Abstract
Broadly neutralizing antibodies (bNAbs) against HIV protect from infection, but their routine elicitation by vaccination has not been achieved. To generate small animal models to test vaccine candidates, we have generated targeted transgenic (“knock-in”) mice expressing, in the physiological immunoglobulin heavy (H) and light (L) chain loci, two well-studied bNAbs: 4E10, which interacts with the membrane proximal external region of gp41, and b12, which binds to the CD4 binding site on gp120. 4E10HL mice are described in the accompanying paper. Here, we describe b12 mice. B cells in b12HL mice, in contrast to the case in 4E10 mice, were abundant and essentially monoclonal, retaining the b12 specificity. In cell culture, b12HL B cells responded avidly to HIV Env gp140 trimers and to BCR ligands, but only weakly to HIV pseudovirions. Upon transfer to wild type recipients, b12HL B cells responded robustly to vaccination with gp140 trimers. Vaccinated b12H mice, while generating abundant precursors and antibodies with affinity for Env, were unable to rapidly generate neutralizing antibodies, highlighting the importance of developing antigen forms that better focus responses to neutralizing epitopes. b12HL and b12H mice should be useful in optimizing HIV vaccine candidates to elicit a neutralizing response while avoiding non-protective specificities.
INTRODUCTION
Broadly neutralizing antibodies to HIV, bNAbs, recognize relatively conserved sites on HIV envelope protein (Env; gp120/gp41) that are similar among many isolates and clades (reviewed in 1, 2, 3). bNAbs to HIV arise in as many as 25% of patients after many months of infection (4), though only ~1% are considered “elite neutralizers” (5). However, single bNAbs are not generally protective in the context of chronic HIV infection. High viral loads and the mutability of HIV lead to escape mutants. Nevertheless, because passive transfer of several bNAbs can prevent infection in animal models, bNAbs are predicted to be protective if elicited prior to infection (2). Therefore, the formulation of vaccines capable of eliciting bNAbs is a high priority in efforts to prevent infection. Here we focus on bNAb b12, which was until recently one of the most potent and broadly neutralizing HIV antibodies known.
b12 was initially identified as one of a group of phage-displayed antibodies generated from bone marrow RNA of an asymptomatic HIV patient who had been infected for 6 years (6). Among 32 gp120-binding phages, b12 belonged to a cohort of 4 sharing CDR3 regions in both H and L chains. b12 was back-engineered to encode a full IgG molecule. Its crystal structure was striking because the putative antigen combining site was marked by extreme protrusion of the H-chain CDR3 (7). The structure was interpreted to indicate that the H-chain CDR3 might make major contacts with the CD4 binding site, consistent with H and L chain shuffling experiments, which indicated that b12 H-chain retained specificity when paired with many different L-chains. However, alteration of 4 specific somatically mutated residues in CDR1 and 3 of the b12 L-chain abolished binding to gp120, suggesting that L-chain contributes contacts (8). By contrast, the b12 L-chain when paired with random H-chains from the same library and selected for gp120 binding reselected the same H-chain CDR3 (9). b12 Fab was subsequently cocrystallized with a truncated, disulfide-stabilized core of gp120, revealing a structure in which all contacts were with H-chain (10).
Surprisingly, in addition to its ability to bind to HIV Env, b12 has been suggested to be an autoantibody (11). This conclusion was based mainly on antibody binding studies. (As we discuss in the accompanying paper, this claim was extended to the gp41 antibodies 4E10 and 2F5.) b12 was found to bind to ribonucleoprotein, double stranded DNA, centromere protein, histones, and HEp-2 cells in a cytoplasmic and nucleolar pattern (11). These data raised the possibility that conserved HIV epitopes might evade the immune system by mimicking self and thereby provoking clonal elimination of reactive B cells. The recessed CD4 binding site might tend to require unusually long, extended CDRH3 regions for antibody neutralization. Long CDRH3s have been associated with polyreactivity/autoreactivity (12–14), and might be counterselected by tolerance. However, thus far, the self-reactivity ascribed to b12 is solely based on antibody binding assays, which are subject to a number of technical caveats and do not necessarily correlate with in vivo reactivity or tolerance.
Here we present data on the generation and analysis of knock-in mice in which the variable portions of b12 were introduced to the physiological mouse Ig H and L loci by gene targeting. We find that B cells in these mice retain the transgene-encoded specificity, and appear not to be negatively regulated by immune tolerance. The b12 H-only and H/L mice should prove useful in assessing vaccine candidates for the ability to drive B cell responses.
MATERIALS AND METHODS
Gene targeting
H-chain constructs were targeted in the C57Bl/6-derived C2 ES cell line (15) at TSRI Mouse Genetics Core facility. The L-chain construct was targeted in C57Bl/6-derived ES cells by InGenious Targeting Laboratories, Inc (Ronkonkoma, New York). Briefly, targeting replaced the DQ52-JH cluster in the IgH locus with b12 H-chain VDJ exon, flanked on the 5′ side by a loxP flanked neo gene, and the mouse VHJ558.85.191 promoter, leader, and intron. Similarly, b12 L-chain VJ element was targeted to replace the Jκ cluster. The light chain gene targeting vector was pVKR2neo and L-chain targeting was carried out essentially as described (16). The L-chain VJ sequences were fused on the 5′ end with a promoter, leader and intronic sequence from mouse Vκ4-53 and introduced as a NotI-SalI fragment. The targeting plasmid was then linearized with ClaI.
