Abstract
Cadherins are Ca2+ dependent cell-cell adhesion proteins that maintain the structural integrity of the epidermis; their principle function is to resist mechanical force. This review summarizes the biophysical mechanisms by which classical cadherins tune adhesion and withstand mechanical stress. We first relate the structure of classical cadherins to their equilibrium binding properties. We then review the role of mechanical perturbations in tuning the kinetics of cadherin adhesion. In particular, we highlight recent studies which show that cadherins form three types of adhesive bonds: catch bonds which become longer lived and lock in the presence of tensile force, slip bonds which become shorter lived when pulled and ideal bonds that are insensitive to tugging.
Introduction
The epidermis serves as a physical barrier that protects organisms from their external environment. This multilayered tissue is composed of keratinocytes bound together by two types of cell-cell adhesion complexes: desmosomes and adherens junctions. (Jensen and Wheelock, 1996). The primary adhesive components of both these structures are the cadherin family of Ca2+ dependent transmembrane proteins (Al-Amoudi and Frangakis, 2008; Green and Simpson, 2007; Gumbiner, 2005; Halbleib and Nelson, 2006; Niessen et al., 2011). Desmosomes are composed of two types of desmosomal cadherins (desmocollin and desmoglein) (Desai et al., 2009; Green and Simpson, 2007), while epidermal adherens junctions contain a single classical type-1 cadherin (either E-cadherin or P-cadherin) (Halbleib and Nelson, 2006; Jensen and Wheelock, 1996). Both desmosomes and adherens junctions act in a coordinated fashion to help the epidermis withstand mechanical stress. While the interactions that mediate desmosomal cadherin binding are not completely understood, the structural basis of classical cadherin adhesion has been extensively characterized.
Classical cadherins share a conserved cytoplasmic domain, and an ectodomain containing five tandem extracellular (EC) repeats. Their expression levels vary within the epidermis; while E-cadherins are present in all keratinocytes, expression of P-cadherins is limited to the basal layer (Halbleib and Nelson, 2006; Takeichi, 1988). Adhesion is mediated by the cadherin ectodomain while the cytoplasmic region binds to adaptor proteins which link cadherins indirectly to the cytoskeleton, regulate cadherin turnover and modulate actin assembly (Nelson and Nusse, 2004; Niessen et al., 2011; Takeichi, 2007). Since the epidermis is a self-renewing tissue with a continuous upward movement of cells, cadherins dynamically tune their adhesive strength in order to preserve epidermal barrier integrity (Niessen, 2007). Epidermal cadherin knockout studies in mice show that loss of E-cadherin correlates with a loss of adherens junctions, altered epidermal differentiation and loss of hair follicles (Tinkle et al., 2004; Young et al., 2003). Similarly, deletion of α-catenin, an adaptor protein associated with the cadherin cytoplasmic domain, results in impaired adhesion and epidermal detachment (Vasioukhin et al., 2001).
Cell-cell adhesion is a dynamic process and classical cadherins tailor their binding kinetics in order to withstand mechanical perturbations. While the equilibrium binding properties of classical cadherins have been extensively characterized (Brasch et al., 2012), the role of mechanical force in altering cadherin binding is only now being measured. Recent studies show that upon being exposed to mechanical perturbation, E-cadherins change their unbinding kinetics (Rakshit et al., 2012). These kinetic changes are not manifested in solution or in the absence of mechanical loading, but are critical for cadherin adhesion.
This brief review summarizes our current understanding of the effect of mechanical force on the kinetics of E-cadherin adhesion. We focus on the ectodomain; the role of the cytoplasmic domain and its associated proteins have been reviewed elsewhere (Gomez et al., 2011; Ladoux and Nicolas, 2012; Leckband et al., 2011; Papusheva and Heisenberg, 2010; Schwartz and DeSimone, 2008). We begin by relating the structure of E-cadherins to their equilibrium binding properties. We then review the role of mechanical perturbations in tuning the kinetics of adhesion. Finally, we discuss major open questions and future directions in this exciting area of research.
Adhesive states of classical cadherins
Classical cadherins adhere via ‘trans’ interactions where ectodomains from opposing cells bridge the inter-membrane gap and interact with each other. Adhesion is strengthened by the cooperative self-assembly of cadherins on the same cell into cis clusters (Brasch et al., 2012).
