Abstract
The objective of this review is to present an argument for performing joint analyses between functional imaging with global gene expression studies. The reason for making this link is that tumor microenvironmental influences on functional imaging can be uncovered. Such knowledge can lead to (1) more informed decisions regarding how to use functional imaging to guide therapy and (2) discovery of new therapeutic targets. As such, this approach could lead to identification of patients who need aggressive treatment tailored toward the phenotype of their tumor vs those who could be spared treatment that carries risk for more normal tissue complications. Only a handful of papers have been published on this topic thus far, but all show substantial promise.
Introduction
It is well known that local control rates following radiotherapy or chemoradiotherapy vary between patient populations, even when matched for clinically important factors, such as stage, grade, and tumor volume. Failure to accurately predict outcome with traditional clinical staging provides strong rationale to search for other characteristics of tumors that can more accurately do so. If such patients could be identified, they could be selected for more aggressive therapy. Alternatively, patients with relatively radiosensitive tumors could be spared treatment that carries increased risk for normal tissue complications. Although the rationale is strong, identification of patients with radioresistant tumors is of little value unless it is possible to use adjuvant therapies that target the source of radioresistance. In this review, we make the case that analyses that combine functional imaging with global gene expression hold the promise to (1) account for intertumoral heterogeneity in terms of prognosis and treatment response and (2) identify novel therapeutic targets that are associated with different outcomes and treatment response. Further, functional imaging may, in some cases, provide complementary information for gene expression, thereby reducing the complexity and cost of making critical treatment decisions.
History of Biomarkers of Radioresistance
Over the past several decades, there have been numerous attempts to identify biomarkers to predict treatment response and local tumor control following radiotherapy. Prior to the development of various genomics tools, many phenomenological assays, such as hypoxic fraction, intrinsic radiosensitivity, deoxyribonucleic acid (DNA) strand break and repair, and fraction of proliferating cells, were examined for their prognostic significance. Others have elegantly reviewed this historical literature1-4; the results have not been discussed in detail in this paper. It should be noted, however, that a fundamental limit to phenomenological assays is that therapeutic strategies to correct them are not clear, because the molecular targets underlying their outcome are likely multifactorial and not always clearly defined. One possible exception to the limitation of phenomenological biomarkers is tumor hypoxia. Although there is strong level 1a evidence that modification of hypoxia influences local tumor control rates and survival,5 there is still no consensus on the standard of care for ameliorating this established cause of radioresist-ance.6 Although hypoxia and other microenvironmental stresses can be directly measured in vivo, these measurements are frequently invasive or require tumors to be snap frozen in a sophisticated laboratory setting. Therefore, these measurements are not easily implemented as a clinical routine.
As for molecular biomarkers, single target genes have been investigated, to some success, to identify relatively resistant tumors2 or tumors that are more likely to metastasize in patients administered radiotherapy.7 However, modern genomic tools, such as microarrays and ribonucleic acid sequencing, have allowed the global profiling of gene expression to capture all the biological activities that may affect their biological and clinical phenotypes. This is based on the assumption that virtually any biological condition and activity can be reflected and captured in the tumor's gene expression, including subtle distinctions in biology. It is then possible to derive multigene signatures associated or predictive of particular phenotypes. Furthermore, the public availability of gene expression data of many tumors and variety of experimental perturbations in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) make it possible to employ gene expression to generate and test biological hypotheses using these data sets. Expression signatures are also portable and provide the capacity to link experimental perturbations in vitro and multiple independent cohorts of tumors in vivo. For example, a hypoxia signature obtained in cultured cells allows the recognition of the molecular features common to multiple cancer types—in turn permitting the identification of patients with strong hypoxia response and high clinical risks. By linking gene expression data with the tumors' response to radiation therapies, such multiple gene predictors (or “metagene” or “signatures”) may provide more robust measures of radio-resistance.2 Such multigene predictors may provide superior predictive ability because of the following reasons: (1) They are derived from multiple genes. This makes them less susceptible to potential errors in classification caused by spurious factors not related to radioresistance that might influence single marker gene expression, and (2) radioresistance might be the consequence of several independent factors that cannot be ascertained from a single test end point. Furthermore, the commercialization of several multiple gene predictors of breast cancer, such as oncotype DX and MammaPrint,8 provide precedents on how to effectively apply these multigene predictors to guide clinical decision.
Linking Gene Expression With Treatment Outcome
Before discussing results, a summary of the terms used is presented in (Table), for those readers who are not familiar with the terminology of gene expression analyses.
Table.