For H-chain targeting we modified the pEASY-FLIRT vector (17). A 3.9 kb 5′ homology arm was cloned into the NotI site, and a 2.6 kb 3′ homology arm was introduced as an AscI-MluI fragment into the AscI site. A 5′ 1540 bp VHJ558.85.191 promoter region including leader exon and intron was generated. The b12 VDJ was then “sewn” onto the VH promoter using overlapping extension PCR with primers carrying MluI sites. The Promoter-VDJ was then cloned into the AscI site of the targeting vector. Plasmid was linearized using an SfiI site 5′ to the upstream arm of homology and used for electroporation of ES cells, followed by G418 selection. ES cells carrying the desired targetings were identified through a series of PCR reactions carried out with genomic DNA from ES cell clones and using Phire polymerase (Thermo), under the recommended conditions and annealing temperatures listed below. Primer locations are shown in Figures S1 and S2 of the accompanying paper. L-chain targeting screening analysis was with primers L1 (5′-CAAGTAGACCTCTCTAATCTTGGTTAG-3′) and L2 (5′-TTCTATCGCCTTCTTGACGA-3′), 58°C annealing temperature. Candidate clones were further identified by a PCR using primers L6 (5′-CCAAGGAGGGATCATGTGTTTGAAT-3′ and L7 (5′-TCAGTGTCACAGGTAGAGATGGGT-3′), with 63°C annealing. H-chain targetings were identified using primers H1 (5′-AGGTAAGGAGCTCAGCAGTTCCAA-3′) and H2 (5′-GCCAGCCTAGTTTAGCTTAGCGGCCCA-3′), with 63.5°C annealing. Candidate clones were confirmed by a PCR using primers H7 (5′-AGGAGTATTCCTGTTCTCCCATGCCTGCATATCA-3′) and H6 (5′-GCTAAAGCGCATGCTCCAGACTG-3′), with 63.5°C annealing. PCR products from ES cells chosen for mouse generation were verified by sequencing. Neor genes were eliminated in the germline by breeding to EIIa-cre expressing mice. Targetings were confirmed by Southern blotting using SacI or BamH1 digestion and a 2 Kb EcoRI 5′ probe in the case of the L-chain gene or BamH1 digestion and a 0.5 kb Xma-BamH1 3′ probe in the case of the H-chain gene.
Mice
All experiments were performed in accordance with relevant institutional and national guidelines and were approved by the TRSI Institutional Animal Care and Use Committee. C57Bl/6J, EIIa-cre, and IgHa mice on the B6 background (B6.Cg-Igha Thy1a Gpi1a/J) were from Jackson Laboratories.
Flow cytometry analyses
Analyses for surface markers and Ca++ flux assay were performed using standard protocols as previously described (18). The following mAbs were used at 1:200 in all experiments described and were from eBioscience unless otherwise indicated: CD19 (1D3; PE/Cy7), CD21 (7E9; FITC), CD23 (B3B4; PE), CD93 (AA4.1; PE or APC), TCRβ (H57; PerCP-Cy5.5), Gr1(PerCP-Cy5.5), StreptAvidin-PE, B220 (RA3-6B2; FITC; BioLegend), Igλ (RLM-42; APC; BioLegend), StreptAvidin-APC (BioLegend). mAbs against mouse IgM (M41; Alexa Fluor 488) and B220 (Pacific blue) were labeled in-house. b12 specific B cells were detected with biotinylated YU2 gp120 monomer or JRFL gp140 (soluble trimers, “gp140-F” in (19)) followed by streptavidin-PE or streptavidin-APC. Samples were read on an LSR-II instrument (BD) and analyzed using the FlowJo program (Tree Star, Inc.).
HIV antigens
YU2 gp120-His6 AviTag, YU2 gp140-F-His6 or JRFL gp140-F-His6 AviTag were prepared as previously described (20). Uncleaved JRFL full-length gene used in Figure 4H carried a pair of R-to-S mutations in the REKR furin cleavage site, which was also a feature of the soluble gp140 trimers used. Supernatants were purified by Galanthus nivalis lectin chromatography followed by size exclusion chromatography.
Figure 4. Maturation status of Env-binding B cells in b12HL and b12H mice.
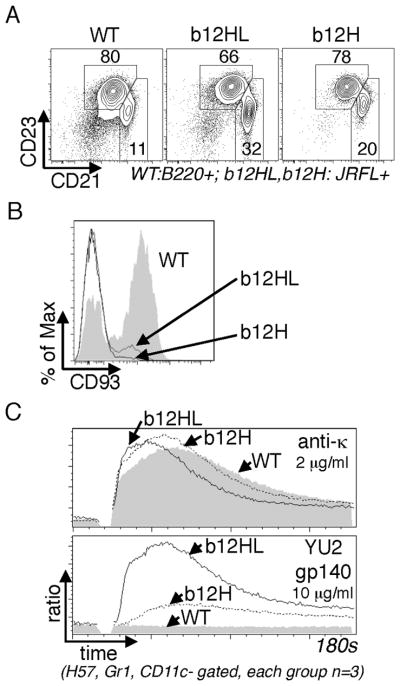
A,B, Flow cytometry analysis of splenic B cell maturation in b12HL and b12H mice. A, Analysis of CD23 and CD21 expression. Plots shown were gated on B220+ (WT) or JRFL+B220+ (b12HL and b12H). Boxes define CD21+CD23+ follicular B cells and CD21++ marginal zone B cells. B, CD93 expression levels on b12HL and b12H CD23+CD21+ B cells (Upper box from A). C, Calcium mobilization analysis of gated splenic B cells of the indicated genotypes upon stimulation with anti-Igκ or soluble Env trimers. Data shown in A and B are representative of at least 6 mice analyzed in three experiments. Data in C are representative of three experiments.
