Abstract
It has been established that the enzyme susceptibility of collagen, the predominant load-bearing protein in vertebrates, is altered by applied tension. However, whether tensile force increases or decreases the susceptibility to enzyme is a matter of contention. It is critical to establish a definitive understanding of the direction and magnitude of the force versus catalysis rate (kC) relationship if we are to properly interpret connective tissue development, growth, remodeling, repair, and degeneration. In this investigation, we examine collagen/enzyme mechanochemistry at the smallest scale structurally relevant to connective tissue: the native collagen fibril. A single-fibril mechanochemical erosion assay with nN force resolution was developed which permits detection of the loss of a few layers of monomer from the fibril surface. Native type I fibrils (bovine) held at three levels of tension were exposed to Clostridium histolyticum collagenase A. Fibrils held at zero-load failed rapidly and consistently (20 min) while fibrils at 1.8 pN/monomer failed more slowly (35–55 min). Strikingly, fibrils at 23.9 pN/monomer did not exhibit detectable degradation. The extracted force versus kC data were combined with previous single-molecule results to produce a “master curve” which suggests that collagen degradation is governed by an extremely sensitive mechanochemical switch.
Keywords: Collagen, ECM (extracellular matrix), Mechanical test, Enzymatic degradation
1 Introduction
A variety of fundamental processes in development, health, and disease depend on the efficient synthesis, modification, and degradation of collagen, the most abundant structural protein in vertebrates. The cleavage and removal of collagen monomers and fibrils is critical for effective wound healing (Beare et al. 2003), reproduction (Hulboy et al. 1997), and growth and remodeling (Inada et al. 2004). Poorly controlled collagen degradation, however, may play a significant role in tumor invasion (McCawley and Matrisian 2000), arthritis (Cawston et al. 1999), ulceration (Baragi et al. 1997), and intervertebral disk degeneration (Kang et al. 1996) as well as fibrosis (Kilian et al. 2007) and scarring. The intact collagen triple-helix, resistant to most enzymes, is cleaved by specific mammalian matrix metalloproteinases (MMPs) and cathepsins, as well as several enzymes produced by bacteria (such as Clostridium histolyticum) which are often associated with diseases (Harrington 1996).
Native collagen fibrils represent the most fundamental continuous structural elements in soft connective tissue and bone. In vivo, soluble collagen monomers are secreted into the extra-cellular matrix (ECM) individually or in microfibril bundles (Kadler 2004) and aggregate to form insoluble fibrils (Perumal et al. 2008) in an energetically driven process that increases collagen stability (Miles and Ghelashvili 1999). In connective tissue structures such as ligament and tendon, collagen fibrils are found generally parallel to load axes, suggesting that collagen accumulates preferentially into fibrils oriented to efficiently resist or transmit the mechanical loads generated by the musculoskeletal system. In more complex loading environments, such as blood vessels and the cornea, fibrils form a weave of quasi-orthogonal layers again suggesting fibrils align to optimize load distribution. While the initial vertebrate template is unquestionably derivative of direct cellular placement and control of extracellular material, we suggest that the molecular design of collagen contributes significantly to the optimization of collagen fibril structure and orientation during connective tissue growth, repair, and remodeling (Flynn et al. 2010; Zareian et al. 2010).
Though collagen molecules are unstable at physiological temperatures (Leikina et al. 2002), incorporation into fibrils stabilizes collagen against thermal denaturation (Miles and Ghelashvili 1999). Moreover, the application of physiologically relevant mechanical tension also increases the thermal stability of collagenous tissue (Bass et al. 2004; Chen et al. 1997). Low levels of mechanical tensile strain (<10 % tissue-level strain) or load (<10 pN/molecule) have also been shown to increase the stability of type I collagen against enzymatic degradation, by bacterial collagenase (BC) from Clostridium histolyticum, at multiple length scales. Mechanical stabilization has been observed in reconstituted collagen tape (Huang and Yannas 1977), in native tissue (Ruberti and Hallab 2005; Nabeshima et al. 1996; Zareian et al. 2010; Wyatt et al. 2009), in reconstituted fibrillar networks (Bhole et al. 2009), and in single, recombinant monomers (Camp et al. 2011). Importantly, the physiological relevance of these observations was supported by the finding that reconstituted type I fibrils also exhibit increased stability against MMP-8 with the application of low loads (Flynn et al. 2010).
While the majority of available literature suggests that mechanical load stabilizes collagen against both enzymatic and thermal degradation, two recently published investigations (Camp et al. 2011; Adhikari et al. 2011) find opposite effects of load on the degradation rate of molecular collagen (though the difference may be ascribed to the nature of the collagen substrate and enzymes used). Because of the numerous and potentially profound implications associated with the magnitude and direction of collagen mechanosensitivity to enzyme, it is critical to determine, unequivocally, whether the application of mechanical force reduces or enhances the stability of collagen-based tissues.
Quantitative characterization of the mechanochemical relationship at whole-tissue scales has been confounded by enzyme diffusion kinetics and network load distribution (Nabeshima et al. 1996; Zareian et al. 2010; Wyatt et al. 2009). Reconstituted collagen fibril networks (Bhole et al. 2009; Flynn et al. 2010) and tapes (Huang and Yannas 1977) are subject to uncontrolled local fibril loads and low maximum loading due to the lack of intermolecular cross-linking. These uncertainties have prevented extraction of a clear relationship between applied load and molecular cleavage rate constants (Zareian et al. 2010; Wyatt et al. 2009; Bhole et al. 2009; Flynn et al. 2010; Nabeshima et al. 1996; Ruberti and Hallab 2005). Massively parallel investigations at the single-molecule scale do permit direct extraction of a relationship between force and cleavage rate for individual collagen monomers (Camp et al. 2011; Adhikari et al. 2011). However, single monomer mechanochemistry methods are force-limited and may not be physiologically relevant because they do not address how molecular packing into fibrils and covalent cross-linking affects enzymatic degradation dynamics (Perumal et al. 2008; Adhikari et al. 2011; Camp et al. 2011).
In this investigation, we develop and employ a single-fibril mechanochemical erosion assay that allows simultaneous measurement of applied force and the resulting strain on a single native collagen fibril and provides a sensitive measure of enzymatic degradation. The assay is a simple, powerful method to study fibril mechanochemistry at a physiologically relevant scale without the confounding effects of diffusion and network complexity. We present data showing a strong relationship between applied mechanical load and collagen fibril degradation. We show force-dependent enzyme cleavage rates and specificities and provide a theoretical model to explain the mechanochemical relationship observed.