Structure and kinetics of trans adhesive states
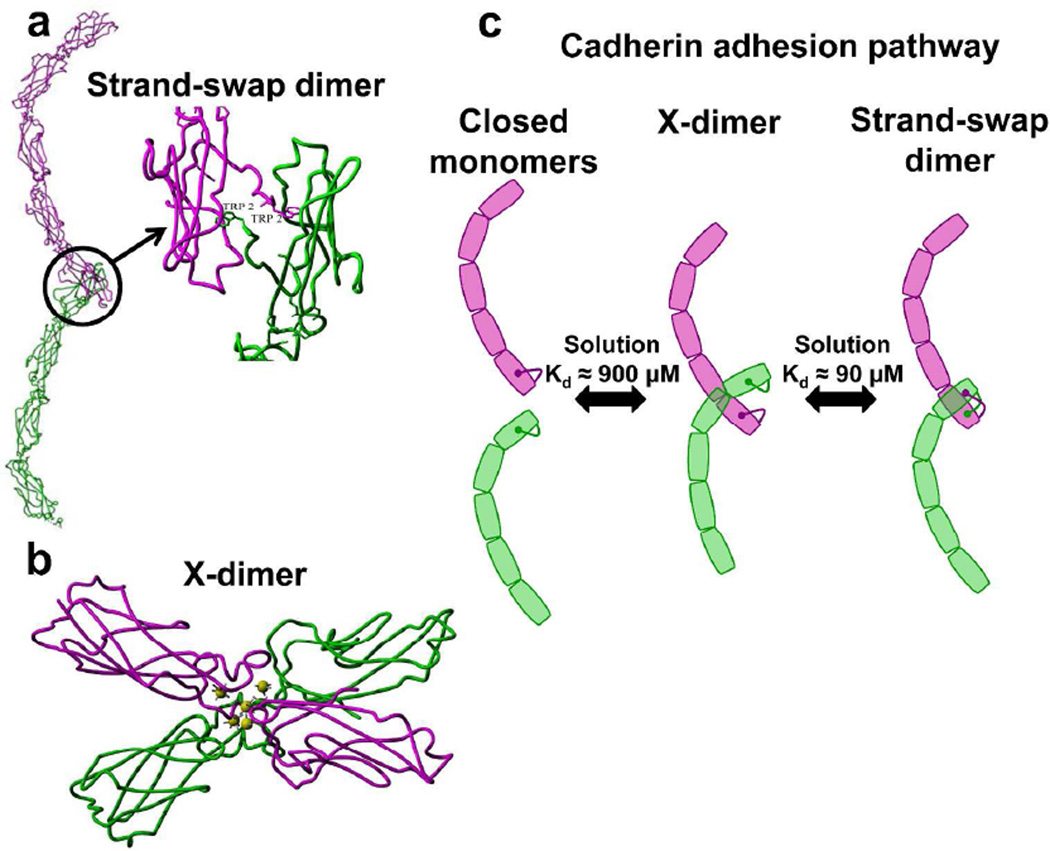
Structural studies of both the complete ectodomain of type I classical C-cadherin (EC1–5) (Boggon et al., 2002) and of smaller fragments of E-Cadherin and N-Cadherin (Harrison et al., 2010; Haussinger et al., 2004; Nagar et al., 1996; Pertz et al., 1999; Shapiro et al., 1995) have identified key interactions that mediate trans adhesion (Fig. 1). The primary adhesive conformation involves the interaction of opposing EC1 domains and is termed the strand-swapped dimer (Fig. 1A). In this structure, N-terminal β-strands between opposing EC1 domains are swapped and the side chain of a conserved Tryptophan at position 2 (W2) is inserted into a pocket on their adhesive partner (Boggon et al., 2002; Haussinger et al., 2004; Parisini et al., 2007; Shapiro et al., 1995) (Fig. 1A). The physiological relevance of this adhesive interface has been confirmed in numerous mutational, structural and cellular studies (Harrison et al., 2005; Pertz et al., 1999; Prakasam et al., 2006a; Shan et al., 2004; Tamura et al., 1998; Troyanovsky et al., 2003). In solution, the affinity for strand-swap dimer formation is low; dissociation constants (Kd) for trans dimers of the full length ectodomain of C-cadherin measured using Analytical Ultra Centrifugation (AUC) is 64 µM (Chappuis-Flament et al., 2001). Similarly, EC1–2 domains of E-cadherin expressed in mammalian and bacterial cells have trans dimer Kd values of 97 µM (Katsamba et al., 2009) and 80 µM (Koch et al., 1997) respectively.
Figure 1. Adhesive states of classical cadherin and the pathway for cadherin binding.
(A) The extracellular region of type-I classical cadherin is composed of five tandem Extracellular (EC) domains. Linkers between successive EC domains are each bound to three Ca2+ ions which give the ectodomain its characteristic curvature. Ectodomains from opposing cells (shown in green and magenta) adhere across the inter-membrane gap via ‘trans’ interactions. The primary trans interface involves the interaction of opposing EC1 domains and is termed the strand-swapped dimer. In this conformation, N-terminal β-strands between opposing EC1 domains are swapped and the side chain of a conserved Tryptophan at position 2 (W2) is inserted into a pocket on their adhesive partner. (B) Prior to strand-swapping, cadherin ectodomains form a non-swapped, intermediate conformation, called an X-dimer. This conformation is formed by extensive surface interactions between the base of the EC1 domain, EC1–EC2 inter-domain linker region and the apex of domain EC2. (C) Cadherin monomers adopt a “closed” conformation where W2 is docked into each monomer’s binding pocket. Monomers from opposing cells interact to form X-dimers and then proceed to swap W2 residues to form a strand-swap dimer. The Kd of the EC1–2 domains of W2A E-cadherin X-dimers is 916 µM (Harrison et al., 2010) while the Kd of the EC1–2 domains of WT E-cadherin strand-swap dimers is 97 µM (Katsamba et al., 2009).