Hybridoma generation
Lymphocytes from b12H or b12HL spleens were cultured for 3 days in Advanced-DMEM supplemented with 10% FCS, 20mM GlutaMax, 55pM 2-mercapthoethanol, IL-4 (20 ng/ml) and anti-CD40 (10 μg/ml) and fused with the SP2/0 myeloma line using polyethelene glycol. Cells from each fusion were then distributed into 96-well plates and hybrids selected with HAT medium. Env-reactive hybrids were selected by ELISA screening of culture supernatants against JRFL gp140.
Serum ELISA
Maxisorp plates (Thermo Fisher Scientific) were coated with 2 μg/ml rat IgG-adsorbed donkey anti-mouse IgG (H+L; Jackson ImmunoResearch Laboratories, Inc.) or M41 (rat anti-mouse IgM) overnight at room temperature, blocked with 1% BSA in buffered saline (ELISA buffer) for 1 h, and incubated for 1 h with mouse sera diluted in ELISA buffer. Purified IgG1 κ (MG1-45; BioLegend) or IgM (C48-6; BD) was used as a standard. For YU2 or JRFL specific ELISA, IgM or IgG1 hybridoma cells obtained from b12HL knock-in mice were used as standards, respectively. Horseradish peroxidase–conjugated anti-mouse Fcγ fragment specific (Jackson ImmunoResearch) or donkey anti-mouse IgM (Jackson ImmunoResearch) diluted in ELISA buffer was used to report signals using 1-Step Ultra-TMB substrate (Thermo Fisher Scientific) per the manufacturer’s instructions. Signals were recorded at 450 nm using a microplate reader (VersaMax; MDS Analytical Technologies).
Neutralization Assay
Neutralization activity of antibodies against pseudovirus in TZM-bl cells was determined as described previously (21, 22). Briefly, TZM-bl cells were seeded in a 96-well flat-bottom plate and infected with pseudovirus in the presence of inhibitors (200 μl, total volume). Viruses were preincubated with the antibody for 1 h at 37°C. Luciferase reporter gene expression was quantified 72 h after infection upon lysis and the addition of luciferase substrate (Promega).
Statistical analysis
Statistical analyses were performed using Prism (GraphPad). Differences between two groups were assessed by the 2-tailed t test; differences between three or more groups were evaluated by one-way ANOVA, followed by Bonerroni’s Multiple Comparison post-test.
RESULTS
Generation of b12 H and L knock-in mice
C57BL/6 ES cells were modified by gene targeting to introduce HIV antibody H- and L-chain variable exons to the natural loci, replacing the respective J clusters, essentially following the strategies described (16, 23–25). Appropriate gene targeting and neo gene removal were carried out as described in Materials and Methods; targeting verification was as described in the supplementary figures of the accompanying manuscript. Mice carrying modified H and L loci of b12 were interbred. We then analyzed B cell development and HIV antigen binding by B cells in mice carrying targetings of H or L, or both Ig loci.
B cells of b12H and b12HL mice are abundant and bind to soluble Env
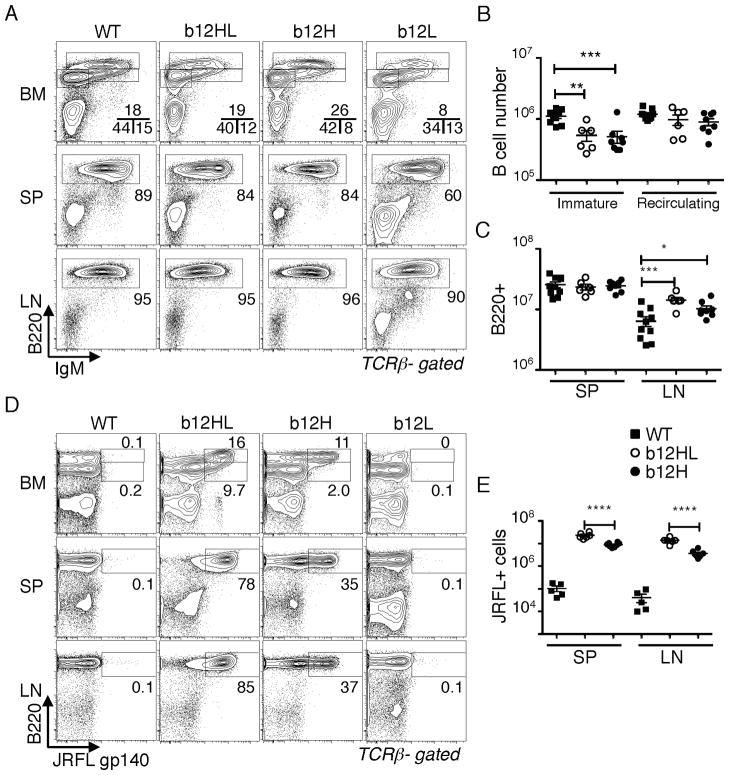
We first compared mice carrying one or both b12 Ig chains. Mice carrying only the H-chain gene (b12H) were bred to IgHa/a mice and expression of the endogenous and transgenic alleles on B cells was measured by flow cytometry. b12H mice displayed excellent allelic exclusion, with >95% of B cells expressing only the “b” allele, indicating successful expression and feedback suppression of endogenous rearrangements (Fig. 1A,B). b12H mice had normal numbers of B lymphocytes in the bone marrow (BM), spleen (SP) and lymph nodes (LN), though immature B cells in BM were reduced (Fig. 2A,B) and B cell numbers in LN were higher than controls (Fig. 2C). Mice carrying both H and L genes (b12HL) had similar B cell numbers compared to b12H mice (Fig. 2B,C).
