Abstract
The default‐mode network (DMN) is a distributed functional‐anatomic network implicated in supporting memory. Current resting‐state functional connectivity studies in humans remain divided on the exact involvement of medial temporal lobe (MTL) in this network at rest. Notably, it is unclear to what extent the MTL regions involved in successful memory encoding are connected to the cortical nodes of the DMN during resting state. Our findings using functional connectivity MRI analyses of resting‐state data indicate that the parahippocampal gyrus (PHG) is the primary hub of the DMN in the MTL during resting state. Also, connectivity of the PHG is distinct from connectivity of hippocampal regions identified by an associative memory‐encoding task. We confirmed that several hippocampal encoding regions lack significant functional connectivity with cortical DMN nodes during resting state. Additionally, a mediation analysis showed that resting‐state connectivity between the hippocampus and posterior cingulate cortex—a major hub of the DMN—is indirect and mediated by the PHG. Our findings support the hypothesis that the MTL memory system represents a functional subnetwork that relates to the cortical nodes of the DMN through parahippocampal functional connections. Hum Brain Mapp 35:1061–1073, 2014. © 2013 Wiley Periodicals, Inc.
Keywords: brain mapping, physiology, human, resting state, functional connectivity, brain networks, Magnetic Resonance Imaging, MTL, Mediation, young adult
INTRODUCTION
The default‐mode network (DMN) (Gusnard and Raichle, 2001; Raichle et al., 2001) is a set of cortical regions identified by reduced activity during many externally oriented tasks (Buckner et al., 2008; Shulman et al., 1997). DMN regions also exhibit coherent fluctuations during resting state (Greicius et al., 2003). The DMN has been implicated in episodic memory function and appears to be particularly vulnerable to the detrimental effects of aging and Alzheimer's disease (AD; e.g., Andrews‐Hanna et al., 2007; Buckner et al., 2005, 2008; Hedden et al., 2009; Sheline et al., 2010a,b; Sperling et al., 2009). Current resting‐state functional connectivity MRI (fcMRI) studies remain divided on the exact involvement of the medial temporal lobe (MTL) in this network at rest. Specifically, it remains unclear if the hippocampus is directly connected to the DMN, or if it forms part of a functional subnetwork that interfaces with the DMN through the parahippocampal gyrus (PHG; Kahn et al., 2008; Vincent et al., 2006). Interestingly, task‐based fMRI studies indicate that MTL subregions can be dynamically coupled and uncoupled with the DMN during memory encoding and retrieval (McCormick et al., 2010; Huijbers et al., 2011; Vannini et al., 2010), implying context dependent, rather than static, interactions between the hippocampus and the DMN. Using resting‐state fcMRI in combination with task‐based fMRI, we test the hypothesis that the PHG is the primary DMN node in the MTL and that the PHG mediates the connectivity between the DMN and MTL structures engaged in memory formation. Notably, this approach allows us to compare MTL activations and DMN connectivity within modality using functional acquisitions collected with identical slice prescriptions and parameters in the same session.
Previous fcMRI studies have reported DMN connectivity with various MTL subregions, including some combination of the hippocampus and PHG (e.g., Greicius et al., 2004; Kahn et al., 2008; Shannon and Buckner, 2004; Vincent et al., 2006, 2007). However, not all studies have found evidence of robust DMN‐MTL connectivity during rest (e.g., Greicius, 2008; Sorg et al., 2007). One potential reason for this discrepancy is that the nodes of the DMN do not display identical intrinsic activity and the strength of the connections between these nodes is not homogenous (Andrews‐Hanna et al., 2010; Uddin et al., 2009). Additionally, prior work indicates that anterior and posterior parts of the hippocampus have different degrees and patterns of connectivity to the DMN (Kahn et al., 2008; Libby et al., 2012), possibly contributing to inconsistencies.
Episodic memory is critically dependent on the MTL and its functional connections to cortex (Milner et al., 1968; Song et al., 2011; Valenstein et al., 1987). Disconnection of the MTL is thought to underlie early memory deficits in AD (Gómez‐Isla et al., 1996; Hyman et al., 1986). Task‐based fMRI has been used to identify the focal subregions within the MTL that are functionally active during episodic memory (Chua et al., 2006; Ranganath et al., 2005; Yassa and Stark, 2008; Zeineh et al., 2003) and altered in the presence of AD pathology (Small et al., 1999; Sperling et al., 2009).
Finally, anatomical studies find few direct connections between hippocampus and cortical DMN regions (Burwell and Amaral, 1998; Suzuki and Amaral, 1994), but many between PHG and the DMN (Burwell, 2000; Furtak et al., 2007; Lavenex and Amaral, 2000; Witter et al., 2010a). Anatomical data also indicate that the entorhinal cortex, part of the PHG, mediates the input and output streams to the hippocampus (Witter et al., 2000ab).
Based on these previous functional and anatomic findings, we predict that the hippocampus is not the locus of functional connectivity between the MTL and the cortical nodes of the DMN during resting state. Instead, we expect to find stronger functional connectivity during resting state between the PHG and cortical nodes of the DMN. Second, given that the PHG has direct connections to both the hippocampus and cortical nodes of the DMN and that the hippocampus can be dynamically coupled and uncoupled with the DMN (Huijbers et al., 2011; Vannini et al., 2010; Young and Mcnaughton, 2009), we predict that functional connectivity between the hippocampus and cortical DMN nodes during resting state is mediated through the PHG.
MATERIAL AND METHODS
Participants
Thirty‐one healthy young adults (11 males, 20 females; age range: 18–28 years; mean age: 23.5 ± 2.3 years) participated in the study. All subjects were right‐handed, native English speakers with normal or corrected‐to‐normal vision. The subjects had no history of psychiatric or neurological disorders, head trauma, and were not using any psychoactive medications. Informed consent was obtained in accordance with guidelines and procedures governed by the institutional review boards of the Massachusetts General Hospital and Brigham and Women's Hospital (Boston, MA). Data from these subjects have not been previously published.
Data Acquisition
Participants underwent functional MRI on a Siemens Trio Tim 3.0 Tesla scanner (Siemens Medical Systems, Erlangen, Germany) equipped with a 12‐channel phased‐array head coil. Visual stimuli for the task were generated using an iBook G4 laptop (Apple Computer, Cupertino, California) running MacStim 2.5 (WhiteAnt Occasional Publishing, Melbourne, Australia) and projected to a screen positioned at the head of the magnet bore and reflected onto a mirror attached to the head coil. Head motion was restrained with extendable foam‐padded clamps. Earplugs and noise‐reduction headphones were used to attenuate scanner noise. Functional data were acquired using a gradient‐echo echo‐planar pulse sequence sensitive to BOLD contrast (Kwong et al., 1992; Ogawa et al., 1992) using the following parameters: TR = 2,000 ms, TE = 30 ms, FA = 90°, 64 × 64 matrix, FOV = 200 mm, 3.125 × 3.125 × 5 skip 1 mm voxels. Thirty interleaved coronal oblique slices aligned perpendicular to the anterior‐posterior commissural plane covered the whole brain. Functional images were acquired in one run of 196 time points for the resting state, and six runs of 127 time points for the encoding task.
Preprocessing
The data were processed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm/; version r4290). Each run was slice‐time corrected, realigned to the first volume of each run with INRIAlign (http://www-sop.inria.fr/epidaure/software/INRIAlign/; Freire and Mangin, 2001; Freire et al., 2002), normalized to the MNI 152 EPI template (Montreal Neurological Institute, Montreal, Canada), and smoothed with an 6 mm FWHM Gaussian kernel. Finally, the data for each subject were manually checked to identify registration errors and signal dropout. No subjects had these errors.