| Abrix | Parameter in DCE-MRI which measures the change in tracer concentration within the tissue of interest over time; also known as amplitude |
| Apparent diffusion coefficient (ADC) | Apparent diffusion coefficient (ADC) measures the magnitude of diffusion (of water molecules) within the tissue during functional MRI. |
| Basal-like breast carcinoma | Molecular subtype of breast cancer; high grade, ER negative or PR negative or HER2 negative; associated with poor prognosis |
| CD31 | Cluster of differentiation 31 is the protein encoded by the gene platelet endothelial cell molecule (PECAM1); CD31 is a member of the immunoglobulin superfamily and is likely involved in leukocyte migration, angiogenesis, and integrin activation; CD31 is used to demonstrate the presence of endothelial cells in histologic tissue sections. |
| Chromogranin A | Chromogranin A is the protein encoded by the gene CHGA; It is a member of the granin family of neuroendocrine secretory proteins; Chromogranin A is the precursor to several functional peptides that negatively modulate neuroendocrine function of cells; Chromogranin A can be used to identify neuroendocrine tumors in histologic tissue sections |
| Claudin-low breast carcinoma | Molecular subtype of breast cancer; claudin-low carcinomas are characterized by low to absent expression of luminal differentiation marks, high epithelial-mesenchymal transition markers, immune response genes, and cancer stem cell like features; clinically, high grade, ER negative or PR negative or HER2 negative; associated with poor prognosis |
| Deep sequencing | The technology that allows a DNA fragment to be repeatedly sequenced in a very short time and delivers great sensitivity and accuracy; deep sequencing of the transcriptome provides the sequence and frequency of RNA molecules that are present at any particular time in a specific cell type, tissue, or organ; quantifying the RNA encoded by individual genes provides an indicator of protein-coding potential, a major contributor to phenotype |
| DCE-MRI | Dynamic contrast-enhanced (DCE)-magnetic resonance imaging (MRI); functional imaging technique in which multiphase MRI scans are taken following the intravenous injection of a contrast agent to noninvasively evaluate tumor vascular characteristics |
| ER; ER alpha and ER beta | Estrogen receptor (ER) is the intracellular protein receptor activated by the hormone estrogen; once activated by estrogen, ER is a DNA-binding transcription factor; ERs are widely expressed in different tissue types; ER alpha is encoded by the gene ESR1 and ER beta is encoded by the gene ESR2 |
| ERBB2 | ERBB2 is the gene that encodes the protein HER2; ERBB2 is amplified and overexpressed in approximately 30% of breast cancers and is strongly associated with increased disease recurrence and worse prognosis |
| 18F-FDG-PET | Positron emission tomography (PET) utilizing the radiopharmaceutical fluorodeoxyglucose (FDG) synthesized with the radioactive isotope fluorine-18 (18F); functional imaging technique that images regions of high glucose metabolism via uptake of the glucose analogue 18F-FDG; high–glucose using cells include cancer cells and normal brain and kidney cells |
| FLT1 | FLT1 is the genethat encodes the protein vascular endothelial growth factor receptor 1 (VEGFR-1); VEGFRs are receptors for the signaling protein vascular endothelial growth factor (VEGF), which is produced by cells that stimulate vasculogenesis and angiogenesis; FLT1 is a member of the src gene family and has tyrosine kinase activity, important for the control of cell proliferation and differentiation |
| Functional imaging | Functional imaging refers to imaging techniques in which a sequence of images follows the distribution of an administered radioactive tracer or contrast agent to delineate one or more physiological processes in the body |
| Gene expression profiling | The measurement of the expression, or activity, of thousands of genes at one time to create a global picture of cellular function |
| Histone deacetylase (HDAC) | Class of enzymes that remove acetyl groups from histones, which allows the histones to wrap DNA more tightly; DNA expression is regulated by acetylation and deacetylation |
| HER2 | Human epidermal growth factor receptor 2 (HER2) is a protein encoded by the ERBB2 gene; HER2 is a member of the epidermal growth factor receptor (EGFR) family which are membrane-bound receptor tyrosine kinases; autophosphorylation of the receptors initiates a variety of signaling pathways |
| HOXC6 | HOXC6 is the gene that encodes the homeobox protein Hox-C6; HOXC6 belongs to the homeobox family, members of which encode a family of transcription factors that play an important role in morphogenesis in multicellular organisms. |
| Hsp90 | Heat shock protein 90 (Hsp90) is a chaperone protein that assists other proteins in proper folding, stabilizes proteins against heat stress, facilitates intracellular transport, cell signaling, and maintenance and degradation of proteins; 5 functional genes encode Hsp90 |
| Inositol trisphosphate receptor 3 | Inositol trisphosphate receptor 3 is a membrane glycoprotein complex in the endoplasmic reticulum acting as a Ca2+ channel activated by inositol trisphosphate; InsP3R3 is encoded by the gene ITPR3 |
| KDR | Kinase insert domain receptor (KDR); also known as vascular endothelial growth factor receptor-2 (VEGFR-2) is a VEGF receptor protein encoded by the gene KDR; KDR is a receptor tyrosine kinase and functions as the main mediator of VEGF-induced endothelial proliferation, survival, migration, tubular morphogenesis, and sprouting |
| Luminal type A breast carcinoma | Molecular subtype of breast cancer; ER positive, low grade |
| Luminal type B breast carcinoma | Molecular subtype of breast cancer; ER positive, but often high grade |
| Metagene, signature, and multigene predictors | A list of genes that is associated with certain biological processes or predictive of biological phenotypes, such as different response to radiotherapy |
| Next generation, massively parallel or deep sequencing, and sequencing | Post-Sanger sequencing methods that generate large amount of data (orders of magnitude more) at incredibly low cost (again, orders of magnitude). This can be accomplished by several platforms, including Illumina, SOLID, Ion Torrent, and those by other companies. This has created amazing opportunities in the biological sciences and enabled amazing new experiments, including the genetic characterization of human cancers. |
| Normal breast-like carcinoma | Molecular subtype of breast cancer; similar expression pattern to normal breast tissue |
| p53 | P53 is a tumor suppressor protein that is encoded by the TP53 gene; regulates the cell cycle and activates DNA repair or initiates apoptosis or both |
| PDGF-B | Platelet-derived growth factor subunit B is a protein encoded by the PDGFB gene; a member of the platelet-derived growth factor family, which are mitogenic factors for cells of mesenchymal orgin and play a significant role in angiogenesis |
| PDK2 | Pyruvate dehydrogenase kinase isoform 2 (PDK2) is a protein that is encoded by the PDK2 gene; acts to inactivate the enzyme pyruvate dehydrogenase, located within the mitochondria, and contributes to the regulation of glucose metabolism |
| PR | Progesterone receptor (PR) is an intracellular protein activated by the hormone progesterone; encoded by the PGR gene; once activated by progesterone, PR is a DNA-binding transcription factor |
| RAD23A | RAD23 homolog A (RAD23)is a protein encoded by the RAD23 geneand is involved in nucleotide excision repair |
| Ras | Ras includes a family of related proteins belonging to a class of protein called small GTPase, and are involved in transmitting signals within cells; the name ras refers to the family of genes encoding these proteins; activates genes involved in cell growth, differentiation, and survival |
