Abstract
The carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII) in mammals undergoes extensive posttranslational modification, which is essential for transcriptional initiation and elongation. Here, we show that the CTD of RNAPII is methylated at a single arginine (R1810) by the coactivator-associated arginine methyltransferase 1 (CARM1). Although methylation at R1810 is present on the hyperphosphorylated form of RNAPII in vivo, Ser2 or Ser5 phosphorylation inhibits CARM1 activity toward this site in vitro, suggesting that methylation occurs before transcription initiation. Mutation of R1810 results in the misexpression of a variety of small nuclear RNAs and small nucleolar RNAs, an effect that is also observed in Carm1−/− mouse embryo fibroblasts. These results demonstrate that CTD methylation facilitates the expression of select RNAs, perhaps serving to discriminate the RNAPII-associated machinery recruited to distinct gene types.
The carboxy-terminal domain (CTD) of the major subunit of RNA polymerase II (RNAPII) in mammals comprises 52 repeats of the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser (1). Although site-specific CTD phosphorylation mediates recruitment of other proteins to RNAPII, how this recruitment facilitates distinct processing events remains poorly understood (2–4). Nonconsensus repeats of the RNAPII CTD contain two arginine and seven lysine substitutions that primarily occur at position seven of the heptad motif. We hypothesized that such arginine and/or lysine residues might be targets for modification of the CTD of RNAPII and, as a consequence, engage activities associated with RNA production.
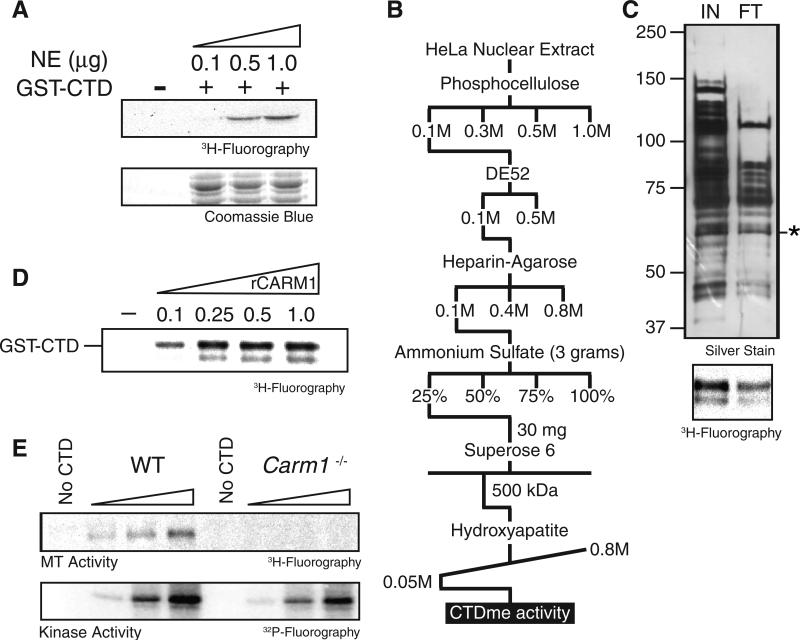
A glutathione S-transferase (GST)–CTD fusion protein containing repeats number 24–52 was not acetylated by HeLa-S3 nuclear extract as a source of enzymes, but specific methylation of the GST-CTD was observed, and its level correlated with increasing amounts of the extract (Fig. 1A). We purified the CTD methyltransferase enzyme from this extract (Fig. 1, B and C), detecting a band at ~65 kD that was crosslinked to a S-adenosyl methionine (SAM) after ultraviolet exposure (fig. S1B) (5). Mass spectrometric analysis revealed the presence of coactivator-associated arginine methyltransferase 1 (CARM1), which migrates at a molecular mass of approximately 65 kD by means of SDS–polyacrylamide gel electrophoresis (SDS-PAGE). To ascertain whether CARM1 was the enzyme that methylates the CTD, we performed methylation reactions using increasing amounts of recombinant CARM1 in the presence of the CTD and confirmed that CARM1 is capable of catalyzing this modification (Fig. 1D). Western blot analysis of the Superose 6 gel filtration fractions derived from conventional purification revealed that CARM1 and the CTD methyltransferase activity co-eluted (fig. S1C) (5). Given that we did not detect any CTD methyltransferase activity that fractionated apart from CARM1 during the purification and that nuclear extracts derived from Carm1−/− mouse embryonic fibroblasts (MEFs) were devoid of this activity (Fig. 1E), we concluded that CARM1 is the enzyme responsible for methylating the CTD. CARM1 is a type I protein arginine methyltransferase (PRMT) that catalyzes a methyltransferase reaction, producing asymmetric dimethylated arginine. Its substrates include histone H3 and p300, and it has been implicated in co-activation of nuclear receptor–directed transcription as well as in mRNA splicing (6,7), although the underlying mechanisms are largely obscure.