Prior to strand-swapping, cadherin monomers are in a “closed” conformation where W2 is docked into each monomer’s binding pocket; the monomers thus act as competitive inhibitors of strand-swapping (Chen et al., 2005). The closed monomeric conformation places a strain on the short swapping strand due to its anchorage at one end by the W2 and at the other by a Ca2+ ion; relieving this conformational strain is the driving force for strand swapping (Vendome et al., 2011). Equilibrium affinity measurements using AUC show that mutations that relieve strain in the swapping strand in E-cadherin monomers, decrease dimerization affinities (Vendome et al., 2011). Single molecule Fluorescence Resonance Energy Transfer (FRET) experiments suggest that prior to swapping N-terminal β-strands, E-cadherin monomers first form a non-swapped, intermediate “encounter complex” (Fig. 1C) (Sivasankar et al., 2009). E-cadherins can be trapped in this encounter complex by mutating W2 (Sivasankar et al., 2009); consequently, W2A fragments weakly adhere to each other (Prakasam et al., 2006a; Sivasankar et al., 2009). Recently, the atomic resolution structure of the encounter complex has been resolved in W2A mutants (Fig. 1B). This conformation, called an X-dimer, is formed by extensive surface interactions between the base of the EC1 domain, EC1–EC2 interdomain linker region and the apex of domain EC2 (Harrison et al., 2010) (Fig. 1B). The affinity for X-dimer formation in solution is significantly weaker than strand-swap dimers; the Kd of W2A cadherin X-dimers is an order of magnitude higher (916 µM) than wild type (WT) cadherin strand-swap dimers (Harrison et al., 2010).
Mutations in the cadherin X-dimer binding interface alter the kinetics of strand-swapping but do not change the structure of the strand-swap dimer. When a key Lys 14 residue in the X-dimer binding interface is mutated to a Glu, the trans-dimers are virtually indistinguishable from WT cadherin strand-swap dimers (Harrison et al., 2010). As measured using Surface Plasmon Resonance, the K14E mutants show no binding in a short–time frame suggesting that their binding rate (on rate) is low. Similarly, sedimentation velocity AUC and size-exclusion chromatography show that the monomer to strand-swap dimer conversion is impeded in these mutants (Harrison et al., 2010). Presumably, lower on-rates are measured since the formation of X-dimers, which serve as kinetic intermediates strand-swapping (Fig. 1C), are impaired in the K14E mutants. In epithelial cells, inactivation of X dimers result in extraordinarily stable cell-cell junctions; this has been interpreted to indicate that X-dimers are an intermediate in the pathway to dissociation of strand-swap dimers (Hong et al., 2011). AUC measurements show that the Kd of the K14E mutants are virtually indistinguishable from WT cadherin which suggests that besides their low on-rate, the dissociation (off-rate) of these mutants is also decreased (Harrison et al., 2010). However, in contrast to these studies, recent single molecule force measurements indicate that the dissociation rate of K14E is similar to WT cadherin (Rakshit et al., 2012). Consequently, the molecular role of X-dimers in the dissociation of strand-swap dimers is unclear.
Structure and kinetics of cis adhesive states
Cadherin adhesion is enhanced by their lateral assembly on the cell surface (Kim et al., 2005; Takeda et al., 1999). However, the biophysical mechanisms by which cis clustering boosts adhesion are just beginning to be understood. Early studies showed that beads decorated with cadherin pairs aggregated to a greater extent than beads with immobilized monomers (Brieher et al., 1996). While this data was interpreted to suggest that cadherin ectodomains form cis dimers, recent single molecule experiments show that ectodomains located adjacent to each cooperatively enhance the probability of adhesion even if they do not associate with each other in a cis geometry (Zhang et al., 2009).
Based on contacts observed in X-ray crystal structures of a range of classical cadherins, it has been proposed that interactions between the apex of EC1 and the base of EC2 of neighboring cadherins mediate dimerization in a cis orientation (Boggon et al., 2002; Harrison et al., 2011). These interactions are however not observed in NMR measurements of EC1–2 (Haussinger et al., 2002), indicating that their Kd exceeds 1mM (Harrison et al., 2011). Similarly, single molecule FRET experiments could not detect cis dimer formation between two cadherin ectodomains that were located adjacent to each other in a configuration that would permit lateral dimerization (Zhang et al., 2009). This discrepancy is explained by recent theoretical studies which predict that the cis assembly of cadherin ectodomains requires prior trans dimerization (Wu et al., 2011; Wu et al., 2010). When trans dimers are formed, the conformational flexibility of ectodomains is dramatically reduced which lowers the entropic penalty associated with cis dimer formation (Wu et al., 2011).