| SAM | Significance analysis of microarrays (SAM) is a statistical technique for determining what genes are statistically significant among different samples from microarrays |
| STAT1 | STAT1 protein is a member of the Signal Transducers and Activators of Transcription family of transcription factors; encoded by STAT1 gene; involved in upregulating genes because of signal by interferons; regulates many aspects of growth, survival, and differentiation in cells; involved in the function of the immune system and maintaining immune tolerance |
| Sialyltransferase-1 | Sialyltransferase-1 is an enzyme that transfers sialic acid to oligosaccharide; membrane protein localized to the Golgi network; expression and activity are associated with tumor metastasis in breast and colon cancers |
| Standard uptake value (SUV) | A ratio of tissue radioactivity concentration at time T and injected dose at the time of injection divided by body weight. |
| TCGA | The Cancer Genome Atlas (TCGA) Project is a large-scale collaborative effort to characterize the genomic changes that occur in cancer. The overarching goal is to comprehensively define the important genomic changes involved in cancer. This knowledge will advance our molecular understanding of the disease and improve our ability to diagnose, treat, and prevent it. |
| TERT | Telomerase reverse transcriptase (TERT) is a catalytic subunit of the enzyme telomerase; telomerase lengthens telomeres in DNA strands, allowing cells that would otherwise become postmitotic and undergo apoptosis to become potentially immortal; TERT is responsible for catalyzing the addition of nucleotides to the ends of a chromosome's telomeres |
| VCAM-1 | Vascular cell adhesion molecule-1 (VCAM-1) is a protein that is encoded by the VCAM1 gene; functions as a cell adhesion molecule, expressed on both large and small blood vessels after the endothelial cells are stimulated by cytokines |
| Vinexin beta | Vinexin is a protein encoded by the SORBS3 gene; the vinexin beta isoform plays a role in cell spreading and enhances the activation of cell growth, differentiation, and survival pathways in response to epidermal growth factor stimulation |
| vWF | Von Willebrand factor (vWF) is a blood glycoprotein crucial to the hemostasis process; encoded by the gene VWF; produced in endothelium, megakaryocytes, and subendothelial connective tissue |
| Zinc finger proteins | A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions to stabilize the fold; their functions are extraordinarily diverse and include DNA recognition, RNA packaging, transcriptional activation, regulation of apoptosis, protein folding and assembly, and lipid binding |
Several reports have been published in which global gene expression analyses have been linked to outcome of radiotherapy. Since the focus of this review is on the subject of linking functional imaging with gene expression studies, we have provided several examples, but this is not comprehensive. A more comprehensive review of this subject was recently reported. Reports that have successfully linked gene expression to treatment outcome can be generally classified into studies that conducted “Bottom-up” and “Top-down” analyses —(1) Bottom-up analyses: these derived gene signatures of particular biological perturbations and biological processes from in vitro studies and then applied them to clinical data sets or (2) Top-down analyses: these focused either on prognosis, where outcome is not linked to particular treatment received, or treatment outcome, where the treatment received is part of the analysis. Examples of these different types of approaches are provided in the following sections.
Bottom-Up Approach: Testing of In Vitro Gene Expression Signatures of Microenvironmental Stresses to Clinical Outcome
Hypoxia and other tumor microenvironmental stresses are postulated to affect the outcomes of radiotherapies. Based on the relevance of these processes, Chi9,10 and Winter et al.11 derived gene signatures for hypoxia response, based on analysis of several cell types. These gene signatures were found to be associated with prognosis in the expression data from patients with a variety of cancers. Although the hypoxia gene signature was successful in predicting the outcome in several tumor types, it did not predict for local tumor control or disease progression in patients with head and neck cancer who were treated with cisplatin and radiotherapy.12 Similar approaches have been extended to other microenvironmental stresses, such as wound healing,13 lactic acidosis,9 and glucose deprivation,14 but these signatures have not yet been extensively tested in cohorts of patients treated with chemoradiotherapy.
Eschnch et al.15 used the NCI-60 cell line genomic data to derive a radiation resistance gene signature, which they then successfully used to predict preoperative chemoradiotherapy downstaging success in patients with rectal and esophageal cancer and 2-year local tumor control in patients with locally advanced head and neck cancer. The gene signature for radiosensitivity was based on clonogenic survival data (surviving fraction at 2 Gy), a well-established measurement of intrinsic radiosensitivity, of 48 cell lines from the NCI-60 cell line set. The analysis also included other factors known to influence radiosensitivity, including p53 and ras status and tissue of origin. Further, the prediction classification was based on gene expression data derived from different analysis platforms (commercial vs single-institution custom arrays). The universality of the result strongly supports the potential robustness and translational potential of this approach.
Eschrich et al.15 recently extended the radiosensitivity gene signature discussed previously, to 5-year relapse-free survival in 2 large data sets of women with breast cancer: n = 159 from Karolinska University Hospital and n = 344 from Erasmus Medical Center.16 In both data sets, patients predicted to be radiosensitive had improved relapse-free survival, but there was no difference in survival for those patients who received surgery alone as the primary treatment modality. Further, the radiosensitivity gene signature was shown to be an independent predictor of outcome in ER+ patients. The robustness of this signature strongly suggests that it be explored in other clinical trial settings. However, it is not known what types of interventions could be used to offset the negative prognosis associated with the signature. Additional research is needed to further test the utility of this very promising prognostic tool, where interventions are included as part of the trial design.
Weichselbaum et al.17 used a similar initial approach to identify an interferon-related DNA damage resistance signature. The original signature came from in vitro studies that identified an interferon signaling pathway associated with DNA damage resistance. This signature was first tested against the NCI-60 cell line data base, showing that it was associated with in vitro radiosensitivity, as assessed by surviving fraction at 2 Gy. They then went on to show that knockdown of the most highly correlated gene within the resistance pathway, STAT1, reduced radioresistance in vitro and in vivo. Expanding on these findings, they demonstrated that the interferon-related DNA damage resistance signature was independently predictive of local tumor control and chemoresponse in several large clinical gene expression databases, involving breast cancer, non—small cell lung cancer (NSCLC), head and neck cancer, and glioblastoma. This was the first study to demonstrate a specific radiation treatment response gene signature that was independent of prognostic gene signatures. The clinical importance of these findings is quite profound in that it suggests that such markers might be used to assist in treatment decisions related to the use of adjuvant chemotherapy. The fact that it is associated with DNA damage resistance provides a clue as to the therapeutic strategies that could be explored.