Figure 1. H-chain allelic exclusion in b12H knock-in B cells.
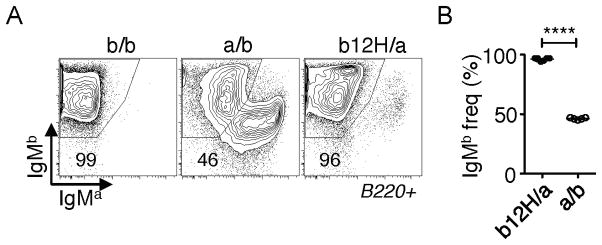
A,B, Flow cytometry analysis of H-chain allelic exclusion in b12H B cells. b12H mice (IgHb) were crossed to IgHa/a mice (“b12H/a”) to allow identification of cells carrying H-chains encoded by the endogenous allele and compared to B6 (b/b) or (B6 x IgHa)F1 (a/b) cells. A, Plot shows expression of IgMa and IgMb on B220+-gated splenocytes from the indicated strains. B, Statistical analysis of IgMb frequency among eight b12H/a and five a/b mice. ****, p < 0.0001.
Figure 2. B cell generation and Env binding by b12 knock-in B cells.
A–C, Comparison of B cell generation in bone marrow (BM), spleen (SP) and lymph nodes (LN) of b12HL, b12H, and b12L mice. A, Flow cytometry analysis of IgM and B220 staining in BM, SP, and LN of the indicated strains. Plots shown were gated on TCRβ negative lymphocytes. Top panels indicate with boxes B220intIgM− immature B cells, B220intIgM+ immature B cells, and B220hi recirculating B cells, respectively. Middle and bottom panels box B220+ B cells. B, Statistical analysis of BM immature vs recirculating B cell numbers from wild type (WT), b12HL and b12H mice. *, p< 0.05; **, p< 0.005; ***, p< 0.0005. C, Analysis of B220+ cell numbers in SP and LN of 9 WT, 6 b12HL and 8 b12H mice. D,E, Binding of soluble Env (JRFL gp140 trimer) by B cells from the indicated strains and tissues. Each data point in E gives the value obtained from an individual mouse.
We next assessed B cells able to bind HIV antigen. A significant subset (~30%) of b12H B cells and >90% of all mature b12HL B cells could bind to soluble trimers of HIV Env (the JRFL isolate) as measured by flow cytometry (Fig. 2D). Absolute numbers of B cells falling into the JRFL+ gate are shown in Fig. 2E. These data indicated that a significant subset of endogenous L-chains is permissive for Env binding when paired with b12H and that in b12HL B cells the transgenic L-chain was paired with b12 H-chain.
Immunoglobulin levels
Analysis of sera from naïve mice revealed that b12H and b12HL mice had normal IgG levels and slightly reduced IgM levels (Fig. 3A,B). b12HL sera, but not b12H sera, contained high titers of HIV Env IgG binding activity (Fig. 3C), which was able to neutralize JRFL (Fig. 3D). As we show in a later section, the low background levels of anti-HIV activity of b12H, compared to b12HL, preimmune sera were likely the result of both the lower frequency and the lower average affinity of B cells reactive to Env. The ability of b12HL B cells to undergo H-chain class switch yet retain b12 specificity supported the notion that the H-chain transgene was appropriately targeted.
Figure 3. Analysis of normal serum Ig levels and HIV reactivity in b12H and HL mice.
