Abstract
Objective
To understand the biological pathways involved in twin-twin transfusion syndrome (TTTS) by performing global gene expression analysis of amniotic fluid (AF) cell-free RNA.
Methods
Prospective whole transcriptome microarray study analyzing cell-free RNA in AF from TTTS recipient twins and singleton controls. Significantly differentially-regulated genes in TTTS cases (N= 8) vs. matched controls (N = 8) were identified and pathways analyses performed. Significant gene expression differences between Stage II TTTS recipients (N = 5) and Stage III TTTS recipients with abnormal Doppler measurements (N = 5) were also analysed.
Results
Analysis of paired data from TTTS cases and controls revealed differential expression of 801 genes, which were significantly enriched for neurological disease and cardiovascular system pathways. We also identified cardiovascular genes and pathways associated with the presence of critically abnormal Doppler measurements in Stage III TTTS recipients.
Conclusions
This study provides the first transcriptome-wide data on the impact of TTTS on fetal development. Our results show that gene expression involving neurological and cardiovascular pathways are altered in recipient fetuses prior to surgical treatment. This has relevance for the origins of long-term complications seen in survivors and for the development of future fetal biomarkers.
Introduction
Twin-twin transfusion syndrome (TTTS) is a unique complication of monochorionic diamniotic (MCDA) twin pregnancy that is associated with very high perinatal mortality rates.1–3 The primary pathophysiological event in TTTS is the net transfer of blood across shared placental vascular anastomoses from one twin (donor) to the other (recipient), leading to complex derangements in fetal growth, fluid balance, and cardiovascular function. The recipient fetus frequently displays cardiomyopathic changes more severe than would be expected from fluid overload alone. Additional mechanisms that have been suggested to explain this phenomenon include alterations in the renin-angiotensin system and other vasoactive mediators, such as endothelin and atrial natriuric factor.4, In the past decade significant improvements in survival of one or both fetuses have been achieved with prenatal laser ablation of the shared placental anastomoses.5, 6
Despite this advance, long-term complications are still considerable in fetuses that have been successfully treated. Of major clinical concern is the persistent risk of abnormal neurological outcomes, with up to 12% of survivors suffering from major neurodevelopmental problems.7,8 As prematurity, low birth weight and treatment modality are important factors contributing to this morbidity, it has been difficult to define the relative contribution of the TTTS disease itself to long-term neurological outcomes.9–12
Another important complication of TTTS is cardiovascular dysfunction, which disproportionately affects recipient twins. More than 50% of recipients demonstrate sonographic features of anatomical or functional cardiac compromise at diagnosis, most commonly right ventricular dysfunction.13, 14 While most recipients regain normal cardiac function after successful laser surgery, they continue to be at increased risk of structural anomalies16, 17 and long term impaired diastolic function.18
Understanding the underlying mechanisms of disease in TTTS is problematic. There are no suitable animal models. Improvements in clinical management have reduced the availability of fetal tissues used in early studies of TTTS.19–21 Postnatal studies are limited by the confounding effects of prenatal treatment and/ or prematurity.16 Recent studies have attempted to identify amniotic fluid (AF) or blood biomarkers of fetal cardiac dysfunction22, 23, 16 and angiogenesis.24 These studies, however, are limited by their reliance on a small number of candidate markers selected from adult or placental studies that may not be directly relevant to fetal pathophysiology.
We have shown that functional analysis of AF cell-free fetal (cff) RNA provides novel insights into human development.25 A prior study of AF cff RNA from TTTS recipients suggested increased expression of an aquaporin water transporter gene.26 We hypothesized that global gene expression analysis of AF cff RNA from TTTS recipients would reveal the molecular mechanisms underlying the recipient's response to untreated disease and identify potential fetal biomarkers that may assist in future prediction of disease, clinical staging or prognosis.
Materials and Methods
Recruitment and sample collection
This was a multi-center prospective study of women having clinically-indicated laser surgery for TTTS. Institutional ethics committee approval was obtained at all participating centers (see Acknowledgments). The diagnosis and staging of TTTS was based on the presence of a MCDA twin pregnancy with AF discordance in accordance with the criteria described by Quintero et al.27 Briefly, Stage II TTTS was defined as AF discordance with non-visualization of the bladder in the donor twin, normal Doppler studies and the absence of hydrops. Stage III TTTS was defined as AF discordance and the presence of critically abnormal Doppler studies in one or both twins in the absence of hydrops. For our analysis, Stage III TTTS cases were categorized as Stage IIIR, IIID or IIIR/D when abnormal Doppler studies were present in the recipient, donor or both twins, respectively.
Informed consent was obtained from women undergoing a clinically-indicated invasive procedure for TTTS, excluding women carrying fetuses with structural malformations unrelated to TTTS, or with a history of a prior invasive procedure. Clinical indications for offering laser therapy were: 16–26 weeks gestational age (GA) with either stage II, III, or IV TTTS or stage I TTTS with severe polyhydramnios causing respiratory compromise or shortened cervix (≤ 1.5 cm length). In order to minimize biological variation due to gestational age, we excluded TTTS cases > 22 weeks GA in our analysis.
AF was collected after entry into the recipient's amniotic sac for laser surgery, but prior to the onset of laser ablation. As the donor sac is not usually entered during this procedure, we only obtained AF from recipient fetuses. All AF samples from the TTTS cases were spun at 350 × g for 10 min at 4°C and the supernatant stored at −80° C. Frozen samples were batched and shipped overnight to Tufts Medical Center on dry ice. Pre-operative ultrasound findings and obstetric outcomes were collected for each case.
Each TTTS case was matched with a singleton control AF sample obtained for routine midtrimester genetic indications. Whole AF was spun at 350 × g for 10 min at 4°C to remove cells for diagnostic testing. The supernatants were de-identified and archived at −80°C for matching to TTTS cases. Cases and controls were matched for GA (+/− 7 days) and fetal sex. Controls were excluded if there was a prenatal diagnosis of major congenital anomaly or abnormal karyotype. As control samples were anonymized, pregnancy outcomes were unavailable for this group.
RNA extraction, amplification and microarray hybridization
RNA was extracted from AF supernatants according to a customized protocol.28 All samples were processed within 6 months of collection. Due to the lower concentration of RNA observed in the TTTS samples, total RNA was extracted from 15–30 ml of AF from TTTS cases and compared with 5 ml AF from singleton controls. Briefly, RNA was extracted using the Qiagen Circulating Nucleic Acid kit (Qiagen Inc; Valencia, CA) with an on-column DNase digestion step to remove genomic DNA. RNA was converted to cDNA and amplified using the Ovation Pico WTA kit (NuGEN Inc; San Carlos, CA). To correct for the different starting volumes of AF supernatant, a standardized quantity of cDNA was loaded onto each microarray. Five micrograms of cDNA from each sample were biotinylated, fragmented and hybridized to a whole human genome expression array (Affymetrix GeneChip Human Genome U133 Plus 2.0; Affymetrix Inc; Santa Clara, CA).
