SUMMARY
Background
Cytochrome P450 2C19 (CYP2C19) is the principle enzyme responsible for converting clopidogrel into its active metabolite and genetic variants have been identified, most notably CYP2C19*2 and CYP2C19*17, that are believed to alter its activity/expression.
Objective
We evaluated whether the consequences of the CYP2C19*2 and CYP2C19*17 variants on clopidogrel response were independent of each other or genetically linked through linkage disequilibrium (LD).
Patients/Methods
We genotyped the CYP2C19*2 and CYP2C19*17 variants in 621 members of the Pharmacogenomics of Anti-Platelet Intervention (PAPI) Study and evaluated the effects of these polymorphisms singly then jointly, taking into account LD, on clopidogrel prodrug level, clopidogrel active metabolite level, and ADP-stimulated platelet aggregation pre- and post-clopidogrel exposure.
Results
The CYP2C19*2 and CYP2C19*17 variants were in LD (|D’|=1.0; r2=0.07). In association analyses that did and did not account for the effects of CYP2C19*17, CYP2C19*2 was strongly associated with levels of clopidogrel active metabolite (beta=−5.24, P=3.0×10−9 and beta=−5.36, P=3.3×10−14, respectively) and post-treatment ADP-stimulated platelet aggregation (beta=7.55, P=2.9×10−16 and beta=7.51, P=7.0×10−15, respectively). In contrast, CYP2C19*17 was associated with clopidogrel active metabolite levels and ADP-stimulated platelet aggregation before (beta=1.57, P=0.04 and beta=−1.98, P=0.01, respectively) but not after (beta=0.40, P=0.59 and beta=−0.13, P=0.69, respectively) adjustment for the CYP2C19*2 variant. Stratified analyses of CYP2C19*2/CYP2C19*17 genotype combinations revealed that CYP2C19*2, and not CYP2C19*17, was the primary determinant in altering clopidogrel response.
Conclusions
Our results suggest that CYP2C19*17 has a small (if any) effect on clopidogrel-related traits and that the observed effect of this variant is due to LD with the CYP2C19*2 loss-of-function variant.
Keywords: Clopidogrel, drug metabolism, poor, CYP2C19-related, linkage disequilibrium, pharmacogenetics, platelets
Clopidogrel therapy is standard of care for treating patients with coronary artery disease (CAD) and/or acute coronary syndrome (ACS) undergoing percutaneous coronary intervention (PCI) [1, 2]. On-treatment variability in clopidogrel efficacy has been well-documented and previous investigations in our laboratory have shown that up to 70% of this variability may be attributed to genetic factors [3]. Indeed, genetic variants have been identified in CYP2C19 that alter clopidogrel active metabolite (CAM) formation, on-clopidogrel adenosine diphosphate (ADP)-stimulated platelet aggregation, and cardiovascular event rates in PCI/ACS patients on clopidogrel therapy [4-6]. As a result of such investigations, the clopidogrel label was updated in March 2010 to inform clinicians that CYP2C19 is important in clopidogrel metabolism, that poor metabolizers of clopidogrel, i.e. patients with 2 CYP2C19 loss-of-function alleles, exhibit higher rates of cardiovascular events, and that alternative treatment or treatment strategies should be considered in these patients [7].
The common loss-of-function CYP2C19*2 (rs4244285) and gain-of-function CYP2C19*17 (rs12248560) variants have been two of the most investigated single nucleotide polymorphisms (SNPs) with regard to clopidogrel efficacy. CYP2C19*2 creates an aberrant splice site that results in a premature stop codon and subsequently a non-functional truncated protein while CYP2C19*17 resides in a regulatory region of the gene and is associated with increased transcriptional activity [3, 8]. These variants have been assumed to operate oppositely and independently primarily on the basis of their presumed functional effects. The goal of this investigation is to determine whether the consequences of the CYP2C19*2 and *17 variants on clopidogrel response are independent of each other or genetically linked through linkage disequilibrium (LD), the non-random association between two or more sequence variants usually due to their close proximity on the same chromosome. To test this, we evaluated the effects of these polymorphisms singly and jointly, taking into account LD, on-clopidogrel prodrug level, the formation of CAM, and ADP-induced platelet aggregation pre- and post-clopidogrel exposure in 621 members of the Amish Pharmacogenomics of Anti-Platelet Intervention (PAPI) Study.