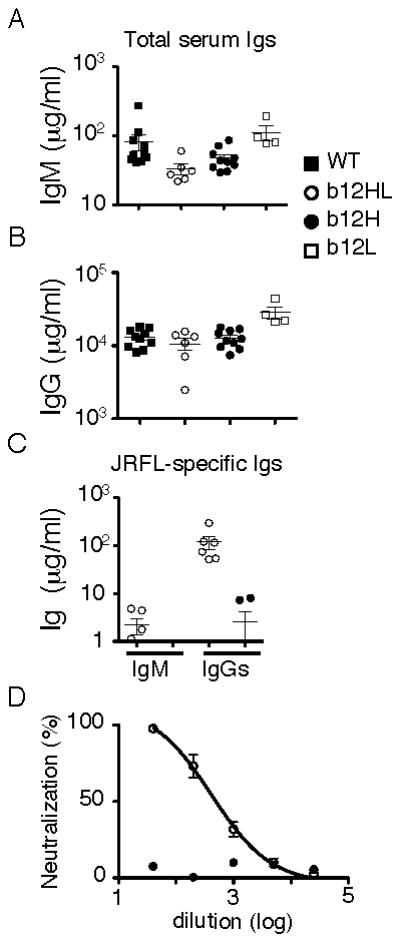
A,B, Statistical analysis of serum IgM and IgG levels in unmanipulated mice. Data from 10 WT, 6 b12HL, 10 b12H and 4 b12L mice. C, Env (JRFL)-specific IgM and IgG concentrations from 6 b12HL and 6 b12H mice. b12 IgM or IgG1 was used to standardize the assay. D, Analysis of neutralization activity against JRFL pseudovirus of normal sera from 4 b12HL and 4 b12H mice. Shown are mean values and SEM. Results were derived from at least three independent experiments except for the serum analyses which were obtained in one experiment.
Maturity and functionality of peripheral B cells
Analysis of B cell maturation revealed that Env-binding B cells in SP of b12H and b12HL mice had mostly a mature follicular phenotype, expressing CD21 and CD23 (Fig. 4A) and mainly lacking expression of CD93 (Fig. 4B), a marker expressed by immature and anergic B cells (26–28). A significant subset of JRFL-binding cells had a MZ B cell phenotype (CD21hiCD23intermediate, Fig. 4A). MZ B cells respond rapidly to blood borne microbes, whereas follicular B cells play a larger role in high affinity secondary responses (29). Furthermore, splenic B cells from b12HL and b12H mice gave normal BCR-triggered Ca++ responses and, unlike wild type B cells, could respond to soluble HIV Env trimers, with the b12HL response being particularly robust (Fig. 4C). Because b12HL B cells showed normal development, were abundant, recognized HIV antigens, triggered normally, and were able to produce high levels of b12 IgG in serum, we conclude that the b12 BCR is “innocuous” — i.e., not autoreactive to a physiologically significant extent.
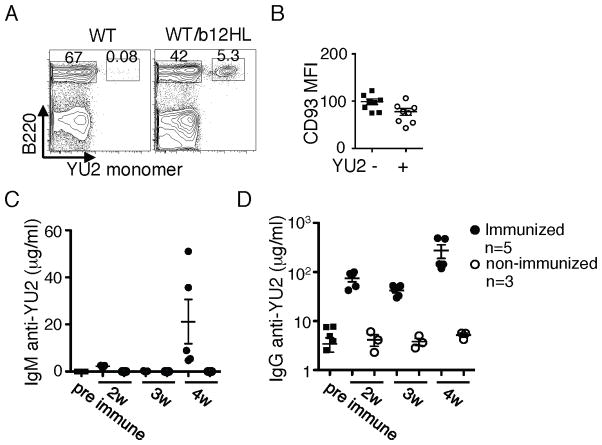
Analysis of b12HL B cells in mixed chimeras
Because B cell monoclonality can rescue survival and function of anergic B cells owing to reduced B cell:B cell competition (30, 31), we tested if b12HL B cells competed well with normal B cells in mixed BM chimeras. b12HL and WT BM were mixed at a 1:1 ratio and used to reconstitute Rag1−/− mice. Six weeks after reconstitution, peripheral blood lymphocytes were analyzed by flow cytometry, revealing an average of 10% of B cells able to bind Env gp120 (YU2) (Fig. 5A). This reduced contribution to the B cell compartment relative to the BM proportion is typical of “innocuous” BCR transgenics owing to accelerated B cell development at the progenitor stage (31, 32). These cells had CD93 levels no higher than did wild type B cells, indicating that they were neither anergic nor immature (Fig. 5B) and the frequency of CD93+ cells was no higher (11.7 +/− 2.4 % versus 16.7 +− 3.5 %; n=8; p=0.2492). The ability of these B cells to respond to immunization was tested by challenging chimeras six weeks after reconstitution with soluble JRFL gp140 trimers in adjuvant, followed by boosting 3 weeks later. The chimeras could rapidly make significant levels of IgG and IgM after boosting that cross-reacted to a heterologous Env isolate, YU2 (Fig. 5C,D). It is not clear why the primary IgM response was low in the immunized mice. As WT mice failed to respond rapidly to this challenge, presumably because the precursor frequency of reactive B cells is too low, we infer that the responses came from the b12HL B cells. We conclude that b12HL B cells compete well with WT cells and can respond to HIV candidate vaccines.
Figure 5. Analysis of b12HL B cell function in mixed chimeras with wild type cells.
Irradiated Rag1−/− mice were reconstituted with a mixture of b12HL and WT BM at a 1:1 ratio. At six weeks post transfer, reconstitution was assessed and immunization initiated. A, Analysis of peripheral blood taken at 6 weeks for the presence of Env-binding (b12HL) and non-binding B cells by gating on TCRβ− lymphocytes and staining for B220 and YU2 gp120 binding. B, CD93 density on B220+YU2− and B220+YU2+ cells. C,D, Serum IgM and IgG responses to Env immunization at various weeks post priming. Chimeric mice were immunized with 40 μg JRFL gp140 in Sigma Adjuvant and boosted three weeks later with antigen in saline. Data are from a total of eight mice as indicated carried out in one experiment.