Resting‐State Analysis
An additional set of preprocessing steps was carried out to enable analysis of functional correlations between regions (see Fox et al., 2005; Vincent et al., 2006). A series of regressors from the resting‐state data were entered into a multiple regression analysis. These were: motion parameters as estimated by the six realignment parameters, average signal from a deep white matter mask, average signal from a ventricle mask, average signal from the whole brain mask, and the first derivative of each of these nuisance regressors. The residuals from this model were then linearly detrended and low‐pass filtered with a second‐order Butterworth filter with a frequency cutoff of 0.08 Hz. These processed data were used as the basis for all seed‐based correlation maps.
As part of hypothesis‐driven analysis to explore DMN connectivity with the MTL, we created seed‐based correlation maps using 10‐mm diameter spherical seeds located in the PCC at MNI coordinates [0 −53 26], and inferior in retrosplenial cortex (RSC) at MNI coordinates [0 −51 15]. The PCC is known to be a key region within the DMN (Fransson and Marrelec, 2008), and has been used as a seed region in multiple publications (e.g., Andrews‐Hanna et al., 2007; Hedden et al., 2009; van Dijk et al., 2010). RSC is known to have strong connectivity with the hippocampal formation (Kobayashi and Amaral, 2003; Yeo et al., 2011), and has been functionally dissociated from the PCC (Huijbers et al., 2010). Seed‐based correlation maps were created by first averaging the BOLD signal across all the voxels within the seed region. Next, this averaged signal was correlated with the preprocessed signal for every voxel in the brain. Finally, the maps were Fisher‐Z transformed with a hyperbolic arc tangent geometric transform to increase normality of the distribution of values for second‐level analyses. We found highly similar patterns of connectivity with similar peaks in the MTL for the PCC and RSP seed maps (Table 1).
Table 1.
Labels, locations, and t values of peaks in the MTL of resting‐state connectivity and task activity
| MNI label | H | Coordinates (MNI) | t(30) |
|---|---|---|---|
| Resting‐state connectivity | |||
| PCC seed | |||
| Parahippocampus | L | [−25 −22 −22] | 10.93 |
| [−15 −28 −16] | 5.24 | ||
| Parahippocampus | R | [27 −19 −25] | 7.96 |
| [18 −7 −37] | 6.92 | ||
| RSC seed | |||
| Parahippocampus | L | [−24 −22 −27] | 6.94 |
| [−18 −10 −34] | 5.75 | ||
| Parahippocampus | R | [24 −19 −28] | 8.1 |
| [30 −34 −13] | 5.61 | ||
| ICA seed | |||
| Parahippocampus | L | [−23 −19 −27] | 9.35 |
| Parahippocampus | R | [27 −19 −22] | 9.24 |
| [27 −34 −13] | 7.95 | ||
| Task activity | |||
| HCH > R | |||
| Hippocampus | L | [−19 −7 −16] | 7.88 |
| [−18 −34 −4] | 6.17 | ||
| Hippocampus | R | [32 −20 −17] | 5.22 |
| Amygdala | L | [−24 2 −25] | 7.00 |
| Amygdala | R | [18 −4 −19] | 9.67 |
| Fusiform | L | [−42 −50 −19] | 8.98 |
| Fusiform | R | [38 −53 −21] | 11.57 |
| MNI label | H | Coordinates (MNI) | ALE (×10−3) |
| ALE activity | |||
| Hippocampus | L | [−22 −10 −16] | 50.0 |
| Hippocampus | R | [18 −8 −16] | 38.0 |
| Fusiform | L | [−42 −46 −22] | 47.5 |
| Fusiform | R | [44 −52 −14] | 53.8 |
H = hemisphere, L = left, R = right, ALE = activation likelihood estimation.
To further test whether the results were a product of the choice of PCC seed location or could be generalized to the DMN as a whole, we used an independent component analysis (ICA) to isolate the DMN. We computed a spatiotemporal ICA using the GIFT (http://mialab.mrn.org/software/gift/; v1.3h; Calhoun et al., 2004) MATLAB (MathWorks, Natick, MA) package. Data that were preprocessed up to and including spatial smoothing were passed into GIFT where they were first intensity normalized. Next, data were reduced in a first‐pass principal component analysis step with 40 principal components. The Infomax ICA algorithm was used to generate 20 independent components, and the GICA3 reconstruction algorithm was used to back‐reconstruct spatial component maps and time courses for each subject. Individual subject component loadings were not normalized post hoc.
The component matching the DMN was identified first through visual inspection by five independent raters. All raters indicated that the ICA produced a single DMN component and selected the same component as the DMN. This component's relationship to the PCC and RSC seed‐derived DMNs was confirmed using a goodness of fit analysis. The goodness of fit metric was computed by first creating a binary mask from the positive values of the group PCC and RSC seed t‐maps, respectively. Goodness of fit for each ICA component was calculated by taking the mean of the value of voxels within the DMN template mask minus the mean of the value of voxels outside the DMN template mask (Greicius et al., 2004). The selected DMN component was an eightfold better match than the next best fitting component for both PCC and RSC seed‐maps, confirming that the DMN did not split into multiple components (see Damoiseaux et al., 2006 for discussion of one versus multiple ICA DMN components).
The ICA produced back‐projected time‐courses for each component for each subject. The individual subject time‐courses for the selected DMN component were used as seed time‐courses to create a new set of connectivity maps. That is, for this analysis, rather than choose a small spherical seed region, we used the signal decomposition of the ICA to isolate the spatiotemporally defined DMN time course for each subject. These time courses were used to generate Fisher‐Z transformed correlation maps for each subject, which allowed for a direct comparison between the ICA results and the seed‐based connectivity maps (see Fig. 1).
Figure 1.
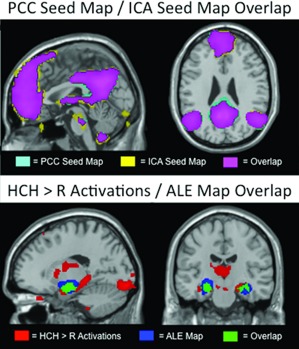
The top panel shows comparison of the PCC (teal) and ICA (yellow) seed maps in a sagittal (left: MNI x = 0) and coronal (right: z = 26) plane. Overlap is shown in purple. The bottom panel shows comparison (at MNI [x = −18, y = −18]) of HCH>R activations (red) and activation likelihood estimation meta‐analytic (Kim, 2011) activations (blue). Overlap is shown in green. All maps are thresholded at α < 0.01 FDR corrected.
To more specifically determine the functional connectivity relationship, if any, between regions of the hippocampus involved in successful memory encoding and the cortical nodes of the DMN, we defined a bilateral “entire” hippocampal seed (all‐hip) based on the High Confidence Hits > Repeated contrast (HCH>R; see “Task‐Based Analysis” section for details). Specifically, the seed was defined as a binary conjunction of the contrast map using an uncorrected threshold of P < 0.001 and the anatomical boundaries of the hippocampus defined by the AAL MNI atlas (Tzourio‐Mazoyer et al., 2002). To compare hippocampus connectivity to PHG connectivity, we created a similar bilateral PHG seed region (para‐hip) from the conjunction of the seed‐derived DMN using an uncorrected threshold of P < 0.001 and the anatomical boundaries of the PHG. These masks allow us to directly compare MTL subregions involved in successful memory formation with MTL subregions that exhibit connectivity with cortical DMN nodes at a liberal threshold while still loosely restricting between the anatomic location of the hippocampus and PHG.