In qualitative agreement with these predictions, micropipette manipulation experiments show that the binding of cadherins from opposing cells occur in two stages: an initial rapid stage ascribed to trans adhesion followed by a second, slower stage interpreted to occur due to cis clustering (Chien et al., 2008). However, while the first stage requires the EC1 domain as expected for trans dimer formation, EC3 is required for the second adhesive state (Chien et al., 2008). Micropipette experiments also demonstrate that hypoglycosylation of EC2 and EC3 enhance the lateral assembly of ectodomains (Langer et al., 2012).
Mechanical tension alters the kinetics of cadherin adhesion
The structural and biophysical studies described above, provide a detailed picture of the kinetic determinants of classical cadherin binding in equilibrium, under force-free conditions. However, the molecular mechanisms by which cadherins alter their binding kinetics in response to mechanical forces are still unclear.
When cadherin trans dimers, are pulled apart, they can form one of three distinct types of bonds (Dembo, 1994; Dembo et al., 1988) (i) Slip bonds which weaken and have a higher off-rate when pulled. (ii) Catch bonds which counter-intuitively strengthen such that their off-rates decrease. (iii) Ideal bonds which are unaffected by mechanical stress. Slip bonds are the most commonly observed interactions in biology. Catch bonds provide a way for the interacting proteins to grip tightly in the presence of tugging forces. Finally, though ideal bonds were theoretically proposed more than a decade ago (Dembo, 1994; Dembo et al., 1988), they had not been experimentally observed in any biological system.
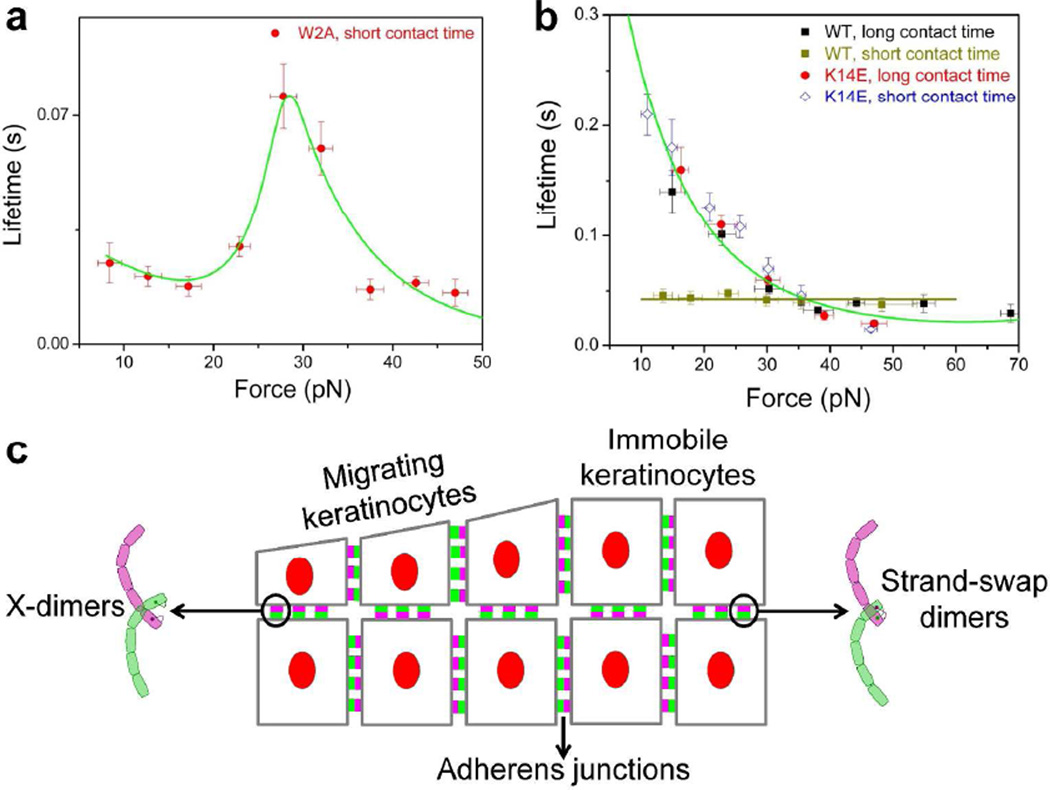
Recently, single molecule Atomic Force Microscope (AFM) force measurements were used to show that E-cadherins form bonds with catch, slip and ideal mechanical properties (Rakshit et al., 2012). The lifetimes of E-cadherin binding conformations were measured as they were subjected to different pulling forces. These experiments showed that while W2A mutant X-dimers formed catch bonds, WT and K14E strand-swap dimers formed slip bonds (Rakshit et al., 2012) (Fig. 2A & 2B). WT cadherins were also shown to form ideal bonds which were hypothesized to arise as X-dimers converted to a strand-swap conformation (Fig. 2B) (Rakshit et al., 2012).