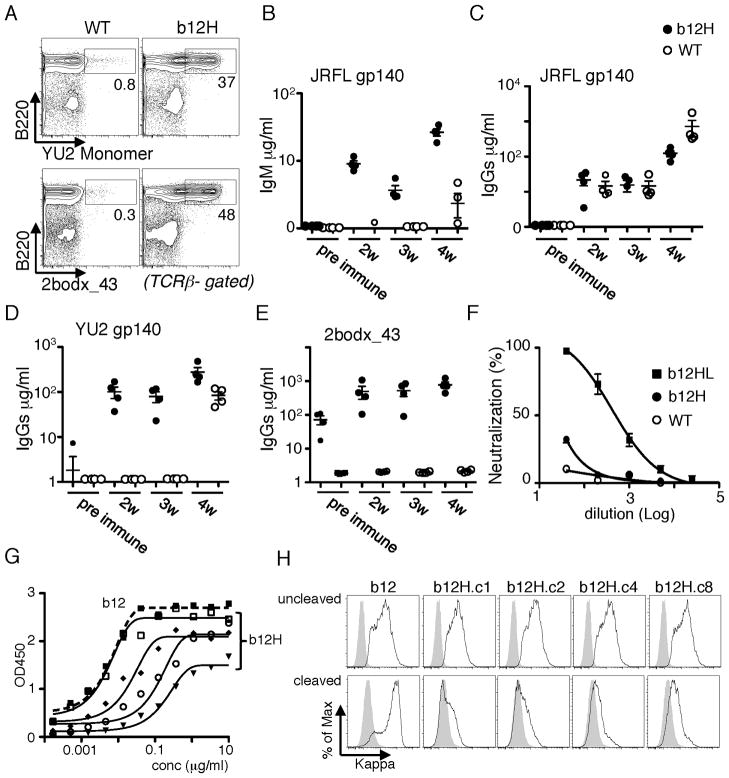
b12H B cells bind to gp140 but do not efficiently neutralize HIV
As normal sera of b12H mice failed to neutralize JRFL pseudovirus, we tested if it was possible to elicit these antibodies by immunization. b12H and WT mice were challenged with soluble JRFL trimers in Ribi adjuvant, then boosted three weeks later. Although b12H mice produced both IgM and IgG JRFL-reactive antibody (Fig. 6B,C), the IgG response was no better than WT. By contrast, an IgG component cross-reactive to YU2 or to the synthetic Env mimetic scaffold 2bodx_43 (33) was rapidly elicited in b12H mice (Fig. 6D,E) but not in WT mice, though at 4 weeks a response arose to YU2 in WT mice. (Fig. 6A demonstrates that 2bodx_43 is recognized by a large proportion of preimmune b12H B cells.) These data indicated that the WT response was not directed to the CD4 binding site of Env. Assays of neutralizing activity indicated that immunized b12H mice made a small but significant JRFL neutralizing response, though much weaker than that present in unmanipulated b12HL mice. The weak, but detectable, response of b12H mice suggests that this model might facilitate a comparison of immunogens designed to elicit a neutralizing response while avoiding production of antibodies to “distracting” epitopes that are non-protective.
Figure 6. Antibody response of b12H mice to soluble Env trimers.
b12H and WT mice were challenged with soluble Env JRFL gp140 trimers and antibody responses measured using the JRFL gp140, YU2 gp120 or 2bodx_43 as the capture reagent in ELISA. The immunization protocol was as in Figure 3, with 4 mice/group. A, Ability of naive b12H B cells to bind to biotinylated YU2 gp120 and 2bodx_43. B,C, JRFL gp140-specific IgM and IgG response. D,E, IgG responses cross-reactive to YU2 gp140 or 2bodx_43. F, Neutralization of JRFL pseudotyped virus by immune sera from four indicated mice. Non-immunized b12HL sera were used as positive control. G,H, Analysis of IgG1 hybridomas from b12H mice. G, ELISA analysis of Env binding by selected hybridoma proteins to plate-bound JRFL gp140. H, Flow cytometry analysis of the binding of the indicated IgG1 hybridoma proteins to 293 cells transfected with full-length JRFL Env (cleaved, lower panels) or JRFL carrying a mutated furin cleavage site (uncleaved, upper panels).
To understand why neutralization activity in sera of b12H mice was weak when gp140-binding B cells were numerous, we studied the specificity of preimmune b12H B cells in more detail. IgG1-producing hybridomas were generated from b12H spleen cells activated in vitro with IL-4 and anti-CD40. As expected, many hybrids produced antibodies reactive to gp140. Their relative binding avidity to JRFL gp140 trimers varied, but could approach that of b12HL antibody (Fig. 6G, “b12”). However, when tested for their ability to bind to cleaved (natural JRFL) or uncleaved JRFLΔCT trimers expressed as full length protein on 293T cells, the b12H hybrids tested bound poorly to cleaved compared to uncleaved versions of Env (Fig. 6H). b12 by contrast bound much better to cleaved trimers. These data confirm that the b12 L-chain not only makes a critical contribution to binding affinity, but also is required for neutralization.
