Abstract
Sensory hair cells are exquisitely sensitive vertebrate mechanoreceptors that mediate the senses of hearing and balance. Understanding the factors that regulate the development of these cells is important, not only for our understanding of ear development and its functional physiology, but also to shed light on how these cells may be replaced therapeutically. In this review, we describe the signals and molecular mechanisms that initiate hair cell development in vertebrates, with particular emphasis on the transcription factor Atoh1 that is both necessary and sufficient for hair cell development. We then discuss recent findings on how microRNAs may modulate the formation and maturation of hair cells. Lastly, we will review recent work on how hair cells are regenerated in many vertebrate groups, and the factors that conspire to prevent this regeneration in mammals.
Keywords: Cochlea, Atoh1, miRNA, Notch, Cell Cycle
A: Intrinsic and extrinsic signals that drive hair cell induction
The inner ear develops from a thickened patch of cranial ectoderm, the otic placode, and then invaginates or cavitates to form a spherical otocyst. At this point, the ear primordium has already received signals that confer regional identity and polarity to the otocyst (Groves and Fekete, 2012). These signals lead to the formation of multiple prosensory patches, each of which will give rise to a particular inner ear sensory organ - the three cristae at the base of each semicircular canal, the two maculae and the auditory epithelium of the cochlea (Alsina et al 2009, Bok et al 2007). These patches are characterized by the expression of the transcription factor Sox2, and mice carrying hypomorphic mutations for Sox2 lack all sensory cells in the inner ear (Kiernan et al 2005). These sensory patches then differentiate to produce the hair cells and supporting cells of each inner ear sensory organ.
A.1: The temporal and spatial regulation of hair cell differentiation
After Sox2+ prosensory tissue has been induced in each sensory organ, the prosensory domain begins to exit the cell cycle and terminally differentiate into hair cells. In most vertebrates, including the vestibular system of mammals, exit from the cell cycle and the appearance of the first markers of hair cells are tightly coupled. In the vestibular system, differentiation typically begins near the center of each prosensory patch, and expands out over an extended period of time. For example, the first hair cells appear in the future striolar region of the mouse utricle at embryonic day 11 (Raft et al 2007), but over half of the total hair cells are generated after birth, with small numbers of hair cells still being generated from mitotic progenitors between postnatal days 12-14 (Burns et al 2012b, Kirkegaard & Nyengaard 2005). In the case of the chicken hearing organ, the basilar papilla, the first hair cells are born in the middle of the superior side of the cochlea beginning at embryonic day 6, spreading both inferiorly and to both the base and apex over the next three days (Katayama & Corwin 1989).
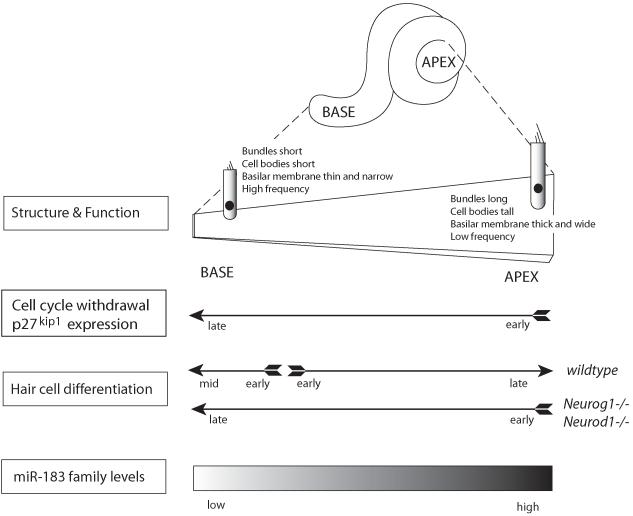
The mammalian organ of Corti has a strikingly different arrangement of hair cells and supporting cells compared to all other vertebrate sensory patches. Instead of a quasi-hexagonal arrangement, where each hair cell is surrounded by between 4-8 supporting cells depending on its position in the sensory epithelium (Goodyear & Richardson 1997), hair cells and supporting cells are arranged in uniform rows and invariant proportions along the length of the cochlear duct (Kelley 2006). This serially repeating pattern is generated by a highly unusual pattern of cell cycle exit and differentiation. In the mouse, the prosensory domain of the future organ of Corti begins to exit the cell cycle in the apical tip of the cochlea at embryonic day 12 (Lee et al 2006, Matei et al 2005, Ruben 1967), and a wave of cell cycle exit then proceeds along the prosensory domain from apex to base over the next 48-60 hours, with some cells in the most basal region still incorporating mitotic labels at E14.5-E15.0 (Lee et al 2006). Starting at about E13.5, cells in the mid-basal region of the cochlea begin to differentiate into hair cells by expressing the transcription factor Atoh1 (Chen et al 2002), and this region of differentiating cells spreads down to the apex over the next 3-4 days. Thus, the first cells to exit the cell cycle in the apex of the cochlear duct are the last ones to terminally differentiate into hair cells five days later, while the last cells to exit the cell cycle in the mid-basal region are some of the first to differentiate into hair cells (Figure 1). This dramatic temporal and spatial uncoupling of cell cycle exit and differentiation has no parallel in any other vertebrate tissue.When maturation is complete, numerous morphological, physiological and molecular properties of the cochlear duct and its resident cells vary systematically along this longitudinal axis and are responsible for the gradient of selectivity to sounds of different frequencies (Figure 1).
Figure 1.
Longitudinal gradients of the mammalian organ of Corti in normal and mutant mice. The cochlea coils from base to apex and exhibits systematic gradients in the dimensions of its fluid-filled chambers, as well as the width and thickness of the basilar membrane (shown uncoiled). Lying on the basilar membrane is the delicate organ of Corti, with hair cells and supporting cells (not shown) also changing systematically in their physical dimensions (shown schematically with one outer hair cell at the extremes). Arrows depict temporal gradients in cell cycle exit, p27kip1 expression and hair cell differentiation in normal and mutant mice. In the mature organ of Corti, the miR-183 family is expressed in a gradient from base (lowest levels) to apex (highest levels).
The mechanisms that propagate the apical-basal gradient of cell cycle exit and the midbasal-apical gradient of differentiation in the mammalian cochlea are poorly understood. The cyclin-dependent kinase inhibitor p27kip1 is up-regulated in the cochlea in an apical-basal gradient concomitant with the gradient of cell cycle exit (Figure 1; Chen & Segil 1999, Lee et al 2006), and is necessary for the correct sequence of cell cycle exit and generation of appropriate numbers of hair cells and supporting cells (Chen & Segil 1999, Kanzaki et al 2006, Lowenheim et al 1999). Its expression is regulated at both the transcriptional and protein levels (Lee et al 2006), but no signals have been identified that directly regulate its expression in the cochlea. The wave of hair cell differentiation does not appear to require contact with the underlying mesenchyme, as it can proceed normally when the embryonic cochlear epithelium is maintained alone in organ culture (Montcouquiol & Kelley 2003). Moreover, there is no absolute requirement for a fully intact cochlear epithelium to propagate differentiation signals in a planar fashion, as the correct basal-apical sequence of differentiation can be recapitulated even when the cochlear epithelium is cut into a series of pieces along its length and cultured separately (Montcouquiol & Kelley 2003). Genetic experiments suggest a role for the transcription factors Neurog1 and Neurod1 in these gradients. The cochlear prosensory region of Neurog1 mutant mice show a premature exit from the cell cycle of about 24-36 hours earlier than normal, and hair cell differentiation, assayed by expression of Atoh1, commences in the apical, not the basal region of the Neurog1 mutant cochlea (Matei et al 2005). This disordered pattern of hair cell differentiation is also seen in Neurod1 mutant mice (Figure 1; Jahan et al 2010). It is important to note that neither Neurod1 nor Neurog1 are expressed in the cochlea at detectable levels during this period of differentiation, suggesting they play an indirect role in triggering the timing of prosensory domain cell cycle exit and differentiation. Both transcription factors are necessary for the differentiation of the spiral ganglion, as both Neurod1 and Neurog1 mutant mice have spiral ganglia that are either greatly reduced or completely absent. This suggests the developing spiral ganglion may regulate the tempo of differentiation of cochlear hair cells.
