Abstract
During B and T cell development, the lymphoid-specific proteins RAG1 and RAG2 act together to initiate antigen receptor gene assembly via a series of site-specific, somatic DNA rearrangements that are collectively termed V(D)J recombination. In the past 20 years, a great deal has been learned about the enzymatic activities of the RAG1-RAG2 complex. Recent studies have revealed several new and exciting regulatory functions of the RAG1-RAG2 complex. Here we discuss some of these functions and suggest that the RAG1-RAG2 complex nucleates a specialized subnuclear compartment that we term the V(D)J recombination factory.
To generate a diverse repertoire of antigen receptors, developing B cells and T cells undergo a complex series of DNA rearrangements collectively termed V(D)J recombination. This process is initiated by the lymphoid-specific proteins RAG1 and RAG2, which function together to generate site-specific DNA double-strand breaks that are then repaired via the classical non-homologous end-joining (NHEJ) pathway. While it is well established that RAG1-RAG2 can function as a recombinase, several recent studies have revealed that RAG1-RAG2 is actually a surprisingly multifaceted enzyme complex that plays an important role in ensuring that V(D)J recombination is faithfully executed and properly regulated in the cell. In this Review, we discuss the role of the RAG1-RAG2 complex in binding to accessible chromatin, mediating allelic pairing during V(D)J recombination, and channeling RAG-generated double-strand breaks towards the classical non-homologous end-joining (NHEJ) pathway (Fig. 1). We conclude by proposing a speculative model in which the RAG1-RAG2 recombinase functions within a specialized subnuclear compartment that we term the V(D)J recombination factory.
Figure 1. Multilayered regulation of V(D)J recombination.
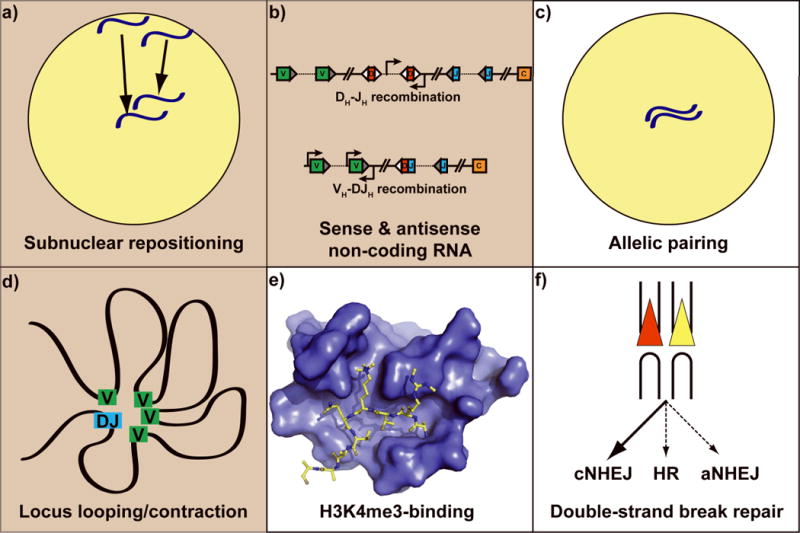
Regulatory mechanisms that are RAG-dependent are shown in white panels (c, e, and f), and those that are RAG-independent are in tan (a, b, and d). a) Subnuclear localization of antigen receptor loci. Within the nucleus (yellow circle) the homologous Igh alleles are depicted in blue. Arrows indicate the centripetal movement of Igh alleles prior to initiation of V(D)J recombination at this locus; b) Sense and antisense non-coding RNA. VH (green), DH (red), JH ( blue), and Cμ (orange) gene segments are shown. Right- and left-facing arrows represent sense and antisense transcription start sites, respectively; c) Allelic pairing. As in a, the nucleus is shown as a yellow circle and homologous Igh alleles are depicted in blue; d) Locus looping and contraction. The curved black line is the Igh locus, with VH gene segments in green and rearranged DJH gene segments in blue. As a result of chromosomal looping, the distance between VH gene segments and the DJH gene segment is decreased; e) Recognition of histone H3 methylation. A space-filling model of the RAG2 PHD finger is shown in blue, bound to the N-terminal tail of histone H3 (shown in yellow), which is trimethylated at lysine 4 and symmetrically dimethylated at arginine 2; f) Preferential usage of the classical NHEJ (cNHEJ) double-strand break repair pathway. The RAG recombinase aids in directing V(D)J recombination intermediates to cNHEJ and away from homologous recombination (HR) and alternative NHEJ (aNHEJ). Signal ends: red and yellow triangles; coding ends: U-shaped black lines.