Kahn et al. (2008) defined two distinct cortical networks that converge on the hippocampal formation. The first network converges on the anterior hippocampus, and includes the anterior temporal lobe, regions of the middle temporal gyrus, and the perirhinal/entorhinal cortices. The second network converges on the posterior hippocampus, and includes the lateral parietal cortex, RSC, PCC, and medial prefrontal cortex—all of which are cortical DMN regions. To test this anterior–posterior split, we constructed two additional seeds. These seeds are subsets of the all‐hip mask. They were created as a binary conjunction mask of a 10‐mm sphere drawn around the most anterior and most posterior HCH>R peaks in the left hippocampus (MNI [−19, −7, −16] and [−18, −34, −4]) and the all‐hip mask. Only the left hippocampus contained both an anterior and posterior peak. These conjunction masks limit our exploration to regions activated during successful memory encoding, while focusing on any difference between anterior and posterior hippocampus. We also used these masks to extract data from task and rest for the purpose of statistical comparisons. These extracted data were normalized using Fisher's r‐to‐z transformation (Zar, 1996). Para‐hip/PCC connectivity was tested against hippocampus/PCC connectivity using a within‐subjects model. We also tested para‐hip task activations against hippocampus task activations using an identical within‐subjects model.
All four of these seeds—entire hippocampus (all‐hip), anterior hippocampus (ant‐hip), posterior hippocampus (post‐hip), and PHG (para‐hip)—were used to create whole‐brain correlation maps to examine patterns of functional connectivity between these regions and the entire cerebral cortex. Each of the hippocampus seed‐based maps was then tested against the PHG seed‐based map with a within‐subjects design to identify regions of significant differing connectivity. To correct for multiple comparisons, we first Bonferroni corrected our initial α < 0.05 to control for multiple tests (Abdi, 2007). The whole‐brain images were then corrected using false discovery rate (FDR; Genovese et al., 2002) correction using the corrected α < 0.01.
Finally, to determine if the interface between the regions of the hippocampus involved in successful memory formation and the DMN are modulated by the PHG, we performed a series of simple and partial correlations. These correlations were based on resting‐state time series data extracted from the previously defined all‐hip, ant‐hip, post‐hip, para‐hip seeds, and the spherical PCC ROI centered at MNI [0 −53 26]. We examined the direct relationship between hippocampus, PHG, and PCC. Additionally, we examined the partial correlations between one MTL structure and the PCC while controlling for the other. This allows us to conduct a mediation analysis. In this analysis, the conditions for mediation are met if: (1) all of the pairwise correlations are significantly greater than zero, (2) the correlation of the hypothesized mediator (B) and dependent variable (C) while controlling for the independent variable (A) is significant, and (3) the correlation of A and C while controlling for B is not significant (Mackinnon et al., 2007). The fulfillment of these conditions indicates a system whereby the connectivity observed between A and C primarily reflects a transitive pathway A‐B‐C, rather than a direct pathway A‐C (see Fig. 5).
Figure 5.
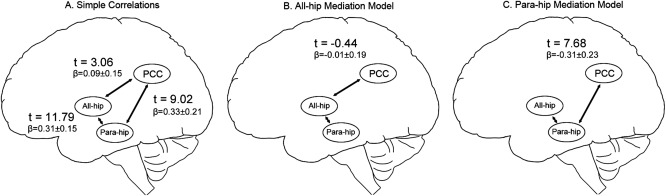
Models of MTL‐DMN mediation.
Task‐Based Analysis
The fMRI memory paradigm was a previously published face‐name associative encoding mixed block‐ and event‐related design (Sperling et al., 2009). Briefly, the task consisted of six runs of four 40 s alternating blocks of novel and repeated face‐name pairs with 25 s fixation between the blocks. Each novel block consisted of seven face‐name pairs with jittered fixation ISI, resulting in 84 total gender‐ and age‐balanced novel face‐name pairs. Repeated blocks consisted of alternating presentation of the same two gender‐balanced face‐name pairs. The two repeated face‐name pairs are never presented in novel blocks, nor are novel face‐name pairs ever repeated. Encoding success of novel face‐name pairs was tested post hoc with a two‐alternative forced‐choice test, followed by a high/low confidence ranking for each pair. The mean correct responses across the subjects were 84.3 ± 8.7%. The mean high confidence correct responses were 62.1 ± 13.0%.
The task analysis removed frequencies with a period of less than 260 s. In addition, bad volume regressors were included to negate volumes that had a global signal value beyond 2.5 standard deviations for the run, translational movement exceeding 0.75 mm per TR, and/or rotational movement exceeding 1.5° per TR. Outlier volume regressors consisted of an additional column in the design matrix for each identified outlier volume that consisted of a value of 1 for the outlier volume and zeros everywhere else. After data screening, no subjects were excluded, and all participants had four good runs that were included in the first‐level analysis.
Contrasts for High Confidence Hits > Repeated (HCH>R) were created for each subject across all runs included in the analysis. A whole brain one‐sample t‐test on the set of 31 participants was used to create a group‐level map. All second‐level analyses were performed with custom in‐house GLM scripts (http://nmr.mgh.harvard.edu/harvardagingbrain/People/AaronSchultz/GLM_Flex.html). These scripts were designed to function identically to SPM8 with the exception that not all data points needed to be present to analyze a particular voxel. If any voxel contained 15 or more values (maximum was 31), it was analyzed. No data were missing in the MTL, PCC, or lateral parietal cortex. Some data were missing in frontal regions, likely due to signal dropout.
RESULTS
Resting‐State Results
The PCC seed maps show resting‐state connectivity with multiple regions of the DMN, including connectivity to medial prefrontal cortex, left and right lateral parietal cortex (LPC), left and right lateral temporal cortex, and MTL. Within the MTL, the connectivity was most significant within the PHG, exhibiting bilateral peaks (Table 1). We did not observe significant (α < 0.01, FDR corrected) map‐level functional connectivity between the PCC seed and hippocampus during resting state. As the lack of significant connectivity at rest between major cortical nodes of the DMN and hippocampus may have been due to the choice of an a priori cortical seed region as hub of the DMN, we also took a data‐driven approach in which we used group ICA to define a DMN time course for each subject. The ICA generated a group‐level map that was virtually identical to the group PCC seed‐based map (Fig. 1, Table 1) suggesting that the findings using the PCC seed were not attributable to idiosyncrasies of the seed location.
Task‐Based Results
The results from the HCH>R task contrast demonstrated significant bilateral activity peaks in hippocampus (Table 1). Peaks were also found bilaterally in the amygdala and fusiform gyrus. No peaks were found in the PHG (see Table 1). Previous work has indicated that structures recruited in support of memory tasks will shift depending on the nature of the task (Davachi, 2006; Ranganath, 2010). To test if our task results differ significantly from frequent activations across many fMRI encoding tasks, we compared our task activations with activation likelihood estimation (ALE; Laird et al., 2005) maps from 74 fMRI studies using a subsequent‐memory approach (Kim, 2011). Activations in the MTL from our task (see red area in Fig. 1) are very similar to the ALE‐maps (blue in Fig. 1, overlap in green), but include a large area of posterior hippocampus that is not significantly activated using the meta‐analytic approach (Fig. 1, Table 1).
Task‐and Resting‐State Locations
A comparison between the group map of HCH>R and the PCC z‐maps (Fig. 2A–C) revealed a distinct difference in the location of task activation in the MTL versus PCC resting‐state connectivity within the MTL. The MTL peaks for memory task‐related activity (MNI [−19, −7, −16]) and PCC connectivity during rest (MNI [−25, −22, −22]) corresponded to the expected atlas coordinates of the hippocampus and PHG, respectively. The overlap of these two maps contained neither peaks of task activation nor peaks of resting‐state functional connectivity, suggesting minimal functional overlap of the two distinct foci.
Figure 2.