2 Materials and methods
2.1 Experimental setup
2.1.1 Fibril isolation and attachment
Individual type I collagen fibrils extracted from bovine sclera were pulled from solution and attached to calibrated glass microneedles (Supplementary Data) by applying epoxy drops (DP-100, 3 M) with a third microneedle. Fibrils and microneedles were observed via 60x differential interference contrast (DIC) microscopy. Microneedles were controlled with micromanipulators (NK2, Eppendorf) with 40 nm resolution. For mechanical testing, one needle was used for actuation, and the other needle was monitored for force deflection. Force was calculated in situ via optical measurement of calibrated needle displacement, with 214 nm resolution, and fibril length and strain were calculated via optical measurement of the distance between epoxy beads (Supplementary Data). For the purposes of this study, microneedles were tuned to tip deflection stiffnesses of 10–60 nN/μm, resulting in fibril force resolution of 2–12 nN (optical deflection resolution × deflection stiffness).
2.1.2 Fibril degradation
Fibrils were washed with 1.85 M NaCl to remove soluble collagen monomers and proteoglycans, submerged in Dulbecco’s Modified Eagle Medium (DMEM), with 15 min to equilibrate hydration, in a thermally controlled (37 °C) microbioreactor, and mechanically tested as shown in Fig. 1. Determining the fibril stiffness allowed calculation of the tensile loads required to reach 2 pN/monomer (low-load), 24 pN/monomer (high-load), and 40 pN/monomer (probe-load) initial load. Each fibril was then loaded to either zero-load (slack), low-load, or high-load and exposed to Clostridium histolyticum bacterial collagenase (BC) type A (C0130, Sigma-Aldrich), 5 μM in DMEM. The enzyme solution and fibril in the microbioreactor were then covered gently, to minimize mixing, with non-drying immersion oil (type-A, Cargille) to minimize dehydration. Fibril load was held constant (load-control), via small manual adjustments using the micromanipulators to maintain in situ, optically measured, force-deflection in the calibrated microneedle, throughout the experiment, except during periodic mechanical probes. To quantify enzymatic degradation rates, via calculated reductions in fibril stiffness, fibrils were mechanically probed in 300 s intervals, up to probe-load values (probing took <30 s). Loading rates were kept below 100 nN/s. It is worth noting the uncertainty (±4 pixels = ±428 nm) in manually measured microneedle deflection at low-load will be higher, 21 %, than the uncertainty for high-load measurements, 0.7 % (See Supplementary Data).
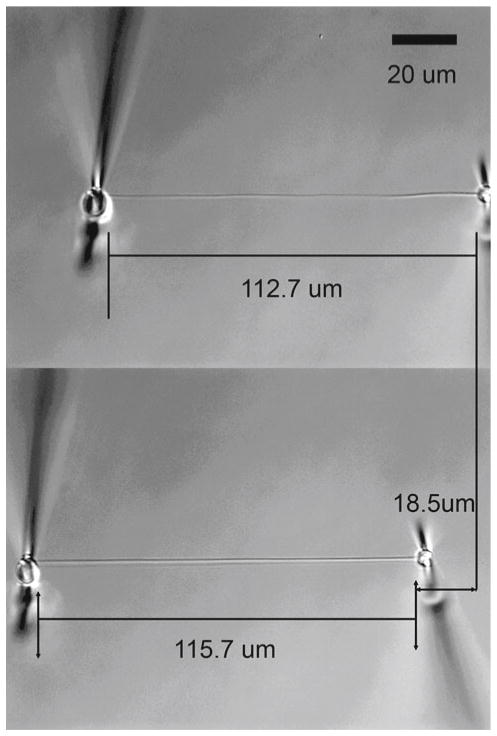
Fig. 1.
DIC image sequence of mechanical testing. An isolated native type I collagen fibril from bovine sclera is epoxied to calibrated (57 ± 8 nN/μm) glass microneedles, submerged in buffer, and mechanically loaded. Top Gauge length (zero-load). Bottom High-load of 1054 ± 149 nN produces 2.6 ± 0.3 % strain
2.1.3 Diameter measurement
Based on estimates of enzyme life 37 °C from supplier Sigma-Aldrich, the maximum experiment length was 4 hours. Fibrils that failed mechanically before experiment end were immediately washed with 2 M NaCl and prepared for scanning electron microscopy (SEM) analysis. Fibrils that did not fail by experiment end (high-load fibrils) were immediately washed in 2 M NaCl, unloaded to zero-load, exposed to a new batch of BC, and mechanically probed as before. These fibrils were also prepared for SEM analysis upon fibril failure. Briefly, microneedles were pressed into carbon tape and fibrils were platinum/palladium coated and then viewed using a Hitachi S4800 SEM. When possible, fibril dry diameters were quantified via SEM and converted to hydrated diameters by multiplying by the ratio of hydrated to dehydrated intermolecular spacing (1.23) (Huang and Meek 1999) to calculate the fibril elastic modulus. Control fibrils followed the zero-load procedure in DMEM without BC and were prepared for SEM analysis.
2.1.4 Force/molecule
We assume a uniform fibril elastic modulus of 0.7 GPa to convert stiffness measurements to radius values. The value 0.70 ± 0.06 GPa was calculated using measured diameters (SEM, n = 3), and agrees with elastic moduli in the literature for single fibrils, determined using atomic force microscopy or MEMS, 0.2–1 GPa for a hydrated cross-sectional area (van der Rijt et al. 2006; Eppell et al. 2006). With a constant fibril-level force, the per-molecule force values will increase as enzymatic degradation reduces cross-sectional area. Thus, we calculated radial degradation rates using stiffness values measured from early mechanical tests, when the fibrils have lost less than 20 % initial diameter. It is worth noting that the transition from continuum scale to the simplified molecular scale used here neglects local differences in packing arrangement, molecule orientation and spacing, and the potential existence of additional non-type I collagens and non-collagenous molecules. While all of these complicating local, molecular heterogeneities likely exist, the variation between fibrils is assumed small, and these contributions are likely averaged out over the large fibril surface areas probed in this study.
2.2 Analysis
2.2.1 1-D Erosion model
The enzymatic degradation of collagen follows a 2-step (binding and cleavage), Michaelis–Menten reaction process (Welgus et al. 1980). Cleavage rate, kC, 0.3–0.6 s−1 for soluble collagen monomers, is considered the degradation-rate-limiting step because the binding rate is faster by a factor of 106 (Mallya et al. 1992). Fibrillar collagen degradation also follows two-step kinetics, but with rates approximately 10x slower, because steric hindrance prevents fibril penetration and limits degradation to surface erosion (Okada et al. 1992; Welgus et al. 1980). Based on the enzyme concentration, molecular binding site density on the fibril surface, and the binding rate (~105 s−1) (Mallya et al. 1992), complete surface binding occurs within seconds of enzyme addition (Supplementary Data). Therefore, we assume all surface collagen molecules are constantly bound by enzyme. Axial and radial symmetry allows the simplification of the degrading fibril to a 1-D line of monomers along the radius.