Top-Down Approach: Identify Gene Expression Signatures Associated With Clinical Outcome
Prostate Cancer
Singh et al.18 performed gene expression analysis from tumor tissues obtained from 52 postprostatectomy patients, to test whether gene signatures could be derived that correlated with known prognostic factors (Gleason score, PSA, positive margins, and capsular invasion). Aside from the Gleason score, they were unable to link a gene expression signature to these other variables. However, they were able to derive a 5-gene signature that independently predicted the outcome in 19 of a subset of 21 patients with sufficient follow-up. The 5 genes were chromogranin A, platelet-derived growth factor beta, HOXC6, inositol triphosphate receptor 3, and sialyltransferase-1. It is not known whether this signature would apply to patients receiving radiotherapy. The study was quite small, which limits the power to detect true differences and increases the likelihood of spurious associations. Thus, the work needs to be reexamined in larger and independent cohorts of patients.
Cervix Cancer
One of the major challenges in the management of locally advanced cervix cancer is to accurately determine lymph node status. Lyng et al.19 performed gene expression analysis on 48 patients with locally advanced cervix cancer; 19 had histologic verification of metastases to regional lymph nodes. Supervised analysis revealed 2 gene sets with significant association with lymph node status: (1) proliferation, oxidative phosphorylation, and invasiveness vs (2) anaerobic metabolism and tolerance to hypoxia. A master regulator of the switch between aerobic and anaerobic metabolism, pyruvate dehydrogenase kinase isoform 2 was found to be independently predictive of lymph node status. This same group also used a combination of copy number alterations (gain or loss of parts of chromosomes) and gene expression analysis, to derive driver genes and biological processes involved in chemoradioresistance of cervix cancer.20
Lung Cancer
Shedden et al.21 examined gene expression profiles in over 400 patients with NSCLC from 4 different institutions. An important aspect of this study was that the gene expression analyses that established the gene signatures were derived from the results of 2 institutions. The validation that followed used independent data from the remaining 2 institutions. The results strongly supported the notion of developing combined gene expression or clinical methods for establishing prognosis. The inclusion of clinical variables was better for classification than gene signatures alone. Second, signatures worked better for patients across all stages, as compared with stage 1 only. Although the treatment received was not included in this report, presumably, a significant proportion of patients with higher stage received radiotherapy as part of their treatment. Validation of the accuracy of this signature in trials with patients receiving radiotherapy would be a logical next step in confirming the predictive power of this signature.
Combination of Gene Expression With Functional Imaging
Functional imaging approaches detect or measure changes in the metabolism, blood flow, regional pressure, and absorption of tumors. These properties may provide independent and complementary information about the tumors. Therefore, the rationale for combining functional imaging with gene expression is multifold: (1) such combinations may reveal the underlying cause for a functional imaging phenotype; (2) it may provide a complementary and independent source of prognostically important information, much in the same way as the combination of clinical variables with gene expression did; (3) such analyses may reveal novel therapeutic targets that cannot be identified with functional imaging or genomic analysis alone; and (4) the addition of functional imaging may provide additional information regarding dose intensification to tumor subvolumes suspected of being relatively radioresistant. In spite of the fact that gene expression signatures and functional imaging have both been shown to correlate with treatment outcome in trials involving radiotherapy, there are only a handful of papers in which these 2 tools have been brought together in the same patient population. These studies have been discussed in more detail later. It should be noted that most of the studies comparing gene expression analyses with functional imaging that have been reported thus far involve small numbers of patients. Thus, results should be considered hypothesis generating and will hopefully spur additional work in larger studies for further validation.
A number of studies have been performed to compare positron emission tomography (PET) tracers, such as fluorine-18 fluorodeoxyglucose-PET (18F-FDG-PET) or hypoxia PET, with treatment outcome without linkages to gene expression analyses. Similar reviews have been written for magnetic resonance (MR)-based parameters. It is outside the purview of this review to discuss these in detail. Readers are referred to review papers on the use of these imaging techniques with radiotherapy. A few relevant reviews are cited here.22-24
18F-FDG-PET
Breast Cancer
Osborne et al.25 examined tumors from 36 patients with locally advanced breast cancer for estrogen receptor (ER) or progesterone receptor and human epidermal growth factor receptor 2 status after 18F-FDG-PET studies were performed (all but 1 patient's tumor was histologic grade III). A subset of 20 biopsies was analyzed for gene expression using Affymetrix microarrays. Standard uptake value (SUV) was significantly and negatively correlated with ER status: ER4+ tumors had lower SUV values than ER— tumors. Unsupervised hierarchical clustering of gene expression data revealed 2 groups of 10, which were largely split by ER status, consistent with other expression study of breast cancer.26 Genes that were differentially expressed between the 2 groups included those involved in glucose metabolism and transport, consistent with strong hypoxia and glycolysis pathways in the ER— tumors. This is similar to the top-down analysis mentioned earlier.14 The investigators went on to validate the gene signature against a separate data set of 99 patients with breast cancer (with known ER status), where gene expression analysis was performed using the same array technology. Given the small sample size of this study, there was no attempt to correlate gene expression patterns with outcome.
NSCLC
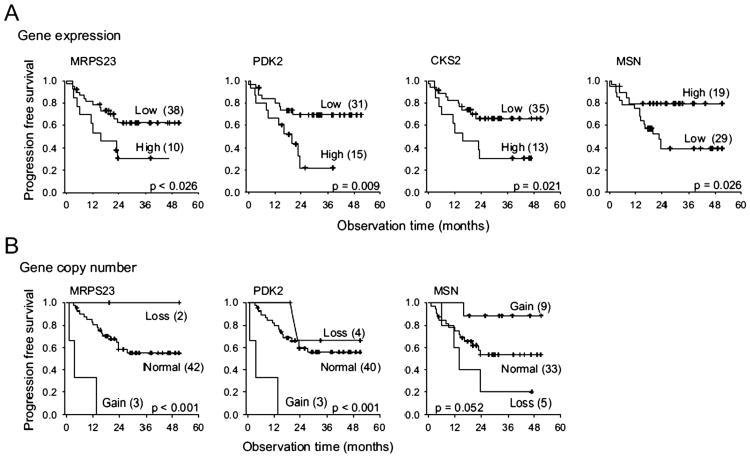
Nair et al.27 performed a detailed analysis of uptake features of 18F-FDG-PET in a set of 25 patients with early-stage NSCLC. This same set of patients also had gene expression analyses performed on tissues acquired at resection. Features of PET tracer uptake were based on standard SUV analysis, but included SUVmin, SUVmax, SUVmean, SUVvariance, SUVskewness, and others. Eight metagenes were identified that correlated with the each of the aforementioned PET features. The PET feature most commonly associated with the metagenes was SUVmax. It was correlated with signatures associated with focal and cell adhesion, protein catabolism, metalloproteinase and collagen, apoptosis, and extracellular matrix or hypoxia. Interestingly, there was no strong correlation with metabolism. Given that higher rates of glucose uptake could be the result of either the Warburg effect (glycolysis in aerobic conditions) or the Pasteur effect (glycolysis in the presence of hypoxia), this is not surprising. Nevertheless, there was a linkage to hypoxia, suggesting that hypoxia may have been a stronger driver of increased glucose uptake in this set of 25 patients. Key PET features of SUVmax, SUVvariance, and SUVPca2 (PCA2—derived from principle component analysis) were found to be associated with survival in 2 independent cohorts (n = 64 and 81) of patients with early-stage NSCLC for whom PET data were available (Fig. 1).