RAG1-RAG2 ‘reads’ marks of accessible chromatin
Given the importance of generating functional B cell antigen receptors (BCRs) and T cell antigen receptors (TCRs) and the need to protect the integrity of the genome from inappropriate RAG-catalyzed double-strand breaks, it is not surprising that the process of V(D)J recombination is strictly regulated at multiple different levels. Specifically, V(D)J recombination exhibits tissue-specificity with recombination only occurring in lymphoid cells, lineage-specificity with complete immunoglobulin (Ig) gene rearrangement restricted to B cells and complete TCR gene rearrangement restricted to T cells, and developmental stage-specificity such that rearrangement only occurs in pro-B, pro-T, pre-B and pre-T cells. Additionally, V(D)J recombination is temporally ordered, such that D-J rearrangement precedes V-DJ rearrangement at the Igh and Tcrb loci, and cell cycle-regulated, with recombination restricted to the G0/G1 stage of the cell cycle. Finally, V(D)J recombination is subject to allelic exclusion, where productive rearrangement on one allele prevents rearrangement on the other allele, and isotypic exclusion of the light chain loci, by which productive rearrangement of Igk prevents rearrangement of Igl and vice versa. As the same RAG1-RAG2 recombinase catalyzes all V(D)J recombination events, and yet only a limited subset of all recombinationally competent gene segments are actually rearranged in a given cell at a given time, V(D)J recombination must be regulated by modulating the accessibility of the various antigen receptor loci to the RAG proteins. Although it is widely believed that alterations in chromatin structure are utilized to achieve this control, mechanisms linking specific aspects of chromatin structure to the function of the RAG recombinase have remained elusive. However, several new studies have provided insight into how chromatin structure may regulate V(D)J recombination.
Recently, an exciting new link between histone methylation and V(D)J recombination was identified. Using a variety of biochemical and biophysical assays, it was demonstrated that RAG2 contains a noncanonical plant homeodomain (PHD) finger that specifically recognizes histone H3 trimethylated at lysine 4 (H3K4me3) 1, 2. Chromatin immunoprecipitation (ChIP) analysis revealed that H3K4me3 is enriched at gene segments poised to undergo V(D)J recombination2, 3. Interestingly, the pattern of H3K4me3 enrichment is consistent with the tissue-specificity, lineage-specificity and temporal ordering of antigen receptor gene rearrangement. For example, H3K4me3 is not present at the Tcrb or Igh loci in fibroblasts, and the Igh locus only exhibits H3K4me3 enrichment in pro-B cells while the Tcrb locus only exhibits H3K4me3 enrichment in pro-T cells. Moreover, the deposition of H3K4me3 and the initiation of recombination are essentially temporally coincident3. A high-resolution crystal structure of the RAG2 PHD finger bound to H3K4me3 revealed the molecular basis of H3K4me3-recognition and facilitated a functional analysis of the RAG2-H3K4me3 interaction (Fig. 1e)2, 4. Strikingly, mutations that abrogate RAG2 recognition of H3K4me3 severely impair V(D)J recombination in vivo1, 2. Similarly, reducing the abundance of H3K4me3 also leads to a marked decrease in V(D)J recombination2. Taken together, the data in these studies demonstrate that recognition of H3K4me3 by RAG2 is crucial for V(D)J recombination.