2.2.2 Molecular cleavage rate, kC
The outermost shell of collagen molecules, each molecule bound at a cleavage site by BC, will degrade at the same cleavage rate, kC, as a single bound molecule (Supplementary Data). In other words, the observed radial degradation rate, converted into a molecular degradation rate by dividing by fibril intermolecular spacing (1.6 nm) (Meek and Leonard 1993), is approximately equal to the molecular cleavage rate, kC. We assume product inhibition, where previously cleaved fragments are continually bound and cleaved by enzyme, though present, is not a significant factor.
2.2.3 Specificity, kC/Km
Specificity (where Michaelis Constant Km = (kC + kr)/kf, kr and kf are reverse and forward binding constants) is calculated, using the reaction-diffusion equation solution for low enzyme and substrate (eq. 111, (Tzafriri et al. 2002), Supplementary Data). Concentration rates are converted to radial degradation rates to yield (1) (Supplementary Data).
| (1) |
where E0 is the initial enzyme concentration, dr/dt is the radial degradation rate, and 1.6 represents the intermolecular spacing within the fibril. It follows that when kr ≪ kC, specificity becomes a measure of the binding rate.
3 Results
3.1 Mechanics
All fibril force-strain curves followed a relatively linear-elastic response in the force range which was probed. Initial calculated fibril diameters were 205–507 nm. Because fibril diameters varied, initial mechanical testing was performed to determine the load required to achieve the desired initial strain and per-molecule force for each fibril.
3.1.1 Fibril ultimate failure
Zero and low-load fibrils were mechanically probed, incrementally up to probe-load (~40 pN/monomer), every 300 s. Zero-load fibrils (slack) failed consistently during the 1,200 s probe (n = 3), low-load fibrils (1.8±0.6 pN/monomer) failed during 2,100–3,300 s probes (n = 3), and high-load fibrils (23.9 ± 2.5 pN/monomer) did not fail or show visible signs of degradation at 14,400 s (n = 3). At test completion, the high-load fibrils, still intact after exposure to BC in the loaded state, were washed in 2 M NaCl to remove degradation products and bound enzyme, then brought to zero-load (slack), and exposed to fresh BC, at which point they degraded in 1,200–1,500 s (n = 3), showing congruence with the no-load fibril group and confirming both minimal degradation in the high-load state and the cleavability of the high-load fibrils.
3.1.2 Fibril diameters
After enzymatic degradation-induced mechanical failure, fibril halves were still visible via DIC and SEM. SEM analysis revealed ~67 nm D-period banding, characteristic of native fibrils (Hulmes et al. 1973), in control fibrils not exposed to enzyme (Fig. 2), and in some fibrils after failure. Banding was not always observed on the remnants of enzymatically degraded fibrils, indicating either loss in surface organization due to degradation or less molecular order in the fibril interior than on the fibril surface. Many fibrils were lost during SEM preparation. SEM diameter measurements, adjusted for dehydration (Huang and Meek 1999), were used to calculate fibril elastic modulus: 0.70 ± 0.06 GPa (n = 3). Though the elastic modulus varies among fibrils, likely due to different cross-link densities, the variation between fibrils is small compared to the differences in degradation rates between zero-load, low-load, and control groups and does not affect the statistical significance of the results. All fibrils were similar at the molecular level; the deviation in mechanical properties and degradation rates would be expected to be quite small. The significant differences found between zero-load, low-load, and high-load degradation rates, despite the expected fibril-to-fibril variation at the molecular scale, indicate the robustness of the differences in degradation rates.
Fig. 2.
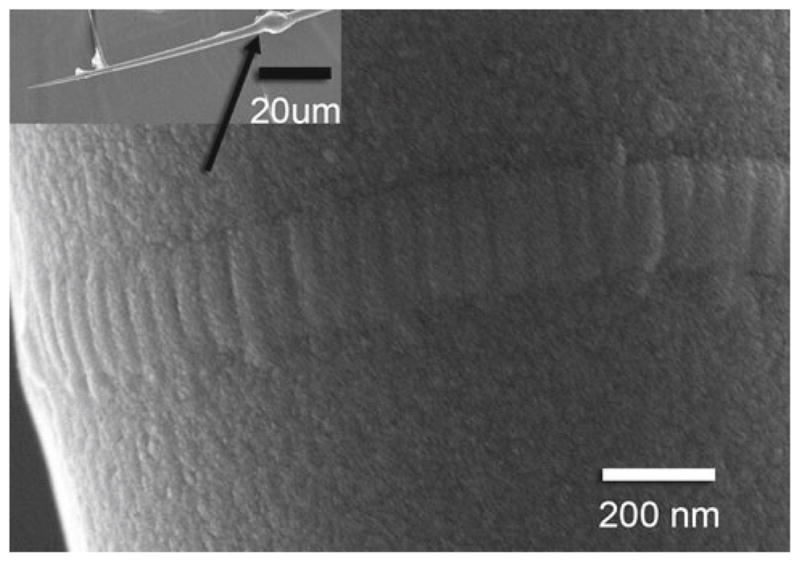
Scanning electron microscope image of a collagen fibril attached to a glass microneedle. Inset Glass microneedle and epoxy bead (arrow). Note the banding pattern typical of native collagen fibrils
3.1.3 Fibril radial degradation rate
Along with observation of ultimate fibril failure, fibril degradation was observed as a decrease in fibril stiffness, calculated from the mechanical probes every 300 s. Fibrils remained relatively linear-elastic throughout enzymatic degradation (Fig. 3) and the radial degradation rates were relatively constant during early stages of digestion (Fig. 4), as predicted by the 1-D radial degradation model. Radial change rates for zero-load, −0.116 ± 0.058 nm s−1 (±SD), and low-load, −0.027±0.009 nm s−1, were significantly different (p ≤ 0.03) from each other and from all other rates, using the t test for unpaired samples with unequal variances. Control, +0.0025 ± 0.0008 nm s−1 (n = 3), and high-load, +0.0023 ± 0.0010 nm s−1, radial rates were not statistically different. Because these mechanochemical erosion tests are load-controlled, there is some concern that the per-monomer load increases as the fibrils degrade. While this concern is not relevant to zero-load fibrils, which are not loaded, or high-load fibrils, which do not significantly degrade, low-load fibrils should experience higher per-monomer loads as they lose load-bearing area through radial degradation. For example, a 50 % loss of fibril radius results in a 75 % loss of load-bearing area, raising the load from 2.5 to 10 pN/monomer for low-load fibrils. To mitigate this effect, radial degradation rates are calculated from the first few mechanical probes, when fibrils have not lost more than 20 % of their initial diameter.