Statistical analysis
Normalization was performed with the three step command from the AffyPLM package in BioConductor, using ideal- mismatch background/signal adjustment, quantile normalization, and the Tukey biweight summary method.29 This summary method included a logarithmic transformation to improve the normality of the data.
We performed two separate analyses of differential gene expression. First, we compared matched TTTS cases and singleton controls, using the dependent t test to identify those genes consistently up or down regulated in all matched pairs. Second, we compared Stage II and Stage IIIR fetuses using the independent t test. The P values from both analyses were adjusted for multiple testing using the Benjamini-Hochberg (BH) correction. We defined genes as significantly differentially regulated if the BH-corrected P value was < 0.05. Our microarray datasets are publicly available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
The independent t test was used to identify any statistically significant differences in the clinical characteristics between the Stage II and IIIR cases using a threshold of 0.05. The variables tested were: GA at surgery, estimated fetal weight of donor and recipient at time of surgery, deepest pool of amniotic fluid prior to surgery, GA at birth, and birth weight of donor and recipient.
Functional analyses
Functional analyses were performed using Ingenuity Pathways Analysis (IPA) Version 9.0 software (Ingenuity; Redwood City, CA). Ingenuity is a manually curated database that identifies over-represented biological processes in a given data set and calculates a significance score for each result using the right tailed Fisher's test. For the comparison between TTTS cases and singleton controls, IPA was used to identify any statistically significantly enriched physiological systems or molecular/cellular functions using a BH correction for multiple pathway testing (BH corrected P value < 0.05). IPA downstream effects analysis was used to predict the activation or inhibition of specific processes based on the direction of differential regulation of genes. Results were considered statistically significant if z –score > 2 (activation in TTTS) or < - 2 (inhibition in TTTS).
For the Stage II vs. IIIR analysis, IPA was used to identify significant genes and functions within the “cardiovascular system development and function” category. This category was preselected as a candidate pathway, based on prior knowledge of the physiological differences between Stage II and IIIR fetuses. Functional annotations within this category with a Fisher's exact test p value < 0.05 were considered statistically significant. As this was a targeted analysis focusing on a system of a priori interest, no BH correction for multiple comparisons in IPA was required.30
Validation of microarray findings by PCR
To validate the findings from the microarray study in an independent dataset, six significantly differentially-regulated genes from the TTTS vs controls analysis were selected for reverse transcription quantitative real-time PCR (RT-qPCR). TTTS cases were matched to singleton controls for the same clinical parameters as in the microarray experiments. The target genes, which were selected on the basis of the magnitude of differential expression and relevance to the pathophysiology of TTTS, were: arginine vasopressin receptor 1A (AVPR1A), endoglin (ENG), fms-related tyrosine kinase 1 (FLT1), neurexin 3 (NRXN3), neurotrophic tyrosine kinase receptor, type3 (NTRK3) and solute carrier family 25, member 37 (SLC25A37). mRNA abundance was assayed in triplicate with TaqMan inventoried gene expression assays using custom TaqMan Array 96-well plates (both from Life Technologies, Inc) on the Applied Biosystems 7900HT sequence detector. Differential expression between cases and controls was compared with the – ΔΔ Ct method utilizing the beta actin gene (ACTB) as the endogenous control. Correlation between the median fold change results from the RT-qPCR and microarray experiments were compared using Pearson's correlation coefficient. For detailed PCR methods see Supplementary data 1.
Results
We performed whole genome microarray analysis on a total of 13 TTTS cases and 8 singleton controls. Figure 1 shows the samples used in each analysis. The clinical characteristics of the TTTS cases are provided in Table 1. An independent sample set of 12 TTTS cases and 12 singleton controls were used for the RT-qPCR experiments (Supplemental data 2).
Figure 1.
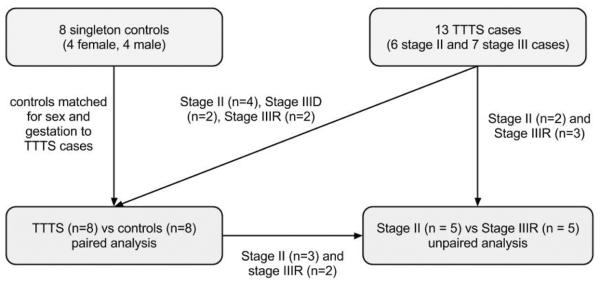
Sample flow chart illustrating the TTTS cases and controls used in each analysis.
Table 1.
Clinical characteristics of TTTS cases
| StudyIDa | GA | TTTS stage | Fetal sex | Recipient twin ultrasound findings | Donor twin ultrasound findings | GA at birth | Mode of birth | Recipient outcome | Donor outcome | Other complications | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EFW (g) | AF DVP (cm) | Doppler abnormalities | EFW (g) | AF DVP (cm) | Doppler abnormalities | Outcome | BW (g) | Outcome | BW (g) | |||||||
| T1 | 17+2 | II | M | 153 | 9.2 | NA | 136 | 0 | NA | 32 | CS | Alive | 1401 | Alive | 1360 | PPROM |
| T2 | 19+6 | II | F | 324 | 8.1 | NA | 219 | 0 | NA | 32 | CS | Alive | 1474 | Alive | 1247 | Preterm labor |
| T3 | 19+6 | II | M | 330 | 12.4 | NA | 230 | 0 | NA | 29 | CS | Alive | 1700 | Alive | 1588 | PPROM |
| T4 | 20+4 | II | F | 394 | 11.8 | NA | 364 | 2.3 | NA | 30 | CS | Alive | 1747 | Alive | 1542 | Preterm labor |
| T5 | 18+0 | IIIR | F | 247 | 9.8 | DV RAW | 174 | 0 | NA | 33 | CS | Alive | 1928 | Alive | 1814 | Preeclampsia |
| T6 | 20+0 | IIIR/D | M | 361 | 9.5 | Pulsatile UV | 324 | 0 | UA AEDF | 21 | VD | NND | 379 | NND | 355 | Preterm labor |
| T7 | 20+6 | IIID | F | 236 | 11.0 | NA | 217 | 0 | UA AEDF, DV RAW, pulsatile UV | 29 | CS | Alive | 1043 | Alive | 816 | Preterm labor |
| T8 | 22+0 | IIID | M | 530 | 10.7 | NA | 350 | 0 | UA AEDF | 38 | VD | Alive | 3402 | Fetal demise | Not recorded | Bipolar reduction of donor twin after laser |
| T9 | 21+2 | II | M | 451 | 16.5 | NA | 312 | 0 | NA | 28 | CS | Alive | 1225 | Alive | 998 | PPROM |
| T10 | 20+1 | II | F | 350 | 8.0 | NA | 212 | 0 | NA | 34 | CS | Alive | 2132 | Alive | 1771 | Preterm labor |
| T11 | 17+1 | IIIR | F | 191 | 9.4 | Absent DV a-wave | 161 | 1.7 | NA | 33 | CS | Alive | 1588 | Alive | 1411 | NA |
| T12 | 19+5 | IIIR | M | 284 | 10.5 | Severe TR, cardiomegaly, pulmonary stenosis | 239 | 0 | NA | 36 | CS | Alive | 2565 | Alive | 2375 | NA |
| T13 | 22+4 | IIIR/D | F | 637 | 8.9 | Pulsatile UV, RA dilatation, TR | 441 | 0 | UA AEDF | 28 | CS | Alive, Normal heart function | 1240 | Alive | 890 | CS for abnormal NST in donor |
Samples T1, T2, T4, T5 and T6 were used in both the TTTS vs. control and the Stage II vs. IIIR analyses
AEDF, absent end diastolic flow; AF, amniotic fluid; Alive, alive at hospital discharge; BW, birth weight; CS, Cesarean section; DV, ductus venosus; DV RAW, ductus venosus reversed a-wave; DVP, deepest vertical pocket; EFW, estimated fetal weight; F, female; GA, gestational age in weeks; M, male; NA, not applicable; NND, neonatal death; NST, non-stress test; PPROM, preterm prelabor rupture of membranes; RA, right atrium; TR, tricuspid regurgitation; UA, umbilical artery; UV, umbilical vein; VD, vaginal delivery
TTTS vs controls microarray results
We analyzed paired data from 8 TTTS cases and 8 matched singleton controls (Table 2). The median GA was 19 weeks (range 17–22 weeks). Only Stage II and III TTTS cases were included due to low numbers of Stage I and IV samples.