Although these studies revealed an intriguing new link between chromatin structure, the RAG recombinase, and V(D)J recombination, many questions remain. For example, given that H3K4me3 is found at many locations throughout the genome, how does the RAG1-RAG2 complex get specifically targeted to the antigen receptor loci? One potential answer is suggested by the crystal structure of the RAG2 PHD finger (RAG2PHD) bound to H3K4me32, 4. Comparing the crystal structures of the PHD finger of RAG2 to the PHD fingers of the transcription factors BPTF and ING2 bound to H3K4me3 suggested that whereas BPTF and ING2 can only recognize histone H3 lysine 4 trimethylation when the nearby arginine 2 residue is unmethylated (H3R2me0K4me3), RAG2 might be able to recognize histone H3 lysine 4 trimethylation when arginine 2 is also methylated. Consistent with this prediction, RAG2PHD not only recognizes histone H3 when it is symmetrically dimethylated at arginine 2 and trimethylated at lysine 4 (H3R2me2sK4me3)4, but it actually binds this dually-modified form of histone H3 more tightly than it binds to H3K4me35. Furthermore, the ability of ING2 to bind H3K4me3 was dramatically impaired by methylation of arginine 24. Therefore, if poised antigen receptor loci are marked by H3R2me2sK4me3 while other portions of the genome are only marked by H3K4me3, then H3R2me2s might serve as a specificity factor that helps recruit the RAG1-RAG2 complex to the antigen receptor loci, and exclude other H3K4me3-binding complexes from these sites. Future experiments will elucidate the functional role of H3R2me2s in V(D)J recombination.
How does RAG2 recognition of H3K4me3 promote V(D)J recombination? There seem to be three general possibilities. First, the RAG2-H3K4me3 interaction may be required for recruiting and/or retaining the RAG1-RAG2 complex at poised antigen receptor loci. This recruitment and/or retention function could be important prior to initial RSS binding and/or during synapsis of a 12/23 RSS pair. Second, the RAG2-H3K4me3 interaction may induce a conformational change in the RAG1-RAG2 complex, thereby allosterically activating the recombinase and stimulating chromosomal V(D)J cleavage. Such activation could occur via a simple conformational change or it could occur via displacement of an inhibitory group. Intriguingly, it has been shown that unlike the PHD fingers of BPTF and ING2, which have closed H3K4me3-binding “cages,” RAG2 has an H3K4me3-binding channel that is open at the top. Furthermore, in the absence of an H3K4me3 peptide, this open channel can be filled from above with another peptide that positions a proline into the aromatic pocket that normally interacts with the trimethylated lysine 4 residue4. This finding suggests the potential for auto-regulation of RAG2 function. Perhaps, in the absence of H3K4me3, a disordered loop of RAG2 folds back upon itself in such a way that the H3K4me3-binding groove is filled from above by a self-peptide. With RAG2 in this conformation, the RAG1-RAG2 complex might exhibit suboptimal catalytic properties. However, in the presence of H3K4me3 (or H3R2me2s/K4me3), the methylated H3 tail would fill the H3K4me3-binding groove from below, displacing the self-peptide, and increasing the rate of V(D)J cleavage. Third, the RAG2-H3K4me3 interaction might stabilize the RAG1-RAG2 post-cleavage complex, helping the recombinase to bind cleaved signal and coding ends, thereby facilitating efficient ‘hand-off’ of V(D)J recombination intermediates to the NHEJ machinery. A great deal of further work will be required to distinguish between these possibilities. Undoubtedly, understanding how the interaction between RAG2 and H3K4me3 regulates chromosomal V(D)J recombination will provide important insights into both the mechanism of V(D)J recombination and the broader question of how chromatin-binding modules affect the function of chromosomal molecular machines.