Figure 1.
Association between 18F-FDG-PET pSUVmax and survival in patients with resected non—small cell lung cancer. Several metagenes were associated with this imaging parameter that related to cell cycle regulation, proliferation, death, and self-recognition. Individual genes associated with pSUVmax were LY6E (encodes a protein that may be involved in cellular differentiation43), RNF149 (encodes a E3 ubiquitin-protern ligase that ubiquitinates the oncogene BRAF f or proteosomal degradation44) , MCM6 (encodes a DNA helicase45), and FAP (encodes or Fibroblast activation protein alpha, a membrane-bound gelatinase involved in cell invasion and motility46). (Data reproduced with permission Nair et al.27)
This paper is important in 3 ways: (1) It shows that careful imaging and gene expression analysis of relatively small cohorts of patients can lead to important lead discoveries regarding the underlying mechanisms driving functional imaging. (2) The principle component analyses revealed methods for describing 18F-FDG-PET data that have biological underpinning. (3) The biological insights have the potential to identify new therapeutic targets.
It remains to be seen whether the gene signatures derived from this study would be applicable to patients with more advanced disease who would receive chemoradiotherapy. Given the wide utilization of 18F-FDG-PET in the diagnosis and management of patients with NSCLC, there is potential for this to be tested in a relatively straightforward manner, however.
Hepatocellular Carcinoma (HCC)
The sensitivity of 18F-FDG-PET to detect HCC is low (50%). However, among those tumors studied, high 18F-FDG-PET uptake is associated with worse outcome. Lee et al.28 compared the 18F-FDG-PET and genomic data from 10 patients with HCC. Of these 10 patients, 4 exhibited high 18F-FDG-PET uptake and the remaining 6 were classified as “low”. Tumors that exhibited high 18F-FDG-PET uptake were classified histologically as Edmonson-Steiner grade III, whereas the tumors classied as low 18F-FDG-PET uptake were relatively lower grade. The high-grade tumors exhibited upregulation of genes involved in invasion or cell spreading, including vascular cell adhesion molecule-1 and vinexin beta. ER beta was among the genes reported to be repressed. The biological significance of this latter finding was not clear, although ER alpha is known to be expressed in normal liver, as opposed to ER beta. The authors suggested that 18F-FDG-PET scans might serve as an important diagnostic tool that would differentiate aggressiveness of these tumors and thereby aid in treatment decisions. This conjecture will need validation in larger patient series.
Gastrointestinal Stromal Tumors (GIST)
Dynamic 18F-FDG-PET scans were performed in 22 patients with GIST.29 These data were compared with gene expression data derived from biopsies taken at the time of surgical resection. GISTs are relatively rare tumors of the gastrointestinal tract. Mutations in the c-KIT gene are found in this tumor, providing rationale for use of imatinib for their treatment. Survival is associated with proliferation, size, and tumor stage. In this study, uptake of 18F-FDG-PET was found to be correlated with the expression of several zinc finger proteins (znf43, znf85, znf91, and znf189). In other work, the expression of zinc finger proteins was associated with sensitivity to imatinib. Thus, 18F-FDG-PET may serve as a screening tool to identify those patients who are most likely to respond favorably to imatinib treatment.
MR Imaging (MRI)
Breast Cancer
Yamamoto et al.30 conducted a combined gene expression and dynamic contrast-enhanced MRI (DCE-MRI) study in 10 patients with breast cancer, combined with gene expression analysis of 353 patients with breast cancer. The image analysis procedures used in this paper were quite extensive and involved detailed analysis of 26 image attributes that reflected tumor vasculature, invasion, 3-dimensional texture, location, edema, and margin assessment. These imaging parameters were compared with gene expression patterns. Unsupervised hierarchical clustering of the 353-patient set revealed the clustering of tumors previously reported, such as luminal types A and B, claudin-low, normal breast-like, basal-like, and ERBB2+/ER- tumors. An interferon subtype was also identified, which was associated with inflammatory genes such as STAT1. As discussed earlier, Weichselbaum associated elevation of STAT1 with a radioresistance gene signature.17 The MRI phenotypes were linked around genes related to immune function and the interferon cluster. Further analysis revealed that 12 MRI characteristics were related to gene expression patterns. These included number of satellite lesions, heterogeneous enhancement, presence of architectural distortion, and absence of regional lymph node involvement. These results suggest a linkage between variation in DCE-MRI parameters and radioresistance that is mediated by differences in gene expression. Comparison of the radiosensitivity gene signature by Eschrich et al.15 and this report might prove fruitful in this regard.