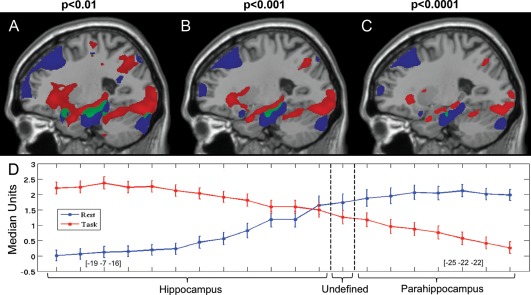
Spatial comparison of task activity and DMN connectivity. Panels A, B, and C display the task and PCC seed maps at varying thresholds in the sagittal plane (MNI x = −25). Panel A is thresholded at P < 0.01, panel B at P < 0.001, and panel C at P < 0.0001 uncorrected. Red areas show significant task activation and blue areas show significant connectivity with the PCC seed. Green areas show overlap between task activation and PCC connectivity. The overlap region is small, runs the border between the hippocampus and parahippocampus, and does not contain any peaks. Panel D displays the median‐normalized values extracted from these maps along a vector running between the hippocampal peak from task activations and the parahippocampal peak from PCC connectivity.
To examine the functional/anatomic boundary between the hippocampus and PHG, we first illustrated the spatial overlap between task and rest in the MTL at three different thresholds for display purposes (P < 0.01, P < 0.001, and P < 0.0001 uncorrected Fig. 2A–C). Then, we evaluated the relative contribution of task‐evoked activity and resting‐state connectivity in the MTL. We took 20 evenly spaced samples from 6 mm ROIs from each subject between the task‐derived left hippocampus peak (MNI [−19, −7, −16]) and the resting state left PHG peak (MNI [−25, −22, −22]). This vector ran linearly from peak to peak. We normalized the extracted values from each dataset by median positive value to allow for visual comparisons of task activity and resting‐state connectivity. The resulting plot (Fig. 2D) displays an anatomic dissociation between task activity and resting‐state connectivity. Most importantly, as we travel along this vector across the hippocampus/PHG anatomic boundary, we observe a shift from task activity‐dominant to resting‐state connectivity dominant. To explicitly test this distinction, we examined the connectivity of the para‐hip, all‐hip, ant‐hip, and post‐hip seeds defined previously. In resting state, Para‐hip/PCC connectivity was significantly greater than any hippocampus seed/PCC connectivity (P < 0.001; Table 2). In task, para‐hip activation was significantly less than any hippocampus seed activation (P < 0.05; Table 2).
Table 2.
Seed‐based analysis of within‐modality differences in connectivity and activity between regions in the MTL
| Seeds | Resting‐state connectivity | Task activations | ||
|---|---|---|---|---|
| Mean (z) | t(60) vs. Para‐hip | Mean (β) | t(60) vs. Para‐hip | |
| Para‐hip | 0.20 ± 0.12 | NA | 0.65 ± 1.13 | NA |
| All‐hip | 0.07 ± 0.12 | 4.19** | 1.51 ± 1.27 | −2.98* |
| Ant‐hip | 0.09 ± 0.14 | 3.47** | 2.57 ± 1.93 | −4.80** |
| Post‐hip | 0.05 ± 0.12 | 4.73** | 1.26 ± 1.14 | −2.11# |
**denotes P < 0.001, *denotes P < 0.01, and #denotes P < 0.05 uncorrected.
Next, we computed whole‐brain seed‐based correlation maps using the all‐hip, para‐hip, ant‐hip, and post‐hip seeds defined previously. As shown in Figure 3A, the para‐hip seed generated a canonical DMN map, whereas none of the task‐derived hippocampal seeds exhibited significant functional connectivity with the standard cortical DMN nodes (Fig. 3B–D; Table 3). Paired samples t‐tests between each hippocampus seed map and the para‐hip seed map (Fig. 4) show these maps to be significantly different (α < 0.01, FDR corrected) in the major regions of cortical DMN connectivity. Notably, neither the ant‐hip nor post‐hip seed exhibited connectivity with the cortical nodes of the DMN with similar magnitude or spatial extent as the para‐hip seed. This suggests that there is no difference in connectivity to cortical DMN nodes within the hippocampus. However, there is a significant difference in connectivity to cortical DMN nodes between the hippocampus and the PHG.
Figure 3.
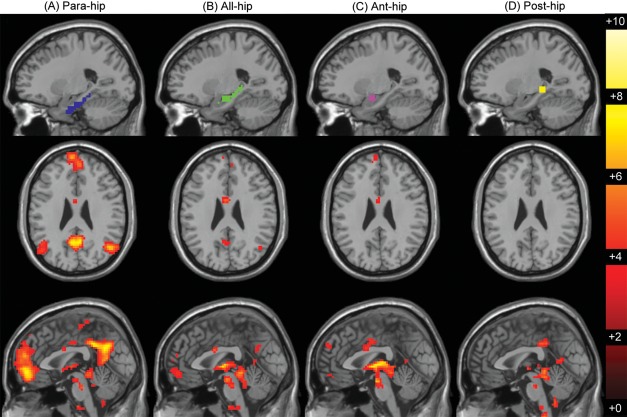
MTL seeds and resulting functional connectivity maps. The panels display connectivity maps seeded from para‐hip (A, blue), all‐hip (B, green), ant‐hip (C, purple), and post‐hip (D, yellow) seeds. Seeds are displayed in the sagittal plane (MNI x = −23). Connectivity maps are displayed below each seed in the axial (MNI z = −26) and sagittal (MNI x = 0) planes and are thresholded at α < 0.01 FDR corrected.
Table 3.
Clusters and peaks from MTL seed‐derived resting‐state correlation maps
| Seed | Lobe | Region | H | Coordinates (MNI) | t(30) | mm3 |
|---|---|---|---|---|---|---|
| All‐hip | Frontal | Orbital middle | L | [−3 59 −6] | 4.97 | 666 |
| Temporal | Middle, inferior, temporal pole, insula, fusiform, olfactory, thalamus, vermis, hippocampus, PHC | L/R | [9 −31 −18] | 8.18 | 33705 | |
| Parietal | Precuneus | L | [−3 −58 26] | 3.94 | 198 | |
| Ant‐hip | Frontal | Orbital middle, superior medial | L | [2 58 20] | 5.75 | 2700 |
| Temporal | Superior, middle, temporal pole, fusiform, caudate, lingual, putamen, amygdala, vermis, hippocampus, PHC | L/R | [21 −7 −19] | 12.68 | 29961 | |
| Parietal | Precuneus | L | [−6 −50 8] | 5.9 | ||
| Post‐hip | Temporal | Fusiform, thalamus, calcarine, hippocampus, PHC | L/R | [−8 −50 4] | 8.02 | 24498 |
| Parietal | PCC | L/R | [−3 −34 35] | 5.97 | 801 |
Figure 4.
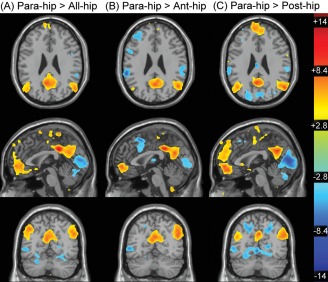
Paired sample t‐tests between the para‐hip seed maps versus all‐hip (A), ant‐hip (B), and post‐hip (C) seed maps. Maps are displayed in the axial (top: z = 26), sagittal (middle: x = 0), and coronal (bottom: y = −63) planes and are thresholded at α < 0.01 FDR corrected.