METHODS
PAPI Study Subjects
Population characteristics, recruitment, and study details of the Amish PAPI Study (NCT00799396) population have been previously described [3]. This report utilized an expanded set of 621 healthy Caucasian individuals recruited from August 2006 to January 2011. Briefly, subjects discontinued all medications, vitamins, and supplements 1 week prior to the initiation of this study and, after an overnight fast, information from medical and family histories, physical examinations, anthropometric measures, blood samples, and other phenotypic measurements were obtained. All participants were given a 300 mg loading dose of clopidogrel and instructed to take 75 mg/day for the following 6 days. Blood was drawn prior to and 1 hour after the last dose of clopidogrel for measures of platelet aggregation and clopidogrel metabolite levels.
Genotyping
Genotyping of the CYP2C19*2 and CYP2C19*17 variants was performed according to the manufacturer's instructions using TaqMan SNP genotyping assays (Applied Biosystems, Foster City, California). The mean genotype concordance rates for these polymorphisms were 100% for both SNPs and the genotype call rates were 99.4% and 98.5%, respectively.
Platelet Aggregation Measurements
Platelet-rich plasma (PRP) was isolated and platelet counts were adjusted to 200,000 platelets/μl using platelet-poor plasma (PPP). Platelet aggregation was evaluated by optical aggregometry using a PAP8E Aggregometer (Bio/Data Corporation, Horsham, Pennsylvania) after stimulation with ADP (20 μmol/L) and was expressed as the maximal percentage change in light transmittance using platelet-poor plasma as a referent [3].
Clopidogrel Prodrug and Active Metabolite Quantification
The methods for quantifying levels of clopidogrel prodrug and its active metabolite have been previously described [9, 10]. We measured both the prodrug and the active metabolite levels in a subset of 475 PAPI Study participants; characteristics of this group were the same as the full PAPI cohort (data not shown). Briefly, plasma levels of clopidogrel and its (E)-2-bromo-3’- methoxyacetophenone (MPB, Sigma Aldrich, St. Louis, Missouri)-derivatized active metabolite were simultaneously assessed using ultra-high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). Chromatographic separation was accomplished using a Waters Acquity UPLC® system (Waters Corporation, Milford, Maryland) and tandem mass spectrometry was performed using an AB Sciex Qtrap® 5500 (AB Sciex, Foster City, California). Parent clopidogrel was selectively identified by the transition of its parent to product ion at m/z 322>212, active metabolite at m/z 504>155, and ticlopidine (internal standard) at m/z 264>154. MRM peak integrations and data analysis were performed using the MultiQuant algorithm from MultiQuant 4.0 (Analyst®, AB Sciex).
Statistical Methods
Summary statistics were generated with SAS version 9.2 (SAS Institute Inc., Cary, NC). Minor allele frequencies, pairwise LD statistics (|D’| and r2), and conformity to Hardy-Weinberg equilibrium (HWE) were assessed using Haploview [11]. To assess generalizability of our results in the Amish to the general population, publically available LD data for the CEU (Utah residents with Northern and Western European ancestry) and YRI (Yoruba in Ibadan, Nigeria) populations were obtained using SNP Annotation and Proxy Search (SNAP) [12] and based on phased genotype data from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) and the 1000 Genomes Project (http://www.1000genomes.org/). Association results for the CYP2C19*17 or the CYP2C19*2 variants were calculated using SOLAR [13], which implements a variance component model accounting for family structure. Relatedness among participants was accounted for by including a polygenic component as a random effect as previously described [3]. Adjusting for family structure is critical in order to minimize type 1 error in genetic analyses of related individuals. The additive effect of genotype on quantitative trait was examined while also simultaneously adjusting for age and sex. The percent variance explained by these variants was estimated by calculating the reduction in residual variance in models that included versus did not include the variant. The effects of haplotypic allele combinations of the CYP2C19*2 and CYP2C19*17 variants on post-treatment levels of clopidogrel prodrug and the active metabolite as well as mean ADP-stimulated platelet aggregation at baseline and after clopidogrel exposure were compared using an analysis of variance approach in SOLAR with a mixed model adjusting for family structure. Likelihood ratio tests were used to determine whether mean trait levels differed between subjects with specific genotype combinations and significance was determined by t-tests.