Table 2.
Paired TTTS cases and controls
| Singleton controls | TTTS cases | ||||||
|---|---|---|---|---|---|---|---|
| Study ID | GA | Fetal sex | Indication for amniocentesis | Study ID | GA | Fetal sex | Quintero Stage |
| N1 | 17+4 | Male | Abnormal serum screening | T1 | 17+2 | Male | II |
| N2 | 18+6 | Female | Abnormal serum screening, echogenic intracardiac focus | T2 | 19+6 | Female | II |
| N3 | 18+6 | Male | Advanced maternal age, echogenic intracardiac focus | T3 | 19+6 | Male | II |
| N4 | 19+6 | Female | Increased nuchal fold | T4 | 20+4 | Female | II |
| N5 | 17+1 | Female | Abnormal serum screening | T5 | 18+0 | Female | IIIR |
| N6 | 19+1 | Male | Choroid plexus cyst | T6 | 20+0 | Male | IIIR/D |
| N7 | 21+5 | Female | Abnormal serum screening | T7 | 20+6 | Female | IIID |
| N8 | 21+5 | Male | Echogenic intracardiac focus | T8 | 22+0 | Male | IIID |
TTTS cases were matched to singleton controls for fetal sex and GA (< or = 7 days difference). Control samples were excluded if there was a diagnosis of aneuploidy or major congenital malformation.
GA, gestational age (weeks + days)
There were 801 genes that had differential regulation in all 8 TTTS cases compared with their matched controls; of these, 472 genes were down-regulated and 329 genes were up-regulated (Supplementary data 3 and 4).
IPA core analysis of the significantly differentially regulated genes identified 14 physiological systems that were statistically significantly over-represented (Table 3). The two systems most affected by long-term complications of TTTS, the nervous and cardiovascular systems, were significantly dysregulated in TTTS fetuses.
Table 3.
Ingenuity Pathways Analysis results - significant physiological systems dysregulated in TTTS vs controls
| Category | Functional Annotationa | BH p-Valueb | No. of Genes |
|---|---|---|---|
| Organ Development | Development of organ | 4.89E-04 | 102 |
| Organogenesis | 4.94E-04 | 103 | |
| Development of epidermis | 8.13E-03 | 17 | |
| Development of liver | 2.62E-02 | 10 | |
| Organismal Survival | Organismal death | 1.22E-03 | 73 |
| Tissue Development | Tissue development | 1.22E-03 | 138 |
| Hair and Skin Development and Function | Skin development | 2.06E-03 | 24 |
| Humoral Immune Response | Immune response of lymphoma cell lines | 6.11E-03 | 4 |
| Nervous System Development and Function | Outgrowth of neurites | 9.87E-03 | 27 |
| Growth of neurites | 2.02E-02 | 29 | |
| Connective Tissue Development and Function | Proliferation of fibroblast cell lines | 2.47E-02 | 20 |
| Hematological System Development and Function | Hematopoiesis | 3.36E-02 | 49 |
| Proliferation of immune cells | 4.48E-02 | 41 | |
| Differentiation of blood cells | 4.92E-02 | 40 | |
| Cardiovascular System Development and Function | Cardiogenesis | 3.90E-02 | 23 |
| Morphogenesis of outflow tract | 4.36E-02 | 5 | |
| Patterning of blood vessel | 4.48E-02 | 6 | |
| Development of outflow tract | 4.48E-02 | 6 | |
| Organ Morphology | Morphogenesis of organ | 4.14E-02 | 25 |
| Organismal Development | Morphology of vessel | 4.21E-02 | 10 |
| Embryonic Development | Gastrulation of embryo | 4.36E-02 | 5 |
| Differentiation of embryonic cells | 4.48E-02 | 14 | |
| Tissue Morphology | Size of vessel | 4.36E-02 | 5 |
| Skeletal and Muscular System Development and Function | Differentiation of muscle cells | 4.48E-02 | 17 |
| Proliferation of muscle cells | 4.48E-02 | 20 |
Functional annotations with < 4 genes not shown
Benjamini-Hochberg corrected P value
There were 13 molecular and cellular functions that were differentially regulated in TTTS cases compared with controls (Table 4). The category “Cell death”, which includes apoptosis, contained 198 genes, including 41 genes annotated to “neuronal cell death” (BH P = 0.022) and 17 genes annotated to “cell death of cerebral cortex cells” (BH P = 0.026).
Table 4.
Ingenuity Pathways Analysis results – significant molecular and cellular pathways dysregulated in TTTS cases vs. controls
| Category | p-value rangea | Number of genes |
|---|---|---|
| Cell Death | 8.23E-07 to 4.48E-02 | 198 |
| Cellular Growth and Proliferation | 3.77E-06 to 4.84E-02 | 201 |
| Cell Cycle | 4.33E-06 to 4.97E-02 | 103 |
| Cellular Movement | 6.64E-06 to 4.48E-02 | 119 |
| Cellular Development | 8.21E-05 to 4.97E-02 | 122 |
| Gene Expression | 7.32E-04 to 4.21E-02 | 129 |
| DNA Replication, Recombination, and Repair | 2.91E-03 | 34 |
| Cell-To-Cell Signaling and Interaction | 6.11E-03 to 4.48E-02 | 8 |
| Protein Trafficking | 6.81E-03 | 25 |
| Cell Morphology | 3.84E-02 to 4.60E-02 | 33 |
| Cellular Assembly and Organization | 9.87E-03 to 4.48E-02 | 90 |
| Cell Signaling | 1.44E-02 to 2.37E-02 | 28 |
| Post-Translational Modification | 3.46E-02 | 20 |
Benjamini-Hochberg corrected P value of individual functional annotations within each category
The IPA downstream effects prediction analysis showed evidence of abnormal nervous system gene expression in TTTS cases, with a predicted increase in the category “neurological disease” (z score = 2.17) (Figure 2). (For details see Supplementary data 5). Other functions that were significantly predicted to be increased in the TTTS fetuses were: cellular growth and proliferation, cellular compromise, and skeletal and muscular system development and function.