Figure 2. Mechanical force tunes the kinetics of cadherin adhesion.
Adapted from (Rakshit et al., 2012). (A) X-dimers form catch bonds which become longer lived and lock in the presence of tensile force. When W2A cadherin X-dimers are pulled, their bond lifetimes increase with force. After reaching a maximum at a critical force of ~ 30 pN, the lifetimes decrease. (B) Strand swap dimers form slip bonds which become shorter lived when pulled. Slip bonds are formed by K14E mutants that interact for short and long periods of time and also by WT cadherins that interact for long periods of time. However, when WT cadherins interact for a short period of time, they form ideal bonds that are insensitive to force. (C) Hypothetical mechanism by which keratinocytes resist tensile forces during skin renewal and wound healing. As skin cells reposition themselves, E-cadherins bind rapidly to form X-dimers that allow cells to grip strongly under load. In immobile keratinocytes, E-cadherins form more robust strand-swap dimers that have a high affinity in the absence of force.
X-dimers form catch bonds
When X-dimers were tugged, their bond lifetimes increased with force, indicative of a catch bond. After reaching a maximum at a critical force of ~ 30 pN, the lifetimes decreased with force (Fig. 2A). A similar behavior was measured when WT cadherins were forced into an X-dimer conformation by competitively inhibiting strand swapping using free W in solution (Rakshit et al., 2012). X-dimer catch bonds are observed because the cadherins reorient when they are pulled such that they form transient, force-induced bonds and lock more tightly.
While this was the first observation of catch bonds in cadherin adhesion, these bonds have previously been measured with other adhesive proteins like selectins (Marshall et al., 2003; Sarangapani et al., 2004; Yago et al., 2004), FimH (Le Trong et al., 2010; Thomas et al., 2002) and integrins (Kong et al., 2009). Although it is tempting to speculate that the physiological role of X-dimer catch bonds is to allow cells to grip tightly and lock in place when pulled; this hypothesis remains to be tested.
Catch bonds resolve discrepancies between solution and surface force measurements
Over a decade ago, Surface Force Apparatus (SFA) measurements of the interactions between cadherin ectodomains immobilized on lipid membranes suggested that classical cadherins bind in three distinct conformations (Sivasankar et al., 2001). The weakest conformation required W2, and corresponded to a strand swapped dimer (Prakasam et al., 2006b; Zhu et al., 2003). The second conformation had an intermediate binding strength and required EC1–2 (Zhu et al., 2003); based on recent structural data, it is likely that this adhesive state corresponds to the X-dimer complex. The third and strongest adhesion required the EC3 domains to interact directly (Zhu et al., 2003); while this adhesive state likely corresponds to a cis-dimer structure, this remains to be confirmed. Single molecule AFM force measurements of the interaction of different classical cadherins confirmed the results of the ensemble SFA measurements (Bayas et al., 2006; Perret et al., 2004; Shi et al., 2008; Shi et al., 2010).
It was initially believed that the SFA measurement of stronger adhesion between X-dimers compared to strand-swap dimers directly contradicted the results of solution affinity measurements which showed that X-dimers have higher off rates than strand-swap dimers. However, the discovery of X-dimer catch bonds resolves this apparent discrepancy (Fig. 2A). Since catch bonds strengthen in the presence of force, X-dimer adhesion which is weak in the absence of force becomes stronger when pulled.
Strand swap dimers form slip bonds
Since strand-swap dimers have a higher binding affinity than X-dimers (Harrison et al., 2010; Katsamba et al., 2009), WT cadherins form strand-swap dimers when they interact for long periods of time. Single molecule AFM force clamp experiments showed that these WT cadherin strand-swap dimers formed slip bonds; their bond lifetimes decreased with increasing tensile force (Fig. 2B). Not surprisingly, identical slip bonds were formed by the K14E strand-swap dimers (Rakshit et al., 2012) (Fig. 2B). The intrinsic off rate of both the WT E-cadherin and K14E strand swap dimers was 1.6 s−1 (Rakshit et al., 2012) which is similar to an off-rate of 0.7 s−1 measured for WT E-cadherins using NMR (Haussinger et al., 2004).