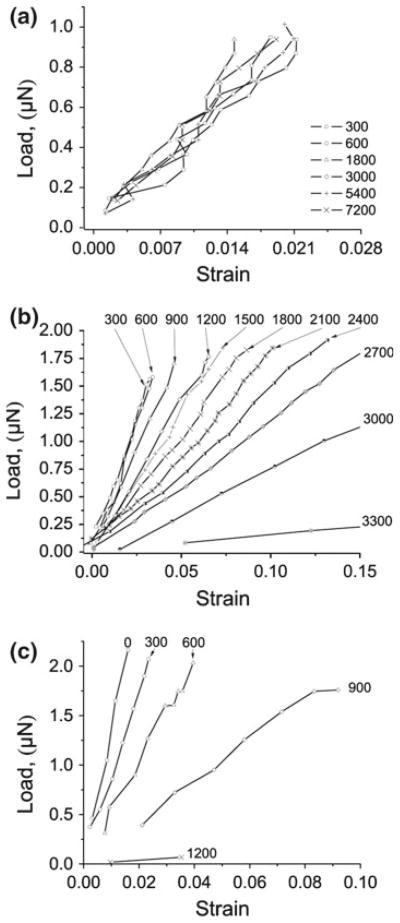
Fig. 3.
Enzyme-mediated mechanical integrity loss of individual collagen fibrils. Representative force versus strain data from mechanical probes performed each 300 s. a Control—no enzyme present. Note the relatively constant mechanical response at low strain values. b Low-load. Note the linearity of the force-strain response throughout enzymatic degradation. Fibril failed during the 3,300 s probe. c Zero-load data. Fibril failed during the 1,200 s probe. Note High-load data are not shown because no degradation was seen and the fibril was not mechanically probed, to avoid allowing degradation during the unloaded phase of mechanical probing
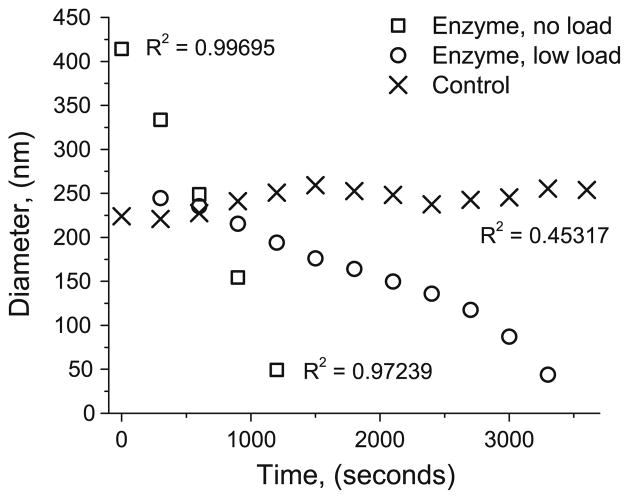
Fig. 4.
Load-modulated enzymatic reduction in fibril diameter for representative individual control, low-load, and no-load experiments. The unloaded fibril fails much sooner (1,200 s) than the low-load fibril (3,300 s) despite a much larger starting diameter. R2 values for linear fits to data for the first 1,200 s were 0.997, 0.985, and 0.917 for no-load, low-load, and control, respectively. The linear nature of diameter reduction at early stages of digestion indicates maximally bound surface erosion for both degradation series. Measurements are highly dependent on accurate determination of the gauge length for each mechanical probe, and slight errors due to vibration are amplified when converting length to strain, which leads to the observed fluctuations. R2 values indicate goodness of linear fit for each dataset
3.2 Enzymatics
3.2.1 Molecular cleavage rate, kC
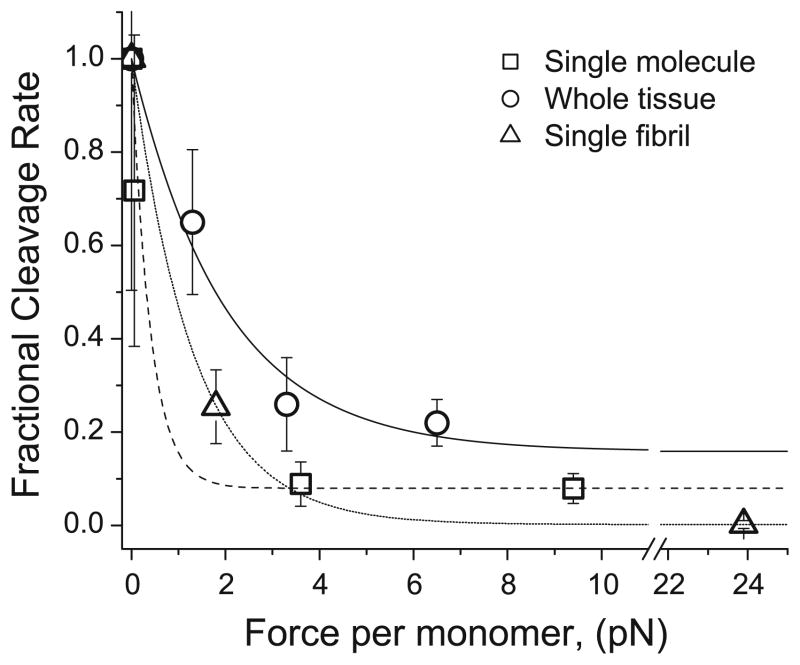
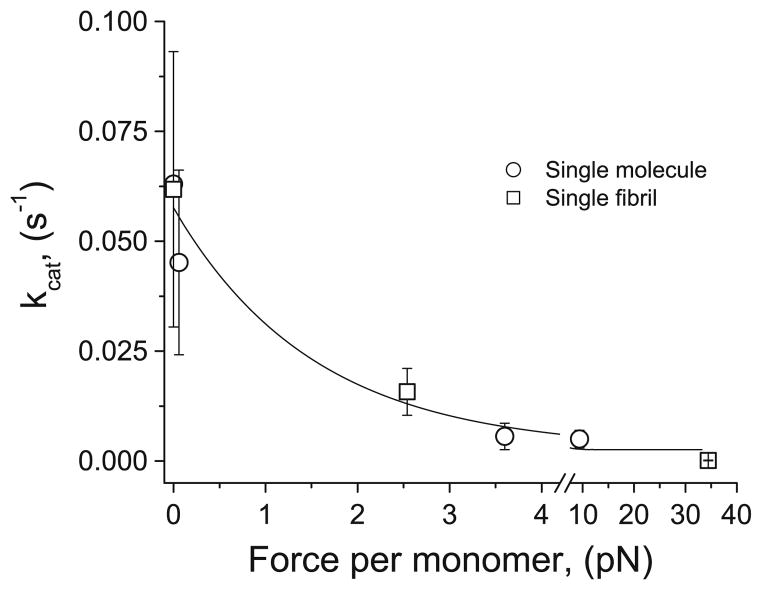
The measured kC for unloaded fibrils (0.074 s−1) is a factor of ~10 lower than published data for soluble collagen monomers (0.3–0.6 s−1), which is in agreement with the previous finding that steric hindrance limits enzyme access to fibrillar collagen and reduces fibrillar collagen cleavage rates to roughly 1/10 of soluble monomer cleavage rates (Welgus et al. 1980; Okada et al. 1992). kC reduces significantly, to 0.018 s−1 for low levels of force per monomer corresponding to 0.1 % strain, and kC is effectively zero for higher forces corresponding to 1.2 % strain (Fig. 5). These strain levels are much lower than those found to reduce enzymatic susceptibility at the whole-tissue scale (4–8 %) (Huang and Yannas 1977). This difference is explained well by findings that molecular sliding and network realignment translates 5 % tissue-level strain to only 0.5–1.5 % molecular-level strain, mainly due to fibril network realignment (Nemetschek et al. 1978). Mechanochemical enzyme-susceptibility data, in the form of degradation rate versus applied force per monomer, from previous single-molecule (Camp et al. 2011) and whole-tissue (Zareian et al. 2010) investigations are reproduced and normalized in Fig. 6 to show the congruence in relative mechanical stabilization across multiple hierarchical scales. In Fig. 7, we combine kC versus force data from previous single-molecule work with the isolated fibril results (we omit whole-tissue results because kC cannot be determined accurately from whole-tissue investigations) and show that the combined data sets are represented well by a single exponential decay (Eq. 2):
| (2) |
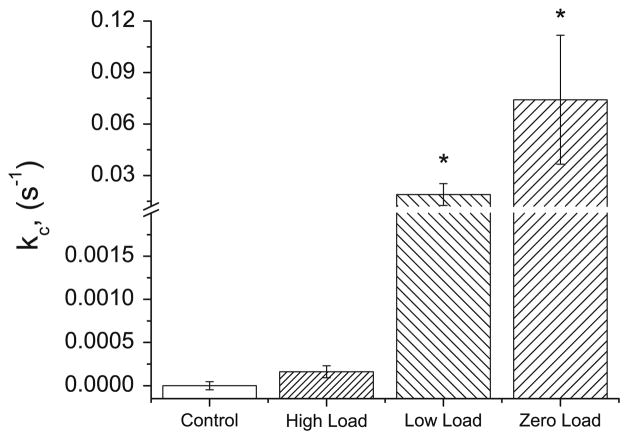
Fig. 5.