In addition to the basal-apical gradient of differentiation in the cochlea, hair cell differentiation also proceeds in a neural-abneural direction at any given position along the length of the cochlear duct, with inner hair cells differentiating before outer hair cells, and pillar cells separating the two differentiating populations. Although birds do not display the same dramatic discontinuity and separation between inner and outer hair cells, they do show a neural-abneural gradient in hair cell length between “tall” and “short” hair cells (Gleich et al 2004). At present it is not clear whether the temporal separation between inner and outer hair cell differentiation in the organ of Corti reflects distinct sets of inductive signals, or a common inducing signal that is interpreted at different times or by different cellular contexts in the inner and outer hair cell regions. For example, recent evidence suggests that a gradient of BMP signaling normally spreads from the abneural to the neural side of the cochlear duct, and that perturbation of this gradient can change the numbers of both inner and outer hair cells (Groves & Fekete 2012, Hwang et al 2010, Ohyama et al 2010, Puligilla et al 2007). However, a number of mouse mutants display specific defects in the differentiation of outer versus inner hair cells. For example, Emx2 mutant mice develop inner hair cells but not outer hair cells or Deiters’ cells (Holley et al 2010) and Jag1 mutant mice develop multiple rows of inner hair cells but also lack outer hair cells and Deiters’ cells along parts of the cochlea (Brooker et al 2006, Kiernan et al 2006). FGF20 mutant mice also lack outer hair cells and Deiters’ cells, a phenotype that seems to be caused by a failure of the prosensory domain to differentiate correctly (Huh et al 2012). These data suggest that regions containing inner versus outer hair cells may differentiate as distinct developmental units.
A.2: Atoh1 – the key initiator of hair cell development
The first transcription factor to be expressed in differentiating hair cell progenitors is the basic helix-loop-helix transcription factor Atoh1. Atoh1 is expressed prior to all other known hair cell genes, and is later down-regulated in maturing hair cells before they begin to participate in hearing and balance (Mulvaney & Dabdoub 2012). Atoh1 mutant mice lack hair cells in all sensory regions of the ear (Ben-Arie et al 1996), with this deficiency being due to hair cell progenitors failing to differentiate and then dying (Chen et al 2002, Pan et al 2011). This suggests Atoh1 is required to initiate the hair cell differentiation program, but is not required for the function of mature hair cells. Atoh1 is also sufficient to generate hair cells when ectopically expressed in some locations in the developing inner ear (Izumikawa et al 2005, Kawamoto et al 2003, Kelly et al 2012, Liu et al 2012b, Zheng & Gao 2000).
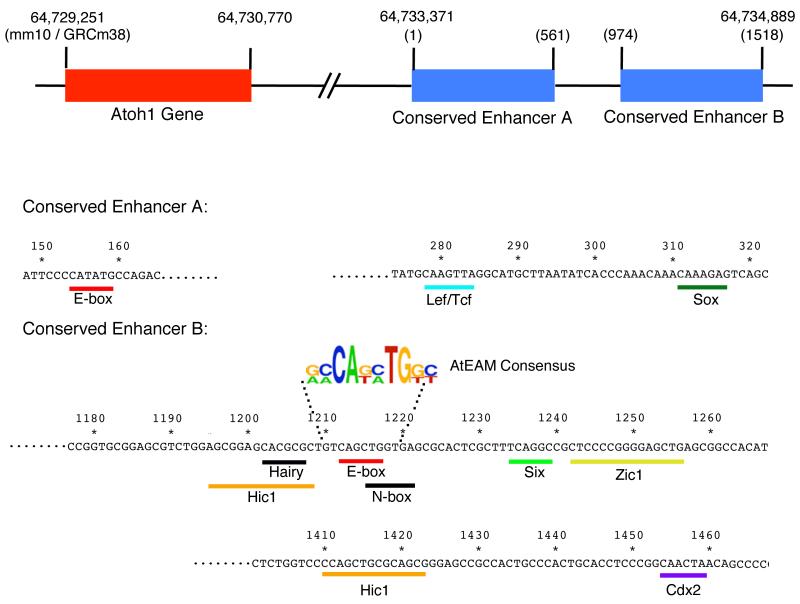
Despite the central role played by Atoh1 in the first steps of hair cell differentiation, surprisingly little is known about the factors that regulate its expression in hair cell progenitors. Manipulation of the BMP, FGF, Shh and Wnt signaling pathways can all alter the numbers of hair cells in inner ear sensory organs (Driver et al 2008, Hayashi et al 2008, Huh et al 2012, Kamaid et al 2010, Li et al 2005, Pirvola et al 2002, Pujades et al 2006, Stevens et al 2003), although it is far less clear to what extent these effects are due to the direct regulation of Atoh1 transcription, to indirect transcriptional regulation by induction of other factors that can themselves regulate Atoh1, or post-translational modification of the Atoh1 protein (Mulvaney & Dabdoub 2012). Far more attention has focused on regions of the Atoh1 locus that regulate its transcription, particularly on two enhancer elements located 3.4kb downstream of the Atoh1 coding region (Figure 2; Helms et al 2000). These enhancers are capable of driving reporter gene expression in the nervous system and in hair cells of transgenic mice (Lumpkin et al 2003), and are strongly conserved in mammals (Helms et al 2000). Atoh1 positively autoregulates its own expression by binding to an E-box motif in this enhancer complex (Helms et al 2000). This autoregulation is necessary for the continued expression of Atoh1 in hair cells, as Atoh1 mutant mice cannot maintain expression in hair cells of a GFP reporter transgene containing the autoregulatory enhancer (Raft et al 2007). However, it is not known whether other regulatory sequences in this enhancer initiate expression of Atoh1 before Atoh1 itself binds to its own enhancer, or whether other enhancers located elsewhere in the Atoh1 locus are responsible for the first burst of Atoh1 transcription.
Figure 2.
A diagram of the mouse Atoh1 locus, showing the position of its 3′ autoregulatory enhancer. Numbers refer to the position on mouse chromosome 6, according to the current build of the mouse genome (mm10/GRCm38). The Atoh1 autoregulatory enhancer consists of two conserved elements, A and B. Transcription factor binding sites that have been experimentally verified are shown in color on relevant regions of the enhancer sequence. Two candidate binding sites, a hairy-preferred motif and an N-box are shown in black; although these have not been experimentally verified, they have been featured in many previous reviews of this regulatory region. The AtEAM consensus sequence for Atoh1 binding identified by Klisch et al. (2011) is shown aligned to the likely E-box site in Enhancer B.
The autoregulatory enhancer region of Atoh1 contains candidate binding sites for a menagerie of transcriptional activators and repressors, some of which have been shown to bind directly to the enhancer by biochemical assays (Figure 2; Akazawa et al 1995, Briggs et al 2008, D’Angelo et al 2010, Ebert et al 2003, Mutoh et al 2006, Shi et al 2010). Some of these transcription factors can also activate or repress Atoh1 expression in vivo or in vitro, although it is not always clear whether these factors work by directly binding the Atoh1 enhancer or are acting indirectly or through other, as-yet unidentified elements in the Atoh1 locus. Recent studies have demonstrated that Six1 and its transcriptional co-activator Eya1, which are expressed in the prosensory domain of the cochlea, can bind directly to the Atoh1 autoregulatory enhancer (Ahmed et al 2012). These two factors are sufficient to induce Atoh1 expression when electroporated into the greater epithelial ridge adjacent to the organ of Corti (Ahmed et al 2012). This activation of Atoh1 can be potentiated by the transcription factor Sox2, which is strongly expressed in all prosensory domains of the inner ear and is necessary for differentiation of each prosensory domain (Kiernan et al 2005). Sox2 has also been reported to be sufficient to activate Atoh1 and induce ectopic hair formation in the chick otocyst (Neves et al 2012). Sox2 is rapidly down-regulated in hair cells as they differentiate, and this down-regulation appears to be required for further maturation of hair cells, since sustained expression of Sox2 in Atoh1-expressing cells blocks the induction of later hair cell markers such as Myosin 7a (Ahmed et al 2012, Puligilla et al 2010).
A.3: The transcriptional control of hair cell differentiation by Atoh1
Despite the generally accepted importance of Atoh1 in hair cell development, we know very little about the molecular basis by which Atoh1 regulates hair cell differentiation. It is conceivable that Atoh1 acts as a “delegator” – sitting atop a large hierarchy of regulatory transcription factors, each devoted to controlling a different aspect of hair cell differentiation. At the other extreme, Atoh1 might act as a “micro-manager”, by directly regulating many different aspects of hair cell development and maturation. Moreover, it is not clear whether Atoh1 simply regulates generic aspects of cell identity shared by hair cells in all inner ear sensory organs, or whether it can regulate genes that distinguish different classes of hair cells, such as inner hair cells versus outer hair cells in the cochlea, or type I versus type II vestibular hair cells.