The dependence of V(D)J recombination on H3K4me3 also raises the question of where the modified nucleosome is positioned with respect to the recombination signal sequence. Does the RSS have to be incorporated into a nucleosome bearing this modification or could it be in close proximity to or even at a considerable linear distance from this modified nucleosome? Although some groups were unable to detect V(D)J cleavage of an RSS within a mononucleosome6, 7, others showed that RAG1-RAG2 can cleave nucleosomal substrates8–10; the latter findings suggest that it is possible for the RSS to be within the modified nucleosome and still participate in V(D)J recombination. On the other hand, the RAG2 PHD finger is separated from the RAG2 core by a flexible hinge which might allow RAG2 to bind H3K4me3 via its PHD finger while the RAG1-RAG2 complex contacts the RSS present in the linker DNA. Of course, depending on what role H3K4me3 plays in the reaction, the time at which H3K4me3 binding is important might be temporally distinct from the actual catalytic steps of the reaction. Future experiments should help to distinguish amongst these possibilities.
The functional importance of the RAG2- H3K4me3 interaction for V(D)J recombination also provides a new insight into the potential role of germline transcription, which is observed at antigen receptor loci and appears to precede V(D)J recombination at these loci (Fig. 1b). This so-called “sterile” transcription has long been known to occur, but its functional importance and relationship to V(D)J recombination has remained unclear. The principal mechanism for trimethylating H3K4 requires transcription, with the MLL-family of methyltransferases being recruited near promoters via recognition of the form of RNA polymerase II whose C-terminal domain is phosphorylated at serine 5. Therefore, the primary function for germline transcription might be to mark these loci with H3K4me3 to allow for efficient V(D)J recombination. If this is the case, an additional question is raised: how is H3K4me3 deposited across a large region of the locus, as opposed to being restricted to the promoter region of the few known germline transcripts that appear prior to V(D)J recombination? At the murine Igh locus, for example, H3K4me3 is present across the 80 kb DJ domain2, even though there is only one well-characterized, active promoter (DQ52) in this region prior to DH-JH recombination11. This discrepancy suggests that there are several, previously unidentified, promoters interspersed throughout the DJ domain (e.g. the poorly-defined promoters driving antisense transcription within this region12, 13) or that there is an additional mechanism that allows H3K4me3 to be deposited at a greater distance from the DQ52 promoter than has been observed for H3K4me3 deposition at other genes. This function of germline transcription in V(D)J recombination remains to be explored.
Finally, this study expands the range of regulatory functions that can be ascribed to the C-terminal, “non-core” portion of RAG2 (aa 384–527). In addition to recognizing methylated histone H3, the C-terminal portion of RAG2 also binds to phosphoinositides14, suppresses RAG transposition15–18, restricts V(D)J recombination to the G0/G1 stage of the cell cycle19, and regulates VH-DJH recombination20–22. We note that RAG2 PHD finger point mutants that are specifically deficient in H3K4me3-binding are more impaired in DH-JH recombination than core RAG2 (aa 1–383), which lacks the entire C-terminal regulatory region2. This apparent paradox may, in part, reflect the fact that core RAG2 lacks the G1/S degradation signal23, and therefore accumulates to much higher quantities in developing B cells than the RAG2 PHD finger mutants2. Thus, if the RAG2-H3K4me3 interaction plays a role in targeting the RAG1-RAG2 recombinase to DH and JH gene segments, core RAG2 might be able to partially overcome its inefficient targeting by simple mass action. Additionally, if RAG2 is subject to PHD finger-dependent auto-inhibition as discussed above, then core RAG2 would be more efficient at V(D)J recombination than PHD finger mutants that are constitutively auto-inhibited due to their inability to bind H3K4me3. This phenomenon, wherein the presence of an impaired regulatory domain is more deleterious than its absence, was also seen in studies of nuclear hormone receptors24 and Src kinase25. In any case, it seems clear that the C-terminal portion of RAG2 is an important regulatory region that affects V(D)J recombination in a variety of interesting ways.