Cleavage rate (kC) for each experimental series, with standard deviation indicated. * No-load and Low-load rates each differed significantly (p < 0.05) from all other series. No significant difference was found between high-load and control
Fig. 6.
Normalized fractional cutting rate versus applied force per monomer for three size scales. Data are normalized against maximum cutting rate, from the zero-load experimental series, and fit to a single exponential curve. Square (Camp et al. 2011), circle (Zareian et al. 2010)
Fig. 7.
Characterization of the mechanochemical switch. Cleavage rate versus applied force per monomer data combined from single-molecule (Camp et al. 2011) and single-fibril work (present study). The combined data are fit using a single exponential (Eq. 2)
The agreement between single-molecule and single-fibril results suggests that the packing of monomers into fibrils does not substantially alter the strain-protection effect.
3.2.2 Specificity, kC/Km
Specificities, kC/Km, were 55.9±29.1, 13.6±4.63, 0.116 ± 0.049 h−1 μm−1 for zero-, low-, and high-load, respectively. These values, for native fibrils, are lower than published values of 200–500 h−1 μm−1 for BC degradation of soluble collagen (Mallya et al. 1992), most likely due to molecular packing and cross-linking (Okada et al. 1992; Welgus et al. 1980).
4 Discussion
Combined with the present study there is mounting evidence that mechanical tension has a stabilizing effect on type I collagen, protecting it against both thermal degradation (Bass et al. 2004) and enzymatic cleavage by two different enzymes, MMP-8 (Flynn et al. 2010) and BC (Bhole et al. 2009; Camp et al. 2011; Huang and Yannas 1977; Nabeshima et al. 1996; Wyatt et al. 2009; Zareian et al. 2010). That tension confers such general stability suggests that the mechanochemical stabilization of collagen we see here is an effect derivative of the structure of the collagen triple-helix and relatively independent of the specific enzyme used.
4.1 Mechanism
The specific mechanism behind mechanochemical stabilization of the triple-helix can be inferred from investigations into the molecular interactions between MMPs and collagen. It has been demonstrated that mammalian collagenases are unable to simultaneously cleave all three alpha chains due to size restrictions in the catalytic cleft (Grams et al. 1995) and thus the triple-helix must partially unwind for collagen degradation to occur. Additionally, collagen triple-helix cleavage is regulated by the local stability of the cleavage sites (Minond et al. 2004) where the alpha chains unwind naturally to form enzyme-accessible loops (Nerenberg et al. 2008; Stultz 2002). The extremely sensitive stabilization effect at 0.1 % strain is difficult to fully explain given the mechanics of BC enzymatic degradation of collagen, which cleaves at multiple locations. While it is possible that the strain is preferentially distributed, causing local deformation at specific cleavage sites, such a small strain distributed over multiple locations would not be expected to alter enzymatic cleavage to the extent measured here. In a recent publication (Chang et al. 2012), we have found that loop structures that form naturally near the MMP cleavage site are pulled back into triple-helical confirmation. Such data suggest that tension may generally pull the collagen alpha chains into a tighter apposition, limiting enzyme access all along the molecule.