DISCUSSION
We generated knock-in mice expressing broadly neutralizing HIV antibodies and analyzed the their B cell development in order to determine if and how B cells carrying these specificities are negatively regulated by immune tolerance processes. b12HL B cells appear not to be negatively selected, as judged by their robust generation, lack of receptor editing, maintenance of b12 specificity, and ability to be triggered by HIV antigens and to secrete antibody. Similarly, b12H B cells are present in large numbers and many bind HIV Env, suggesting that a large fraction of endogenous L-chains are permissive for binding. This is consistent with crystal structure and chain swapping data indicating that the H-chain is largely responsible for b12 specificity (8–10).
The suggestion that b12 might be autoreactive (11) was not supported by our studies. Not only could these cells be triggered in vitro by HIV ligands, they responded robustly to soluble gp140 trimers in vivo. By contrast, as we show in the accompanying paper, 4E10 appears to be autoreactive and regulated by tolerance. Our findings illustrate the importance of bioassays to identify physiologically significant autoreactivity.
The b12H chain has a long CDR3 region. Long CDRH3s have been documented in some studies to promote polyreactivity, and to present a barrier to B cell development owing to problems with immune tolerance, pairing with surrogate L-chain components at the precursor stage, and, possibly, pairing with L-chains at the small preB stage (12, 13, 34–37). It is therefore significant that the b12H mouse demonstrates no evidence of B cell developmental defects. B cells carrying the transgene specificity were abundant in the peripheral lymphoid organs and apparently fully functional.
Because of the functionality of their Env-reactive B cells, b12H and b12HL mice shouldbe useful models for HIV vaccine development. In vitro assays can be carried out with primary Env-reactive B cells. Many B cells produced by these mice recognize Env, allowing one to easily monitor ongoing responses, to rank the efficacy of vaccine candidates, and to detect weak or even abortive responses.
The results from immunization of b12H mice with experimental vaccine candidates illustrates the daunting challenge of HIV vaccination and might contribute to our understanding of how best to formulate and design vaccines. b12H mice contain a huge number of B cells with Env reactivity but lacking in neutralizing capacity. We presume that a very small subset of these cells might have appropriate L-chains that promote or permit eventual neutralizing capacity either prior to or after appropriate somatic mutation. Cleaved Env trimers on the surface of virions are required for infection and are the targets of neutralizing antibodies such as b12. But randomly selected B cells from b12H mice captured as hybridomas preferentially recognize the uncleaved form. This result emphasizes the importance of cleavage for the generation of a conformation of Env trimer accurately mimicking that of the functional trimer on the virion surface of on transfected/infected cells (38). The only recombinant Env trimers that are cleaved and currently available are based on the SOSIP framework (39, 40). Most recombinant Env constructs are uncleaved and many use a heterologous trimerization motif to impose trimer symmetry (41, 42). b12H mice should be useful to assess vaccine candidates that selectively recruit B cells recognizing the most relevant epitope.
A future challenge will be to learn how to train germline B cells to adopt specificity to achieve both high affinity and the ability to neutralize HIV. In this effort, mice carrying germline versions of b12 HL and other bNAbs will be of special value. However, these B cells lack detectable affinity for HIV (20, 43). Recent progress on rational immunogen design has suggested a pathway to recruit such cells (44). However, our study of b12H mice suggests that we still have much to learn from this model about how to extend moderate binding energy of BCRs to promote production of antibody able to neutralize HIV.
Acknowledgments
The authors thank the TSRI Mouse Genetics facility and Patrick Skog for excellent technical assistance, and Roberta Pelanda (NJH) for the pVKR2neo targeting vector.
Abbreviations used
- BM
bone marrow
- bNAb
broadly neutralizing antibody
- Env
envelope
- HIV
human immunodeficiency virus
- LN
lymph nodes
- SP
spleen
Footnotes
This work was supported by NIH research grant R01AI073148 and UO1AI078224, and by the International AIDS Vaccine Initiative Neutralizing Antibody Center and the Ragon Institute.