Fig. 1.
CARM1 methylates the CTD of RNAPII. (A) (Top) Fluorography after SDS-PAGE analysis of GST-CTD protein methylated by increasing amounts of nuclear extract. (Bottom) Coomassie blue staining as a loading control for the substrate. (B) Schematic showing the chromatographic strategy that was used to identify the CTD methyltransferase activity. (C) (Top) Silver stain of the input and flow-through fractions of the hydroxyapatite chromatographic step. (Bottom) CTD methyltransferase activity of the fractions indicated above. The flow-through fraction was subjected to mass spectrometry, and CARM1 was identified as the CTD methyltransferase activity. The asterisk denotes the molecular weight of CARM1. (D) GST-CTD methylation assay performed in vitro using increasing amounts (micrograms) of recombinant CARM1 (rCARM1). (E) (Top) GST-CTD methylation assay using nuclear extract from WT or Carm1−/− MEF cells. (Bottom) GST-CTD phosphorylation assay with the same extracts.
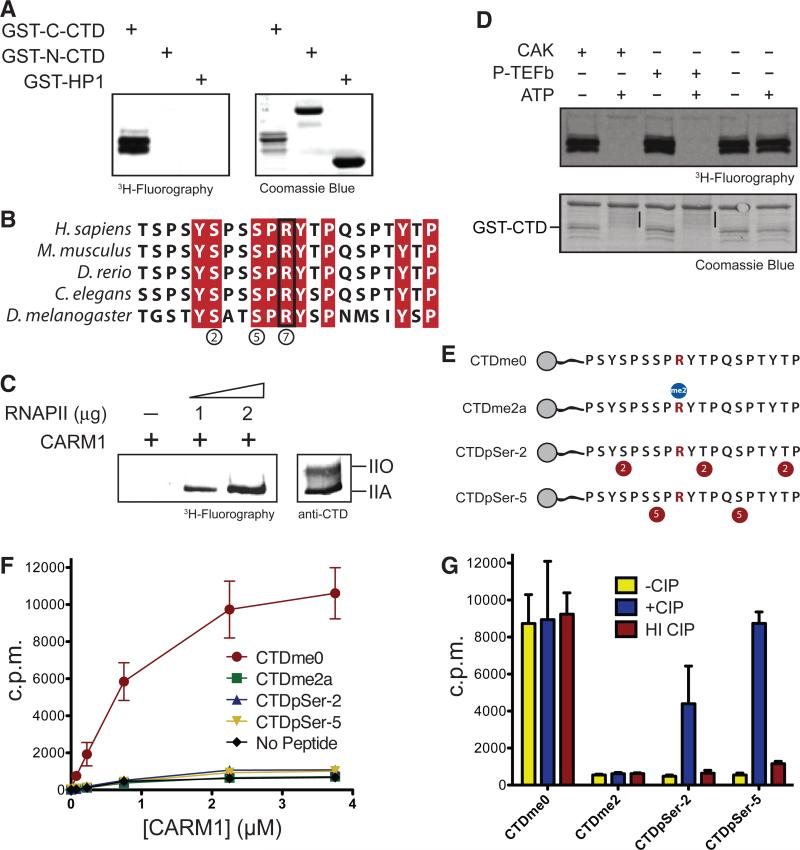
The largest subunit of RNAPII contains two arginine residues within the CTD; one is present within the N-terminal half of the CTD (R1603, second repeat), and the other within the C-terminal half (R1810, repeat number 31) (8). We tested whether both arginines are targets of CARM1 using methylation assays with GST fusion proteins containing either the N-terminal (GST-N-CTD) or C-terminal (GST-C-CTD) portions of the CTD. Only the GST-C-CTD substrate was methylated by recombinant CARM1, indicating that CARM1 targets R1810 (Fig. 2A). An alignment of the residues surrounding R1810 from different species suggests that CARM1 methylation may be conserved throughout evolution (Fig. 2B).