Figure 2.
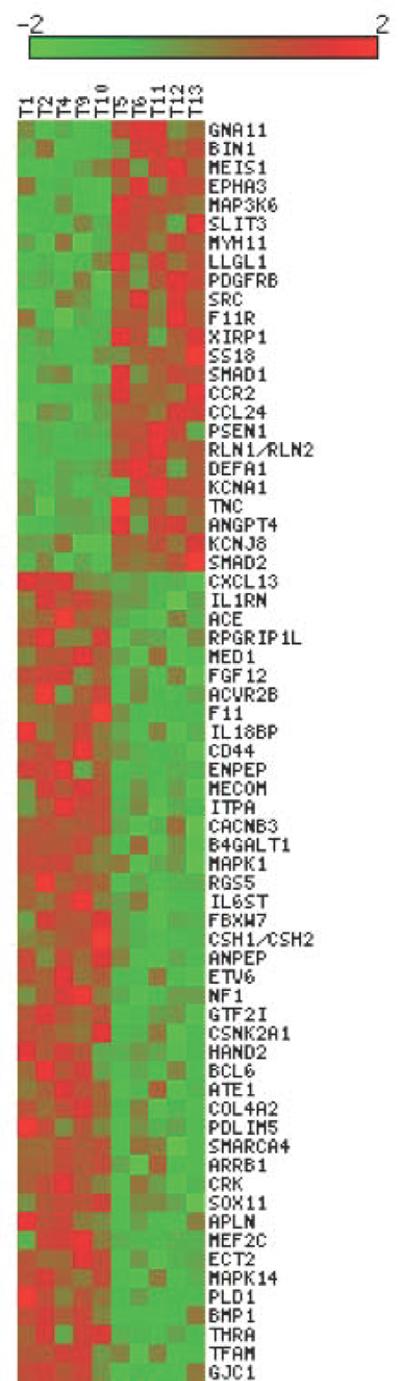
Heat map illustration of the 79 differentially regulated genes in the TTTS vs controls analysis annotated to “neurological disease” by Ingenuity Pathways Analysis. Each column represents an individual sample. The color scale represents normalized gene expression levels, with data zero-centered by rows (genes) and sorted in descending order according the fold change value between TTTS and control groups. Up- and down-regulation of gene expression is shown in red and green respectively. (Images were created using matrix2png by Pavlidis, P. and Noble W.S. (2003) Matrix2png: A Utility for Visualizing Matrix Data. Bioinformatics 19: 295–296)
Many individual differentially regulated genes appeared biologically relevant to TTTS recipients because of their role in fluid homeostasis, blood pressure regulation system, or angiogenesis (Table 5).31
Table 5.
Selected differentially regulated genes of interest in analysis of TTTS cases vs. controls
| Gene symbol | Gene name | Fold changea | Gene functionb |
|---|---|---|---|
| SLC25A37 | solute carrier family 25, member 37 | 6.9 | Mitochondrial iron transporter that specifically mediates iron uptake in developing erythroid cells, thereby playing an essential role in heme biosynthesis. |
| ENG | endoglin | 4.3 | Major glycoprotein of vascular endothelium. May play a critical role in the binding of endothelial cells to integrins and other receptors |
| ADRA1D | adrenergic, alpha-1D-, receptor | 3.5 | Alpha1-adrenoceptors are widely distributed in both the central and peripheral nervous system where they play a major role in smooth muscle contraction. |
| SYNGR1 | synaptogyrin 1 | 3.3 | Involved in the regulation of short-term and long-term synaptic plasticity |
| AVPR1A | arginine vasopressin receptor 1A | 3.1 | Receptor for arginine vasopressin found in high density on smooth muscle cells. It is involved in regulation of blood pressure through arterial vasoconstriction and has been found to stimulate VEGF secretion. |
| FLT1 | Fms-related tyrosine kinase 1 | 3.1 | Tyrosine-protein kinase that acts as a cell-surface receptor for VEGFA, VEGFB and PGF, and plays an essential role in the development of embryonic vasculature, the regulation of angiogenesis, cell survival, cell migration, macrophage function, chemotaxis, and cancer cell invasion. May play an essential role as a negative regulator of embryonic angiogenesis by inhibiting excessive proliferation of endothelial cells. |
| CHRNB2 | cholinergic receptor, nicotinic, beta 2 (neuronal) | 2.3 | Member of the superfamily of ligand-gated ion channels which allow the flow of sodium and potassium across the plasma membrane in response to ligands such as acetylcholine and nicotine. |
| AQP6 | aquaporin 6, kidney specific | 2.0 | The protein encoded by this gene is an aquaporin protein, which functions as a water channel in cells. This protein is specific for the kidney. |
| NTRK3 | neurotrophic tyrosine kinase, receptor, type 3 | −2.3 | Receptor for neurotrophin-3, which induces neuronal differentiation and survival. Cardiac expression of this receptor is also required for normal development of the outflow tracts. |
| BASP1 | brain abundant, membrane-attached signal protein 1 | −2.7 | Associated with the membranes of growth cones that form the tips of elongating axons. |
| MEIS1 | Meis homeobox 1 | −3.1 | Homeobox genes play a crucial role in normal development. Required for hematopoiesis, megakaryocyte lineage development and vascular patterning. |
| AMOTL2 | angiomotin like 2 | −3.5 | The protein encoded by this gene is related to angiomotin and is a member of the motins protein family. Angiomotin is a protein that binds angiostatin, a circulating inhibitor of angiogenesis. |
| EGFR | epidermal growth factor receptor | −4.2 | This protein is a receptor for members of the epidermal growth factor family. many cellular responses, including changes in gene expression, cytoskeletal rearrangement, anti-apoptosis and increased cell proliferation. |
| GJA1 | gap junction protein, alpha 1, 43kDa | −5.9 | The encoded protein is the major protein of gap junctions in the heart that are thought to have a crucial role in the synchronized contraction of the heart and in embryonic development. |
| NRXN3 | neurexin 3 | −8.5 | Neuronal cell surface protein that may be involved in cell recognition and cell adhesion. May play a role in angiogenesis. |
| GCH1 | GTP cyclohydrolase 1 | −20.6 | Positively regulates nitric oxide synthesis in umbilical vein endothelial cells. |
Positive and negative fold changes indicate up- and downregulation in TTTS cases relative to controls respectively
Gene functions summarized from public databases via The Human Gene Compendium (www.genecards.org) with particular focus on relevance to the nervous and cardiovascular systems
GTP, guanosine 5'-triphosphate; VEGFA, vascular endothelial growth factor A; VEGFB, vascular endothelial growth factor B; PGF, placental growth factor
Stage II vs. IIIR TTTS microarray results
The gene expression profiles of five recipient fetuses with abnormal Doppler measurements (from 3 Stage IIIR and 2 Stage IIIR/D cases) were compared with five stage II cases. There were no statistically significant differences in the clinical characteristics between the two groups (p > 0.05 for all individual variables). A total of 611 genes were significantly differentially regulated in stage II vs. IIIR fetuses (Supplementary data 6 and 7).