4.2 Implications
4.2.1 Developmental biology
The results presented here complete a 3-tier hierarchical investigation on collagen mechanochemistry and strongly support the theory that mechanical load stabilizes collagen against enzymatic degradation. There are numerous implications for our understanding of connective tissue development, growth, and remodeling (Grytz et al. 2011; Sander et al. 2011) provided by the characterization and quantification of the general mechanochemical switch operating in collagenous fibrils, molecules, and tissues. One of the more important implications is that force triggered stability of collagen is simplifying. Though cells certainly play a direct role in the initial formation and deposition of the embryonic connective tissue structural template, control of subsequent epigenetic adaptation of connective tissue is likely to be exerted from a distance by resident fibroblasts. It has been shown that fibroblasts transduce mechanical loads to effect optimized modeling and remodeling of the matrix (Arokoski et al. 2000; Cowin 2004; Humphrey 2001; Michna and Hartmann 1989) via physical tensioning of fibrils (Hormann 1982; Lyubimov and Vasiliev 1982; Bell et al. 1979) and secretion of matrix molecules including collagen and collagenolytic enzymes (Prajapati et al. 2000; Koskinen et al. 2004; Blain et al. 2001).
We have suggested that mechanical tension is a critical controlling parameter for both collagen degradation and assembly (Bhole et al. 2009) and we propose that the fundamental role of the fibroblast is to synthesize and secrete matrix and signaling molecules in response to mechanical and chemical stimulation, and to generate local tension to control matrix remodeling from a distance. This postulate significantly alters the traditionally accepted view of ECM control from one of micromanagement attributed to the fibroblast (Canty et al. 2004) to one where the mechanical state of the matrix drives the accumulation and removal of collagen. Mechanochemical stabilization also explains the potential conundrum whereby collagenolytic enzymes are ubiquitous in the ECM, yet the structural integrity of the musculoskeletal system is maintained and the accumulation of excess, unloaded collagen is generally avoided.
4.2.2 Connective tissue disease
The characterization of the collagen mechanochemical switch also provides new insight for research in the prevention and management of connective tissue diseases. In osteoarthritis and intervertebral disk disease, the collagen network may lose its protective tension with age, through reduced proteoglycan quality and synthesis, or injury resulting in unsustainable ECM degradation rates. In cases where collagen accumulation is detrimental, such as fibrosis, the strain-stabilization theory suggests the converse problem: undesired collagen is being loaded and over-stabilized such that local collagenolytic activity is not high enough to maintain balance. Numerous therapeutic approaches can be envisioned which could modulate the tension on connective tissue to increase or decrease the rate of fibril accumulation and degradation. The strain-stabilization theory also suggests new ways to engineer synthetic load-bearing tissue constructs. The judicious application of collagenolytic enzymes, monomeric collagen, and mechanical tension could be utilized to refine acellular collagen matrices into more natural, organized scaffolds in a process somewhat akin to lithography (albeit force-directed).
4.2.3 Single-fibril mechanochemical assay
Finally, the development of the mechanochemical erosion assay in this investigation provides a new and highly sensitive method to determine mechanical properties and enzyme kinetics for native collagen fibrils as well as other fiber-forming proteins. Most available data on collagenase activity values have been obtained with collagen monomers in free solution (Mallya et al. 1992) with a few enzyme kinetics rates extracted from experiments using reconstituted fibrils (Okada et al. 1992; Welgus et al. 1980). Neither assay, however, is physiologically accurate. Native fibrils in real tissue comprise packed arrays of crosslinked collagen monomers whose aggregate structure may alter the rate at which enzymes hydrolyze the constituent collagen molecules (Perumal et al. 2008). There are very little available data on the enzyme kinetics in the context of intact, native collagen fibrils. The assay we introduce in this manuscript is capable of detecting the loss of a few layers of monomer from the surface of degrading fibrils. In addition to the high sensitivity of the method, the ability to simultaneously measure enzyme kinetics while applying mechanical force permits extraction of the strain dependency of the enzyme activity. Combined with fluorescence techniques, the assay can be used to extract binding and degradation rates as a function of applied load for numerous native collagen fibril/enzyme pairs and to study the force dependence of interactions between different non-enzymatic ECM proteins.
4.3 Future work
A theoretical, force-dependent degradation model was developed based on the force-degradation exponential fit for comparison. While the model accurately reproduces the results for zero- and high-load fibrils, the low-load fibrils do not fail when expected (Supplementary Data). It is worth noting again that the force per monomer should increase with degradation of load-bearing area when fibril load is held constant. So we should expect the low-load fibrils to degrade until they reach the high-load per-monomer force, and then degradation should slow or stop. We cannot fully explain why the low-load fibrils fail. This discrepancy indicates either 1) a more complex relationship between force and degradation rate than can be determined from three force groups or 2) collagen fibrils are not as uniform through their diameter as we assume. Additional investigation is required to answer this question.
5 Conclusion
A single-fibril mechanochemical erosion assay has been developed and used to probe the relationship between applied force and enzymatic degradation rate for individual, native collagen fibrils. The results strongly support the hypothesis that mechanical tension stabilizes collagen against enzymatic degradation. The base (zero-load) enzymatic rates found in this investigation agree with previously published rates, and the force-rate relationship follows the same exponential function recently found at the single-molecule level. The data suggest an extremely sensitive mechanochemical switch (Fig. 7, Eq. 2) with significant reductions in cleavage rates occurring at fibril strains as small as 0.1 %. We suggest that the prevalence of collagen as the dominant structural protein in vertebrates is derivative of this incredibly sensitive mechanochemical switch mechanism which reduces its susceptibility to both enzymatic and thermal denaturation at physiologically relevant tensile loads. Such a mechanism elevates collagen from a simple structural element, which is positioned by cells, to a more active player which participates in load-bearing structure formation and optimization via preferential retention, through enhanced stability, in locations of elevated tensile load.
Supplementary Material
Acknowledgments
This work was funded in part by NSF IGERT Nanomedicine Award, DGE-0504331 and by NIH NEI-R01015500.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10237-012-0399-2) contains supplementary material, which is available to authorized users.