Cervix Cancer
An important study was conducted by Halle et al.31 who examined a series of patients with locally advanced cervix cancer who were treated with chemoradiotherapy. This is the only paper we identified that provided a linkage analysis between functional imaging and gene expression for human patients treated with chemoradiotherapy. A total of 78 patients had DCE-MRI studies performed before therapy. Radiotherapy consisted of external radiotherapy to 50 Gy to the tumor, parametria, and adjacent pelvic wall + 45 Gy to remaining pelvis, followed by brachytherapy of 21 Gy to point A. Cisplatin (40 mg/m2) was administered weekly during radio-therapy. The Brix model was used to fit the DCE-MRI data.32
Of the parameters used to fit the DCE-MRI contrast uptake curve, amplitude (Abrix) was most strongly correlated with progression-free survival. Gene expression analysis was then performed on the initial set of 46 patients. In this cohort, Abrix correlated most strongly with genes associated with HIF-1 and hypoxia. Other published gene sets for intrinsic radioresistance and DNA repair pathways were included in the analysis, but were not significant. A signature for cell cycle checkpoints was significant, but the most highly ranked genes in that signature were also known to be hypoxia regulated. Hierarchical clustering showed that this group of patients had 2 main clusters, ranked by high and low Abrix. Kaplan-Meier analysis demonstrated that progression-free survival was shorter for patients with a high hypoxia scores (Fig. 2). The prognostic value of the hypoxia gene signature was validated in an independent cohort of 109 patients with locally advanced cervix cancer. In a multivariate analysis including other known prognostic variables, the hypoxia gene signature maintained independent predictive value for progression-free survival (relative risk = 2.5) and local-regional control (relative risk = 3.7).
Figure 2.
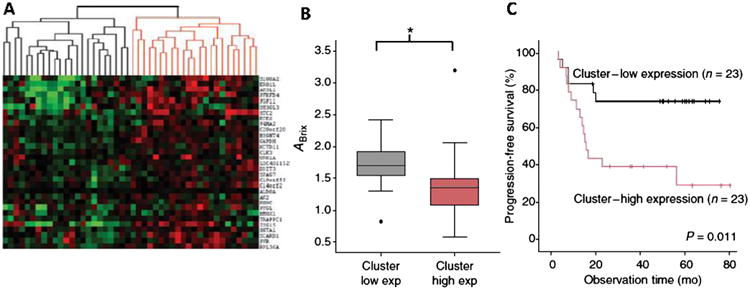
Key results from gene expression analysis of patients with locally advanced cervix cancer treated with chemoradiotherapy. (A) Unsupervised clustering of expression of 31 genes identified in a training dataset consisting of 78 patients with variable DCE-MRI-derived Abrix scores that was associated with treatment outcome. The clustering reveals a significant split by Abrix value in an independent and validation cohort (46 patients) (B). (C) In the same set of 46 patients, the signature for low expression reveals better prognosis. This independent validation is an important step toward a truly beneficial imaging biomarker as therapeutic intervention to reduce hypoxia may prove valuable in the subset with low Abrix score. (Data reproduced with permission from Halle et al.31) (Color version of figure is available online.)
In summary, this publication demonstrates that there is a strong linkage between DCE-MRI contrast enhancement and hypoxia for locally advanced cervix cancer. Given the relative importance of hypoxia in controlling the outcome in this disease,33-36 this finding is useful. Currently, there is no direct method for measuring hypoxia in cervix cancer, as the Eppendorf electrode system is no longer commercially available. The widespread availability of MRI means that it is feasible to consider examination of DCE-MRI in cooperative group trials as a surrogate for hypoxia for cervix cancer.
Soft Tissue Sarcoma
Chi et al.37 conducted genomic analysis on 22 companion dogs with spontaneous soft tissue sarcomas that were treated with thermoradiotherapy. Analyses were done before therapy and at 24 hour after the first hyperthermia treatment. One to two fractions of radiotherapy (2.25 Gy/fraction) would have been given by that time. Apparent diffusion coefficient of water (ADC) was measured by MRI in 17 of these subjects before and at the end of therapy. Approximately 2700 gene sets were found to exhibit at least a 2-fold change associated with the thermoradiotherapy. Commonly induced genes included those involved in cell cycle control and DNA damage response and heat shock proteins (HSP).
Unsupervised analysis was performed on these gene sets, revealing 2 primary clusters. These 2 clusters were significantly different in their change in ADC in response to treatment (Fig. 3). Tumors that exhibited an increase in ADC after treatment showed increases in genes associated with tissue remodeling and inflammation. This is consistent in that one would expect an increase in tissue water content (and therefore ADC) in response to an inflammatory stimulus. There appeared to be an angiogenic stimulus as well, as expression of both FLT1 and kinase insert domain receptor were positively correlated with change in ADC. Tumors that exhibited less change in ADC in response to treatment exhibited genes associated with a more mature vasculature, such as CD31 and von Willebrand factor. Additional genes found to be associated with volume change at the end of therapy vs before treatment included telomerase reverse transcriptase and the DNA repair gene, RAD23A.
Figure 3.
(A) Global analysis of change in gene expression (24 hour after thermoradiotherapy treatment) vs change in diffusion coefficient of water (ADC) from baseline to end of treatment course in 22 dogs with spontaneous soft tissue sarcomas. Unsupervised analysis revealed 2 gene clusters (B) that were significantly different (red and blue boxes), based on the change in ADC (C). Genes that were differentially expressed were associated with inflammation (IL1b, IL6, IL8, IL10, and MARCO) and tissue remodeling (MMP-1, MMP-2A, MMP-3, and TGF-β). The linkage between change in ADC and change in gene expression in the same subjects reveals biological insight into how the tumors respond to this therapy. MMP, matrix metalloproteinase; TGF-β, transforming growth factor beta. (Data reproduced with permission from Chi et al.37) (Color version of figure is available online.)
The expression data were also used to identify compounds with potential to enhance the effects of thermoradiotherapy. Significance analysis of microarrays was used to identify probe sets that varied significantly between the 2 clusters. These data were compared with “connectivity map,” which contains gene signatures for response to thousands of drugs. Inhibitors of histone deacetylase and HSP90 were highly correlated with the gene expression data from this study. To validate the connection, clonogenic studies of thermoradiotherapy response ± geldanamycin (an inhibitor of HSP90) were conducted. Geldanamycin significantly enhanced toxicity of radiation and hyperthermia when used alone and in a combination.
In summary, this report shows how one can link changes in tumor physiology, as assessed by MRI, to changes in gene expression. Further, a method for translating such information to potential new therapeutic targets was demonstrated.