RAG1-RAG2 mediates allelic pairing
While it is clear that localized molecular events such as histone methylation play an important role in regulating V(D)J recombination, it is becoming increasingly apparent that V(D)J recombination is also regulated by large-scale chromosome dynamics. In pro-B cells, which are poised to undergo V(D)J recombination, the Igh loci are repositioned from the peripheral nuclear lamina to a central region of the nucleus (Fig. 1a)26, where these loci then undergo large-scale contraction26 via a DNA looping mechanism(Fig. 1d)27, 28. A similar process of locus contraction also occurs at the Tcra and Tcrb loci in developing thymocytes29. Igh locus contraction is dependent upon the interleukin 7 receptor26 and the transcription factors Pax530, YY131, and Ikaros32, and may play a role in regulating the accessibility of distal VH gene segments during VH-DJH recombination. Although several factors have been implicated in Igh locus contraction, little is known about the initial process of subnuclear repositioning of the Igh locus. Nonetheless, it seems clear that large-scale chromosome dynamics play an important role in regulating antigen receptor loci accessibility during lymphoid development.
A very recent study extends this theme, demonstrating that during B cell development, homologous immunoglobulin alleles pair up in a lineage-specific and developmental stage-specific manner that correlates with the recombinational activity of the immunoglobulin loci (Fig. 1c)33. That is, in pre-pro-B and pro-B cells, which are undergoing DH-JH and VH-DJH recombination respectively, the two Igh alleles pair with each other, while in pre-B and immature B cells, which are undergoing Vκ-Jκ recombination, the two Igk alleles pair with each other. Surprisingly, this allelic pairing seems to be dependent upon RAG1 expression, but independent of the RAG1-RAG2-catalyzed V(D)J cleavage because it still occurs with a catalytically inactive form of RAG1. Thus, the RAG1-RAG2 complex seems to play a role in mediating allelic pairing during B cell development.
Of note, the role of RAG2 in this pairing, or more specifically, the histone-binding2 and phosphoinositide-binding14 activities of the RAG2 PHD finger, have not yet been explored. The question of how the RAG1-RAG2 complex influences allelic pairing also remains. Does it directly bind and synapse the homologous Ig alleles? Or does it play an indirect role, perhaps by serving as a molecular scaffold that interacts with other factors once these factors have bound to the Ig loci? Complementing Rag1−/− cells with binding-deficient RAG1 mutants34–36 could help to distinguish between these possibilities. Also, what other factors are involved in the allelic pairing process? And finally, if the purpose of allelic pairing is to allow the two alleles to communicate with each other, thereby ensuring that productive recombination only occurs on one of the two immunoglobulin alleles, how is information transferred from one allele to the other? That is, how does allelic pairing enable one allele to assess the recombinational status of the other allele? Although many questions remain, the finding that the RAG1-RAG2 complex plays a role in mediating allelic pairing of immunoglobulin loci during B cell development is very exciting and certainly expands our view of how the RAG1-RAG2 complex helps to regulate V(D)J recombination.
RAG1-RAG2 promotes classical non-homologous end-joining
During lymphoid development, the RAG1-RAG2 complex initiates V(D)J recombination by binding to a pair of recombination signal sequences (RSS) and generating DNA double-strand breaks at the RSS-coding joint junctions. These double-strand breaks are then repaired via non-homologous end-joining to form a precise signal joint and an imprecise coding joint. It is well-established that the classical NHEJ (cNHEJ) machinery is required to properly repair the programmed double-strand breaks generated during V(D)J recombination. It has also been shown that mutations in RAG135, 37, 38 and RAG239 can impair the repair of signal ends and coding ends. However, it has been rather unclear why RAG-generated double-strand breaks are repaired by the classical NHEJ pathway as opposed to the alternative NHEJ (aNHEJ) pathway, which relies on microhomology between the broken ends that are being repaired.
A recent report suggests that RAG2 may play an important role in channeling V(D)J recombination intermediates towards the classical NHEJ repair pathway (Fig. 1f)40. Using a cellular V(D)J recombination assay that distinguishes between classical non-homologous end-joining and microhomology-driven alternative non-homologous end-joining, it was demonstrated that a RAG2 frameshift mutant (FS361) that is missing 22 residues more than are absent from RAG2core exhibits a greatly increased frequency of alternative NHEJ-mediated repair of signal ends and coding ends. This mutant allows V(D)J recombination intermediates to access the alternative NHEJ pathway in wild-type cells, where the classical NHEJ machinery is intact, and it also rescues the end-joining defects found in cNHEJ-deficient cells. The fact that this mutation allows broken ends to be repaired by the alternative NHEJ pathway, even when the classical NHEJ machinery is fully functional, suggests that the C-terminal portion of RAG2 plays a key role in channeling RAG-generated DNA double-strand breaks to the classical NHEJ pathway. Moreover, the observation that the FS361 mutant exhibits significantly more aNHEJ-mediated repair of signal ends and coding ends than does RAG2core suggests that residues 362–383 of RAG2 may play an especially important role in directing RAG-generated double-strand breaks to the classical NHEJ pathway.