References
- 1.Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol. 2010;22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hessell AJ, Haigwood NL. Neutralizing antibodies and control of HIV: moves and countermoves. Curr HIV/AIDS Rep. 2012;9:64–72. doi: 10.1007/s11904-011-0105-5. [DOI] [PubMed] [Google Scholar]
- 3.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 4.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat Med. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 5.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton DR, Barbas CF, III, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 8.Zwick MB, Parren PW, Saphire EO, Church S, Wang M, Scott JK, Dawson PE, Wilson IA, Burton DR. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:5863–5876. doi: 10.1128/JVI.77.10.5863-5876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbas CF, 3rd, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR, et al. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 10.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 12.Aguilera I, Melero J, Nunez-Roldan A, Sanchez B. Molecular structure of eight human autoreactive monoclonal antibodies. Immunology. 2001;102:273–280. doi: 10.1046/j.1365-2567.2001.01159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichiyoshi Y, Casali P. Analysis of the structural correlates for antibody polyreactivity by multiple reassortments of chimeric human immunoglobulin heavy and light chain V segments. J Exp Med. 1994;180:885–895. doi: 10.1084/jem.180.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, Nussenzweig MC. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gertsenstein M, Nutter LM, Reid T, Pereira M, Stanford WL, Rossant J, Nagy A. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PloS one. 2010;5:e11260. doi: 10.1371/journal.pone.0011260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelanda R, Schaal S, Torres RM, Rajewsky K. A prematurely expressed Ig(kappa) transgene, but not V(kappa)J(kappa) gene segment targeted into the Ig(kappa) locus, can rescue B cell development in lambda5-deficient mice. Immunity. 1996;5:229–239. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- 17.Casola S. Conditional gene mutagenesis in B-lineage cells. Methods Mol Biol. 2004;271:91–109. doi: 10.1385/1-59259-796-3:091. [DOI] [PubMed] [Google Scholar]
- 18.Ota T, Ota M, Duong BH, Gavin AL, Nemazee D. Liver-expressed Ig{kappa} superantigen induces tolerance of polyclonal B cells by clonal deletion not {kappa} to {lambda} receptor editing. The Journal of Experimental Medicine. 2011;208:617–629. doi: 10.1084/jem.20102265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, McKee K, Tran K, O’Dell S, Schmidt SD, Phogat A, Forsell MN, Karlsson Hedestam GB, Mascola JR, Wyatt RT. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site. J Biol Chem. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ota T, Doyle-Cooper C, Cooper AB, Huber M, Falkowska E, Doores KJ, Hangartner L, Le K, Sok D, Jardine J, Lifson J, Wu X, Mascola JR, Poignard P, Binley JM, Chakrabarti BK, Schief WR, Wyatt RT, Burton DR, Nemazee D. Anti-HIV B Cell Lines as Candidate Vaccine Biosensors. J Immunol. 2012;189:4816–4824. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 12. Chapter 12. 2005. p. 11. [DOI] [PubMed] [Google Scholar]
- 23.Taki S, Meiering M, Rajewsky K. Targeted insertion of a variable region gene into the immunoglobulin heavy chain locus [see comments] Science. 1993;262:1268–1271. doi: 10.1126/science.8235657. [DOI] [PubMed] [Google Scholar]
- 24.Luning Prak E, Weigert M. Light chain replacement: a new model for antibody gene rearrangement. J Exp Med. 1995;182:541–548. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Prak EL, Weigert M. Editing disease-associated autoantibodies. Immunity. 1997;6:97–105. doi: 10.1016/s1074-7613(00)80673-1. [DOI] [PubMed] [Google Scholar]
- 26.Rolink AG, Andersson J, Melchers F. Characterization of immature B cells by a novel monoclonal antibody, by turnover and by mitogen reactivity. Eur J Immunol. 1998;28:3738–3748. doi: 10.1002/(SICI)1521-4141(199811)28:11<3738::AID-IMMU3738>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Merrell KT, Benschop RJ, Gauld SB, Aviszus K, Decote-Ricardo D, Wysocki LJ, Cambier JC. Identification of anergic B cells within a wild-type repertoire. Immunity. 2006;25:953–962. doi: 10.1016/j.immuni.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Allman D, Lindsley RC, DeMuth W, Rudd K, Shinton SA, Hardy RR. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- 29.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulcher DA, Basten A. Reduced life span of anergic self-reactive B cells in a double-transgenic model. J Exp Med. 1994;179:125–134. doi: 10.1084/jem.179.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cyster JG, Hartley SB, Goodnow CC. Competition for follicular niches excludes self-reactive cells from the recirculating B-cell repertoire [see comments] Nature. 1994;371:389–395. doi: 10.1038/371389a0. [DOI] [PubMed] [Google Scholar]
- 32.Spanopoulou E, Roman CA, Corcoran LM, Schlissel MS, Silver DP, Nemazee D, Nussenzweig MC, Shinton SA, Hardy RR, Baltimore D. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 33.Azoitei ML, Correia BE, Ban YE, Carrico C, Kalyuzhniy O, Chen L, Schroeter A, Huang PS, McLellan JS, Kwong PD, Baker D, Strong RK, Schief WR. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. [DOI] [PubMed] [Google Scholar]
- 34.Ivanov, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr Development of the expressed Ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, and charge that are first established in early B cell progenitors. J Immunol. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 35.Vettermann C, Jack HM. The pre-B cell receptor: turning autoreactivity into self-defense. Trends Immunol. 2010;31:176–183. doi: 10.1016/j.it.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Arnold LW, Spencer DH, Clarke SH, Haughton G. Mechanisms that limit the diversity of antibody: three sequentially acting mechanisms that favor the spontaneous production of germline encoded anti-phosphatidyl choline. Int Immunol. 1993;5:1365–1373. doi: 10.1093/intimm/5.11.1365. [DOI] [PubMed] [Google Scholar]
- 37.Ye J, McCray SK, Clarke SH. The transition of pre-BI to pre-BII cells is dependent on the VH structure of the mu/surrogate L chain receptor. EMBO J. 1996;15:1524–1533. [PMC free article] [PubMed] [Google Scholar]
- 38.Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 39.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakrabarti BK, Pancera M, Phogat S, O’Dell S, McKee K, Guenaga J, Robinson J, Mascola J, Wyatt RT. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res Hum Retroviruses. 2011;27:877–887. doi: 10.1089/aid.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsell MN, Schief WR, Wyatt RT. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Current opinion in HIV and AIDS. 2009;4:380–387. doi: 10.1097/COH.0b013e32832edc19. [DOI] [PubMed] [Google Scholar]
- 43.Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, Macpherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science. 2013 doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]