All statistical tests were 2-sided. Individuals with missing data were omitted from analyses. Power estimates in this population (n = 621), calculated using the Quanto software program [14], indicated 80% power at α = 0.05 to detect SNP associations accounting for at least 1.3% of the residual trait variance.
RESULTS
Characteristics of the PAPI participants are shown in Table 1. Minor allele frequencies of CYP2C19*2 (0.17) and CYP2C19*17 (0.26) were similar to those reported in non-Amish Caucasians of the CEU HapMap Project cohort (CYP2C19*2 = 0.14, CYP2C19*17 = 0.23) and in prior studies of Caucasians investigating the effect of these SNPs on clopidogrel efficacy [15, 16]. Both SNPs evaluated in this study conformed to expectations of HWE (P value > 0.05).
Table 1.
Characteristics of Amish PAPI Study Participants.
| Characteristic, Units | Males (n = 302) | Females (n = 319) | Total (n = 621) |
|---|---|---|---|
| Mean age ± SD | 43.4 ± 12.3 | 46.9 ± 13.8 | 45.2 ± 13.2 |
| Mean body mass index* ± SD | 25.9 ± 3.5 | 28.4 ± 5.4 | 27.2 ± 4.7 |
| Systolic blood pressure ± SD (mm Hg) | 116.7 ± 11.4 | 117.8 ± 14.1 | 117.3 ± 12.8 |
| Diastolic blood pressure ± SD (mm Hg) | 70.9 ± 7.3 | 69.9 ± 7.4 | 70.4 ± 7.3 |
| Hypertension, No. (%) | 17 (5.6) | 24 (7.5) | 41 (6.6) |
| Total cholesterol ± SD (mg/dl) | 206.2 ± 42.7 | 214.4 ± 51.1 | 210.4 ± 47.3 |
| LDL-C ± SD (mg/dl) | 137.4 ± 39.7 | 137.3 ± 47.4 | 137.4 ± 43.8 |
| HDL-C ± SD (mg/dl) | 55.1 ± 14.5 | 61.9 ± 15.1 | 58.6 ± 15.2 |
| Triglycerides ± SD (mg/dl) | 68.7 ± 39.3 | 75.7 ± 42.3 | 72.3 ± 40.9 |
| Hypercholesterolemia, No. (%) | 54 (17.9) | 64 (20.1) | 118 (19.0) |
| Taking aspirin, No. (%) | 6 (2.0) | 2 (0.6) | 8 (1.3) |
| Self-reported diabetes, No. (%) | 1 (0.3) | 2 (0.6) | 3 (0.5) |
| Mean hematocrit ± SD, % | 41.6 ± 2.4 | 37.7 ± 2.3 | 39.6 ± 3.0 |
| Median (IQR) white blood cell count, X 1000/μL | 5.9 (4.3 -7.5) | 5.9 (4.3 -7.5) | 5.9 (4.3 -7.5) |
| Mean platelet count ± SD, X 100,000/μL | 234.9 ± 45.3 | 246.9 ± 51.7 | 240.6 ± 48.9 |
| Current smoker, No. (%) | 62 (20.5) | 0 (0.0) | 62 (10.0) |
Abbreviations: HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; PAPI, Pharmacogenomics of Anti-platelet Intervention; SD, standard deviation.