Current efforts to identify direct targets of transcription factors typically involve chromatin immunoprecipitation of the transcription factor bound to DNA in the cells of interest, followed by high throughput sequencing of the DNA (ChIP-sequencing). The application of these approaches to finding Atoh1 targets in hair cells have been confounded by the relatively small number of hair cells in mammalian sensory organs, and by the large number of other cell types in the cochlea. However, ChIP-sequencing has recently been used to identify Atoh1 targets in cerebellar granule cells, which can be obtained in large numbers and which represent about 50% of the total cells in a neonatal mouse cerebellum (Klisch et al 2011). This work supports the idea that Atoh1 is more of a micro-manager than a delegator – it not only regulates genes associated with transcription, but also cell division, chromosomal organization, metabolism, cell migration and cell adhesion (Klisch et al 2011). Clearly many of these genes are unlikely to be expressed in post-mitotic and non-migratory hair cells, and indeed, a recent comparison of the ChIP-sequencing data from the cerebellum with gene expression data sets from cerebellar granule neurons, dorsal spinal interneurons and hair cells have identified only three genes – Rab15, Selm and Atoh1 itself – that are candidates to be direct targets of Atoh1 in all three cell types (Lai et al 2011). However, the ChIP-sequencing results from the cerebellum have also led to the identification of a consensus Atoh1 binding motif (G/A,C/A,CA,G/T,C/A,TG,G/T,C/T) that is longer and more specific than the generic E-box sequence (CANNTG) bound by many bHLH transcription factors (Figure 2; Klisch et al 2011). This Atoh1 E-box associated motif, or AtEAM, is present in the majority of sequences bound by Atoh1 in the cerebellum, and thus may be of use in predicting candidate Atoh1 targets in hair cells. The significance of the specificity of the Atoh1-binding AtEAM site was recently demonstrated by replacing the Atoh1 coding sequence with that of the closely related bHLH factor Neurog1, which is expressed in precursors of inner ear neurons, but not hair cells. Mice homozygous for this replacement allele have a severely disrupted organ of Corti containing very few hair cells (Jahan et al 2012). The surviving hair cells were extremely immature and lacked significant stereocilia, although this phenotype was not as severe as that seen in Atoh1 null mice. This suggests that bHLH transcription factors that are closely related to Atoh1 are unable to correctly activate sufficient Atoh1 target genes to promote correct hair cell differentiation and survival, even when expressed in the correct cells at the correct time.
B: Modulation of hair cell gene expression through microRNAs (miRNAs)
MicroRNAs (miRNAs) are widely appreciated for their role in post-transcriptional repression of gene expression (Hobert 2007, Hobert 2008, Kloosterman & Plasterk 2006, Takacs & Giraldez 2010). miRNAs are single-stranded RNAs, usually 21-23 nucleotides in length, that are generated from long RNA polymerase II transcripts and then processed through a series of double-stranded-specific ribonuclease III enzymes such as Drosha and Dicer (Esquela-Kerscher & Slack 2006, Kosik 2006, Winter et al 2009). Mature miRNAs ultimately bind to target messenger RNA (mRNA) transcripts, which either destabilizes the targets or lowers their translation efficiency (Filipowicz et al 2008, Guo et al 2010, Lewis & Steel 2010, Liu et al 2008). The resulting repression in protein levels rarely exceeds 50%, at least for the small number of miRNAs that have been tested to date (Baek et al 2008, Bartel 2009, Selbach et al 2008). Thus, miRNAs are modulators of protein expression levels, rather than “off” switches. Several comprehensive reviews have recently highlighted the literature related to inner ear miRNAs (Friedman et al 2009, Rudnicki & Avraham 2012, Soukup 2009, Weston & Soukup 2009). In this section, we narrow the focus to the expression and manipulation of miRNAs and their targets in hair cells and hearing. ..
B.1: Manipulating miRNA levels in developing hair cells
To evaluate the role of miRNAs in hair cell development and maintenance, the miRNA processing enzyme, Dicer, has been conditionally deleted in the mouse inner ear. Several different Cre driver lines have been used to eliminate a floxed Dicer allele at different developmental time points. Foxg1-Cre and Pax2-Cre lines excise Dicer at the otocyst stages (Soukup et al 2009), Atoh1-Cre recombines the locus about the time the hair cells are born (Weston et al 2011), and Pou4f3-Cre shortly thereafter (Friedman et al 2009). These conditional mutants each show defects in hair cell development, with the severity dependent upon the stage of Dicer excision, and probably also on the amount and duration of residual miRNA expression that persists after Cre onset (see Discussion by Soukup, 2009). In general, however, deletion of Dicer in hair cells leads to disrupted hair bundle morphology and subsequent hair cell death.
A major caveat of this approach is that phenotypes resulting from Dicer knockouts may not necessarily reflect a “worst-case scenario” that reveals the full role played by miRNAs. This is because some miRNAs, as well the transcripts that they repress, may act in direct opposition to others that are co-expressed at the same time and place. As a result, the loss of a single species of miRNA (or miRNA family) may throw the system further out of equilibrium than the loss of two opposing miRNAs, or the blockade of all miRNA processing. Such a scenario is seen in the wing imaginal disc of the fly larvae, where the absence of miR-9 alone is more severe than deletion of Dicer (Bejarano et al 2010).
With this in mind, let us consider the necessity of miRNA processing for proper hair cell development and maturation. The miR-183 family includes 3 members (miR-96, miR-182 and miR-183) that are expressed at high levels in young hair cells, are undetectable in supporting cells, and are also present in the inner ear ganglion neurons (Li et al 2010b, Weston et al 2006). There is a discrepancy in the literature about whether these miRNAs remain confined to hair cells in the postnatal organ of Corti (Sacheli et al 2009, Weston et al 2011). Applying whole mount in situ hybridization methods to the cochlear epithelium, Weston and colleagues reported that all 3 miR-183 family members behave similarly and remain restricted to hair cells. Expression is stronger in the base than the apex at P0, but then disappears from the base to establish a gradient from apex (high) to base (low) that persists into adulthood (Figure 1; Sacheli et al 2009, Weston et al 2011).
It is thus tempting to speculate that the molecular basis of anatomical variation and frequency selectivity that is systematically arrayed along the length of the cochlea (shown schematically in Figure 1) might be modulated, and perhaps even enforced, by systematic differences in the levels of the miR-183 family and its targets. If so, then loss of this gene family should lead to weaker longitudinal gene expression gradients. A counterintuitive result is obtained when comparing the expression patterns of the miR-183 family with the phenotype observed when Dicer is deleted selectively in hair cells using the Atoh1-Cre driver line. Here, Dicer deletion using Atoh1-Cre enhances transcript gradients (Sacheli et al 2009, Weston et al 2011). This leads to the conclusion that, in toto, hair-cell-expressed miRNAs serve to repress longitudinal transcript gradients, rather than to promote them. Resolution of this paradox must await experiments that selectively delete only the miR-183 family to discover whether or not they share this en masse effect of miRNA loss. So far, studies in zebrafish have revealed that the number of hair cells in developing macular sensory organs is affected by levels of miR-183 family members: over-expression of miR-96 or -182 initially yields more hair cells, whereas knockdown of miR-96, -182 and/or -183 generates fewer hair cells (Li et al 2010b). While these studies emphasize the impact of the miR-183 family on hair cell fate specification, they cannot address the question of longitudinal maturation gradients, since zebrafish do not have a cochlea.
B.2: miR-96 mutations underlie inherited hearing loss in mice and humans
Progressive, non-syndromic autosomal semi-dominant or dominant hearing loss has been associated with miR-96 mutations in Diminuendo mice and in DFNA50 human families, respectively (Lewis et al 2009, Mencia et al 2009). Diminuendo hair cells are born and initiate differentiation, but become stalled at an early postnatal stage and fail to differentiate fully into inner and outer hair cells (Kuhn et al 2011). It is uncertain to what degree this phenotype reflects the loss-of-function of the wild type allele versus a gain-of-function effect of the novel allele in repressing a new set of target genes. One fact arguing in favor of the former mechanism is that two human families have unique point mutations in the seed region of miR-96 (and thus presumably could interact with unique sets of potential new targets; Mencia et al 2009), while a third family has a mutation in the precursor miRNA transcript that results in poor expression levels of the mature miR-96 (Solda et al 2012). Despite these allele differences among the human DFNA50 families and the Diminuendo mice, progressive hearing loss is a common outcome. To address the underlying cause of the hair cell phenotype in Diminuendo homozygous mice, cochlear tissue was subjected to gene expression profiling in an effort to identify target transcripts (Lewis et al 2009). As might be expected, genes with 3′UTRs carrying heptamer sequences complementary to the seed region of miR-96 were enriched in the pool of transcripts that were up-regulated in the organ of Corti of Diminuendo homozygotes. Likewise, the binding site for the Diminuendo allele of miR-96 was enriched in the pool of down-regulated transcripts. However, binding sites from this latter group of mouse genes were not conserved in human and rat orthologs. Ultimately, distinguishing gain-of-function from loss-of-function mechanisms may be revealed by genetic rescue of Diminuendo hair cells.