Fig. 2.
The hypophosphorylated form of the RNAPII CTD is methylated on a single arginine residue, and phosphorylated CTD cannot be methylated in vitro. (A) In vitro methylation assays with rCARM1 using GST-C-CTD and GST-N-CTD, with GST-HP1 as control. The reaction products were detected with fluorography (left). (B) Alignment of human CTD-R1810 with CTD sequences from the species indicated. Red columns indicate conserved residues. (C) (Left) Methylation assay using recombinant CARM1 and highly purified RNAPII from HeLa-S3 nuclei. (Right) Western blot using highly purified RNAPII and an antibody to the CTD that recognizes both IIA and IIO forms. (D) Methylation assays on GSTCTD phosphorylated by the indicated kinases. (Bottom) Black bars denote the change in mobility when the CTD is phosphorylated. (E) Schematic of biotinylated CTD peptides used. Red circles indicate phosphorylation sites, and the blue circle indicates the methylation site. (F) Methylation of CTD peptides shown in (E) in the presence of increasing amounts of rCARM1, as scored with scintillation counting. Points represent the mean T SD of three different experiments. (G) Scintillation counting of CTD peptide methylation as in (F), after treatment with calf intestine phosphatase (CIP) or heatinactivated (HI) CIP. Bars represent the mean T SD of three different experiments.
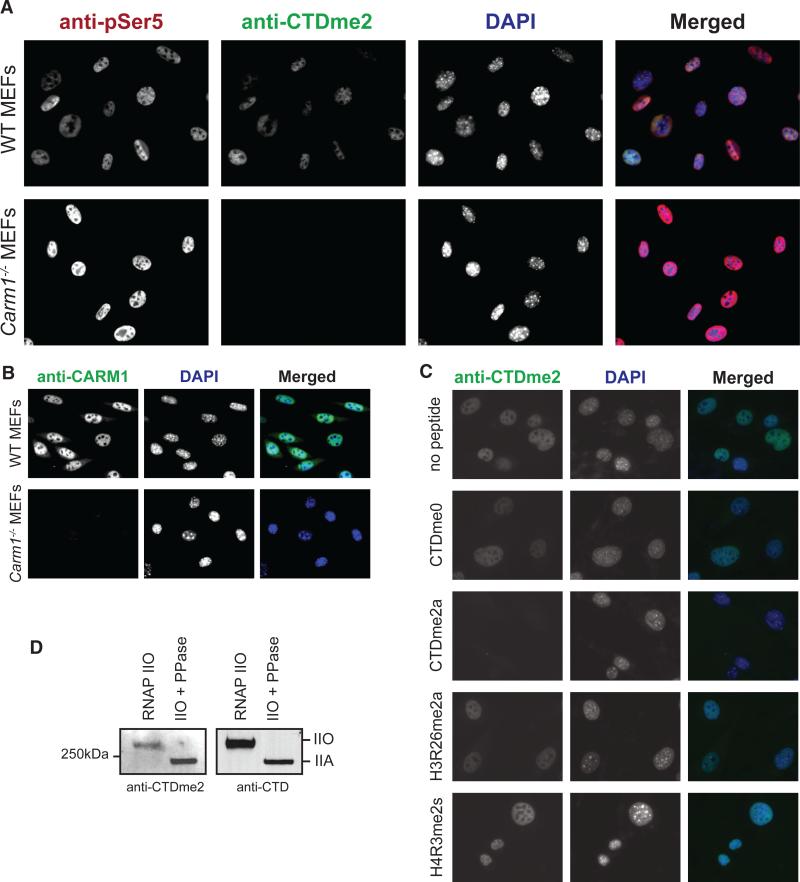
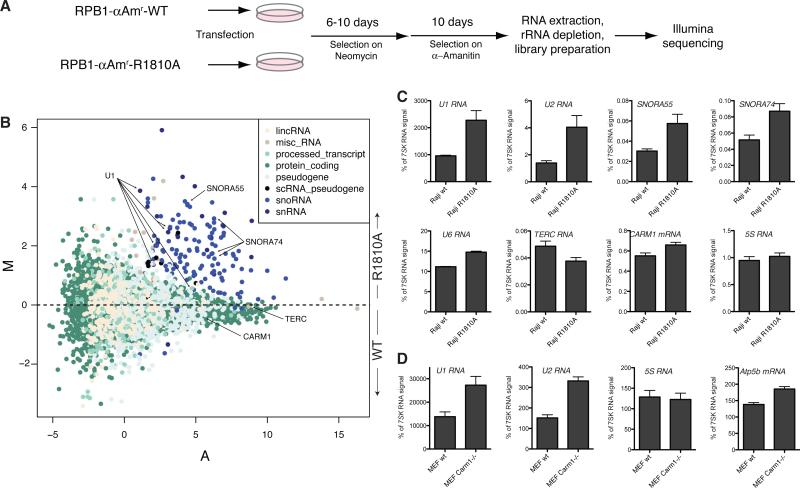
CARM1 methylated highly purified RNAP II derived from HeLa-S3 cells (Fig. 2C): Only the hypophosphorylated (IIA) form of RNAPII was methylated; the hyperphosphorylated form of RNAPII (IIO) was not. Methylation experiments by use of the CTD fusion protein that was prephosphorylated by Ser5- (CAK) or Ser2-specific (P-TEFb) kinases suggest that CARM1 methylation is sensitive to Ser2 and Ser5 phosphorylation (Fig. 2D). Furthermore, CARM1 was ineffectual when provided with synthetic peptides that were phosphorylated at Ser2 or Ser5 as substrates (Fig. 2, E and F). Phosphatase treatment of these peptides restored their ability to be methylated by CARM1 (Fig. 2G). The CTD of RNAPII is phosphorylated during the initiation of transcription; CARM1-mediated methylation probably takes place before phosphorylation, which is consistent with its recruitment during the early phases of transcriptional activation (9, 10). Immunofluorescence analyses performed on MEF cells by using a polyclonal antibody against a CTD peptide containing R1810me2a (fig. S2, A and B) showed that this modification is localized in the nuclei, which is similar to the case of phosphorylated CTD (anti-pSer5, clone 4H8) (Fig. 3A). Carm1−/− MEFs were devoid of CARM1, as expected (Fig. 3B) and also devoid of methylated CTD, as evidenced by the absence of signal using antibodies to CTDRme2a (anti-CTDRme2a) (Fig. 3A), which supports the conclusion that the CTD is methylated in vivo