SI conversion factors: To convert HDL-cholesterol, LDL-cholesterol, and total cholesterol values to mmol/L, multiply by 0.0259; triglycerides to mmol/L, multiply by 0.0113.
Calculated as weight in kilograms divided by height in meters squared
In the PAPI Study, LD analysis revealed that CYP2C19*2 and CYP2C19*17 were in complete LD (|D’|=1). These results are consistent with |D’| values reported in non-Amish Caucasian and African populations (HapMap CEU (|D’|=1) and YRI (|D’|=1) populations, respectively). Since the minor rs12248560T allele (*17) is always carried on the major rs4244285G allele (*1), r2 estimates in our sample, as well as HapMap CEU and YRI populations, were considerably lower (r2=0.07, 0.04, and 0.08, respectively). The strong LD among these two variants is reflected by the absence of the *2 — *17 (rs4244285A — rs12248560T) haplotype. As a result, three of the eight possible haplotypes, namely CYP2C19*2/CYP2C19*17 genotype combinations *1*2/*17*17, *2*2/*1*17 and *2*2/*17*17 are non-existent in our sample (P = 3.2 × 10−10 for observed genotype combination frequencies vs. expected genotype frequencies assuming independent assortment, Table 2). These data affirm that these two variants are genetically linked due to their close physical proximity and the lack of significant recombination in the populations studied.
Table 2.
Observed number of CYP2C19*2/CYP2C19*17 genotype combinations in Amish PAPI Study participants
| CYP2C19*2 Genotype | ||||
|---|---|---|---|---|
| *1/*1 (G/G) | *1/*2 (G/A) | *2/*2 (A/A) | ||
| CYP2C19*17 Genotype | *1/*1 (C/C) | 195 | 132 | 14 |
| *1/*17 (C/T) | 183 | 57 | 0 | |
| *17/*17 (T/T) | 40 | 0 | 0 | |
We performed association analyses with each SNP separately followed by analyses in which both SNPs were included in the model as covariates to account for the non-independence of the variants. Results of single and multi-SNP association analyses evaluating the effect of CYP2C19*2 and CYP2C19*17 on CAM levels are shown in Table 3. In the single SNP analysis, CYP2C19*2 was significantly associated with decreased CAM levels (beta = −5.36, P = 3.3 × 10−14) and the effect size was virtually unchanged with adjustment for CYP2C19*17 (beta = −5.24, P = 3.0 × 10−9), with this SNP accounting for 8.5% and 7.4% of the variation observed in this trait, respectively. As expected, CYP2C19*2 was not associated with clopidogrel prodrug levels in either the single SNP or multi-SNP analyses (P = 0.19 and P = 0.18, respectively).
Table 3.
Association analysis results of the CYP2C19*2 and CYP2C19*17 variants with clopidogrel active metabolite level and post-clopidogrel ADP-stimulated platelet aggregation*
| Single SNP association analysis | Multi-SNP association analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Clopidogrel active metabolite level | ADP-stimulated platelet aggregation | Clopidogrel active metabolite level | ADP-stimulated platelet aggregation | |||||
| beta | p | beta | p | beta | p | beta | p | |
| CYP2C19*2 (rs4244285) | −5.36 | 3.3 × 10−14 | 7.55 | 2.9 × 10−16 | −5.24 | 3.0 × 10−9 | 7.51 | 7.0 × 10−15 |
| CYP2C19*17 (rs12248560) | 1.57 | 0.04 | −1.98 | 0.01 | 0.40 | 0.59 | −0.13 | 0.69 |
Multi-SNP analysis model includes both CYP2C19*2 and CYP2C19*17 in the model
Before adjusting for CYP2C19*2, association results suggested that the CYP2C19*17 variant was marginally associated with increased CAM levels (beta = 1.57, P = 0.04). However, when CYP2C19*2 genotype was added to the model as a covariate in the multi-SNP analysis, the association with CYP2C19*17 was markedly attenuated and no longer associated with CAM (beta = 0.40, P = 0.59). Correspondingly, the CYP2C19*17 variant accounted for 0.09% of the variation in CAM levels prior to adjusting for the CYP2C19*2 variant and 0.01% after the effects of CYP2C19*2 were taken into consideration. CYP2C19*17 was not associated with clopidogrel prodrug levels in either the single SNP or multi-SNP analyses (P = 0.92 and 0.81, respectively).
