synapses and HIV-1 pathogenesis. Immunologic research. 2012;54:133–139. doi: 10.1007/s12026-012-8320-8. [DOI] [PubMed] [Google Scholar]
- 44.Sharova N, et al. Macrophages archive HIV-1 virions for dissemination in trans. The EMBO journal. 2005;24:2481–2489. doi: 10.1038/sj.emboj.7600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carr JM, et al. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology. 1999;265:319–329. doi: 10.1006/viro.1999.0047. [DOI] [PubMed] [Google Scholar]
- 46.Cameron PU, et al. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science. 1992;257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 47.Nath A, et al. Infection of human fetal astrocytes with HIV-1: viral tropism and the role of cell to cell contact in viral transmission. Journal of neuropathology and experimental neurology. 1995;54:320–330. doi: 10.1097/00005072-199505000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Piguet V, Sattentau Q. Dangerous liaisons at the virological synapse. J Clin Invest. 2004;114:605–610. doi: 10.1172/JCI22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett AE, et al. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS pathogens. 2009;5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mothes W, et al. Virus cell-to-cell transmission. Journal of virology. 2010;84:8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen P, et al. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. Journal of virology. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimitrov DS, et al. Quantitation of human immunodeficiency virus type 1 infection kinetics. Journal of virology. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aquaro S, et al. Long-term survival and virus production in human primary macrophages infected by human immunodeficiency virus. Journal of medical virology. 2002;68:479–488. doi: 10.1002/jmv.10245. [DOI] [PubMed] [Google Scholar]
- 54.Brown A, et al. In vitro modeling of the HIV-macrophage reservoir. J Leukoc Biol. 2006;80:1127–1135. doi: 10.1189/jlb.0206126. [DOI] [PubMed] [Google Scholar]
- 55.Lekkerkerker AN, et al. Viral piracy: HIV-1 targets dendritic cells for transmission. Curr HIV Res. 2006;4:169–176. doi: 10.2174/157016206776055020. [DOI] [PubMed] [Google Scholar]
- 56.Wang JH, et al. Functionally distinct transmission of human immunodeficiency virus type 1 mediated by immature and mature dendritic cells. Journal of virology. 2007;81:8933–8943. doi: 10.1128/JVI.00878-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arrighi JF, et al. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. The Journal of experimental medicine. 2004;200:1279–1288. doi: 10.1084/jem.20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gringhuis SI, et al. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat Immunol. 2010;11:419–426. doi: 10.1038/ni.1858. [DOI] [PubMed] [Google Scholar]
- 59.Vasiliver-Shamis G, et al. HIV-1 Virological Synapse is not Simply a Copycat of the Immunological Synapse. Viruses. 2010;2:1239–1260. doi: 10.3390/v2051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDonald D, et al. Recruitment of HIV and its receptors to dendritic cell- T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 61.Felts RL, et al. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13336–13341. doi: 10.1073/pnas.1003040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nikolic DS, et al. HIV-1 activates Cdc42 and induces membrane extensions in immature dendritic cells to facilitate cell-to-cell virus propagation. Blood. 2011;118:4841–4852. doi: 10.1182/blood-2010-09-305417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong P, et al. Cell-to-cell transmission of viruses. Current opinion in virology. 2013;3:44–50. doi: 10.1016/j.coviro.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blumenthal R, et al. HIV entry and envelope glycoprotein-mediated fusion. The Journal of biological chemistry. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sougrat R, et al. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS pathogens. 2007;3:e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobrowsky TM, et al. Organization of cellular receptors into a nanoscale junction during HIV-1 adhesion. PLoS computational biology. 2010;6:e1000855. doi: 10.1371/journal.pcbi.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lifson JD, Haigwood NL. Lessons in nonhuman primate models for AIDS vaccine research: from minefields to milestones. Cold Spring Harbor perspectives in medicine. 2012;2:a007310. doi: 10.1101/cshperspect.a007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flynn NM, et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. The Journal of infectious diseases. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 69.Pitisuttithum P, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. The Journal of infectious diseases. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 70.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. The New England journal of medicine. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 71.O'Connell RJ, et al. Human immunodeficiency virus vaccine trials. Cold Spring Harbor perspectives in medicine. 2012;2:a007351. doi: 10.1101/cshperspect.a007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karasavvas N, et al. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS research and human retroviruses. 2012;28:1444–1457. doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]