Finally, to explore whether the pattern of connectivity relationships support a model in which PHG mediates the relationship between the hippocampus and the DMN, we computed a series of partial correlations for each subject and tested whether these correlation values were significant (i.e., >0 connectivity). First, we correlated the all‐hip with the PCC, the para‐hip with the PCC, and the all‐hip with the para‐hip (Fig. 5A). All of these correlations were models significantly greater than zero [t(30) > 3]. However, all‐hip was not significantly correlated with the PCC when controlling for para‐hip (Fig. 5B), whereas the para‐hip was highly significantly correlated with the PCC while controlling for the all‐hip seed (Fig. 5C). This mediation analysis indicates that the majority of the variance shared between the hippocampus and PCC is contained within the PHG–PCC relationship, consistent with a model where the connection between hippocampus and PCC is indirect and mediated by the PHG. To determine whether the same pattern was present in other hippocampal seeds, we repeated this analysis using ant‐hip and post‐hip seeds, and the results statistically equivalent. Overall, these patterns are consistent with the hypothesis of the PHG as a major node of the DMN.
DISCUSSION
We found evidence for functional connectivity between major cortical nodes of DMN and specific subregions in the MTL, consistent with previous fcMRI studies (e.g., Andrews‐Hanna et al., 2007; Buckner et al., 2008; Hedden et al., 2009; Vincent et al., 2006). We extended these findings by using a functional localizer that activated the hippocampus, and showed a clear dissociation within the MTL between the locations of hippocampal activity during memory encoding and PHG connectivity with the DMN during rest. This dissociation of activation and connectivity was robust across multiple seeds and analysis methods. Finally, we extend findings that PHG and hippocampus have different patterns of cortical connectivity (Kahn et al., 2008, Libby et al., 2012) by demonstrating that the PHG mediates the connectivity between the hippocampus and the posterior cingulate cortex (PCC), the posterior cortical hub of the DMN.
Agreement With Previous Anatomic Connectivity Studies
Our functional connectivity findings in humans parallel anatomic studies of connectivity conducted in macaque and rat. Anatomical studies have established that PHG has many reciprocal connections with cortex (Burwell, 2000; Furtak et al., 2007; Lavenex and Amaral, 2000; Witter et al., 2010a). However, the hippocampus itself has few direct cortical connections (Burwell and Amaral, 1998; Suzuki and Amaral, 1994; van Strien et al., 2009). It is important to note that these studies of monosynaptic connectivity may not entirely reflect the complicated polysynaptic connections that underlie human functional connectivity. However, the relevance of those structural findings are supported by fcMRI analyses showing that macaques and rats have fcMRI‐derived homologs to major posterior DMN nodes (Lu et al., 2012; Margulies et al., 2009; Rilling et al., 2007; Vincent et al., 2007). Additionally, Catani et al. (2003) have also shown similarities between human and macaque MTL anatomy using diffusion tensor imaging. These anatomical findings implicate the PHG as a DMN node, which we have confirmed in humans with functional connectivity fMRI.
The MTL Memory System is Distinct from, but Interfaces with the DMN
When using the task‐defined hippocampal seed regions (Fig. 3C–E), we see very little functional connectivity with cortex, but a high degree of bilateral local coherence. Notably, this is true for both the anterior‐ and posterior‐specific hippocampus seeds. This parallels previous findings that indicate that the hippocampus is highly correlated across hemispheres (Buckner et al., 2008; Kahn et al., 2008; Vincent et al., 2006; Wang et al., 2010). Given the weak cortical functional connectivity found when seeding the hippocampus, our findings imply that the regions of the hippocampus engaged in encoding are not functionally connected to major cortical DMN nodes during rest, but rather are part of a functional subnetwork whose interface with the DMN is mediated through the PHG. Additionally, connectivity measures have been shown to be dynamic during resting‐state scans (Chang and Glover, 2010; Jones et al., 2012), possibly due to spontaneous memory processes, which in turn might also reflect coupling and decoupling of the DMN and MTL memory system.
The MTL memory system is likely composed of several subnetworks that support distinct memory processes (Eichenbaum et al., 2007; Ranganath and Richey 2012; Yassa and Stark 2008). Multiple task‐based fMRI studies have confirmed the role of the hippocampus during memory encoding, particularly in associative or relational encoding (e.g., Hannula and Ranganath, 2008; Zeineh et al., 2003). Other memory tasks, including spatial memory or recollected retrieval also activate MTL regions, although the foci differ (e.g., Poppenk and Moscovitch, 2011; Spaniol et al., 2009). Previous work using resting‐state fcMRI has described the MTL regions involved in memory as a subnetwork of the DMN (Vincent et al., 2008), but did not address this hypothesis within a memory task paradigm. In our view, the DMN exhibits condition‐dependent dynamic functional connectivity with the MTL memory systems. Specifically, in the context of memory encoding and retrieval, a change in the relationship between activity in the MTL and cortical nodes of the DMN has been observed. This is typically characterized by inversely correlated activity patterns during encoding (MTL will activate and DMN will deactivate) and positively correlated activity patterns during retrieval (both MTL and DMN will activate) (Daselaar et al., 2009; Huijbers et al., 2011; Vannini et al., 2010).
Further support for the distinction between MTL‐based memory systems and the DMN can be found in the multiple pathologies that show DMN disruption but not memory impairment. Changes in DMN connectivity have been widely reported in a range of neurological disorders, including depression (Greicius et al., 2007; Sheline et al., 2004b), autism spectrum disorders (Assaf et al., 2010; Cherkassky et al., 2006; Kennedy and Courchesne, 2008), schizophrenia (Bluhm et al., 2007; Rotarska‐Jagiela et al., 2010; Zhou et al., 2007), and obsessive‐compulsive disorder (Jang et al., 2010). However, many of these disorders do not manifest episodic memory impairment as the most salient clinical feature. Additionally, these results focus on differences in functional connectivity among cortical nodes of the DMN. The majority of functional connectivity studies in AD and mild cognitive impairment have reported specific evidence of altered connectivity between the DMN and the MTL (Celone et al., 2006; Greicius et al., 2004; Petrella et al., 2011; Rombouts et al., 2005; Sorg et al., 2007), suggesting that disconnection between these two systems may be more specific for amnestic disorders. Because cortical DMN dysfunction does not appear to be specific to amnestic disorders, it may be a more general indicator of synaptic pathology (Buckner et al., 2008).
Previous studies have indicated that lesions to the PHG that spare the hippocampus can also cause major memory deficits (Suzuki et al., 1993; Zola‐Morgan et al., 1989). A study of anatomic connectivity indicated that the input and output streams of the hippocampus are mediated through the superficial and deep layers, respectively, of the entorhinal cortex (Witter et al., 2000ab). Recent work in rats indicates that chemical inhibition of neural activity in the PHG disrupts memory retrieval of previously conditioned behavior (Morrissey et al., 2012). Additionally, fcMRI studies of patients with damage to the hippocampus find reduced connectivity in the ipsilesional regions of the MTL, but not with the cortical nodes of the DMN (Frings et al., 2009). Further, patients with specific hippocampal damage showed no difference in cortical thickness in DMN regions when compared with controls (Bernhardt et al., 2008), consistent with intact connections between the PHG and DMN. These findings indicate that previous observations of hippocampus/DMN connectivity (e.g., Andrews‐Hanna et al., 2010; Poppenk and Moscovitch, 2011) may reflect connectivity mediated through the PHG. This, collectively, supports our finding that the PHG serves as the interface between the MTL memory system and cortical nodes of the DMN and suggests that memory deficits caused by direct insult to PHG may be related to disruption of critical connections between the MTL memory system and the DMN.