How does the C-terminal portion of RAG2 determine whether V(D)J recombination intermediates are repaired via classical or alternative NHEJ? Perhaps the C-terminal portion of RAG2 plays a role in stabilizing the RAG1-RAG2 post-cleavage complex, as has been demonstrated for other joining-deficient RAG mutants37, 41. Alternatively, as it has recently been found that the N-terminal portion of RAG1 forms protein-protein interactions (either directly or indirectly) with two components of the cNHEJ machinery (Ku70 and Ku80)42, perhaps the C-terminal portion of RAG2 also interacts with components of the cNHEJ machinery. Such an interaction could promote the efficient transfer of signal ends and coding ends from the RAG1-RAG2 post-cleavage complex to the cNHEJ pathway. Future studies will determine the mechanism(s) by which the C-terminal portion of RAG2 influences the fate of RAG-generated double-strand breaks.
Model
Given the myriad functions required to effect the complex chromosomal gymnastics that occur during antigen receptor gene rearrangement, including subnuclear repositioning26, allelic pairing and unpairing33, reversible locus contraction26, 29, dynamic chromosomal looping27, 28, and long-range 12/23 synapsis and double-strand break repair, one might wonder how these various functions are integrated within the nucleus to ensure the faithful execution of V(D)J recombination. We hypothesize that antigen receptor gene assembly may occur within a specialized subnuclear compartment that we will term the V(D)J recombination factory (Fig. 2). This specialized compartment, which likely exists within a central, euchromatic region of the nucleus26, would localize the factors required to alter the large-scale chromosomal architecture of the antigen receptor loci (e.g. Pax530, YY131, Ikaros32, ATM43, and potentially many others), the factors required to alter the local chromatin structure of the antigen receptor loci (e.g. ATP-dependent chromatin remodeling enzymes9, 10, 44, 45, H3/H4 lysine-acetyltransferases (KATs)9, 45–48, and H3K4 lysine-methyltransferases (KMTs)1, 2), as well as the RAG1-RAG2 recombinase and the cNHEJ machinery. We hypothesize that the V(D)J recombination factory exists as a single subnuclear compartment that is uniquely equipped to carry out the process of chromosomal V(D)J recombination. Only a single compartment would be required since typically, only a single antigen receptor locus is undergoing V(D)J recombination at any given point in time. Such a unique site would be in contrast to transcription factories, which seem to exist at multiple sites within the nucleus likely reflecting that, at any point in time, many different genes are being transcribed from several different chromosomes. In light of the many ways that RAG1-RAG2 complex seems to influence other processes involved in V(D)J recombination including allelic pairing and unpairing and choice of double-strand break repair pathways, and given that the RAG1-RAG2 recombinase is the central protein machine driving V(D)J recombination, we suggest that the RAG1-RAG2 complex is the organizing factor that nucleates the V(D)J recombination factory. Other factors would then be recruited to and retained within this subnuclear compartment. By physically bringing together all of the factors that influence chromosomal V(D)J recombination, and concentrating them within a V(D)J recombination factory, the cell could functionally couple the numerous processes that occur during V(D)J recombination, thereby enhancing both the efficiency and the regulation of antigen receptor gene rearrangement. If the cell excludes non-antigen receptor loci from this subnuclear compartment, then the V(D)J recombination factory might also help to minimize the formation of spurious RAG-generated double-strand breaks at cryptic RSSs49, 50 and non-B-form DNA51 throughout the genome, as well as decrease the opportunity for deleterious interchromosomal translocations52–55.