Discussion
In this review we first presented an overview of how gene expression can be used to understand differences in radio-sensitivity and how such information can be translated to prediction of outcome in human trials. Two important gene signatures from studies by Eschrich et al.15 and Weichselbaum et al.17 require further testing to determine the general applicability for chemoradiotherapy trials for a variety of tumor sites.
The main focus of the review, however, was on how the integration of functional imaging with gene expression can reveal novel insights into tumor pathophysiology and treatment responses. Several examples were discussed that involved 18F-FDG-PET. Unfortunately, none of these studies involved the use of this PET tracer in groups of patients who received chemoradiotherapy as part of their management. Suggestions were made as to how such investigations could be made.
Finally, 2 examples were shown linking MRI parameters to gene expression. The paper by Halle et al.31 provides strong rationale for further examining DCE-MRI measurements in patients with cervix cancer enrolled in therapeutic trials involving chemoradiotherapy. The paper by Chi et al. presents an analysis that focuses on changes in gene expression early in the course of thermoradiotherapy that predict for treatment response. The changes in gene expression revealed differential expression of important gene sets associated with inflammation and angiogenesis as well as DNA damage repair. Importantly, the results suggest that changes in ADC may also have prognostic value. With the knowledge of the underlying pathophysiology that accompanies changes in ADC, one might have opportunities to further target the tumor.
The integration of genome-level characterization and functional imaging will undoubtedly be improved by the enormous technical advances in these areas. For example, the genetic characterization of tumors has benefited tremendously from next generation sequencing technology. These technical advances triggered the coordinated efforts of The Cancer Genome Atlas to genetically characterize the somatic mutations, copy number alterations, epigenetic alterations, and gene expression for a large number of tumors and has allowed better integration of these complementary and highly interdependent layers of genetic information to produce a comprehensive picture of the dysregulated processes that contribute to oncogenesis. These characterizations have identified many somatic mutations associated with novel subtypes of tumors with distinct phenotypes. If information on the microenvironmental stresses or functional imaging data is available for these Cancer Genome Atlas tumors, this will present an unprecedented opportunity to further integrate the genetic information to better predict prognosis and treatment response. Recently, point mutations at Arg132 of the cytoplasmic nicotinamide adenine dinucleotide phosphate+-dependent isocitrate dehydrogenase 1 (IDH1) and IDH2 were found to occur fre-quently38 in glioma and several other tumor types. These mutations result in a gain of function to produce the “oncometabolite” 2-hydroxyglutarate.39 It is now possible to use in vivo MR spectroscopy to detect high levels of tumor 2- hydroxyglutarate.40 Therefore, it is possible to use functional imaging to indirectly detect the somatic mutations of glioma and other tumors bearing IDH1 mutations.
One of the challenges in genomic analysis (and deep sequencing) is how to statistically account for the large numbers of genes involved. Statistically, one must correct for the likelihood of false positives. Here we show that functional imaging can help to tame this problem by allowing one to focus on genes that reflect the tumor microenvironment. As a cautionary note, many of the more contemporary imaging methods are still in development and there is no consensus on standardization.2 Standardization is essential for future investigations of these methods, particularly as they move from single-institution to multi-institutional trials. Furthermore, the significant intratumoral heterogeneity in both genetic landscape41 and tumor microenvironmental stress42 may present challenges in extrapolating the results from the sampled areas to the clinical phenotypes of whole tumors.
Acknowledgments
We recognize research support from the NIH (CA 40355) and (CA42745) to M.W.D. and NIH (NCI R01CA125618) and USAMRMC (BC111548) to J.T.C. The authors appreciate the editorial assistance of Dr. Keara Boss.
The funding sources have no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; or in the decision to submit the manuscript for publication.
References
- 1.Torres-RocaJF, Stevens CW. Predicting response to clinical radiotherapy: Past, present, and future directions. Cancer Control. 2008;15(2):151–156. doi: 10.1177/107327480801500207. [DOI] [PubMed] [Google Scholar]
- 2.Lambin P, van Stiphout R, Starmans MHW, et al. Predicting outcomes in radiation oncology-multifactorial decision support systems. Nat Rev Clin Oncol. 2013;10(1):27–40. doi: 10.1038/nrclinonc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg AC. Predicting recurrence after radiotherapy in head and neck cancer. Semin Radiat Oncol. 2012;22(2):108–118. doi: 10.1016/j.semradonc.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Begg AC. Predicting response to radiotherapy: Evolutions and revolutions. Int J Radiat Biol. 2009;85(10):825–836. [PubMed] [Google Scholar]
- 5.Overgaard J. Hypoxic modification of radiotherapy in squamous cell carcinoma of the head and neck—A systematic review and meta-analysis. Radiother Oncol. 2011;100(1):22–32. doi: 10.1016/j.radonc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Harada H. How can we overcome tumor hypoxia in radiation therapy? J Radiat Res. 2011;52(5):545–556. doi: 10.1269/jrr.11056. [DOI] [PubMed] [Google Scholar]
- 7.Le QT, Harris J, Magliocco AM, et al. Validation of lysyl oxidase as a prognostic marker for metastasis and survival in head and neck squamous cell carcinoma: Radiation therapy oncology group trial 90-03. J Clin Oncol. 2009;27(26):4281–4286. doi: 10.1200/JCO.2008.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross JS, Hatzis C, Symmans WF, et al. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist. 2008;13(5):477–493. doi: 10.1634/theoncologist.2007-0248. [DOI] [PubMed] [Google Scholar]