As predicted, cardiovascular system development and function was significantly enriched on IPA analysis of the differentially regulated gene list (p < 0.03) (Figure 3). Cardiogenesis was the most statistically significant functional annotation within this category (P = 0.0002, 24 genes). Morphology of cardiovascular system (P = 0.001, 35 genes) and angiogenesis (P = 0.005, 32 genes) were also statistically significant (Supplementary data 8). Selected genes from the Stage II vs. IIIR analysis are listed in Table 6.
Figure 3.
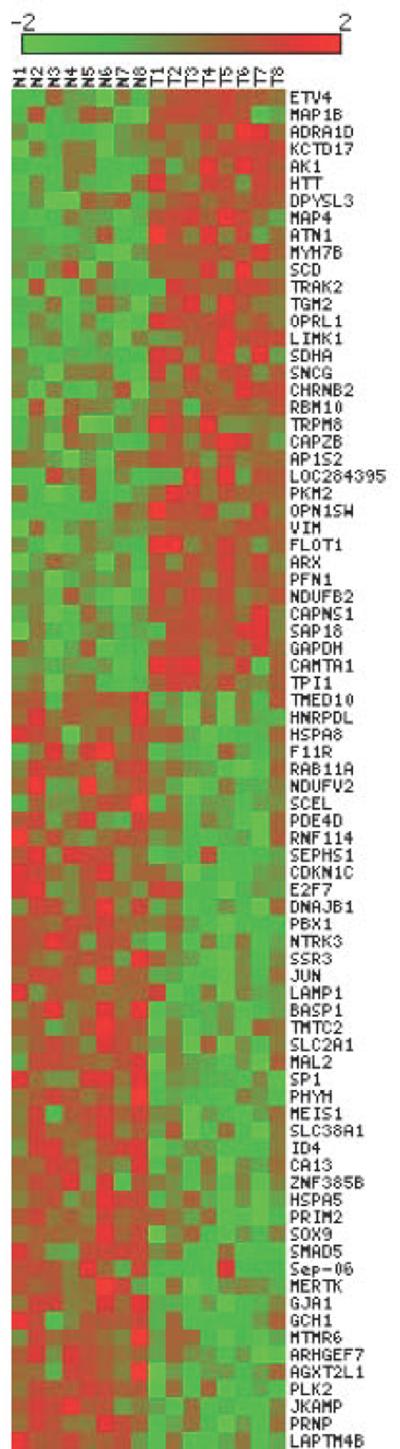
Heat map illustration of the 67 differentially regulated genes in the Stage II vs IIIR TTTS analysis annotated to “cardiovascular system development and function” by Ingenuity Pathways Analysis. Each column represents an individual sample. The color scale represents normalized gene expression levels, with data zero-centered by rows (genes) and sorted in descending order according the fold change value between the Stage II and IIIR groups. Up and down-regulation of gene expression is shown in red and green respectively. (Images were created using matrix2png by Pavlidis, P. and Noble W.S. (2003) Matrix2png: A Utility for Visualizing Matrix Data. Bioinformatics 19: 295–296)
Table 6.
Selected differentially regulated genes of interest in analysis of Stage II vs. Stage IIIR TTTS
| Gene symbol | Gene Name | Fold Changea | Gene functionb |
|---|---|---|---|
| TACR1 | tachykinin receptor 1 | 6.7 | Also known as the neurokinin (NK) 1 receptor, this member of the tachykinin family is localized in high concentrations in the CNS and peripheral tissues. NK1 receptors are thought to mediate central stress reactions, mood control, excitatory neurotransmission, immune modulation, and airway and lung function. |
| MYH11 | myosin, heavy chain 11, smooth muscle | 5.6 | Found predominantly in myocytes and mediates muscle contraction through cyclic interactions with actin-rich thin filaments, creating a contractile force. |
| PDGFRB | platelet-derived growth factor receptor, beta polypeptide | 5.2 | Plays an essential role in blood vessel development by promoting proliferation, migration and recruitment of pericytes and smooth muscle cells to endothelial cells. Plays a role in the migration of vascular smooth muscle cells and the formation of neointima at vascular injury sites. Required for normal development of the cardiovascular system. |
| TSHR | thyroid stimulating hormone receptor | 5.1 | Receptor for thyrothropin. Plays a central role in controlling thyroid cell metabolism. |
| NCDN | neurochondrin | 5.0 | Probably involved in signal transduction in the nervous system. Required for the spatial learning process. May be involved in bone metabolism and neurite outgrowth. |
| RLN1/RLN2 | relaxin 2 | 3.2 | Relaxin is an ovarian hormone that acts with estrogen to produce dilatation of the birth canal in many mammals. May be involved in remodeling of connective tissues during pregnancy, promoting growth of pubic ligaments and ripening of the cervix |
| NPDC1 | neural proliferation, differentiation and control, 1 | 3.1 | Suppresses oncogenic transformation in neural and non-neural cells and down-regulates neural cell proliferation. |
| AGRN | agrin | 3.0 | Plays a central role in the formation and the maintenance of the neuromuscular junction. |
| CHRM4 | cholinergic receptor, muscarinic 4 | 2.7 | The muscarinic acetylcholine receptor mediates various cellular responses, including inhibition of adenylate cyclase, breakdown of phosphoinositides and modulation of potassium channels through the action of G proteins. |
| ANGPT4 | angiopoietin 4 | 2.6 | Angiopoietins are proteins with important roles in vascular development and angiogenesis. This gene product is an agonist that promotes endothelial cell survival, migration and angiogenesis. |
| BASP1 | brain abundant, membrane attached signal protein 1 | −2.2 | Associated with the growth cones that form the tips of elongating axons. |
| ACE | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | −2.3 | Converts angiotensin I to angiotensin II, resulting in an increase of the vasoconstrictor activity of angiotensin. Also able to inactivate bradykinin, a potent vasodilator. |
| MTPN | myotrophin | −2.4 | Promotes dimerization of NF-kappa-B subunits and regulates NF-kappa-B transcription factor activity. Plays a role in the regulation of the growth of actin filaments. Promotes growth of cardiomyocytes, but not cardiomyocyte proliferation. Promotes cardiac muscle hypertrophy. |
| CASK | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | −4.0 | Multidomain scaffolding protein with a role in synaptic transmembrane protein anchoring and ion channel trafficking. Contributes to neural development and regulation of gene expression. Binds to cell-surface proteins, including amyloid precursor protein, neurexins and syndecans. |
| HAND2 | heart and neural crest derivatives expressed 2 | −5.5 | Essential for cardiac morphogenesis, particularly for the formation of the right ventricle and the aortic arch arteries. Required for vascular development and regulation of angiogenesis, possibly through a VEGF signaling pathway. Plays an important role in limb development, and the development of branchial arches. |
| SOX11 | SRY (sex determining region Y)-box 11 | −8.1 | Probably important in the developing nervous system. May also have a role in tissue modeling during development. |
| APLN | apelin | −8.7 | Endogenous ligand for the apelin receptor that has considerable sequence homo logy to the angiotensin receptor. The apelin receptor and peptides act as mediators of cardiovascular function, fluid homeostasis and adipocyte endocrine secretion. |