Ideal bonds are formed as X-dimers transition to a strand-swap conformation
Besides forming catch and slip bonds, cadherins also form ideal bonds that behave like mechanical dampers and prevent the abrupt jolting of cells. When WT cadherin interaction time was decreased, the lifetimes of their interactions were independent of force; they formed ideal bonds (Rakshit et al., 2012) (Fig. 2B). It was hypothesized that ideal bonds correspond to an intermediate state which is formed when X-dimers transition to strand-swap binding (Rakshit et al., 2012). However, the structure of the intermediate state and the molecular contacts responsible for ideal bond formation still need to be resolved.
Future Directions
Catch, slip and ideal bonds suggest a physical mechanism that E-cadherins use to resist tensile force as cells rearrange during skin renewal and wound healing. It is tempting to speculate the as keratinocytes reposition themselves, E-cadherins bind rapidly to form X-dimer catch bonds that allow cells to grip strongly under load (Rakshit et al., 2012) (Fig. 2C). Over time, the X-dimers proceed to form more robust strand-swap dimers that have a high affinity in the absence of force (Fig. 2C); this conversion is facilitated by an intermediate conformation that is insensitive to tensile force (Rakshit et al., 2012). However, it is currently unclear if keratinocytes utilize such a mechanism to tune adhesive properties. Studying cadherin bond mechanics in living cells will be a crucial first step to addressing this question.
Besides mediating robust adhesion, classical cadherins play a key role in mechanotransduction by sensing physical stimuli at cell-cell junctions, transmitting them to the cytoplasm and activating a biochemical response (Ladoux et al., 2010; le Duc et al., 2010; Liu et al., 2010; Weber et al., 2012). It is believed that cadherins along with their adaptor proteins, β-catenin and α-catenin form the core force-bearing unit in the transmission of mechanical signals (Leckband et al., 2011). To accomplish this, it is critical that the interactions between cadherins and catenins remain intact when exposed to force. However, the force dependent binding kinetics of these interactions have not yet been studied. Furthermore, the role of these adapter proteins in altering cadherin mechanical properties is still an open question. For instance, it is known that the adapter protein α-catenin plays an important role in strengthening cadherin bonds following initial adhesion (Bajpai et al., 2008). Whether α-catenin and other adapter proteins alter the force dependent kinetics of cadherin bonds needs to be investigated.
Some of the discrepancies in cadherin binding measured using solution affinity measurements and force measurements arise due to differences between cadherin interactions in solution, under force independent conditions, and cadherin adhesion in the presence of mechanical stress. The discovery that cadherins vary their lifetimes in response to force reconciles some of these differences (Rakshit et al., 2012). However, several open questions remain. For instance, the molecular interactions by which cadherins form catch bonds are not known. Furthermore, the hypothesis that ideal bonds correspond to an intermediate state which is formed as cadherin X-dimers transition to a strand-swap conformation needs to be tested at the molecular level. Finally, the role that X-dimers play in the dissociation of strand-swap dimers is unclear.
Acknowledgments
The author thanks Dr. Sabyasachi Rakshit for his help in preparing Figure 2. This work was supported in part by a Basil O’Connor Starter Scholar Award from the March of Dimes Foundation and an American Heart Association National Scientist Development Grant.
Footnotes
The author declares no conflict of interest.
References
- Al-Amoudi A, Frangakis AS. Structural studies on desmosomes. Biochem Soc Trans. 2008;36:181–187. doi: 10.1042/BST0360181. [DOI] [PubMed] [Google Scholar]
- Bajpai S, Correia J, Feng YF, Figueiredo J, Sun SX, Longmore GD, et al. alpha-Catenin mediates initial E-cadherin-dependent cell-cell recognition and subsequent bond strengthening. Proc Natl Acad Sci U S A. 2008;105:18331–18336. doi: 10.1073/pnas.0806783105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayas MV, Leung A, Evans E, Leckband D. Lifetime measurements reveal kinetic differences between homophilic cadherin bonds. Biophys J. 2006;90:1385–1395. doi: 10.1529/biophysj.105.069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher WM, Yap AS, Gumbiner BM. Lateral dimerization is required for the homophilic binding activity of C-cadherin. J Cell Biol. 1996;135:487–496. doi: 10.1083/jcb.135.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappuis-Flament S, Wong E, Hicks LD, Kay CM, Gumbiner BM. Multiple cadherin extracellular repeats mediate homophilic binding and adhesion. Journal of Cell Biology. 2001;154:231–243. doi: 10.1083/jcb.200103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Posy S, Ben-Shaul A, Shapiro L, Honig BH. Specificity of cell-cell adhesion by classical cadherins: Critical role for low-affinity dimerization through beta-strand swapping. Proc Natl Acad Sci U S A. 2005;102:8531–8536. doi: 10.1073/pnas.0503319102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien YH, Jiang N, Li F, Zhang F, Zhu C, Leckband D. Two stage cadherin kinetics require multiple extracellular domains but not the cytoplasmic region. J Biol Chem. 2008;283:1848–1856. doi: 10.1074/jbc.M708044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M. Lectures on Mathematics in the Life Sciences, Some Mathematical Problems in Biology. Vol. 24. Providence, Rhode Island: American Mathematical Society; 1994. On peeling an adherent cell from a surface; pp. 51–77. [Google Scholar]