B.3: Evaluation of putative targets for hair-cell-enriched miRNAs
Although the miR-183 family holds particular interest for understanding hair cell biology, the members of this family are also expressed in developing retina and olfactory epithelium (Wienholds et al 2005, Xu et al 2007). Furthermore, they are mis-regulated in certain tumor tissues and cancer cell lines (Guttilla & White 2009, Li et al 2010c, Lin et al 2010, Liu et al 2012c, Lowery et al 2010, Mihelich et al 2011, Sarver et al 2010, Segura et al 2009, Wang et al 2012a, Yan et al 2012, Yu et al 2010). These discoveries have sparked efforts to identify target transcripts and explore their potential functions in other systems, thereby generating lists of genes that could eventually be evaluated as targets in hair cells. Bioinformatic analysis is useful as a preliminary target screening tool. Target-search algorithms, such as PicTar, TargetScan and Microcosm, favor transcripts containing sequences in their 3′ untranslated regions (3′UTRs) that are complementary to the seed regions of miRNAs, particularly nucleotides 2 through 7 or 8 (Lewis et al 2005, Lewis et al 2003). Other criteria can include the number of putative miRNA binding sites as well as their evolutionary conservation. Each of the common target-prediction programs can generate lists of genes numbering in the hundreds for a single miRNA. Ultimately, though, the translational repression of potential targets requires validation.
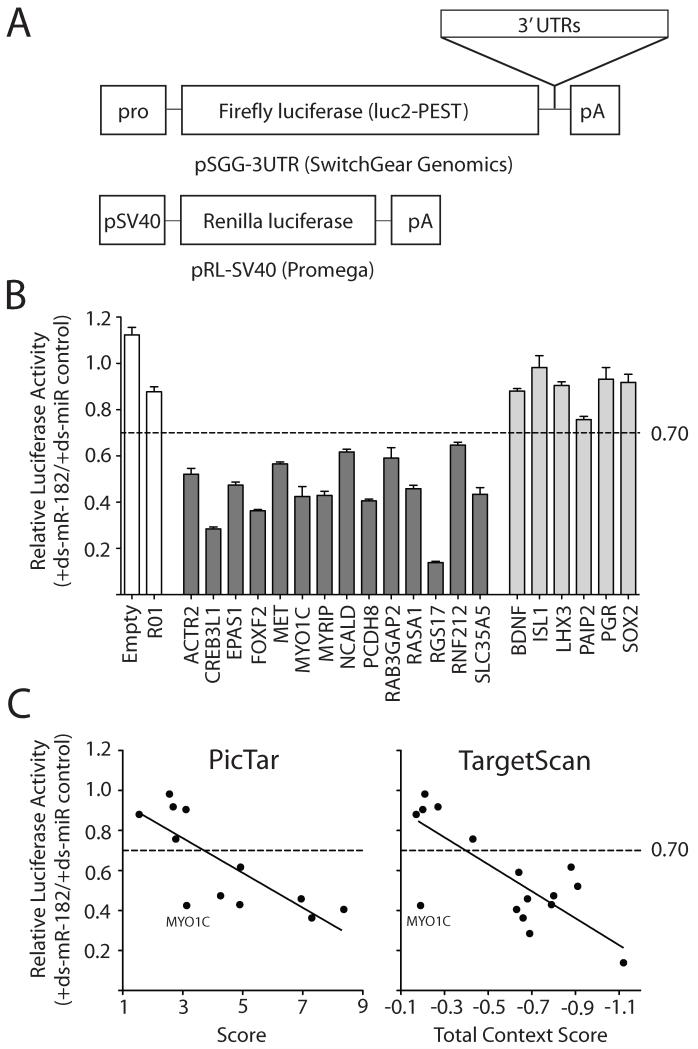
In vitro assays can be employed to evaluate protein levels directly (such as with immunostains or Western blots) or to monitor translational activity indirectly using luciferase reporters. By way of offering an example of the latter method, Figure 3 shows results from luciferase reporter assays performed in human embryonic kidney cells (HEK293T cells) for 20 human 3′ UTRs that varied widely in their score values as predicted targets for miR-182. Normalized luciferase activity was reduced by at least 30% in the presence of ds-miR-182 mimic for 14 tested targets. Interestingly, the strength of the scores generated from both TargetScan and PicTar algorithms showed moderately good correlations with the magnitude of the knockdowns in the luciferase assays. An exception was seen for the myosin1C (myo1C) 3′UTR reporter, which showed relatively greater repression by miR-182 in comparison with the other potential targets (Figure 3C, D).
Figure 3.
Luciferase assays for validating potential targets of miR-182. (A) Plasmids for in vitro luciferase assays. Human 3′UTRs were placed downstream of the coding region for destabilized Firefly luciferase (purchased from SwitchGear Genomics) and these test plasmids were then co-transfected with Renilla luciferase plasmids into HEK293T cells, along with double-stranded (ds) miRNA mimics (Thermo Scientific Dharmacon). (B) Relative luciferase activity. At 24 hours post-transfection, luminescence originating from Renilla protein was used to normalize the transfection efficiency in each well of a 96-well plate using the Dual-Glo Luciferase Assay System (Promega). Then, the luminescence from Firefly luciferase in the presence of miRIDIAN double-stranded (ds) miRNA-182 mimic was compared that obtained in the presence of a miRIDIAN ds-miRNA mimic negative control #1 (5′-UCACAACCUCCUAGAAAGAGUAGA-3′) provided by the manufacturer. Values are shown as means plus standard errors from at least six replicates performed over at least two independent experiments. White bars are negative controls. Dark bars are constructs with luminescence ratios below 0.70 (dashed line); the knockdowns by miR-182 are statistically significant in comparison to a plasmid carrying the R01 3′UTR that has no predicted binding sites for miR-182 (t-test, p<0.05). Grey bars are constructs that were not significantly different from the R01. (C) Relative luciferase activity shown as a function of target prediction scores for PicTar and TargetScan. Note that each program outputs a score for only a subset of the 20 tested 3′UTRs. Linear regression analysis of the data gives modest correlation coefficients for TargetScan (R2 = 0.5961) and PicTar (R2 = 0.5851)
Combining these results with other assays reported in the literature, we compiled a list of validated targets of miR-96, miR-182 and miR-183 (Table 1). A number of these transcripts are expressed in hair cells, as determined by deep sequencing of purified hair cells (SHIELD; Shared Harvard Inner-Ear Laboratory Database; https://shield.hms.harvard.edu/gene_search.html). For these genes, the miR-183 family likely acts to modulate target protein levels, possibly to initiate or maintain a mature hair cell phenotype. Others have speculated that while miR-183 may promote hair cell differentiation, miR-96 and miR-182 may instead promote a proliferative state (Hei et al 2011). This idea is based on their association with oncogenesis in other cells types, as well as temporal changes in their expression levels as sphere-forming cochlear progenitors are grown under differentiation conditions for two weeks: miR-183 peaks at 6 days as the other two fall precipitously. However, the percentage of cells differentiating into hair-cell-like cells was not evaluated in these cultures, and is likely to be below 0.3% (Diensthuber et al 2009). A resolution of the potentially conflicting activities of miR-183 family members will require a more comprehensive accounting and verification of their direct targets, as well as in vivo validations such as miR-specific knockouts.