| BMP1 | bone morphogenetic protein 1 | −17.1 | Induces cartilage and bone formation. May participate in dorsoventral patterning during early development by cleaving chordin. |
| THRA | thyroid hormone receptor, alpha | −26.0 | Nuclear hormone receptor. High affinity receptor for triiodothyronine. |
| STX3 | syntaxin 3 | −45.5 | Potentially involved in docking of synaptic vesicles at presynaptic active zones. |
positive and negative fold change values indicate up- and down-regulation in the Stage IIIR group compared to the Stage II group respectively
Gene functions summarized from public databases via The Human Gene Compendium (www.genecards.org) with particular focus on relevance to the nervous and cardiovascular systems
VEGF, vascular endothelial growth factor
RT-qPCR Amplification Results
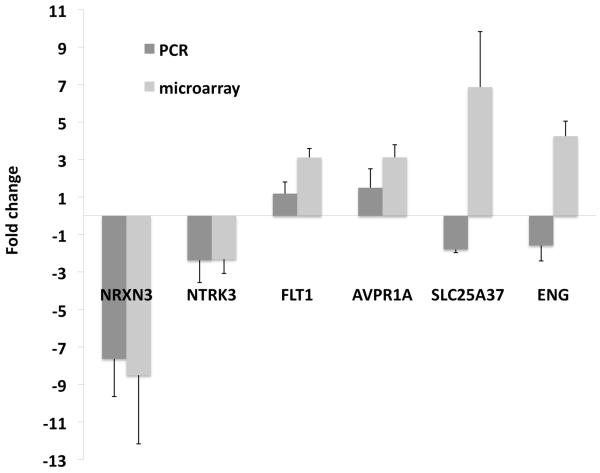
Six genes that were significantly differentially regulated in the TTTS vs. control analysis were tested in an independent sample set of 12 TTTS cases and 12 matched controls using RT-qPCR amplification. The Pearson's correlation coefficient showed a positive correlation between the microarray and the PCR fold changes (r = 0.78). Four of the six target genes (NTRK3, NRXN3, FLT1, AVPR1A) showed a similar direction of differential regulation to the microarray results (Figure 4). Further details are provided in Supplementary data 9.
Figure 4.
Comparison of median fold changes in gene expression between TTTS cases and controls for six genes based on microarray and RT-qPCR analyses using an independent sample set of 12 TTTS cases and 12 matched controls. Standard error bars are shown. Pearson correlation coefficient showed a positive correlation between the microarray and RT-qPCR amplification results (r = 0.78). NTRK3 and NRXN3 were statistically significantly downregulated on RT-qPCR (paired 2-tailed t test on normalized mean cycle threshold difference, P = 0.02 and 0.005 respectively)
Discussion
The pathophysiology of TTTS poses great challenges for researchers due to the absence of a suitable animal model and the complexity of the disease. We attempted to overcome these barriers to in vivo research by performing a whole transcriptome analysis of cell-free RNA using AF from live fetuses with TTTS before treatment. Our results suggest that neurological and cardiovascular abnormalities are present prior to laser ablation. These dysregulated pathways and genes may be involved in the long-term morbidity observed in successfully treated survivors.
The pathogenesis of the cerebral injury in TTTS is heterogeneous and ill defined. In this study, functional analysis indicated that the process of neurite outgrowth is specifically impaired in TTTS recipients compared with controls. We also found that the neurological disease predicted to be increased in TTTS was “movement disorder”, which may have relevance for the increased risk of cerebral palsy in survivors. In addition, a gene essential for normal synapse development, neurexin 3, was significantly downregulated in the TTTS fetuses by gene expression microarrays and confirmed by RT-qPCR experiments in an independent sample set.
The significant cardiovascular annotations in the TTTS vs. control analysis were notable for their specificity to recipient twins, namely, structural abnormalities of the outflow tracts. This is despite only one of the TTTS cases having documented pulmonary stenosis. This suggests that normal cardiac development is still affected in recipients, even in the absence of overt structural anomalies. We also observed significant downregulation of the NTRK3 gene, which was confirmed by RT-qPCR. This gene codes for a nerve growth factor receptor that is involved in cardiac as well as neurological development. Animal models have shown that cardiac underexpression of NTRK3 results in severe congenital cardiac anomalies involving structures of neural crest origin, in particular pulmonary stenosis.31
Another dysregulated gene of interest that has not been previously described in TTTS was AVPR1A. The arginine vasopressin (AVP) type 1A receptor is highly expressed in vascular smooth muscle and is responsible for the vasoconstrictive effects of vasopressin. Fetal plasma vasopressin levels appear to be positively correlated with increased disease severity in recipients.32 The type IA receptor is also expressed in cardiac myocytes, where overexpression causes ventricular hypertrophy, dilatation, and upregulation of natriuretic peptide expression.33 All these gene functions are biologically consistent with the pathophysiology observed in recipients.
Novel results from the TTTS Stage II vs. IIIR analysis include the significant differential regulation of apelin, a potent endogenous stimulator of cardiac contractility. Apelin reduces left ventricular preload and afterload, and increases contractile reserve without causing hypertrophy.34 It also decreases central vasopressin release. Apelin is crucial to maintaining cardiac contractility in pressure overload and its eight-fold downregulation in fetuses with critically abnormal Doppler measurements suggest an important role in hemodynamic deterioration.
These findings illustrate the advantages of global gene expression studies over a traditional gene-by-gene approach. Its unbiased nature overcomes our substantial knowledge gap regarding the choice of candidate genes to study in the human fetus. The upregulation of AVPR1A in TTTS recipients is an example of how discovery-driven research can suggest candidate drugs for future fetal therapies. AVP receptor antagonists are used to treat hyponatremia and decompensated heart failure in adults.35–37 While more research is required before this information can be used clinically, these results allow potential adjunctive medical therapies such as AVP receptor antagonists to be considered for future investigation.