by CARM1. The anti-CTDme2a signal observed by using MEF cells was blocked by CTDme2a peptide but not by other synthetic peptides that are analog to CARM1 products, namely H3R26me2a or a symmetrically methylated peptide H4R3me2s (Fig. 3C). Using this antibody on Western blots, we detected methylation on hyperphosphorylated RNAPII purified from HeLa cells (Fig. 3D). Thus, CARM1 methylates the CTD of RNAPII in vivo and does so before the CTD is phosphorylated. Yet given that the methylated CTD is a substrate for subsequent phosphorylation and that phosphorylated RNAPII exhibits this methylation state (Fig. 3D), it is highly probable that R1810me is preserved on the transcribing polymerase, not having a major impact on CTD phosphorylation. To address the function of RNAPII methylation, we generated cell lines expressing an RNAPII resistant to a-amanitin and carrying either wildtype (WT) R1810 or an arginine-to-alanine substitution at that same residue, abolishing R1810 methylation of the CTD (Fig. 4A). In cells cultured in aamanitin, the a-amanitin–resistant mutants fully replaced the functions of endogenous RNAPII, allowing us to study whether gene expression is affected by the absence of R1810me (11). Our transcriptome analysis results suggest that steady state levels of small nuclear RNAs (snRNAs) as well as small nucleolar RNAs (snoRNAs) are affected in a general manner (Fig. 4B and fig. S3). We confirmed these results for a number of candidate transcripts through quantitative reverse transcription polymerase chain reaction (RT-PCR) using total RNA derived from WT or R1810A cells (Fig. 4C). Because a large number of snRNAs and snoRNAs were affected, we asked whether there was a general correlation between the size of the RNA species whose expression was affected as a consequence of the absence of R1810me (fig. S4). The effect appeared specific to snRNAs and snoRNAs, however, because transcripts of comparable size with that of snRNAs and snoRNAs but belonging to different classes of RNA were not upregulated. Also, expression of RNAPIII-transcribed small RNAs, such as 5S and U6, was not affected (Fig. 4B, bottom plots). The misregulation of snRNA expression was also detected in Carm1−/− MEFs through quantitative RT-PCR (Fig. 4D), suggesting that the functions of CARM1 and R1810me are related and that they are conserved, at least in mammals. The observed de-repression of snRNAs and snoRNAs in the absence of R1810me suggests that this CTD modification functions in opposition to the activating effect of Ser phosphorylation (12–14). In addition to the direct function of CARM1 in splicing, in which it modulates the recognition of 5′ splice sites through its interaction with U1 small nuclear ribonucleoprotein complex (15), CARM1 may also contribute to splicing indirectly through modulating the levels of spliceosome components, such as U1 and U2 (Fig. 4).