TABLE 1. Validated targets of hair-cell-enriched miRNAs based on bioactivity assays.
| miR-182 Target | Reference |
|---|---|
| ACTR2 (h) | Fig. 3 |
| ADCY6 (h) | Xu et al., 2007; (%) Jalvy-Delvaille et al., 2012 |
| ARRDC3 (m) | Zhu et al., 2011 |
| BCL2 (h) | Yan et al., 2012 |
| BRCA1 (h) | Moskwa et al., 2011 |
| CASP2 (m) | Zhu et al., 2011 |
| CCND2 (h) | Yan et al., 2012 |
| CLOCK (h) | Saus et al., 2010 |
| CREB3L1 (h) | Fig. 3 |
| EPAS1 (h) | Fig. 3 |
| FOXF2 (h) | Fig. 3 |
| FOXO1 (h) | Guttilla et al., 2009; (%) not a target in Jalvy-Delvaille et al., 2012 |
| FOXO3 (h) | Segura et al., 2009 |
| MET (h) | Fig. 3 |
| MITF (h) | Xu et al., 2007; Segura et al., 2009; Yan et al., 2012 |
| MTSS1 (h) | Wang et al., 2012 (a) |
| MYO1C (h) | Fig. 3 |
| MYRIP (h) | Fig. 3 |
| NCALD (h) | Fig. 3 |
| PCDH8 (h) | Fig. 3 |
| RAB3GAP2 (h) | Fig. 3 |
| RASA1 (h) | Fig. 3 |
| RGS17 (h) | Sun et al., 2010; Fig. 3 |
| RNF212 (h) | Fig. 3 |
| SLC30A1 (h) % | Mihelich et al., 2011 |
| SLC30A7 (h) % | Mihelich et al., 2011 |
| SLC35A5 (h) | Fig. 3 |
| SLC39A1 (h) % | Mihelich et al., 2011 |
| SLC39A7 (h) % | Mihelich et al., 2011 |
| SOX2 (h) # | Weston et al., 2011 |
| SPAST (h) % | Henson et al., 2012 |
| TBX1 (m) | Wang et al., 2012 (b) |
| miR-183 Target | Reference |
|---|---|
| EGR1 (h) | Sarver et al., 2010 |
| ITGB1 (h) | Li et al., 2010 (a) |
| KIF2A (h) | Li et al., 2010 (a) |
| PDCD4 (h) | Li et al., 2010 (c) |
tested as a potential target for hs-mR-96(13G>A), but also repressed by wildtype hs-miR-96
tested as a potential target for hs-mR-96(14C>A), but also repressed by wildtype hs-miR-96
miR-96 binding site is present in the coding region, not in the 3′UTR
not validated by luciferase assay
these may not be valid targets because they were repressed less than 30% in the presence of the miRNA
BOLDgenes expressed in cochlear hair cells (GFP+) at greater than 50 reads at P0, P4 and/or P7 in Pou4f3-GFP mice; SHIELD database
italicsgenes repressed at least 30% in P0 cochlear hair cells (GFP+) vs. other cochlear cells (GFP−) in Pou4f3-GFP mice; SHIELD database
m, h mouse, human
C: The genetic mechanisms and constraints of hair cell regeneration
All non-mammalian vertebrates examined to date have the capacity to regenerate their hair cells. This can occur either as part of normal turnover (for example, in the constant replacement of lateral line hair cells in bony fish or in the vestibular organs of birds; (Ma & Raible 2009, Roberson et al 1992), or following loss of hair cells after injury in fish, amphibians, lizards and birds (Avallone et al 2008, Corwin & Cotanche 1988, Ma & Raible 2009, Ryals & Rubel 1988, Straube & Tanaka 2006). Supporting cells are capable of directly trans-differentiating into hair cells following injury – this has been best characterized in birds and begins in the first 24 hours after hair cell loss (Cafaro et al 2007). The potential depletion of supporting cells by this regenerative strategy is offset by a robust proliferation of supporting cells that begins several days after damage, and it is likely that at least some of these cell divisions are asymmetric, generating both hair cells and supporting cells (Stone & Cotanche 2007). Mammals, in contrast, have a very limited capacity for hair cell regeneration (Groves 2010). Although a very small amount of proliferation can be seen in the mature vestibular system of mammals following damage, together with a small number of morphologically immature hair cells (Forge et al 1993, Forge et al 1998, Kawamoto et al 2009), the degree of vestibular hair cell regeneration seen in mammals is a fraction of that seen in other vertebrates. Moreover, there is almost no evidence of proliferation or hair cell recovery following damage to the mature organ of Corti (Roberson & Rubel 1994, Yamasoba & Kondo 2006). Nevertheless, the widespread occurrence of hair cell loss in aging humans has prompted many comparisons between mammals and non-mammalian vertebrates in the hopes of identifying genetic or biochemical pathways that underlie regeneration but which may lie dormant in mammals.
C.1: To what extent is hair cell regeneration a recapitulation of hair cell development?
As discussed in section A, Atoh1 is both necessary and in some circumstances sufficient for hair cell differentiation. Its role appears to be confined to establishing rather than maintaining hair cell fate, as it is rapidly down-regulated in hair cells as they mature. A small number of Atoh1-expressing cells can be observed in sensory organs of non-mammalian vertebrates that exhibit normal hair cell turnover, such as fish lateral line organs (Ma et al 2008) or bird vestibular organs (Cafaro et al 2007), This suggests that Atoh1 is re-activated in supporting cells during the process of regeneration, an idea borne out by the observation that Atoh1 is rapidly and broadly up-regulated in fish and bird supporting cells following damage (Cafaro et al 2007, Lewis et al 2012, Ma & Raible 2009, Ma et al 2008). Recent experiments using adenovirally-transduced reporters of Atoh1 enhancer activity in cultures of the chicken basilar papilla treated with ototoxic antibiotics suggest that only about 50% of supporting cells that re-activate the Atoh1 enhancer go on to differentiate into hair cells as defined by the expression of Myosin6 (Lewis et al 2012). It is possible that some cells that begin to express Atoh1 are eventually diverted back to a supporting cell fate by processes such as Notch-mediated lateral inhibition. Supporting this, the Notch ligand Delta1 is one of the first genes to be re-activated during hair cell regeneration in birds and fish (Ma et al 2008, Stone & Rubel 1999). Moreover, pharmacological inhibition of Notch signaling in drug-damaged chick basilar papilla cultures increased the proportion of hair cells that derived from cells activating the Atoh1 enhancer from about 50% to 75% (Lewis et al 2012).
Although only very limited hair cell regeneration is seen in the vestibular system of mammals, recent work suggests that the potential of mammalian sensory tissue to respond to damage may be greater than previously realized. Transduction of Atoh1 reporter constructs into cultures of the drug-damaged adult mouse utricle showed that an average of almost 200 cells/utricle activated the Atoh1 enhancer after 18 days of culture (Lin et al 2011). However, very few (less than 5%) of these cells expressed Atoh1 protein or hair cell markers such as Myosin7a. Similar results have also been observed an in vivo ototoxic lesion to the vestibular system (Kawamoto et al 2009). As in birds, the efficiency of hair cell generation in mouse utricular cultures could be significantly increased – by almost 30-fold - by blocking Notch signaling (Lin et al 2011), although the numbers of hair cells generated by the damaged mouse utricle is still far less than those seen in birds even after Notch inhibition.
These results suggest that hair cell regeneration has at least some superficial similarities with hair cell development, in that Atoh1 is activated rapidly and in a manner that is at least partly subject to regulation by the Notch pathway (Puligilla & Kelley 2009). Moreover, mammalian vestibular tissue is able to initiate at least some of these regenerative steps, albeit at a much lower efficiency than in other vertebrates. One significant difference is that regenerating hair cells are derived from supporting cells rather than prosensory progenitors. At present, the lack of unambiguous markers to distinguish these two cell populations confounds the question of whether supporting cells undergo partial de-differentiation into a prosensory cell state during the course of regeneration, or are able to directly activate Atoh1 and become hair cells from their mature state.
C.2: Age-dependent obstacles to hair cell regeneration in mammals
A number of studies suggest that young mammals possess at least some of the molecular components to respond to damage by replacing hair cells or triggering proliferation of supporting cells, but that this potential is rapidly lost with age. For example, cochlear hair cells can be replaced in the embryonic organ of Corti after laser ablation, but not in neonatal mice (Kelley et al 1995). Genetic ablation of neonatal mouse utricular hair cells with diphtheria toxin leads to significant mitotic replacement of hair cells, most likely by supporting cells, but this phenomenon is no longer observed in five day old mice (Burns et al 2012a). Supporting cells purified from the neonatal mouse cochlea are capable of re-entering the cell cycle and generating Atoh1+ hair cells in culture, but this capacity is lost by two weeks after birth (White et al 2006). Similarly, the ability of cochlear or vestibular sensory tissue to generate proliferative non-adherent sphere cultures also declines with age (Oshima et al 2007).