- 9.Chen JLY, Lucas JE, Schroeder T, et al. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS Genet. 2008;4(12) doi: 10.1371/journal.pgen.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi JT, Wang Z, Nuyten DSA, et al. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3(3):395–409. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter SC, Buffa FM, Silva P, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67(7):3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 12.Pramana J, Van den Brekel MWM, Van Velthuysen MLF, et al. Gene expression profiling to predict outcome after chemoradiation in head and neck cancer. Int J Radiat Oncol Biol Phys. 2007;69(5):1544–1552. doi: 10.1016/j.ijrobp.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 13.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004;2(2):206–214. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatza ML, Kung HN, Blackwell KL, et al. Analysis of tumor environmental response and oncogenic pathway activation identifies distinct basal and luminal features in HER2-related breast tumor subtypes. Breast Cancer Res. 2011;13(3) doi: 10.1186/bcr2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eschrich SA, Pramana J, Zhang HL, et al. A gene expression model of intrinsic tumor radiosensitivity: Prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75(2):489–496. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eschrich SA, Fulp WJ, Pawitan Y, et al. Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res. 2012;18(18):5134–5143. doi: 10.1158/1078-0432.CCR-12-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weichselbaum RR, Ishwaran H, Yoon T, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105(47):18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh D, Febbo PG, Ross K, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1(2):203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 19.Lyng H, Svendsrud DH, Brovig RS, et al. Molecular markers for lymph node involvement in advanced cervical carcinomas. Radiother Oncol. 2006;78:S60–S61. [Google Scholar]
- 20.Lando M, Holden M, Bergersen LC, et al. Gene dosage, expression, and ontology analysis identifies driver genes in the carcinogenesis and chemoradioresistance of cervical cancer. PLoS Genet. 2009;5(11) doi: 10.1371/journal.pgen.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shedden K, Taylor JMG, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahra MA, Hollingsworth KG, Sala E, et al. Dynamic contrast-enhanced MRI as a predictor of tumour response to radiotherapy. Lancet Oncol. 2007;8(1):63–74. doi: 10.1016/S1470-2045(06)71012-9. [DOI] [PubMed] [Google Scholar]
- 23.Heijmen L, Verstappen M, ter Voert E, et al. Tumour response prediction by diffusion-weighted MR imaging: Ready for clinical use? Crit Rev Oncol Hematol. 2012;83(2):194–207. doi: 10.1016/j.critrevonc.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Bollineni VR, Wiegman EM, Pruim J, et al. Hypoxia imaging using positron emission tomography in non-small cell lung cancer: Implications for radiotherapy. Cancer Treat Rev. 2012;38(8):1027–1032. doi: 10.1016/j.ctrv.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Osborne JR, Port E, Gonen M, et al. F-18-FDG PET of locally invasive breast cancer and association of estrogen receptor status with standardized uptake value: Microarray and immunohistochemical analysis. J Nucl Med. 2010;51(4):543–550. doi: 10.2967/jnumed.108.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 27.Nair VS, Gevaert O, Davidzon G, et al. Prognostic PET F-18-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res. 2012;72(15):3725–3734. doi: 10.1158/0008-5472.CAN-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JD, Yun MJ, Lee JM, et al. Analysis of gene expression profiles of hepatocellular carcinomas with regard to F-18-fluorodeoxyglucose uptake pattern on positron emission tomography. Eur J Nucl Med Mol Imaging. 2004;31(12):1621–1630. doi: 10.1007/s00259-004-1602-1. [DOI] [PubMed] [Google Scholar]
- 29.Strauss LG, Dimitrakopoulou-Strauss A, Koczan D, et al. Correlation of dynamic PET and gene array data in patients with gastrointestinal stromal tumors. Sci World J. 2012 doi: 10.1100/2012/721313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto S, Maki DD, Korn RL, et al. Radiogenomic analysis of breast cancer using MRI: A preliminary study to define the landscape. Am J Roentgenol. 2012;199(3):654–663. doi: 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- 31.Halle C, Andersen E, Lando M, et al. Hypoxia-induced gene expression in chemoradioresistant cervical cancer revealed by dynamic contrast-enhanced MRI. Cancer Res. 2012;72(20):5285–5295. doi: 10.1158/0008-5472.CAN-12-1085. [DOI] [PubMed] [Google Scholar]
- 32.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7(1):91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 33.Moon EJ, Brizel DM, Chi JTA, et al. The potential role of intrinsic hypoxia markers as prognostic variables in cancer. Antioxid Redox Signal. 2007;9(8):1237–1294. doi: 10.1089/ars.2007.1623. [DOI] [PubMed] [Google Scholar]
- 34.Hockel M, Schlenger K, Aral B, et al. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996;56(19):4509–4515. [PubMed] [Google Scholar]
- 35.Vaupel P, Kelleher DK, Hockel M. Oxygenation status of malignant tumors: Pathogenesis of hypoxia and significance for tumor therapy. Semin Oncol. 2001;28(2):29–35. doi: 10.1016/s0093-7754(01)90210-6. [DOI] [PubMed] [Google Scholar]
- 36.Fyles AW, Milosevic M, Wong R, et al. Oxygenation predicts radiation response and survival in patients with cervix cancer. Radiother Oncol. 1998;48(2):149–156. doi: 10.1016/s0167-8140(98)00044-9. [DOI] [PubMed] [Google Scholar]
- 37.Chi JT, Thrall DE, Jiang C, et al. Comparison of genomics and functional imaging from canine sarcomas treated with thermoradiotherapy predicts therapeutic response and identifies combination therapeutics. Clin Cancer Res. 2011;17(8):2549–2560. doi: 10.1158/1078-0432.CCR-10-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi C, Ganji SK, DeBerardinis RJ, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreso A, O'Brien CA, van Galen P, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339(6119):543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bamezai A. Mouse Ly-6 proteins and their extended family: Markers of cell differentiation and regulators of cell signaling. Arch Immunol Ther Exp. 2004;52(4):255–266. [PubMed] [Google Scholar]
- 44.Hong SW, Jin DH, Shin JS, et al. Ring finger protein 149 is an E3 ubiquitin ligase active on wild-type v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) J Biol Chem. 2012;287(28):24017–24025. doi: 10.1074/jbc.M111.319822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harvey CB, Wang YX, Darmoul D, et al. Characterisation of a human homologue of a yeast cell division cycle gene, MCM6, located adjacent to the 5′‘ end of the lactase gene on chromosome 2q21. FEBS Lett. 1996;398(2-3):135–140. doi: 10.1016/s0014-5793(96)01189-1. [DOI] [PubMed] [Google Scholar]
- 46.Kelly T, Huang Y, Simms AE, et al. Fibroblast activation protein-alpha: A key modulator of the microenvironment in multiple pathologies. Int Rev Cell Mol Biol. 2012;297:83–116. doi: 10.1016/B978-0-12-394308-8.00003-0. [DOI] [PubMed] [Google Scholar]