- Dembo M, Torney DC, Saxman K, Hammer D. The Reaction-Limited Kinetics of Membrane-to-Surface Adhesion and Detachment. Proc R Soc Lond B Biol Sci. 1988;234:55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- Desai BV, Harmon RM, Green KJ. Desmosomes at a glance. J Cell Sci. 2009;122:4401–4407. doi: 10.1242/jcs.037457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez GA, McLachlan RW, Yap AS. Productive tension: force-sensing and homeostasis of cell-cell junctions. Trends Cell Biol. 2011;21:499–505. doi: 10.1016/j.tcb.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Green KJ, Simpson CL. Desmosomes: New perspectives on a classic. J Invest Dermatol. 2007;127:2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Bahna F, Katsamba PS, Jin XS, Brasch J, Vendome J, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17 doi: 10.1038/nsmb.1784. 348-U121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Corps EM, Berge T, Kilshaw PJ. The mechanism of cell adhesion by classical cadherins: the role of domain 1. J Cell Sci. 2005;118:711–721. doi: 10.1242/jcs.01665. [DOI] [PubMed] [Google Scholar]
- Harrison OJ, Jin XS, Hong SJ, Bahna F, Ahlsen G, Brasch J, et al. The Extracellular Architecture of Adherens Junctions Revealed by Crystal Structures of Type I Cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, Grzesiek S. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 2004;23:1699–1708. doi: 10.1038/sj.emboj.7600192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussinger D, Ahrens T, Sass HJ, Pertz O, Engel J, Grzesiek S. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation and mapping of contact regions. J Mol Biol. 2002;324:823–839. doi: 10.1016/s0022-2836(02)01137-3. [DOI] [PubMed] [Google Scholar]
- Hong SJ, Troyanovsky RB, Troyanovsky SM. Cadherin exits the junction by switching its adhesive bond. J Cell Biol. 2011;192:1073–1083. doi: 10.1083/jcb.201006113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Wheelock MJ. The relationships among adhesion, stratification and differentiation in keratinocytes. Cell Death and Differentiation. 1996;3:357–371. [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci U S A. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Johnson KR, Wheelock MJ. N-cadherin-mediated cell motility requires cis dimers. Cell Commun Adhes. 2005;12:23–39. doi: 10.1080/15419060500305971. [DOI] [PubMed] [Google Scholar]
- Koch AW, Pokutta S, Lustig A, Engel J. Calcium binding and homoassociation of E-cadherin domains. Biochemistry. 1997;36:7697–7705. doi: 10.1021/bi9705624. [DOI] [PubMed] [Google Scholar]
- Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoux B, Anon E, Lambert M, Rabodzey A, Hersen P, Buguin A, et al. Strength Dependence of Cadherin-Mediated Adhesions. Biophys J. 2010;98:534–542. doi: 10.1016/j.bpj.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoux B, Nicolas A. Physically based principles of cell adhesion mechanosensitivity in tissues. Reports on Progress in Physics. 2012;75 doi: 10.1088/0034-4885/75/11/116601. [DOI] [PubMed] [Google Scholar]
- Langer MD, Guo HB, Shashikanth N, Pierce JM, Leckband DE. N-glycosylation alters cadherin-mediated intercellular binding kinetics. J Cell Sci. 2012;125:2478–2485. doi: 10.1242/jcs.101147. [DOI] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, et al. Structural Basis for Mechanical Force Regulation of the Adhesin FimH via Finger Trap-like beta Sheet Twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband DE, le Duc Q, Wang N, de Rooij J. Mechanotransduction at cadherin-mediated adhesions. Curr Opin Cell Biol. 2011;23:523–530. doi: 10.1016/j.ceb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, et al. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci U S A. 2010;107:9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen CM. Tight junctions/adherens junctions: Basic structure and function. J Invest Dermatol. 2007;127:2525–2532. doi: 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Leckband D, Yap AS. Tissue Organization by Cadherin Adhesion Molecules: Dynamic Molecular and Cellular Mechanisms of Morphogenetic Regulation. Physiol Rev. 2011;91:691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papusheva E, Heisenberg C-P. Spatial organization of adhesion: force-dependent regulation and function in tissue morphogenesis. EMBO J. 2010;29:2753–2768. doi: 10.1038/emboj.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisini E, Higgins JMG, Liu JH, Brenner MB, Wang JH. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J Mol Biol. 2007;373:401–411. doi: 10.1016/j.jmb.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret E, Leung A, Feracci H, Evans E. Trans-bonded pairs of E-cadherin exhibit a remarkable hierarchy of mechanical strengths. Proc Natl Acad Sci U S A. 2004;101:16472–16477. doi: 10.1073/pnas.0402085101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O, Bozic D, Koch AW, Fauser C, Brancaccio A, Engel J. A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 1999;18:1738–1747. doi: 10.1093/emboj/18.7.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakasam A, Chien YH, Maruthamuthu V, Leckband DE. Calcium site mutations in cadherin: impact on adhesion and evidence of cooperativity. Biochemistry. 2006a;45:6930–6939. doi: 10.1021/bi060213m. [DOI] [PubMed] [Google Scholar]