Figure 2. Model depicting a putative V(D)J recombination factory.
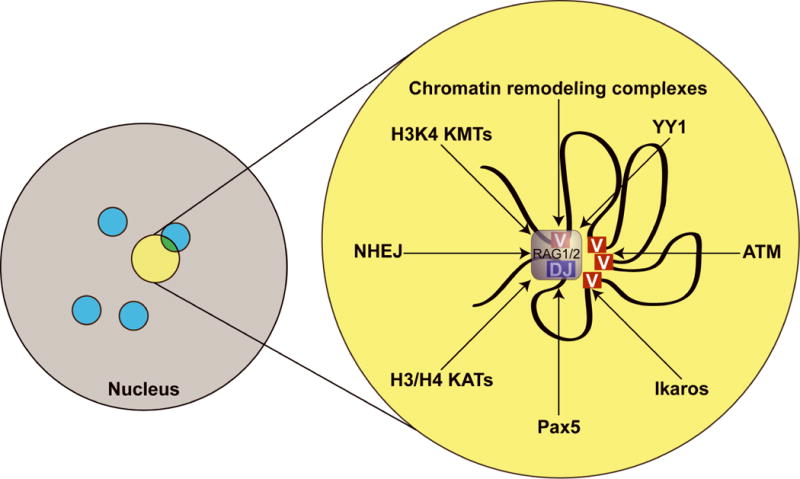
Within the nucleus (gray circle), the V(D)J recombination machinery may exist as a specialized, centrally-located, subnuclear compartment (yellow circle). This compartment is likely to be distinct from transcription factories (blue). An expanded view of the V(D)J recombination factory is shown on the right. Igh locus: curved black line; VH gene segments: red, a rearranged DJH gene segment: blue. The RAG1-RAG2 recombinase (light purple) is shown synapsing the DJH gene segment and one of the VH gene segments just prior to VH-DJH recombination. Factors that are likely to be enriched within the V(D)J recombination factory are indicated. The central position of the RAG recombinase within this figure reflects the hypothesis that the RAG1-RAG2 complex nucleates the factory, while arrows pointing towards the RAG1-RAG2 complex represent the recruitment of additional regulatory factors by the RAG1-RAG2 complex.
Conclusion
This is an exciting time in the field of V(D)J recombination. Recent studies have demonstrated that the RAG1-RAG2 complex is a talented and versatile molecular machine that does much more than simply catalyze the phosphoryl transfer reactions required to generate DNA double-strand breaks. In addition to cutting DNA, the RAG1-RAG2 complex also interacts with H3K4me3, plays a role in immunoglobulin allelic pairing, and helps determine whether the broken ends generated during V(D)J recombination are repaired via classical NHEJ, alternative NHEJ, or homologous recombination. A great deal of future research will be required to unravel the mechanisms underlying these processes and to test the hypothesis that antigen receptor gene assembly occurs within specialized V(D)J recombination factories. Nonetheless, it seems clear that in addition to its central role in the development of the vertebrate adaptive immune system, V(D)J recombination is also becoming a valuable system to study many fundamental questions pertaining to chromatin accessibility, nuclear dynamics, long-range chromosomal interactions, and DNA double-strand break repair.
Acknowledgments
We apologize to those whose work we were unable to cite because of space limitations. Supported by the US National Institutes of Health (M.A.O.). A.G.W.M. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1981-08).