Caveats
The fMRI data were relatively low resolution (3.125 × 3.125 × 6) to achieve whole‐brain coverage in the oblique coronal plane, and the analyses were conducted in common MNI atlas space. Low‐resolution images may make precise anatomic localization difficult, and nonlinear normalization to group space may result in localization errors. However, no localization errors were observed. Additionally, our functional slices sample perpendicular to the anterior‐posterior commissural plane and we are able to use the task analysis as a within‐modality functional localizer. Our slice prescription is optimized to observe MTL activity during either task or rest. This allows excellent separation in the coronal plane as well as a direct comparison with resting‐state connectivity using known hippocampal engagement during task.
Clinical Implications
Our finding that PHG serves as mediator between the cortical DMN nodes and the hippocampus proper supports the hypothesis that early pathological changes within the PHG may isolate the hippocampus from the DMN via changes in PHG‐DMN connectivity, rather than direct changes in hippocampus‐DMN connections (Gómez‐Isla et al., 1997; Hyman et al., 1986). It is possible that disrupted connectivity between these subnetworks leads to hippocampal hyperactivation and associated failure to deactivate posteromedial cortices during memory encoding (Miller et al., 2008; Sperling et al., 2009). A recent finding of CA3/dentate hyperactivity and entorhinal hypoactivity in patients with amnestic mild cognitive impairment suggests that this interface is critically important for memory tasks (Yassa et al., 2010b). We have also recently found evidence that hippocampal hyperactivity is associated with thinning in the entorhinal cortex and DMN regions and with the degree of memory impairment in patients with early mild cognitive impairment (Putcha et al., 2011). Finally, recent work indicating that tau pathology can propagate from one neuron to another along network connections (de Calignon et al., 2012; Liu et al., 2012), as well as evidence that neurodegeneration proceeds along functionally connected networks (de Calignon et al., 2012; Liu et al., 2012; Pievani et al., 2011; Seeley, 2011; Seeley et al., 2009), may link early focal MTL pathology, such as neurofibrillary tangle formation and neuronal loss, to dysfunction of large‐scale networks distributed throughout the cortex.
CONCLUSIONS
Our finding that the PHG, rather than the hippocampus proper, is functionally coupled to the DMN at rest has several implications. First, it suggests that the hippocampus is part of a distinct MTL memory system that interfaces with, but is not directly part of, the DMN. Second, the PHG appears to modulate functional connectivity between cortical DMN nodes and the hippocampus, which is consistent with previously published patterns of anatomical connectivity. Finally, our findings may have implications for the role of early PHG pathology in the disconnection of the DMN from the hippocampus proper in AD. If the PHG is in fact the nexus linking the MTL and cortical nodes of the DMN, then sensitive measures of PHG connectivity may prove to be a particularly promising biomarker of early AD‐related network dysfunction.
ACKNOWLEDGMENTS
The authors thank Hongkeun Kim for providing the ALE maps.
REFERENCES
- Abdi H (2007): Bonferroni and Šidák corrections for multiple comparisons In: Salkind NJ, editor.Encyclopedia of Measurement and Statistics. Thousand Oaks, CA:Sage; p. 284–290. [Google Scholar]
- Andrews‐Hanna JR, Snyder AZ, Vincent JL, Lustig C, Head D, Raichle ME, Buckner RL (2007): Disruption of large‐scale brain systems in advanced aging. Neuron 56:924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews‐Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL (2010): Functional‐anatomic fractionation of the brain's default network. Neuron 65:550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf M, Jagannathan K, Calhoun VD, Miller L, Stevens MC, Sahl R, O'boyle JG, Schultz RT, Pearlson GD (2010): Abnormal functional connectivity of default mode sub‐networks in autism spectrum disorder patients. NeuroImage 53:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt BC, Worsley KJ, Besson P, Concha L, Lerch JP, Evans AC, Bernasconi N (2008): Mapping limbic network organization in temporal lobe epilepsy using morphometric correlations: Insight on the relation between mesiotemporal connectivity and cortical atrophy. NeuroImage 42:515–524. [DOI] [PubMed] [Google Scholar]
- Bluhm RL, Miller J, Lanius RA, Osuch EA, Boksman K, Neufeld RWJ, Théberge J, Schaefer B, Williamson P (2007): Spontaneous low‐frequency fluctuations in the BOLD signal in schizophrenic patients: anomalies in the default network. Schizophr Bull 33:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, Larossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA (2005): Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci 25:7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Andrews‐Hanna JR, Schacter DL (2008): The brain's default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci 1124:1–38. [DOI] [PubMed] [Google Scholar]
- Burwell RD (2000): The parahippocampal region: corticocortical connectivity. Ann N Y Acad Sci 911:25–42. [DOI] [PubMed] [Google Scholar]
- Burwell RD, Amaral DG (1998): Cortical afferents of the perirhinal, postrhinal, and entorhinal cortices of the rat. J Comp Neurol 398:179–205. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pekar JJ (2004): A method for comparing group fMRI data using independent component analysis: application to visual, motor and visuomotor tasks. J Magn Reson Imaging 22:1181–1191. [DOI] [PubMed] [Google Scholar]
- Catani M, Jones DK, Donato R, Ffytche DH (2003): Occipito‐temporal connections in the human brain. Brain 126:2093–2107. [DOI] [PubMed] [Google Scholar]
- Celone K, Calhoun VD, Dickerson BC, Atri A, Chua EF, Miller SL, Depeau K, Rentz DM, Selkoe DJ, Blacker D, Albert MS, Sperling RA (2006): Alterations in memory networks in mild cognitive impairment and Alzheimer's disease: an independent component analysis. J Neurosci 26:10222–10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH (2010): Time‐frequency dynamics of resting‐state brain connectivity measured with fMRI. NeuroImage 50:81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkassky VL, Kana RK, Keller TA, Just MA (2006): Functional connectivity in a baseline resting‐state network in autism. Neuroreport 17:1687–1690. [DOI] [PubMed] [Google Scholar]
- Chua EF, Schacter DL, Rand‐Giovannetti E, Sperling RA (2006): Understanding metamemory: neural correlates of the cognitive process and subjective level of confidence in recognition memory. NeuroImage 29:1150–1160. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF (2006): Consistent resting‐state networks across healthy subjects. Proc Natl Acad Sci U S A 103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daselaar SM, Prince SE, Dennis NA, Hayes SM, Kim H, Cabeza R (2009): Posterior midline and ventral parietal activity is associated with retrieval success and encoding failure. Front Hum Neurosci 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davachi L (2006): Item, context and relational episodic encoding in humans. Curr Opin Neurobiol 16:693–700. [DOI] [PubMed] [Google Scholar]
- de Calignon A, Polydoro M, Suárez‐Calvet M, William C, Adamowicz DH, Kopeikina KJ, Pitstick R, Sahara N, Ashe KH, Carlson GA, Spires‐Jones TL, Hyman BT (2012): Propagation of tau pathology in a model of early Alzheimer's disease. Neuron 73:685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy HM (2005): The Human Hippocampus: Functional Anatomy, Vascularization, and Serial Sections with MRI. New York:Springer. [Google Scholar]
- Eichenbaum H, Yonelinas AR, Ranganath C (2007): The medial temporal lobe and recognition memory. Annu Rev Neurosci 30:123–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, van Essen DC, Raichle ME (2005): The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A 102:9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P, Marrelec G (2008): The precuneus/posterior cingulate cortex plays a pivotal role in the default mode network: Evidence from a partial correlation network analysis. NeuroImage 42:1178–1184. [DOI] [PubMed] [Google Scholar]