Consistent with the results of the CAM analysis, CYP2C19*2 was significantly associated with increased ADP-stimulated platelet aggregation following clopidogrel exposure in both single SNP (beta = 7.55, P = 2.9 × 10−16) and multi-SNP (beta = 7.51, P = 7.0 × 10−15) analyses (Table 3), with this SNP accounting for 13.1% and 11.8% of the variation in ADP-stimulated platelet aggregation post-clopidogrel treatment in single SNP and multi-SNP analyses, respectively. No association was observed pre-clopidogrel treatment using either model (P = 0.87 and 0.84 for single and multi-SNP analysis, respectively). Of note, the association between CYP2C19*2 and post-clopidogrel ADP-stimulated platelet aggregation as well as the percentage of trait variation explained by this SNP have been previously reported in a subset (n=429) of the current Amish sample population (P = 4.3 × 10−11 and 12%, respectively) [3].
In association analyses of ADP-stimulated platelet aggregation, CYP2C19*17 was statistically significant in single SNP analyses (beta= −1.98, P = 0.01); however, this association was also markedly attenuated and no longer statistically significant in the multi-SNP model that included CYP2C19*2 as a covariate (beta = −0.13, P = 0.69). The variation in ADP-stimulated platelet aggregation explained by the CYP2C19*17 variant decreased considerably in the multi-SNP model compared to the single SNP analysis (0.1% vs. 1.0%, respectively).
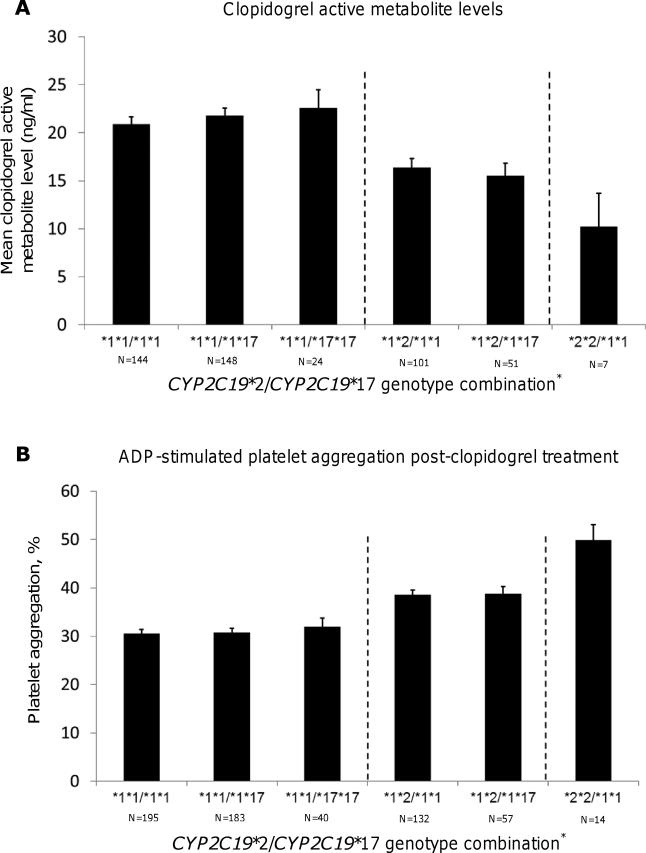
Figures 1 shows mean CAM levels and ADP-stimulated platelet aggregation values stratified by CYP2C19*2/CYP2C19*17 genotype combinations. Among subjects who did not carry the CYP2C19*2 allele, CAM levels were not statistically different with increasing copies of the CYP2C19*17 allele (P = 0.38) Similarly, while individuals who were CYP2C19*2 heterozygotes had decreased CAM levels compared to those who did not carry the CYP2C19*2 allele (P = 0.0003), addition of the CYP2C19*17 allele in CYP2C19*2 heterozygotes did not affect CAM levels (P = 0.57). Finally, individuals homozygous for the CYP2C19*2 allele had decreased mean levels of CAM as compared to those with 0 or 1 copy of this allele (P = 1.3 × 10−5).