Another strength of our study was that we avoided the confounders of antenatal surgery and premature birth by using samples taken prior to surgical correction of TTTS. Reliance on neonatal samples has the potential to miss important cardiovascular changes because cardiac function in the recipient often normalizes after laser surgery. Postnatal assessment of neurological function also suffers from the confounding effects of premature birth, a common obstetric complication of TTTS that affected all of our cases.
One of the limitations of our study was the use of singleton controls. Other investigators studying AF from TTTS recipients have experienced this requirement.23 We were unable to obtain AF from uncomplicated MCDA controls due to the rarity of GA-matched specimens. The results from our TTTS vs. control analysis could therefore be influenced by inherent properties of monochorionic pregnancies, rather than, or in addition to, the TTTS disease process itself. However, our Stage II vs. IIIR analysis consisted of TTTS recipient cases only, allowing us to confidently determine the disease-specific molecular mechanisms involved in increasing cardiac dysfunction. Further research may allow us to identify differential gene expression before cardiovascular anomalies become clinically detectable, potentially improving stratification and patient selection.
Our study presumes that the cell-free RNA obtained from the AF of recipient twins originates from only the recipient fetus. It is possible that some blood-borne cell-free transcripts from the donor fetus may enter the circulation of the recipient via placental anastomoses. Any donor cell-free RNA in the blood circulation of the recipient, however, would be unlikely to exist in quantities sufficient to significantly influence the overall gene expression profile ascertained via its AF..
The field of prenatal diagnosis is now exploring high dimensional biology approaches to better understand disease and develop new approaches to fetal treatment.38 This study provides new molecular data on the impact of TTTS on the developing nervous and cardiovascular system of the recipient fetus and responds to the call for a better understanding of TTTS pathophysiology.4 The dysregulated genes and pathways identified this study have relevance for the long-term sequelae of TTTS, both for understanding the molecular mechanisms and for suggesting novel medical therapeutic strategies.
Supplementary Material
What is already known about this topic?
Twin-twin transfusion syndrome (TTTS) survivors are at increased risk for neurological and cardiovascular complications even after prenatal therapy
What does this study add?
This study provides the first transcriptome-wide data on the impact of TTTS on fetal development
The results show that gene expression involving neurological and cardiovascular pathways are altered in recipient fetuses prior to surgical treatment
This has relevance for postnatal complications and the development of future biomarkers
Acknowledgments
The authors thank the programs listed here, along with the specific individuals in parentheses, for their help in sample acquisition and data collection: Texas Fetal Center, University of Texas School of Medicine, Houston, TX (Elisa Garcia); Texas Children's Fetal Center, Baylor College of Medicine & Texas Children's Hospital, Houston, TX; Fetal Treatment Program of New England, Brown University, RI (Debra Watson-Smith, RN); Royal Hospital for Women, Randwick NSW Australia; Department of Cytogenetics, Tufts Medical Center, Boston (Janet Cowan, Ph.D. and Karen Krajewski). In addition we thank Professor Jonathan Morris from the University of Sydney for critical review of the manuscript.
Financial sources
Supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development grants R01 HD42053-09 and R01 HD058880; the University of Sydney Medical School (Albert S. McKern Research Scholarship to Dr Hui); and the Royal Australian and New Zealand College of Obstetricians and Gynaecologists (Fotheringham Fellowship to Dr Hui).
Footnotes
Disclosures
The authors report no conflicts of interest.
References
- 1.Gonsoulin W, Moise KJ, Jr., Kirshon B, et al. Outcome of twin-twin transfusion diagnosed before 28 weeks of gestation. Obstet Gynecol. 1990;75:214–6. [PubMed] [Google Scholar]
- 2.Saunders NJ, Snijders RJ, Nicolaides KH. Therapeutic amniocentesis in twin-twin transfusion syndrome appearing in the second trimester of pregnancy. Am J Obstet Gynecol. 1992;166:820–4. doi: 10.1016/0002-9378(92)91340-g. [DOI] [PubMed] [Google Scholar]
- 3.Shah DM, Chaffin D. Perinatal outcome in very preterm births with twin-twin transfusion syndrome. Am J Obstet Gynecol. 1989;161:1111–3. doi: 10.1016/0002-9378(89)90644-3. [DOI] [PubMed] [Google Scholar]
- 4.Fisk NM, Duncombe GJ, Sullivan MHF. The basic and clinical science of twin-twin transfusion syndrome. Placenta. 2009;30:379–90. doi: 10.1016/j.placenta.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Roberts D, Gates S, Kilby M, Neilson JP. Interventions for twin-twin transfusion syndrome: a Cochrane review. Ultrasound Obstet Gynecol. 2008;31:701–11. doi: 10.1002/uog.5328. [DOI] [PubMed] [Google Scholar]
- 6.Senat MV, Deprest J, Boulvain M, et al. Endoscopic laser surgery versus serial amnioreduction for severe twin-to-twin transfusion syndrome. N Engl J Med. 2004;351:136–44. doi: 10.1056/NEJMoa032597. [DOI] [PubMed] [Google Scholar]
- 7.Salomon LJ, Ortqvist L, Aegerter P, et al. Long-term developmental follow-up of infants who participated in a randomized clinical trial of amniocentesis vs laser photocoagulation for the treatment of twin-to-twin transfusion syndrome. Am J Obstet Gynecol. 2010;203:444, e1–7. doi: 10.1016/j.ajog.2010.08.054. [DOI] [PubMed] [Google Scholar]
- 8.Rossi AC, Vanderbilt D, Chmait RH. Neurodevelopmental outcomes after laser therapy for twin-twin transfusion syndrome: a systematic review and meta-analysis. Obstet Gynecol. 2011;118:1145–50. doi: 10.1097/AOG.0b013e318231827f. [DOI] [PubMed] [Google Scholar]
- 9.Kowitt B, Tucker R, Watson-Smith D, et al. Long-term morbidity after fetal endoscopic surgery for severe twin-to-twin transfusion syndrome. J Pediatr Surg. 2012;47:51–6. doi: 10.1016/j.jpedsurg.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Lopriore E, van Wezel-Meijler G, Middeldorp JM, et al. Incidence, origin, and character of cerebral injury in twin-to-twin transfusion syndrome treated with fetoscopic laser surgery. Am J Obstet Gynecol. 2006;194:1215–20. doi: 10.1016/j.ajog.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Quarello E, Molho M, Ville Y. Incidence, mechanisms, and patterns of fetal cerebral lesions in twin-to-twin transfusion syndrome. J Matern Fetal Neonatal Med. 2007;20:589–97. doi: 10.1080/14767050701449638. [DOI] [PubMed] [Google Scholar]