What are the age-dependent constraints on supporting cell proliferation and hair cell replacement in mammals? Recent work from Corwin and colleagues has established a very strong correlation between the decline in the ability of supporting cells to spread and proliferate in normal or damaged utricular explants with the establishment of thick bands of F-actin and E-cadherin in the apical junctions between supporting cells (Burns et al 2008). In contrast, the cortical actin bands in chicken utricular supporting cells remain thin throughout the life of the animal. In the absence of perturbation studies, it is not clear at present whether the establishment of thick F-actin/E-cadherin-containing bands in mammals directly regulates the capacity of supporting cells to divide, or whether it is simply an indicator of more global changes occurring in supporting cells with age that might also include their ability to generate hair cells. In this regard, it is interesting to note that supporting cells in the striolar region of the utricle – which has the capacity to activate Atoh1 expression in the adult mouse after damage and inhibition of Notch signaling – contain significantly less E-cadherin and have thinner actin bands than their extra-striolar counterparts (Collado et al 2011).
In addition to changes in cell structure, it is also likely that the mature sensory epithelium of mammals may exhibit changes in responsiveness to cell-cell signaling. In particular, several recent studies suggest the Notch signaling pathway may no longer be instrumental in maintaining the balance between hair cells and supporting cells in older animals. For example, blocking Notch signaling in the undamaged chicken basilar papilla or the undamaged zebrafish larval lateral line has no effect on supporting cell proliferation or hair cell generation (Daudet et al 2009, Ma et al 2008), suggesting that either Notch signaling is not necessary to maintain the mature array of hair cells and supporting cells, or that other factors are able to compensate for the loss of Notch activity. Moreover, although pharmacological or genetic inhibition of Notch signaling can rapidly up-regulate Atoh1 and promote the expression of hair cell markers such as Myosin6 and Myosin7a in perinatal mouse cochlear or utricular cultures, these effects decline rapidly with age, both in vitro and in vivo (Doetzlhofer et al 2009, Hori et al 2007, Lin et al 2011).
As described above, some of the age-dependent changes in the ability of mammalian sensory epithelium to regenerate may be due to a failure to re-activate expression of Atoh1 after it is down-regulated in early postnatal life. In an attempt to bypass these age-dependent limitations, a number of studies have used adenoviral or transgenic expression of Atoh1 to generate new hair cells (e.g. Izumikawa et al., 2005). Two recent studies used transgenic techniques to activate Atoh1 expression either throughout the inner ear (Kelly et al 2012) or specifically in sub-populations of supporting cells (Liu et al 2012b). In either case, expression of Atoh1 in supporting cells or in non-sensory cochlear epithelium was able to induce new hair cells, some of which assembled stereociliary bundles and displayed voltage-dependent currents (Kelly et al 2012, Liu et al 2012b). However, in both studies, the ability of Atoh1 to promote ectopic hair cell formation in all regions of the cochlea declined rapidly and was severely compromised by two weeks of age, the time at which hearing begins in mice (Kelly et al 2012, Liu et al 2012b). This failure was also seen when Atoh1 was activated in supporting cells of adult mice in which hair cells had been ablated with ototoxic drugs (Liu et al 2012b).
There are many possible reasons why the mature mammalian ear becomes refractory to over-expression of Atoh1. In a recent review of Atoh1 function, Mulvaney and Dabdoub (2012) discuss a number of ways in which Atoh1 activity and binding to its E-box DNA targets might be regulated at the post-translational level, and each of these might contribute to the failure of Atoh1 to activate hair cell-specific gene expression in the adult mammal. For example, the C-terminus of Atoh1 has a conserved serine-rich domain that contains potential phosphorylation sites that could regulate its activity (Mulvaney & Dabdoub 2012). Basic helix-loop-helix proteins like Atoh1 require bHLH binding partners such as E47 to correctly bind to their E-box targets. Competition for the partners of Atoh1 by inhibitory Id HLH family members, or by other bHLH proteins, might reduce or abolish the transcriptional activity of Atoh1 in adult tissue. Atoh1 activity can also be modulated in different cell types by forming complexes with different bHLH factors that promote binding to different sets of E-boxes. The expression of such factors in adult supporting cells might direct Atoh1 away from the promoters of hair cell genes and hence block its hair cell promoting activity. Finally, it is possible that other classes of transcription factors co-operate with Atoh1 in young sensory tissue but are no longer expressed in older tissue, thus depriving Atoh1 of transcriptional partners.
A final possibility for why Atoh1 is unable to promote hair cell formation in the mature mammalian inner ear is that direct transcriptional targets of Atoh1 become epigenetically modified in supporting cells with age and are rendered unavailable for transcription, even in the presence of exogenous Atoh1. In such a model, some form of epigenetic reprogramming would be required to render mammalian supporting cells competent to respond to Atoh1 again. Whether this could be achieved by altering the activity of histone-modifying complexes or would require defined reprogramming factors (for example, similar to those required to reprogram fibroblasts into neurons; (Lujan et al 2012, Vierbuchen et al 2010, Yang et al 2011) is not clear. A better understanding of the epigenetic regulation of Atoh1 targets in the inner ear is hampered by a lack of good candidates for genes directly regulated by Atoh1. As discussed above, two recent studies used ChIP-seq, RNA-seq and mining of microarray databases to identify and validate a set of potential direct targets of Atoh1 in cerebellar granule cells and dorsal spinal cord interneurons (Klisch et al 2011, Lai et al 2011). However, of these, only three – Rab15, Selm and Atoh1 itself – have been shown to be expressed in hair cells.
ACRONYMS
- ChIP-Seq
Chromatin immunoprecipitation and deep sequencing
- GFP
Green Fluorescent Protein
- miRNA
MicroRNA
REFERENCES
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012;22:377–90. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Alsina B, Giraldez F, Pujades C. Patterning and cell fate in ear development. Int J Dev Biol. 2009;53:1503–13. doi: 10.1387/ijdb.072422ba. [DOI] [PubMed] [Google Scholar]
- Avallone B, Fascio U, Balsamo G, Marmo F. Gentamicin ototoxicity in the saccule of the lizard Podarcis Sicula induces hair cell recovery and regeneration. Hear Res. 2008;235:15–22. doi: 10.1016/j.heares.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Smibert P, Lai EC. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol. 2010;338:63–73. doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Arie N, McCall AE, Berkman S, Eichele G, Bellen HJ, Zoghbi HY. Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis. Hum Mol Genet. 1996;5:1207–16. doi: 10.1093/hmg/5.9.1207. [DOI] [PubMed] [Google Scholar]
- Bok J, Chang W, Wu DK. Patterning and morphogenesis of the vertebrate inner ear. Int J Dev Biol. 2007;51:521–33. doi: 10.1387/ijdb.072381jb. [DOI] [PubMed] [Google Scholar]
- Briggs KJ, Eberhart CG, Watkins DN. Just say no to ATOH: how HIC1 methylation might predispose medulloblastoma to lineage addiction. Cancer Res. 2008;68:8654–6. doi: 10.1158/0008-5472.CAN-08-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–86. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Burns J, Christophel JJ, Collado MS, Magnus C, Carfrae M, Corwin JT. Reinforcement of cell junctions correlates with the absence of hair cell regeneration in mammals and its occurrence in birds. J Comp Neurol. 2008;511:396–414. doi: 10.1002/cne.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Cox BC, Thiede BR, Zuo J, Corwin JT. In vivo proliferative regeneration of balance hair cells in newborn mice. J Neurosci. 2012a;32:6570–7. doi: 10.1523/JNEUROSCI.6274-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, On D, Baker W, Collado MS, Corwin JT. Over Half the Hair Cells in the Mouse Utricle First Appear After Birth, with Significant Numbers Originating from Early Postnatal Mitotic Production in Peripheral and Striolar Growth Zones. J Assoc Res Otolaryngol. 2012b doi: 10.1007/s10162-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro J, Lee GS, Stone JS. Atoh1 expression defines activated progenitors and differentiating hair cells during avian hair cell regeneration. Dev Dyn. 2007;236:156–70. doi: 10.1002/dvdy.21023. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126:1581–90. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- Collado MS, Thiede BR, Baker W, Askew C, Igbani LM, Corwin JT. The postnatal accumulation of junctional E-cadherin is inversely correlated with the capacity for supporting cells to convert directly into sensory hair cells in mammalian balance organs. J Neurosci. 2011;31:11855–66. doi: 10.1523/JNEUROSCI.2525-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240:1772–4. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- D’Angelo A, Bluteau O, Garcia-Gonzalez MA, Gresh L, Doyen A, et al. Hepatocyte nuclear factor 1alpha and beta control terminal differentiation and cell fate commitment in the gut epithelium. Development. 2010;137:1573–82. doi: 10.1242/dev.044420. [DOI] [PubMed] [Google Scholar]