Fig. 3.
The CTD of RNAPII is methylated in vivo. (A) Immunofluorescenceanalysis of WT and Carm1−/− MEFs using the indicated antibodies. (B) Comparison of WT and Carm1−/− MEFs using antibody to CARM1. (C) The specificity of antibody to CTDme2a is shown as a function of its incubation with the indicated blocking peptides (5 mg/mL) followed by immunofluorescence. (D) Western blot of purified hyperphosphorylated RNAPII from HeLa-S3 nuclei as a function of phosphatase treatment as indicated on top, using the indicated antibodies.
Fig. 4.
R1810A mutation affects snRNA and snoRNA expression. (A) Schematic of the generation of Raji cells containing R1810A mutation and RNA-seq library preparation. (B) MA plot of genome-wide expression changes as determined with RNA-seq. Log2 fold-changes (M) in normalized number of reads are plotted on the y axis, and average intensities (A) are plotted on the x axis. Each dot represents a gene from ENSEMBL59. Colors indicate gene biotypes; only the eight most numerous are depicted here. (C) Quantitative RT-PCR validation on Raji cells carrying WT or R1810A a-amanitin–resistant RNAPII. Top four plots show affected snRNAs and snoRNAs. Bottom four plots show expression levels of RNAs that were unaffected. Bars represent mean + SEM from two (WT) or three (R1810A) biological replicates and two independent cDNA preparations. (D) Quantitative RT-PCR analysis of WT and Carm1−/− MEF cells. Bars represent mean + SEM from two independent RNA preparations from MEFs at different passages.
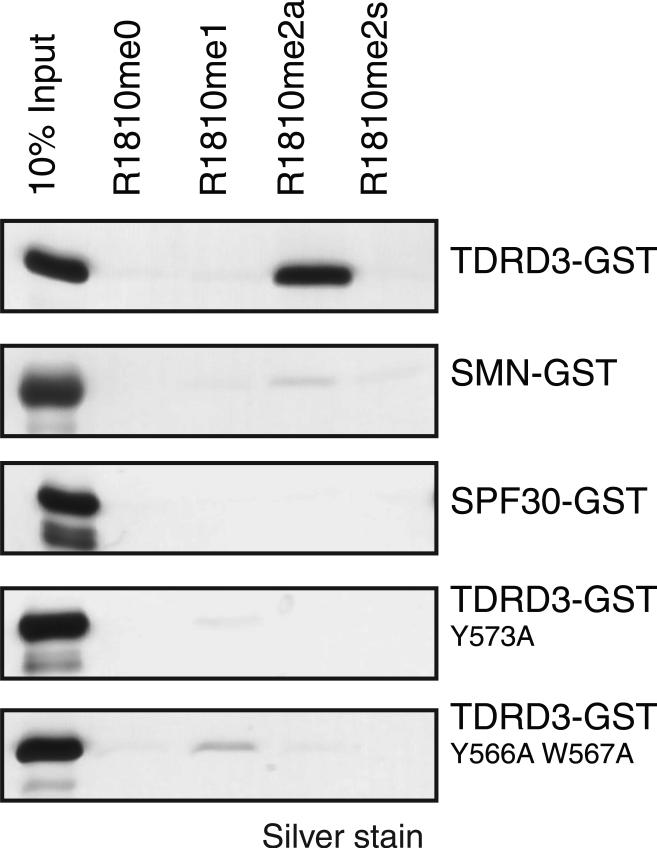
The Tudor domains of several proteins specifically associate with methylated arginine residues (16–18). Tudor domain–containing proteins SMN, SPF30, and TDRD3 bind preferentially to both symmetric and asymmetric dimethyl arginine in the context of Gly-Arg patches (GR motifs). TDRD3 displayed a specific interaction with the R1810me2a-containing CTD peptide, as opposed to the unmodified CTD or mono- or symmetric-dimethylated R1810 (Fig. 5). In contrast, SMN and SPF30 did not exhibit affinity for either the unmodified CTD or any methylated version thereof. TDRD3 can be recruited to chromatin in a CARM1-dependent manner, although it is not clear whether histones or other substrates of CARM1 are recognized by this Tudor domain–containing protein (19). On the basis of the crystal structure of the TDRD3 tudor domain (Protein Data Bank ID 2D9T), we mutated residues typically responsible for binding to methylated arginines (Fig. 5, bottom). The relevant TDRD3 mutants did not bind R1810me2a, demonstrating that TDRD3 interaction with dimethyl-R1810 probably depends on an aromatic cage structure within its tudor domain. However, knockdown of TDRD3 did not result in the same genome-wide alterations of snRNA and snoRNA expression as in the case of either the R1810A mutant or Carm1−/− cells (fig. S5), suggesting that TDRD3 may be involved in a different function of CTD methylation, unrelated to snRNA and snoRNA regulation. We have shown here that the CTD of RNAPII is methylated at R1810 by CARM1 in vitro and in vivo. This modification plays a role in the regulation of snRNA and snoRNA expression. In addition, the tudor domain of TDRD3 specifically recognizes CTD-R1810me2a. Although TDRD3 does not play a role in the regulation of snoRNA expression, it could very well participate in different downstream functions of CTD-R1810me2a.