References
- Adhikari AS, Chai J, Dunn AR. Mechanical load induces a 100-fold increase in the rate of collagen proteolysis by MMP-1. J Am Chem Soc. 2011 doi: 10.1021/ja109972p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arokoski JP, Jurvelin JS, Vaatainen U, Helminen HJ. Normal and pathological adaptations of articular cartilage to joint loading. Scand J Med Sci Sports. 2000;10(4):186–198. doi: 10.1034/j.1600-0838.2000.010004186.x. [DOI] [PubMed] [Google Scholar]
- Baragi VM, Qiu L, Gunja-Smith Z, Woessner JF, Jr, Lesch CA, Guglietta A. Role of metalloproteinases in the development and healing of acetic acid-induced gastric ulcer in rats. Scand J Gastroenterol. 1997;32(5):419–426. doi: 10.3109/00365529709025075. [DOI] [PubMed] [Google Scholar]
- Bass EC, Wistrom EV, Diederich CJ, Nau WH, Pellegrino R, Ruberti J, Lotz JC. Heat-induced changes in porcine annulus fibrosus biomechanics. J biomech. 2004;37(2):233–240. doi: 10.1016/j.jbiomech.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Beare AH, O’Kane S, Krane SM, Ferguson MW. Severely impaired wound healing in the collagenase-resistant mouse. J Invest Dermatol. 2003;120(1):153–163. doi: 10.1046/j.1523-1747.2003.12019.x. doi:10.1046/j.1523-1747. 2003.12019.x. [DOI] [PubMed] [Google Scholar]
- Bell E, Ivarsson B, Merrill C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc Natl Acad Sci USA. 1979;76(3):1274–1278. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhole AP, Flynn BP, Liles M, Saeidi N, Dimarzio CA, Ruberti JW. Mechanical strain enhances survivability of collagen micronetworks in the presence of collagenase: implications for load-bearing matrix growth and stability. Philos Trans A Math Phys Eng Sci. 2009;367(1902):3339–3362. doi: 10.1098/rsta.2009.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain EJ, Gilbert SJ, Wardale RJ, Capper SJ, Mason DJ, Duance VC. Up-regulation of matrix metalloproteinase expression and activation following cyclical compressive loading of articular cartilage in vitro. Arch Biochem Biophys. 2001;396(1):49–55. doi: 10.1006/abbi.2001.2575. [DOI] [PubMed] [Google Scholar]
- Camp RJ, Liles M, Beale J, Saeidi N, Flynn BP, Moore E, Murthy SK, Ruberti JW. Molecular mechanochemistry: low force switch slows enzymatic cleavage of human type I collagen monomer. J Am Chem Soc. 2011 doi: 10.1021/ja110098b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty EG, Lu Y, Meadows RS, Shaw MK, Holmes DF, Kadler KE. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J Cell Biol. 2004;165(4):553–563. doi: 10.1083/jcb.200312071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawston T, Billington C, Cleaver C, Elliott S, Hui W, Koshy P, Shingleton B, Rowan A. The regulation of MMPs and TIMPs in cartilage turnover. Ann NY Acad Sci. 1999;878:120–129. doi: 10.1111/j.1749-6632.1999.tb07678.x. [DOI] [PubMed] [Google Scholar]
- Chang S-W, Flynn BP, Ruberti JW, Buehler MJ. Molecular mechanism of force induced stabilization of collagen against enzymatic breakdown. Biomaterials. 2012;33(15):3852–3859. doi: 10.1016/j.biomaterials.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Wright NT, Humphrey JD. Heat-induced changes in the mechanics of a collagenous tissue: isothermal free shrinkage. J Biomech Eng. 1997;119(4):372–378. doi: 10.1115/1.2798281. [DOI] [PubMed] [Google Scholar]
- Cowin SC. Tissue growth and remodeling. Annu Rev Biomed Eng. 2004;6:77–107. doi: 10.1146/annurev.bioeng.6.040803.140250. [DOI] [PubMed] [Google Scholar]
- Eppell SJ, Smith BN, Kahn H, Ballarini R. Nano measurements with micro-devices: mechanical properties of hydrated collagen fibrils. J R Soc Interface. 2006;3(6):117–121. doi: 10.1098/rsif.2005.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn BP, Bhole AP, Saeidi N, Liles M, Dimarzio CA, Ruberti JW. Mechanical strain stabilizes reconstituted collagen fibrils against enzymatic degradation by mammalian collagenase matrix metalloproteinase 8 (MMP-8) PLoS One. 2010;5(8) doi: 10.1371/journal.pone.0012337. doi:10. 1371/journal.pone.0012337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grams F, Reinemer P, Powers JC, Kleine T, Pieper M, Tschesche H, Huber R, Bode W. X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design. Eur J Biochem. 1995;228(3):830–841. doi: 10.1111/j.1432-1033.1995.tb20329.x. [DOI] [PubMed] [Google Scholar]
- Grytz R, Meschke G, Jonas JB. The collagen fibril architecture in the lamina cribrosa and peripapillary sclera predicted by a computational remodeling approach. Biomech Model Mechanobiol. 2011;10(3):371–382. doi: 10.1007/s10237-010-0240-8. [DOI] [PubMed] [Google Scholar]
- Harrington DJ. Bacterial collagenases and collagen-degrading enzymes and their potential role in human disease. Infect Immun. 1996;64(6):1885–1891. doi: 10.1128/iai.64.6.1885-1891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormann H. Fibronectin–mediator between cells and connective tissue. Klin Wochenschr. 1982;60(20):1265–1277. doi: 10.1007/BF01727483. [DOI] [PubMed] [Google Scholar]
- Huang C, Yannas IV. Mechanochemical studies of enzymatic degradation of insoluble collagen fibers. J Biomed Mater Res Symp. 1977;11(1):137–154. doi: 10.1002/jbm.820110113. [DOI] [PubMed] [Google Scholar]
- Huang Y, Meek KM. Swelling studies on the cornea and sclera: the effects of pH and ionic strength. Biophys J. 1999;77(3):1655–1665. doi: 10.1016/S0006-3495(99)77013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulboy DL, Rudolph LA, Matrisian LM. Matrix metalloproteinases as mediators of reproductive function. Mol Hum Reprod. 1997;3(1):27–45. doi: 10.1093/molehr/3.1.27. [DOI] [PubMed] [Google Scholar]
- Hulmes DJ, Miller A, Parry DA, Piez KA, Woodhead-Galloway J. Analysis of the primary structure of collagen for the origins of molecular packing. J Mol Biol. 1973;79(1):137–148. doi: 10.1016/0022-2836(73)90275-1. [DOI] [PubMed] [Google Scholar]
- Humphrey JD. Stress, strain, and mechanotransduction in cells. J Biomech Eng. 2001;123(6):638–641. doi: 10.1115/1.1406131. [DOI] [PubMed] [Google Scholar]
- Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, Lopez-Otin C, Krane SM. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci USA. 2004;101(49):17192–17197. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadler K. Matrix loading: assembly of extracellular matrix collagen fibrils during embryogenesis. Birth Defects Res C Embryo Today. 2004;72(1):1–11. doi: 10.1002/bdrc.20002. [DOI] [PubMed] [Google Scholar]
- Kang JD, Georgescu HI, McIntyre-Larkin L, Stefanovic-Racic M, Donaldson WF, III, Evans CH. Herniated lumbar intervertebral discs spontaneously produce matrix metalloproteinases, nitric oxide, interleukin-6, and prostaglandin E2. Spine (Phila Pa 1976) 1996;21(3):271–277. doi: 10.1097/00007632-199602010-00003. [DOI] [PubMed] [Google Scholar]
- Kilian O, Pfeil U, Wenisch S, Heiss C, Kraus R, Schnettler R. Enhanced alpha 1(I) mRNA expression in frozen shoulder and dupuytren tissue. Eur J Med Res. 2007;12(12):585–590. [PubMed] [Google Scholar]
- Koskinen SO, Heinemeier KM, Olesen JL, Langberg H, Kjaer M. Physical exercise can influence local levels of matrix metalloproteinases and their inhibitors in tendon-related connective tissue. J Appl Physiol. 2004;96(3):861–864. doi: 10.1152/japplphysiol.00489.2003. [DOI] [PubMed] [Google Scholar]
- Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA. 2002;99(3):1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AV, Vasiliev JM. Distribution of fibronectin-containing structures on the surface of lamelloplasm and endoplasm of fibroblasts; hypothesis of receptor-mediated assembly of fibronectin structures. Cell Biol Int Rep. 1982;6(2):105–112. doi: 10.1016/0309-1651(82)90086-8. [DOI] [PubMed] [Google Scholar]
- Mallya SK, Mookhtiar KA, Van Wart HE. Kinetics of hydrolysis of type I, II, and III collagens by the class I and II Clostridium histolyticum collagenases. J Protein Chem. 1992;11(1):99–107. doi: 10.1007/BF01025096. [DOI] [PubMed] [Google Scholar]
- McCawley LJ, Matrisian LM. Matrix metalloproteinases: multifunctional contributors to tumor progression. Mol Med Today. 2000;6(4):149–156. doi: 10.1016/s1357-4310(00)01686-5. S1357-4310(00)01686-5. [DOI] [PubMed] [Google Scholar]
- Meek KM, Leonard DW. Ultrastructure of the corneal stroma: a comparative study. Biophys J. 1993;64:273–280. doi: 10.1016/S0006-3495(93)81364-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michna H, Hartmann G. Adaptation of tendon collagen to exercise. Int Orthop. 1989;13(3):161–165. doi: 10.1007/BF00268040. [DOI] [PubMed] [Google Scholar]
- Miles CA, Ghelashvili M. Polymer-in-a-box mechanism for the thermal stabilization of collagen molecules in fibers. Biophys J. 1999;76(6):3243–3252. doi: 10.1016/S0006-3495(99)77476-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minond D, Lauer-Fields JL, Nagase H, Fields GB. Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry. 2004;43(36):11474–11481. doi: 10.1021/bi048938i. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y, Grood ES, Sakurai A, Herman JH. Uniaxial tension inhibits tendon collagen degradation by collagenase in vitro. J Orthop Res. 1996;14:123–130. doi: 10.1002/jor.1100140120. [DOI] [PubMed] [Google Scholar]
- Nemetschek T, Jonak R, Nemetschek-Gansler H, Riedl H, Rosenbaum G. On the determination of changes in the large periodic structure of collagen (author’s transl) Z Naturforsch C. 1978;33(11–12):928–936. [PubMed] [Google Scholar]
- Nerenberg PS, Salsas-Escat R, Stultz CM. Do collagenases unwind triple-helical collagen before peptide bond hydrolysis? Reinterpreting experimental observations with mathematical models. Proteins. 2008;70(4):1154–1161. doi: 10.1002/prot.21687. [DOI] [PubMed] [Google Scholar]
- Okada T, Hayashi T, Ikada Y. Degradation of collagen suture in vitro and in vivo. Biomaterials. 1992;13:448–454. doi: 10.1016/0142-9612(92)90165-k. [DOI] [PubMed] [Google Scholar]
- Perumal S, Antipova O, Orgel JP. Collagen fibril architecture, domain organization, and triple-helical conformation govern its proteolysis. Proc Natl Acad Sci USA. 2008;105(8):2824–2829. doi: 10.1073/pnas.0710588105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati RT, Chavally-Mis B, Herbage D, Eastwood M, Brown RA. Mechanical loading regulates protease production by fibroblasts in three-dimensional collagen substrates. Wound Repair Regen. 2000;8(3):226–237. doi: 10.1046/j.1524-475x.2000.00226.x. [DOI] [PubMed] [Google Scholar]
- Ruberti JW, Hallab NJ. Strain-controlled enzymatic cleavage of collagen in loaded matrix. Biochem Biophys Res Commun. 2005;336(2):483–489. doi: 10.1016/j.bbrc.2005.08.128. [DOI] [PubMed] [Google Scholar]
- Sander EA, Barocas VH, Tranquillo RT. Initial fiber alignment pattern alters extracellular matrix synthesis in fibroblast-populated fibrin gel cruciforms and correlates with predicted tension. Ann Biomed Eng. 2011;39(2):714–729. doi: 10.1007/s10439-010-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stultz CM. Localized unfolding of collagen explains collagenase cleavage near imino-poor sites. J Mol Biol. 2002;319(5):997–1003. doi: 10.1016/S0022-2836(02)00421-7. [DOI] [PubMed] [Google Scholar]
- Tzafriri AR, Bercovier M, Parnas H. Reaction diffusion model of the enzymatic erosion of insoluble fibrillar matrices. Biophys J. 2002;83:776–793. doi: 10.1016/S0006-3495(02)75208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Rijt JA, Van der Werf KO, Bennink ML, Dijkstra PJ, Feijen J. Micromechanical testing of individual collagen fibrils. Macromol Biosci. 2006;6(9):697–702. doi: 10.1002/mabi.200600063. [DOI] [PubMed] [Google Scholar]
- Welgus HG, Jeffrey JJ, Stricklin GP, Roswit WT, Eisen AZ. Characteristics of the action of human skin fibroblast collagenase on fibrillar collagen. J Biolo Chem. 1980;255:6808–6813. [PubMed] [Google Scholar]
- Wyatt KE, Bourne JW, Torzilli PA. Deformation-dependent enzyme mechanokinetic cleavage of type I collagen. J Biomech Eng. 2009;131(5):051004. doi: 10.1115/1.3078177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareian R, Church KP, Saeidi N, Flynn BP, Beale JW, Ruberti JW. Probing collagen/enzyme mechanochemistry in native tissue with dynamic, enzyme-induced creep. Langmuir. 2010;26(12):9917–9926. doi: 10.1021/la100384e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.