- Daudet N, Gibson R, Shang J, Bernard A, Lewis J, Stone J. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev Biol. 2009;326:86–100. doi: 10.1016/j.ydbio.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diensthuber M, Oshima K, Heller S. Stem/progenitor cells derived from the cochlear sensory epithelium give rise to spheres with distinct morphologies and features. J Assoc Res Otolaryngol. 2009;10:173–90. doi: 10.1007/s10162-009-0161-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver EC, Pryor SP, Hill P, Turner J, Ruther U, et al. Hedgehog signaling regulates sensory cell formation and auditory function in mice and humans. J Neurosci. 2008;28:7350–8. doi: 10.1523/JNEUROSCI.0312-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert PJ, Timmer JR, Nakada Y, Helms AW, Parab PB, et al. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 autoregulation. Development. 2003;130:1949–59. doi: 10.1242/dev.00419. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Forge A, Li L, Corwin JT, Nevill G. Ultrastructural evidence for hair cell regeneration in the mammalian inner ear. Science. 1993;259:1616–9. doi: 10.1126/science.8456284. [DOI] [PubMed] [Google Scholar]
- Forge A, Li L, Nevill G. Hair cell recovery in the vestibular sensory epithelia of mature guinea pigs. J Comp Neurol. 1998;397:69–88. [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, Tenne T, Toren G, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009;106:7915–20. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleich O, Fischer FP, Köppl C, Manley GA, Manley GA, Popper AN, Fay RR. Evolution of the Vertebrate Auditory System. Springer Verlag; New York: 2004. Hearing organ evolution and specialization: Archosaurs; pp. 224–55. [Google Scholar]
- Goodyear R, Richardson G. Pattern formation in the basilar papilla: evidence for cell rearrangement. J Neurosci. 1997;17:6289–301. doi: 10.1523/JNEUROSCI.17-16-06289.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 2010;235:434–46. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–57. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–16. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci. 2008;28:5991–9. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hei R, Chen J, Qiao L, Li X, Mao X, et al. Dynamic changes in microRNA expression during differentiation of rat cochlear progenitor cells in vitro. Int J Pediatr Otorhinolaryngol. 2011;75:1010–4. doi: 10.1016/j.ijporl.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–96. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Henson BJ, Zhu W, Hardaway K, Wetzel JL, Stefan M, et al. Transcriptional and post-transcriptional regulation of SPAST, the gene most frequently mutated in hereditary spastic paraplegia. PLoS One. 2012;7:e36505. doi: 10.1371/journal.pone.0036505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. miRNAs play a tune. Cell. 2007;131:22–4. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Holley M, Rhodes C, Kneebone A, Herde MK, Fleming M, Steel KP. Emx2 and early hair cell development in the mouse inner ear. Dev Biol. 2010;340:547–56. doi: 10.1016/j.ydbio.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori R, Nakagawa T, Sakamoto T, Matsuoka Y, Takebayashi S, Ito J. Pharmacological inhibition of Notch signaling in the mature guinea pig cochlea. Neuroreport. 2007;18:1911–4. doi: 10.1097/WNR.0b013e3282f213e0. [DOI] [PubMed] [Google Scholar]
- Huh SH, Jones J, Warchol ME, Ornitz DM. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 2012;10:e1001231. doi: 10.1371/journal.pbio.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CH, Guo D, Harris MA, Howard O, Mishina Y, et al. Role of bone morphogenetic proteins on cochlear hair cell formation: analyses of Noggin and Bmp2 mutant mice. Dev Dyn. 2010;239:505–13. doi: 10.1002/dvdy.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–6. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Calisto LE, Morris KA, et al. Expression of Neurog1 instead of Atoh1 can partially rescue organ of Corti cell survival. PLoS One. 2012;7:e30853. doi: 10.1371/journal.pone.0030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One. 2010;5:e11661. doi: 10.1371/journal.pone.0011661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalvy-Delvaille S, Maurel M, Majo V, Pierre N, Chabas S, et al. Molecular basis of differential target regulation by miR-96 and miR-182: the Glypican-3 as a model. Nucleic Acids Res. 2012;40:1356–65. doi: 10.1093/nar/gkr843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KP, Covault J. Human miR-1271 is a miR-96 paralog with distinct non-conserved brain expression pattern. Nucleic Acids Res. 2010;39:701–11. doi: 10.1093/nar/gkq798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KP, Covault J, Conner TS, Tennen H, Kranzler HR, Furneaux HM. A common polymorphism in serotonin receptor 1B mRNA moderates regulation by miR-96 and associates with aggressive human behaviors. Mol Psychiatry. 2009;14:381–9. doi: 10.1038/mp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaid A, Neves J, Giraldez F. Id gene regulation and function in the prosensory domains of the chicken inner ear: a link between Bmp signaling and Atoh1. J Neurosci. 2010;30:11426–34. doi: 10.1523/JNEUROSCI.2570-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki S, Beyer LA, Swiderski DL, Izumikawa M, Stover T, et al. p27(Kip1) deficiency causes organ of Corti pathology and hearing loss. Hear Res. 2006;214:28–36. doi: 10.1016/j.heares.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Katayama A, Corwin JT. Cell production in the chicken cochlea. J Comp Neurol. 1989;281:129–35. doi: 10.1002/cne.902810110. [DOI] [PubMed] [Google Scholar]
- Kawamoto K, Ishimoto S, Minoda R, Brough DE, Raphael Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J Neurosci. 2003;23:4395–400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto K, Izumikawa M, Beyer LA, Atkin GM, Raphael Y. Spontaneous hair cell regeneration in the mouse utricle following gentamicin ototoxicity. Hear Res. 2009;247:17–26. doi: 10.1016/j.heares.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–49. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- Kelley MW, Talreja DR, Corwin JT. Replacement of hair cells after laser microbeam irradiation in cultured organs of corti from embryonic and neonatal mice. J Neurosci. 1995;15:3013–26. doi: 10.1523/JNEUROSCI.15-04-03013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal Mammalian cochlea in vivo. J Neurosci. 2012;32:6699–710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–5. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard M, Nyengaard JR. Stereological study of postnatal development in the mouse utricular macula. J Comp Neurol. 2005;492:132–44. doi: 10.1002/cne.20736. [DOI] [PubMed] [Google Scholar]
- Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci U S A. 2011;108:3288–93. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–20. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kuhn S, Johnson SL, Furness DN, Chen J, Ingham N, et al. miR-96 regulates the progression of differentiation in mammalian cochlear inner and outer hair cells. Proc Natl Acad Sci U S A. 2011;108:2355–60. doi: 10.1073/pnas.1016646108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–71. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Liu F, Segil N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development. 2006;133:2817–26. doi: 10.1242/dev.02453. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–8. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MA, Steel KP. MicroRNAs in mouse development and disease. Semin Cell Dev Biol. 2010;21:774–80. doi: 10.1016/j.semcdb.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RM, Hume CR, Stone JS. Atoh1 expression and function during auditory hair cell regeneration in post-hatch chickens. Hear Res. 2012;289:74–85. doi: 10.1016/j.heares.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J Biol Chem. 2010a;285:5461–71. doi: 10.1074/jbc.M109.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Corrales CE, Wang Z, Zhao Y, Wang Y, et al. BMP4 signaling is involved in the generation of inner ear sensory epithelia. BMC Dev Biol. 2005;5:16. doi: 10.1186/1471-213X-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010b;30:3254–63. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Fu H, Xu C, Tie Y, Xing R, et al. miR-183 inhibits TGF-beta1-induced apoptosis by downregulation of PDCD4 expression in human hepatocellular carcinoma cells. BMC Cancer. 2010c;10:354. doi: 10.1186/1471-2407-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Dai T, Xiong H, Zhao X, Chen X, et al. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5:e15797. doi: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin V, Golub JS, Nguyen TB, Hume CR, Oesterle EC, Stone JS. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J Neurosci. 2011;31:15329–39. doi: 10.1523/JNEUROSCI.2057-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Liang XH, Su RW, Lei W, Jia B, et al. Combined analysis of microRNome and 3′-UTRome reveals a species-specific regulation of progesterone receptor expression in the endometrium of rhesus monkey. J Biol Chem. 2012a;287:13899–910. doi: 10.1074/jbc.M111.301275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–21. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dearman JA, Cox BC, Walters BJ, Zhang L, et al. Age-dependent in vivo conversion of mouse cochlear pillar and deiters’ cells to immature hair cells by atoh1 ectopic expression. J Neurosci. 2012b;32:6600–10. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu J, Segura MF, Shao C, Lee P, et al. MiR-182 overexpression in tumourigenesis of high-grade serous ovarian carcinoma. J Pathol. 2012c doi: 10.1002/path.4000. [DOI] [PubMed] [Google Scholar]