Figure 1. Effect of CYP2C19*2/CYP2C19*17 genotype combination on the formation of clopidogrel active metabolite level (A) and ADP-stimulated platelet aggregation (B) in members of the Pharmacogenomics of Anti-Platelet Intervention (PAPI) Study.
Vertical dashed lines are used to separate PAPI participants based on CYP2C19*2 genotype. * Due to linkage disequilibrium, there were no individuals with CYP2C19*2/CYP2C19*17 *1*2/*17*17, *2*2/*1*17, and *2*2/*17*17 genotype combinations.
As described above, CYP2C19*2 was significantly associated with measures of ADP-stimulated platelet aggregation. However, among individuals who carried no copies of the CYP2C19*2 allele, levels of platelet aggregation were not associated with presence of the CYP2C19*17 allele (P = 0.96) (Fig 1B). Similarly, no effect of the CYP2C19*17 allele on ADP-stimulated platelet aggregation could be observed among CYP2C19*2 heterozygotes (P = 0.90).
DISCUSSION
We evaluated whether the effects of the CYP2C19*2 and CYP2C19*17 variants on clopidogrel prodrug and active metabolite levels as well as ADP-stimulated platelet aggregation pre- and post-clopidogrel treatment are independent of each other or are correlated due to LD. LD is calculated using the r2 and D’ statistics and allows for assessment of how well alleles are correlated with each other and whether alleles are inherited over time together, respectively. We observed that while the r2 statistic was low given the difference in allele frequencies between these variants, |D’| estimates show that CYP2C19*2 and CYP2C19*17 are in complete LD with each other and thus these variants are inherited together on the same haplotype. Furthermore, our data regarding measures of LD are consistent with HapMap and 1,000 Genomes Project data and strengthen our confidence in the generalizability of our results. Thus, the CYP2C19*17 variant is always on the haplotype lacking the CYP2C19*2 variant. Therefore, it is important to consider the effect one variant has on the other in terms of clopidogrel efficacy and to examine the nature of haplotypic diversity that exists in study populations.
When measures of LD were not taken into account, the CYP2C19*17 variant was significantly associated with increased levels of CAM and decreased ADP-stimulated platelet aggregation. However, when taking into account the effects of the CYP2C19*2 locus, the results of these analyses changed substantially showing no independent effect of the CYP2C19*17 variant on either CAM levels or ADP-stimulated platelet aggregation. In contrast, the CYP2C19*2 variant was significantly associated with both CAM levels and ADP-stimulated platelet aggregation with or without regard to the CYP2C19*17 variant. Together, these data suggest that the CYP2C19*2 variant is the main determinant of clopidogrel efficacy and that the observed influence of the CYP2C19*17 variant on clopidogrel-related traits is driven primarily, and perhaps entirely, by non-independence with the CYP2C19*2 variant.