- 12.Spruijt M, Steggerda S, Rath M, et al. Cerebral injury in twin-twin transfusion syndrome treated with fetoscopic laser surgery. Obstet Gynecol. 2012;120:15–20. doi: 10.1097/AOG.0b013e31825b9841. [DOI] [PubMed] [Google Scholar]
- 13.Habli M, Michelfelder E, Cnota J, et al. Prevalence and progression of recipient-twin cardiomyopathy in early-stage twin-twin transfusion syndrome. Ultrasound Obstet Gynecol. 2012;39:63–8. doi: 10.1002/uog.10117. [DOI] [PubMed] [Google Scholar]
- 14.Stirnemann JJ, Mougeot M, Proulx F, et al. Profiling fetal cardiac function in twin-twin transfusion syndrome. Ultrasound Obstet Gynecol. 2010;35:19–27. doi: 10.1002/uog.7488. [DOI] [PubMed] [Google Scholar]
- 15.Karatza AA, Wolfenden JL, Taylor MJ, et al. Influence of twin-twin transfusion syndrome on fetal cardiovascular structure and function: prospective case-control study of 136 monochorionic twin pregnancies. Heart. 2002;88:271–7. doi: 10.1136/heart.88.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopriore E, Bokenkamp R, Rijlaarsdam M, et al. Congenital heart disease in twin-to-twin transfusion syndrome treated with fetoscopic laser surgery. Congenit Heart Dis. 2007;2:38–43. doi: 10.1111/j.1747-0803.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- 17.Herberg U, Gross W, Bartmann P, et al. Long term cardiac follow up of severe twin to twin transfusion syndrome after intrauterine laser coagulation. Heart. 2006;92:95–100. doi: 10.1136/hrt.2004.057497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halvorsen CP, Bilock SL, Pilo C, et al. Childhood cardiac function after twin-to-twin transfusion syndrome--a 10-year follow up. Acta Paediatr. 2009;98:1468–74. doi: 10.1111/j.1651-2227.2009.01376.x. [DOI] [PubMed] [Google Scholar]
- 19.Bajoria R, Ward S, Chatterjee R. Natriuretic peptides in the pathogenesis of cardiac dysfunction in the recipient fetus of twin-twin transfusion syndrome. Am J Obstet Gynecol. 2002;186:121–7. doi: 10.1067/mob.2002.118845. [DOI] [PubMed] [Google Scholar]
- 20.Mahieu-Caputo D, Dommergues M, Delezoide AL, et al. Twin-to-twin transfusion syndrome. Role of the fetal renin-angiotensin system. Am J Pathol. 2000;156:629–36. doi: 10.1016/S0002-9440(10)64767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajoria R, Sullivan M, Fisk NM. Endothelin concentrations in monochorionic twins with severe twin-twin transfusion syndrome. Hum Reprod. 1999;14:1614–8. doi: 10.1093/humrep/14.6.1614. [DOI] [PubMed] [Google Scholar]
- 22.Habli M, Cnota J, Michelfelder E, et al. The relationship between amniotic fluid levels of brain-type natriuretic peptide and recipient cardiomyopathy in twin-twin transfusion syndrome. Am J Obstet Gynecol. 2010;203:404, e1–7. doi: 10.1016/j.ajog.2010.06.070. [DOI] [PubMed] [Google Scholar]
- 23.Van Mieghem T, Done E, Gucciardo L, et al. Amniotic fluid markers of fetal cardiac dysfunction in twin-to-twin transfusion syndrome. Am J Obstet Gynecol. 2010;202:48, e1–7. doi: 10.1016/j.ajog.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Fox CE, Lash GE, Pretlove SJ, et al. Maternal plasma and amniotic fluid angiogenic factors and their receptors in monochorionic twin pregnancies complicated by twin-to-twin transfusion syndrome. Ultrasound Obstet Gynecol. 2010;35:695–701. doi: 10.1002/uog.7515. [DOI] [PubMed] [Google Scholar]
- 25.Hui L, Slonim DK, Wick HC, et al. The amniotic fluid transcriptome: a source of novel information about human fetal development. Obstet Gynecol. 2012;119:111–8. doi: 10.1097/AOG.0b013e31823d4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larrabee PB, Johnson KL, Lai C, et al. Global gene expression analysis of the living human fetus using cell-free messenger RNA in amniotic fluid. JAMA. 2005;293:836–42. doi: 10.1001/jama.293.7.836. [DOI] [PubMed] [Google Scholar]
- 27.Quintero RA, Morales WJ, Allen MH, et al. Staging of twin-twin transfusion syndrome. J Perinatol. 1999;19:550–5. doi: 10.1038/sj.jp.7200292. [DOI] [PubMed] [Google Scholar]
- 28.Dietz JA, Johnson KL, Massingham LJ, et al. Comparison of extraction techniques for amniotic fluid supernatant demonstrates improved yield of cell-free fetal RNA. Prenat Diagn. 2011;31:598–9. doi: 10.1002/pd.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentleman R. Reproducible research: a bioinformatics case study. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1034. Article2. [DOI] [PubMed] [Google Scholar]
- 30.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28:323–32. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tessarollo L, Tsoulfas P, Donovan MJ, et al. Targeted deletion of all isoforms of the trkC gene suggests the use of alternate receptors by its ligand neurotrophin-3 in neuronal development and implicates trkC in normal cardiogenesis. Proc Natl Acad Sci USA. 1997;94:14776–81. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bajoria R, Ward S, Sooranna SR. Influence of vasopressin in the pathogenesis of oligohydramnios-polyhydramnios in monochorionic twins. Eur J Obstet Gynecol Reprod Biol. 2004;113:49–55. doi: 10.1016/S0301-2115(03)00318-X. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Chan TO, Myers V, et al. Controlled and cardiac-restricted overexpression of the arginine vasopressin V1A receptor causes reversible left ventricular dysfunction through Galphaq-mediated cell signaling. Circulation. 2011;124:572–81. doi: 10.1161/CIRCULATIONAHA.111.021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashley EA, Powers J, Chen M, et al. The endogenous peptide apelin potently improves cardiac contractility and reduces cardiac loading in vivo. Cardiovasc Res. 2005;65:73–82. doi: 10.1016/j.cardiores.2004.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arai Y, Fujimori A, Sudoh K, Sasamata M. Vasopressin receptor antagonists: potential indications and clinical results. Curr Opin Pharmacol. 2007;7:124–9. doi: 10.1016/j.coph.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Valania G, Singh M, Slawsky MT. Targeting hyponatremia and hemodynamics in acute decompensated heart failure: is there a role for vasopressin antagonists? Curr Heart Fail Rep. 2011;8:198–205. doi: 10.1007/s11897-010-0035-3. [DOI] [PubMed] [Google Scholar]
- 37.Thibonnier M. Vasopressin receptor antagonists in heart failure. Curr Opin Pharmacol. 2003;3:683–7. doi: 10.1016/j.coph.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Bianchi DW. From prenatal genomic diagnosis to fetal personalized medicine: progress and challenges. Nat Med. 2012;18:1041–51. doi: 10.1038/nm.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.