- Lowenheim H, Furness DN, Kil J, Zinn C, Gultig K, et al. Gene disruption of p27(Kip1) allows cell proliferation in the postnatal and adult organ of corti. Proc Natl Acad Sci U S A. 1999;96:4084–8. doi: 10.1073/pnas.96.7.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109:2527–32. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–95. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- Ma EY, Raible DW. Signaling pathways regulating zebrafish lateral line development. Curr Biol. 2009;19:R381–6. doi: 10.1016/j.cub.2009.03.057. [DOI] [PubMed] [Google Scholar]
- Ma EY, Rubel EW, Raible DW. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J Neurosci. 2008;28:2261–73. doi: 10.1523/JNEUROSCI.4372-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–50. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–13. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- Mihelich BL, Khramtsova EA, Arva N, Vaishnav A, Johnson DN, et al. miR-183-96-182. cluster is overexpressed in prostate tissue and regulates zinc homeostasis in prostate cells. J Biol Chem. 2011;286:44503–11. doi: 10.1074/jbc.M111.262915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Kelley MW. Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J Neurosci. 2003;23:9469–78. doi: 10.1523/JNEUROSCI.23-28-09469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41:210–20. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvaney J, Dabdoub A. Atoh1, an Essential Transcription Factor in Neurogenesis and Intestinal and Inner Ear Development: Function, Regulation, and Context Dependency. J Assoc Res Otolaryngol. 2012 doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H, Sakamoto H, Hayakawa H, Arao Y, Satoh K, et al. The intestine-specific homeobox gene Cdx2 induces expression of the basic helix-loop-helix transcription factor Math1. Differentiation. 2006;74:313–21. doi: 10.1111/j.1432-0436.2006.00074.x. [DOI] [PubMed] [Google Scholar]
- Neves J, Uchikawa M, Bigas A, Giraldez F. The prosensory function of Sox2 in the chicken inner ear relies on the direct regulation of Atoh1. PLoS One. 2012;7:e30871. doi: 10.1371/journal.pone.0030871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, Groves AK. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J Neurosci. 2010;30:15044–51. doi: 10.1523/JNEUROSCI.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Grimm CM, Corrales CE, Senn P, Martinez Monedero R, et al. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol. 2007;8:18–31. doi: 10.1007/s10162-006-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Kopecky B, Santi P, et al. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear Res. 2011;275:66–80. doi: 10.1016/j.heares.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–80. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Pujades C, Kamaid A, Alsina B, Giraldez F. BMP-signaling regulates the generation of hair-cells. Dev Biol. 2006;292:55–67. doi: 10.1016/j.ydbio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30:714–22. doi: 10.1523/JNEUROSCI.3852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, et al. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn. 2007;236:1905–17. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Kelley MW. Building the world’s best hearing aid; regulation of cell fate in the cochlea. Curr Opin Genet Dev. 2009;19:368–73. doi: 10.1016/j.gde.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, et al. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–15. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- Riester A, Issler O, Spyroglou A, Rodrig SH, Chen A, Beuschlein F. ACTH-dependent regulation of microRNA as endogenous modulators of glucocorticoid receptor expression in the adrenal gland. Endocrinology. 2012;153:212–22. doi: 10.1210/en.2011-1285. [DOI] [PubMed] [Google Scholar]
- Roberson DF, Weisleder P, Bohrer PS, Rubel EW. Ongoing production of sensory cells in the vestibular epithelium of the chick. Hear Res. 1992;57:166–74. doi: 10.1016/0378-5955(92)90149-h. [DOI] [PubMed] [Google Scholar]
- Roberson DW, Rubel EW. Cell division in the gerbil cochlea after acoustic trauma. Am J Otol. 1994;15:28–34. [PubMed] [Google Scholar]
- Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. 1967;Suppl 220:1–44. [PubMed] [Google Scholar]
- Rudnicki A, Avraham KB. microRNAs: the art of silencing in the ear. EMBO Mol Med. 2012 doi: 10.1002/emmm.201100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240:1774–6. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- Sacheli R, Nguyen L, Borgs L, Vandenbosch R, Bodson M, et al. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr Patterns. 2009;9:364–70. doi: 10.1016/j.gep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–80. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- Saus E, Soria V, Escaramis G, Vivarelli F, Crespo JM, et al. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum Mol Genet. 2010;19:4017–25. doi: 10.1093/hmg/ddq316. [DOI] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shi F, Cheng YF, Wang XL, Edge AS. Beta-catenin up-regulates Atoh1 expression in neural progenitor cells by interaction with an Atoh1 3′ enhancer. J Biol Chem. 2010;285:392–400. doi: 10.1074/jbc.M109.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda G, Robusto M, Primignani P, Castorina P, Benzoni E, et al. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet. 2012;21:577–85. doi: 10.1093/hmg/ddr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA. Little but loud: small RNAs have a resounding affect on ear development. Brain Res. 2009;1277:104–14. doi: 10.1016/j.brainres.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, et al. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–41. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CB, Davies AL, Battista S, Lewis JH, Fekete DM. Forced activation of Wnt signaling alters morphogenesis and sensory organ identity in the chicken inner ear. Dev Biol. 2003;261:149–64. doi: 10.1016/s0012-1606(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Stone JS, Cotanche DA. Hair cell regeneration in the avian auditory epithelium. Int J Dev Biol. 2007;51:633–47. doi: 10.1387/ijdb.072408js. [DOI] [PubMed] [Google Scholar]
- Stone JS, Rubel EW. Delta1 expression during avian hair cell regeneration. Development. 1999;126:961–73. doi: 10.1242/dev.126.5.961. [DOI] [PubMed] [Google Scholar]
- Straube WL, Tanaka EM. Reversibility of the differentiated state: regeneration in amphibians. Artif Organs. 2006;30:743–55. doi: 10.1111/j.1525-1594.2006.00296.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fang R, Li C, Li L, Li F, et al. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Commun. 2010;396:501–7. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- Takacs CM, Giraldez AJ. MicroRNAs as genetic sculptors: fishing for clues. Semin Cell Dev Biol. 2010;21:760–7. doi: 10.1016/j.semcdb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012a;12:227. doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XR, Zhang XM, Du J, Jiang H. MicroRNA-182 regulates otocyst-derived cell differentiation and targets T-box1 gene. Hear Res. 2012b;286:55–63. doi: 10.1016/j.heares.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Huang JW, Calses P, Kemp CJ, Taniguchi T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res. 2012c doi: 10.1158/0008-5472.CAN-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, et al. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011;240:808–19. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Weston MD, Soukup GA. MicroRNAs sound off. Genome Med. 2009;1:59. doi: 10.1186/gm59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441:984–7. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–66. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Yamasoba T, Kondo K. Supporting cell proliferation after hair cell injury in mature guinea pig cochlea in vivo. Cell Tissue Res. 2006;325:23–31. doi: 10.1007/s00441-006-0157-9. [DOI] [PubMed] [Google Scholar]
- Yan D, Dong XD, Chen X, Yao S, Wang L, et al. Role of MicroRNA-182 in Posterior Uveal Melanoma: Regulation of Tumor Development through MITF, BCL2 and Cyclin D2. PLoS One. 2012;7:e40967. doi: 10.1371/journal.pone.0040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Ng YH, Pang ZP, Sudhof TC, Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell. 2011;9:517–25. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Lu Z, Liu C, Meng Y, Ma Y, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–25. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3:580–6. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Sun W, Okano K, Chen Y, Zhang N, et al. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J Biol Chem. 2011;286:31749–60. doi: 10.1074/jbc.M111.259028. [DOI] [PMC free article] [PubMed] [Google Scholar]