In order to further evaluate the independent effects, or lack thereof, of the CYP2C19*2 and CYP2C19*17 variants on clopidogrel efficacy, we compared mean traits values of CAM levels and ADP-stimulated platelet aggregation for each observed 2-SNP haplotype (Figures 1A and 1B). Consistent with the results of the single- and multi-SNP association analyses, in individuals who carried 0 copies of the CYP2C19*2 allele, increasing the number of CYP2C19*17 alleles did not result in a significant increase in CAM or a corresponding decrease in ADP-stimulated platelet aggregation. Furthermore, increasing the number of CYP2C19*17 alleles in CYP2C19*2 heterozygotes yielded similar results. In contrast, increasing the number of the CYP2C19*2 allele resulted in reduced CAM formation and a subsequent increase in ADP-stimulated platelet aggregation regardless of the number of CYP2C19*17 alleles present. Due to complete LD, the CYP2C19*17 variant is non-existent or very uncommon in CYP2C19*2 homozygotes, and thus could not be evaluated in this group. Overall, these results are not consistent with the work of Sibbing and colleagues who showed that co-inheritance of the CYP2C19*2 and CYP2C19*17 alleles resulted in an intermediate level of platelet aggregation similar to what is observed in individuals who are homozygous for the wild-type alleles of both these SNPs [17]. However, these results do support the claims of Gurbel et al. that the “gain-of-function” effect attributed to the CYP2C19*17 variant may instead be due to the fact that patients who have increasing copies of the CYP2C19*17 allele are less likely to carry the CYP2C19*2 risk allele due to LD [18].
Several reports have been published evaluating the CYP2C19*17 variant on clopidogrel efficacy with mixed results. While some of these studies suggest that this variant significantly decreases on-treatment platelet reactivity and/or occurrence of cardiovascular events, several investigations have observed no such effect (Table 4). Furthermore, data from recently performed meta-analyses are also inconsistent. For example, independent meta-analyses performed by Li et al. [19] and Zabalza et al. [20] suggest that clopidogrel-treated patients who carry the CYP2C19*17 variant have decreased rates of cardiovascular events (OR = 0.82, 95%CI 0.72-0.94 and HR = 0.75, 95%CI 0.66-0.87, respectively) and increased rates of adverse bleeding (OR = 1.25, 95%CI 1.07-1.47 and HR = 1.26, 95%CI 1.05-1.50, respectively). In contrast, a systematic review and meta-analysis performed by Bauer and colleagues [21] showed that clopidogrel-treated patients who carried at least one copy of the CYP2C19*17 allele did not have an increased risk of experiencing stent thrombosis (OR = 0.99, 95%CI 0.60-1.62) or other cardiovascular events (OR = 0.93, 95%CI 0.75-1.14). Differences in study design, patient populations, and statistical power may explain the mixed results between studies. Alternatively, adjustment for the CYP2C19*2 variant or lack thereof may explain, at least in part, these discrepancies.
Table 4.
Summary of genetic association studies with CYP2C19*17 and clopidogrel response traits
| Study* | Patients | Outcome(s) | Results of association studies |
|---|---|---|---|
| Geisler 2008 | CAD | ADP-induced platelet aggregation (LTA) | no association |
| Frere 2009 | ACS | ADP-induced platelet aggregation (LTA) | no association |
| VASP phosphorylation | decreased platelet reactivity | ||
| ADP-induced platelet aggregation (LTA) | no association | ||
| Gurbel 2009 | CAD | VASP phosphorylation | no association |
| TEG | no association | ||
| Mega 2009 | ACS | MACE | no association |
| bleeding | no association | ||
| Shuldiner 2009 | PCI/healthy | ADP-induced platelet aggregation (LTA) | no association |
| MACE | no association | ||
| Simon 2009 | MI | MACE | no association |
| ACS | MACE | decreased event rate | |
| Pare 2010 | bleeding | no association | |
| AF | MACE | no association | |
| ADP-induced platelet aggregation (LTA) | decreased platelet aggregation | ||
| Sibbing 2010 | CAD | MACE | no association |
| bleeding | increased risk | ||
| Tiroch 2010 | MI | TLR | decreased event rate |