- Freire L, Mangin JF (2001): Motion correction algorithms may create spurious brain activations in the absence of subject motion. NeuroImage 14:709–722. [DOI] [PubMed] [Google Scholar]
- Freire L, Roche A, Mangin J (2002):What is the best similarity measure for motion correction in fMRI time series? IEEE Trans Med Imaging 2 1:470–484. [DOI] [PubMed] [Google Scholar]
- Frings L, Schulze‐Bonhage A, Spreer J, Wagner K (2009): Remote effects of hippocampal damage on default network connectivity in the human brain. J Neurol 256:2021–2029. [DOI] [PubMed] [Google Scholar]
- Furtak S, Wei S, Agster K (2007): Functional neuroanatomy of the parahippocampal region in the rat: The perirhinal and postrhinal cortices. Hippocampus 722:709–722. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T (2002): Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage 15:870–880. [DOI] [PubMed] [Google Scholar]
- Gómez‐Isla T, Price JL, Mckeel DW Jr, Morris JC, Growdon JH, Hyman BT (1996): Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer's disease. J Neurosci 16:4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Isla T, Hollister R, West H, Mui S, Growdon JH, Petersen RC, Parisi JE, Hyman BT (1997): Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Ann Neurol 41:17–24. [DOI] [PubMed] [Google Scholar]
- Greicius MD (2008): Resting‐state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol 21:424. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V (2003): Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A 100:253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V (2004): Default‐mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A 101:4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, Reiss AL, Schatzberg AF (2007): Resting‐state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry 62:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnard DA, Raichle ME (2001): Searching for a baseline: functional imaging and the resting human brain. Nat Rev Neurosci 2:685–694. [DOI] [PubMed] [Google Scholar]
- Hannula DE, Ranganath C (2008): Medial temporal lobe activity predicts successful relational memory binding. J Neurosci 28:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden T, van Dijk KRA, Becker JA, Mehta A, Sperling RA, Johnson KA, Buckner RL (2009): Disruption of functional connectivity in clinically normal older adults harboring amyloid burden. J Neurosci 29:12686–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers W, Pennartz CMA, Daselaar SM (2010): Dissociating the “retrieval success” regions of the brain: effects of retrieval delay. Neuropsychologia 48:491–497. [DOI] [PubMed] [Google Scholar]
- Huijbers W, Pennartz CMA, Cabeza R, Daselaar SM (2011): The hippocampus is coupled with the default network during memory retrieval but not during memory encoding. PLoS One 6:e17463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, van Hoesen GW, Kromer LJ, Damasio AR (1986): Perforant pathway changes and the memory impairment of Alzheimer's disease. Ann Neurol 20:472–481. [DOI] [PubMed] [Google Scholar]
- Jang JH, Kim J‐H, Jung WH, Choi J‐S, Jung MH, Lee J‐M, Choi C‐H, Kang D‐H, Kwon JS (2010): Functional connectivity in fronto‐subcortical circuitry during the resting state in obsessive‐compulsive disorder. Neurosci Lett 474:158–162. [DOI] [PubMed] [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, Przybelski SA, Gregg BE, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR (2012): Non‐stationarity in the “Resting Brain's” modular architecture. PLoS One 7:e39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn I, Andrews‐Hanna JR, Vincent JL, Snyder AZ, Buckner RL (2008): Distinct cortical anatomy linked to subregions of the medial temporal lobe revealed by intrinsic functional connectivity. J Neurophysiol 100:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DP, Courchesne E (2008): The intrinsic functional organization of the brain is altered in autism. NeuroImage 39:1877–1885. [DOI] [PubMed] [Google Scholar]
- Kim H (2011): Neural activity that predicts subsequent memory and forgetting: a meta‐analysis of 74 fMRI studies. NeuroImage 54:2446–2461. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Amaral DG (2003): Macaque monkey retrosplenial cortex: II. Cortical afferents. J Comp Neurol 466:48–79. [DOI] [PubMed] [Google Scholar]
- Kwong KK, Belliveau JW, Chesler DA, Goldberg IE, Weisskoff RM, Poncelet BP, Kennedy DN, Hoppel BE, Cohen MS, Turner R (1992): Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci U S A 89:5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Price CJ, Glahn DC, Uecker AM, Lancaster JL, Turkeltaub PE, Kochunov P, Fox PT (2005): ALE meta‐analysis: controlling the false discovery rate and performing statistical contrasts. Hum Brain Mapp 25:155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenex P, Amaral DG (2000): Hippocampal‐neocortical interaction: a hierarchy of associativity. Hippocampus 10:420–430. [DOI] [PubMed] [Google Scholar]
- Libby LA, Ekstrom AD, Ragland JD, Ranganath C (2012): Differential connectivity of perirhinal and parahippocampal cortices within human hippocampal subregions revealed with high‐resolution functional imaging. J Neurosci 32:6550–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, Duff K (2012): Trans‐synaptic spread of tau pathology in vivo. PLoS One 7:e31302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zou Q, Gu H, Raichle ME, Stein EA, Yang Y (2012): Rat brains also have a default mode network. Proc Natl Acad Sci 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon DP, Fairchild AJ, Fritz MS (2007): Mediation analysis. Annu Rev Psychol 58:593–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Vincent JL, Kelly C, Lohmann G, Uddin LQ, Biswal BB, Villringer A, Castellanos FX, Milham MP, Petrides M (2009): Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci U S A 106:20069–20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Moscovitch M, Protzner AB, Huber CG, Mcandrews MP (2010): Hippocampal‐neocortical networks differ during encoding and retrieval of relational memory: functional and effective connectivity analyses. Neuropsychologia 48:3272–3281. [DOI] [PubMed] [Google Scholar]
- Miller SL, Celone K, Depeau K, Diamond E, Dickerson BC, Rentz DM, Pihlajamäki M, Sperling RA (2008): Age‐related memory impairment associated with loss of parietal deactivation but preserved hippocampal activation. Proc Natl Acad Sci U S A 105:2181–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner B, Corkin S, Teuber H‐L (1968): Further analysis of the hippocampal amnesic syndrome: 14‐year follow‐up study of H. M. Neuropsychologia 6:215–234. [Google Scholar]
- Morrissey MD, Maal‐Bared G, Brady S, Takehara‐Nishiuchi K (2012): Functional dissociation within the entorhinal cortex for memory retrieval of an association between temporally discontiguous stimuli. J Neurosci 32:5356–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Tank DW, Menon R, Ellermann JM, Kim SG, Merkle H, Ugurbil K (1992): Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci U S A 89:5951–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella JR, Sheldon FC, Prince SE, Calhoun VD, Doraiswamy PM (2011): Default mode network connectivity in stable vs progressive mild cognitive impairment. Neurology 75:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pievani M, de Haan W, Wu T, Seeley WW, Frisoni GB (2011): Functional network disruption in the degenerative dementias. Lancet Neurol 10:829–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenk J, Moscovitch M (2011): A hippocampal marker of recollection memory ability among healthy young adults:contributions of posterior and anterior segments. Neuron,72:931–937. [DOI] [PubMed] [Google Scholar]
- Putcha D, Brickhouse M, O'keefe K, Sullivan C, Rentz D, Marshall G, Dickerson B, Sperling R (2011): Hippocampal hyperactivation associated with cortical thinning in Alzheimer's disease signature regions in non‐demented elderly adults. J Neurosci 31:17680–17688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Macleod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001): A default mode of brain function. Proc Natl Acad Sci U S A 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath C (2010): A unified framework for the functional organization of the medial temporal lobes and the phenomenology of episodic memory. Hippocampus 20:1263–1290. [DOI] [PubMed] [Google Scholar]