- Prakasam AK, Maruthamuthu V, Leckband DE. Similarities between heterophilic and homophilic cadherin adhesion. Proc Natl Acad Sci U S A. 2006b;103:15434–15439. doi: 10.1073/pnas.0606701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S. Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci U S A. 2012;109:18815–18820. doi: 10.1073/pnas.1208349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani KK, Yago T, Klopocki AG, Lawrence MB, Fieger CB, Rosen SD, et al. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J Biol Chem. 2004;279:2291–2298. doi: 10.1074/jbc.M310396200. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, DeSimone DW. Cell adhesion receptors in mechanotransduction. Curr Opin Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Yagita Y, Wang Z, Koch A, Fex Svenningsen A, Gruzglin E, et al. The minimal essential unit for cadherin-mediated intercellular adhesion comprises extracellular domains 1 and 2. J Biol Chem. 2004;279:55914–55923. doi: 10.1074/jbc.M407827200. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, et al. Structural Basis of Cell-Cell Adhesion by Cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- Shi QM, Chien YH, Leckband D. Biophysical properties of cadherin bonds do not predict cell sorting. J Biol Chem. 2008;283:28454–28463. doi: 10.1074/jbc.M802563200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi QM, Maruthamuthu V, Li F, Leckband D. Allosteric Cross Talk between Cadherin Extracellular Domains. Biophys J. 2010;99:95–104. doi: 10.1016/j.bpj.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar S, Gumbiner B, Leckband D. Direct measurements of multiple adhesive alignments and unbinding trajectories between cadherin extracellular domains. Biophys J. 2001;80:1758–1768. doi: 10.1016/S0006-3495(01)76146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar S, Zhang Y, Nelson WJ, Chu S. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17:1075–1081. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Shimoyama Y, Nagafuchi A, Hirohashi S. E-cadherin functions as a cis-dimer at the cell-cell adhesive interface in vivo. Nat Struct Biol. 1999;6:310–312. doi: 10.1038/7542. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The Cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639. doi: 10.1242/dev.102.4.639. -&. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- Tamura K, Shan WS, Hendrickson WA, Colman DR, Shapiro L. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 1998;20:1153–1163. doi: 10.1016/s0896-6273(00)80496-1. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- Tinkle CL, Lechler T, Pasolli HA, Fuchs E. Conditional targeting of E-cadherin in skin: Insights into hyperproliferative and degenerative responses. Proc Natl Acad Sci U S A. 2004;101:552–557. doi: 10.1073/pnas.0307437100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky RB, Sokolov E, Troyanovsky SM. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol Cell Biol. 2003;23:7965–7972. doi: 10.1128/MCB.23.22.7965-7972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- Vendome J, Posy S, Jin X, Bahna F, Ahlsen G, Shapiro L, et al. Molecular design principles underlying Beta-strand swapping in the adhesive dimerization of cadherins. Nat Struct Mol Biol. 2011;18:693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. A Mechanoresponsive Cadherin-Keratin Complex Directs Polarized Protrusive Behavior and Collective Cell Migration. Dev Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Vendome J, Shapiro L, Ben-Shaul A, Honig B. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature. 2011;475:510–513. doi: 10.1038/nature10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YH, Jin XS, Harrison O, Shapiro L, Honig BH, Ben-Shaul A. Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc Natl Acad Sci U S A. 2010;107:17592–17597. doi: 10.1073/pnas.1011247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yago T, Wu JH, Wey CD, Klopocki AG, Zhu C, McEver RP. Catch bonds govern adhesion through L-selectin at threshold shear. J Cell Biol. 2004;166:913–923. doi: 10.1083/jcb.200403144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Boussadia O, Halfter H, Grose R, Berger P, Leone DP, et al. E-cadherin controls adherens junctions in the epidermis and the renewal of hair follicles. EMBO J. 2003;22:5723–5733. doi: 10.1093/emboj/cdg560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sivasankar S, Nelson WJ, Chu S. Resolving cadherin interactions and binding cooperativity at the single molecule level. Proc Natl Acad Sci U S A. 2009;106:109–114. doi: 10.1073/pnas.0811350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Chappuis-Flament S, Wong E, Jensen IE, Gumbiner BM, Leckband D. Functional analysis of the structural basis of homophilic cadherin adhesion. Biophys J. 2003;84:4033–4042. doi: 10.1016/S0006-3495(03)75129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]