Fig. 5.
The methylated CTD is recognized selectively by TDRD3. Shown are silver staining of fractions eluted from the immobilized biotinylated peptides containing different modifications at R1810, as indicated on top. TDRD3 tudor domain selectively recognized the R1810me2a peptide. Mutations at the indicated TDRD3 aromatic residues abolished binding.
Supplementary Material
Acknowledgments
These studies were supported by the HHMI (to D.R.) and grants from NIH (GM-37120 to D.R. and GM-71166 to R.J.S.). R.B. is a fellow of the Helen Hay Whitney foundation. D.E. was supported by Deutsche Forschungsgemeinschaft (SFB/Transregio-5, SFB684) and José Carreras LeukämieStiftung eV. We thank L. Vales, E. Lecona, and P. Voigt for careful reading of the manuscript; R. D. Chapman and K. Burger for technical support; and members of the Reinberg lab for critical commentaries during the development of this project. RNA-seq data have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus as GEO series GSE27315.
References and Notes
- 1.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Fong N, Bentley DL. Capping, splicing, and 3. Genes Dev. 2001;15:1783. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell. 1999;3:697. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 4.Ryan K, Murthy KGK, Kaneko S, Manley JL. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3. Mol. Cell. Biol. 2002;22:1684. doi: 10.1128/MCB.22.6.1684-1692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Materials and methods are available as supporting material on Science Online.
- 6.Cheng D, Côté J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell. 2007;25:71. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. J. Biol. Chem. 2005;280:28927. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- 8.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. In the mutants, other amino acids were substituted at certain locations; for example, H134R indicates that histidine at position 134 was replaced by arginine.
- 9.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Métivier R, et al. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 11.Meininghaus M, Chapman RD, Horndasch M, Eick D. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J. Biol. Chem. 2000;275:24375. doi: 10.1074/jbc.M001883200. [DOI] [PubMed] [Google Scholar]
- 12.Baillat D, et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Egloff S, et al. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem. 2010;285:20564. doi: 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egloff S, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J. Biol. Chem. 2005;280:28927. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- 16.Côté J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 17.Shaw N, et al. The multifunctional human p100 protein “hooks’ methylated ligands. Nat. Struct. Mol. Biol. 2007;14:779. doi: 10.1038/nsmb1269. [DOI] [PubMed] [Google Scholar]
- 18.Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: Conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 2003;327:507. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 19.Yang YZ, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell. 2010;40:1016. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.