- Ranganath C, Heller A, Cohen MX, Brozinsky CJ, Rissman J (2005): Functional connectivity with the hippocampus during successful memory formation. Hippocampus 15:997–1005. [DOI] [PubMed] [Google Scholar]
- Ranganath C, Ritchey M (2012): Two cortical systems for memory‐guided behaviour. Nat Rev Neurosci 13:713–726. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Barks SK, Parr LA, Preuss TM, Faber TL, Pagnoni G, Bremner JD, Votaw JR (2007): A comparison of resting‐state brain activity in humans and chimpanzees. Proc Natl Acad Sci U S A 104:17146–17151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts SARB, Barkhof F, Goekoop R, Stam CJ, Scheltens P (2005): Altered resting state networks in mild cognitive impairment and mild Alzheimer's disease: an fMRI study. Hum Brain Mapp 26:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotarska‐Jagiela A, van de Ven V, Oertel‐Knöchel V, Uhlhaas PJ, Vogeley K, Linden DEJ (2010): Resting‐state functional network correlates of psychotic symptoms in schizophrenia. Schizophr Res 117:21–30. [DOI] [PubMed] [Google Scholar]
- Seeley WW (2011): Divergent network connectivity changes in healthy APOE ε4 carriers:disinhibition or compensation? Arch Neurol 68:1107–1108. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD (2009): Neurodegenerative diseases target large‐scale human brain networks. Neuron 62:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon BJ, Buckner RL (2004): Functional‐anatomic correlates of memory retrieval that suggest nontraditional processing roles for multiple distinct regions within posterior parietal cortex. J Neurosci 24:10084–10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Morris JC, Snyder AZ, Price JL, Yan Z, D'angelo G, Liu C, Dixit S, Benzinger T, Fagan A, Goate A, Mintun MA (2010a): APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Aβ42. J Neurosci 30:17035–17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Raichle ME, Snyder AZ, Morris JC, Head D, Wang S, Mintun MA (2010b): Amyloid plaques disrupt resting state default mode network connectivity in cognitively normal elderly. Biol Psychiatry 67:584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GL, Fiez JA, Corbetta M, Buckner RL, Miezin FM, Raichle ME, Petersen SE (1997): Common blood flow changes across visual tasks: II. Decreases in cerebral cortex. J Cogn Neurosci 9:648–663. [DOI] [PubMed] [Google Scholar]
- Small SA, Perera GM, Delapaz R, Mayeux R, Stern Y (1999): Differential regional dysfunction of the hippocampal formation among elderly with memory decline and Alzheimer's disease. Ann Neurol 466–472. [DOI] [PubMed] [Google Scholar]
- Song Z, Wixted JT, Hopkins RO, Squire LR (2011): Impaired capacity for familiarity after hippocampal damage. Proc Natl Acad Sci U S A 108:9655–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg C, Riedl V, Mühlau M, Calhoun VD, Eichele T, Läer L, Drzezga A, Förstl H, Kurz A, Zimmer C, Wohlschläger AM (2007): Selective changes of resting‐state networks in individuals at risk for Alzheimer's disease. Proc Natl Acad Sci U S A 104:18760–18765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaniol J, Davidson PSR, Kim ASN, Han H, Moscovitch M, Grady CL (2009): Event‐related fMRI studies of episodic encoding and retrieval: Meta‐analyses using activation likelihood estimation. Neuropsychologia 47:1765–1779. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Laviolette PS, O'keefe K, O'boyle JG, Rentz DM, Pihlajamäki M, Marshall G, Hyman BT, Selkoe DJ, Hedden T, Buckner RL, Becker JA, Johnson KA (2009): Amyloid deposition is associated with impaired default network function in older persons without dementia. Neuron 63:178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki WA, Amaral DG (1994): Perirhinal and parahippocampal cortices of the macaque monkey: cortical afferents. J Comp Neurol 350:497–533. [DOI] [PubMed] [Google Scholar]
- Suzuki WA, Zola‐Morgan S, Squire LR, Amaral DG (1993): Lesions of the perirhinal and parahippocampal cortices in the monkey produce long‐lasting memory impairment in the visual and tactual modalities. J Neurosci 13:2430–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Kelly AM, Biswal BB, Xavier Castellanos F, Milham MP (2009): Functional connectivity of default mode network components: correlation, anticorrelation, and causality. Hum Brain Mapp 30:625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenstein E, Bowers D, Verfaellie M, Heilman KM, Day A, Watson RT (1987): Retrosplenial amnesia. Brain 110( Pt 6):1631–1646. [DOI] [PubMed] [Google Scholar]
- van Dijk KRA, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL (2010): Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol 103:297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien NM, Cappaert NLM, Witter MP (2009): The anatomy of memory: an interactive overview of the parahippocampal‐hippocampal network. Nat Rev Neurosci 10:272–282. [DOI] [PubMed] [Google Scholar]
- Vannini P, O'boyle JG, O'keefe K, Pihlajamäki M, Laviolette P, Sperling RA (2010): What goes down must come up: Role of the posteromedial cortices in encoding and retrieval. Cereb Cortex 21:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini P, Hedden T, Becker JA, Sullivan C, Putcha D, Rentz D, Johnson KA, Sperling RA (2011): Age and amyloid‐related alterations in default network habituation to stimulus repetition. Neurobiol Aging 33:1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Snyder AZ, Fox MD, Shannon BJ, Andrews‐Hanna JR, Raichle ME, Buckner RL (2006): Coherent spontaneous activity identifies a hippocampal‐parietal memory network. J Neurophysiol 96:3517–3531. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME (2007): Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447:83–86. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Kahn I, Snyder AZ, Raichle ME, Buckner RL (2008): Evidence for a frontoparietal control system revealed by intrinsic functional connectivity. J Neurophysiol 100:3328–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Negreira A, Laviolette PS, Bakkour A, Sperling RA, Dickerson BC (2010): Intrinsic interhemispheric hippocampal functional connectivity predicts individual differences in memory performance ability. Hippocampus 20:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter MP, Naber PA, van Haeften T, Machielsen WC, Rombouts SARB, Barkhof F, Scheltens P, Lopes da Silva FH (2000a): Cortico‐hippocampal communication by way of parallel parahippocampal‐subicular pathways. Hippocampus 10:398–410. [DOI] [PubMed] [Google Scholar]
- Witter MP, Wouterlood FG, Naber PA, van Haeften T (2000b): Anatomical organization of the parahippocampal‐hippocampal network. Ann N Y Acad Sci 911:1–24. [DOI] [PubMed] [Google Scholar]
- Yassa MA, Muftuler LT, Stark CEL (2010): Ultrahigh‐resolution microstructural diffusion tensor imaging reveals perforant path degradation in aged humans in vivo. Proc Natl Acad Sci U S A 107:12687–12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa MA, Stark CEL (2008): Multiple signals of recognition memory in the medial temporal lobe. Hippocampus 18:945–954. [DOI] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, Fischl B, Liu H, Buckner RL (2011): The organization of the human cerebral cortex estimated by functional connectivity. J Neurophysiol 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CK, Mcnaughton N (2009): Coupling of theta oscillations between anterior and posterior midline cortex and with the hippocampus in freely behaving rats. Cereb Cortex 19:24–40. [DOI] [PubMed] [Google Scholar]
- Zar JH (1996): Biological Analysis. Upper Saddle River, NJ:Prentice‐Hall. [Google Scholar]
- Zeineh MM, Engel SA, Thompson PM, Bookheimer SY (2003): Dynamics of the hippocampus during encoding and retrieval of face‐name pairs. Science 299:577–580. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liang M, Tian L, Wang K, Hao Y, Liu H, Liu Z, Jiang T (2007): Functional disintegration in paranoid schizophrenia using resting‐state fMRI. Schizophr Res 97:194–205. [DOI] [PubMed] [Google Scholar]
- Zola‐Morgan S, Squire LR, Amaral DG, Suzuki WA (1989): Lesions of perirhinal and parahippocampal cortex that spare the amygdala and hippocampal formation produce severe memory impairment. J Neurosci 9:4355–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]