References
- 1.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perkins EJ, Kee BL, Ramsden DA. Histone 3 lysine 4 methylation during the pre-B to immature B-cell transition. Nucleic acids research. 2004;32:1942–1947. doi: 10.1093/nar/gkh523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramon-Maiques S, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews AG, Oettinger MA. unpublished observation. [Google Scholar]
- 6.Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. Embo J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. Embo J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Molecular cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 9.Kwon J, Morshead KB, Guyon JR, Kingston RE, Oettinger MA. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Molecular cell. 2000;6:1037–1048. doi: 10.1016/s1097-2765(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 10.Patenge N, Elkin SK, Oettinger MA. ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. The Journal of biological chemistry. 2004;279:35360–35367. doi: 10.1074/jbc.M405790200. [DOI] [PubMed] [Google Scholar]
- 11.Alessandrini A, Desiderio SV. Coordination of immunoglobulin DJH transcription and D-to-JH rearrangement by promoter-enhancer approximation. Molecular and cellular biology. 1991;11:2096–2107. doi: 10.1128/mcb.11.4.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolland DJ, et al. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Molecular and cellular biology. 2007;27:5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty T, et al. Repeat organization and epigenetic regulation of the DH-Cmu domain of the immunoglobulin heavy-chain gene locus. Molecular cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Elkin SK, et al. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. The Journal of biological chemistry. 2005;280:28701–28710. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 15.Matthews AG, Elkin SK, Oettinger MA. Ordered DNA release and target capture in RAG transposition. Embo J. 2004;23:1198–1206. doi: 10.1038/sj.emboj.7600131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkin SK, Matthews AG, Oettinger MA. The C-terminal portion of RAG2 protects against transposition in vitro. Embo J. 2003;22:1931–1938. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai CL, Schatz DG. Regulation of RAG1-RAG2-mediated transposition by GTP and the C-terminal region of RAG2. Embo J. 2003;22:1922–1930. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson PC, Volkmer D, Wang L. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. The Journal of biological chemistry. 2004;279:4034–4044. doi: 10.1074/jbc.M311100200. [DOI] [PubMed] [Google Scholar]
- 19.Lin WC, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci U S A. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akamatsu Y, et al. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc Natl Acad Sci U S A. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang HE, et al. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 22.Kirch SA, Rathbun GA, Oettinger MA. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. Embo J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 24.Jurutka PW, et al. Molecular endocrinology. Vol. 14. Baltimore, Md: 2000. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB; pp. 401–420. [DOI] [PubMed] [Google Scholar]
- 25.Hubbard SR, Mohammadi M, Schlessinger J. Autoregulatory mechanisms in protein-tyrosine kinases. The Journal of biological chemistry. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]
- 26.Kosak ST, et al. Science. Vol. 296. New York, N.Y: 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development; pp. 158–162. [DOI] [PubMed] [Google Scholar]
- 27.Sayegh CE, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes & development. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jhunjhunwala S, et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skok JA, et al. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nature immunology. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 30.Fuxa M, et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes & development. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes & development. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynaud D, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nature immunology. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hewitt SL, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nature immunology. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 35.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Molecular and cellular biology. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spanopoulou E, et al. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 37.Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes & development. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarnell Schultz H, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Molecular cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 39.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Molecular cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 40.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 41.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 42.Raval P, Kriatchko AN, Kumar S, Swanson PC. Evidence for Ku70/Ku80 association with full-length RAG1. Nucleic acids research. 2008;36:2060–2072. doi: 10.1093/nar/gkn049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hewitt SL, et al. RAG1 and ATM coordinate mono-allelic recombination and nuclear positioning of immunoglobulin loci. Nature immunology. doi: 10.1038/ni.1735. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osipovich O, et al. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nature immunology. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- 45.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Molecular and cellular biology. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMurry MT, Krangel MS. Science. Vol. 287. New York, N.Y: 2000. A role for histone acetylation in the developmental regulation of VDJ recombination; pp. 495–498. [DOI] [PubMed] [Google Scholar]
- 48.McBlane F, Boyes J. Stimulation of V(D)J recombination by histone acetylation. Curr Biol. 2000;10:483–486. doi: 10.1016/s0960-9822(00)00449-8. [DOI] [PubMed] [Google Scholar]
- 49.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Molecular and cellular biology. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marculescu R, Le T, Simon P, Jaeger U, Nadel B. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. The Journal of experimental medicine. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 52.Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes & development. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Molecular and cellular biology. 2006;26:1558–1568. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Messier TL, O’Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. Embo J. 2003;22:1381–1388. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marculescu R, et al. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA repair. 2006;5:1246–1258. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]