| MACE | decreased event rate | ||
| Wallentin 2010 | ACS | MACE | no association |
| bleeding | increased risk | ||
| Bouman 2011 | PCI | stent thrombosis | no association |
| Platelet reactivity (VerifyNow P2Y12) | decreased platelet reactivity | ||
| Campo 2011 | PCI | MACE | no association |
| bleeding | increased risk | ||
| Gurbel 2011 | CAD | ADP-induced platelet aggregation (LTA) | no association |
| HPR | decreased rate | ||
| Trenk 2011 | PCI | ADP-induced platelet aggregation (LTA) | decreased platelet aggregation |
| Bauer 2011 | Meta-Analysis | MACE | no association |
| stent thrombosis | no association | ||
| Rideg 2011 | PCI | ADP-induced platelet aggregation (LTA) | decreased platelet aggregation |
| VASP phosphorylation | decreased platelet reactivity | ||
| Chan 2012 | PCI | VASP phosphorylation | decreased platelet reactivity |
| Dai 2012 | PCI | ADP-induced platelet aggregation (LTA) | decreased platelet aggregation |
| bleeding | increased risk | ||
| ADP-induced platelet aggregation (LTA) | decreased platelet aggregation | ||
| Harmze 2012 | PCI | Platelet reactivity (VerifyNow P2Y12) | decreased platelet reactivity |
| bleeding | increased risk of bleeding | ||
| ADP-induced platelet aggregation (LTA) | no association | ||
| Tello-Montoliu 2012 | ACS | VASP phosphorylation | decreased platelet reactivity |
| TRAP (LTA) | no association | ||
| MACE | no association | ||
| HPR | decreased rate | ||
| Li 2012 | Meta-Analysis | MACE | decreased event rate |
| stent thrombosis | no association | ||
| bleeding | increased risk | ||
| Zebalza 2012 | Meta-Analysis | MACE | decreased event rate |
| bleeding | increased risk | ||
| Kassimis 2012 | PCI | Platelet reactivity (VerifyNow P2Y12) | no association |
| Subraja 2012 | CAD | ADP-induced platelet aggregation (LTA) | no association |
Abbreviations: ACS, acute coronary syndrome; ADP, adenosine diphosphate; AF, atrial fibrillation; CAD, coronary artery disease; CYP2C19, cytochrome P450 2C19; HPR, high platelet reactivity; LTA, light transmission aggregometry; MACE, major adverse cardiovascular event; MI, myocardial infarction; PCI, percutaneous coronary intervention; TEG, thromboelastography; TLR, target lesion revascularization; VASP, vasodilator-stimulated phosphoprotein.
Full references for all investigations are shown in the online Supporting Information.
There are some limitations of this study that we would like to acknowledge. The method used to quantitate CAM in this study does not differentiate between isomers generated by CYP2C19 and paraoxonase (PON1); however, it has been previously shown that the major isomer found in plasma of clopidogrel-treated patients derives from the CYP2C19-dependent pathway and thus we believe that this does not significantly affect our results [22]. Furthermore, quantification of clopidogrel prodrug and active metabolite was measured at one time point in this investigation. Therefore, we suggest using caution when interpreting our metabolite data in the context of the overall clopidogrel exposure.
The use of genetic information to help clinicians personalize anti-platelet treatment offers great potential to reduce recurrent cardiovascular events and improve patient care. Indeed, studies of implementation of CYP2C19 genotype-directed anti-platelet therapy have been reported [23] and others are currently underway [24]. Furthermore, guidelines and treatment algorithms have been developed to help guide clinicians in choosing the most effective anti-platelet regimen based on CYP2C19*2 genotype [25]. Such guidelines, primarily driven by the current evidence base, categorize patients who carry the CYP2C19*17 variant as being “ultrametabolizers” of clopidogrel. However, results of this investigation suggest that the CYP2C19*17 variant has a small (if any) independent effect on clopidogrel metabolism, but rather the observed effect of this variant on clopidogrel-related traits is dependent on the CYP2C19*2 variant due to LD.
ACKNOWLEDGEMENTS
We gratefully acknowledge our Amish liaisons and field workers and the extraordinary support of the Amish community, without which these studies would not have been possible.
FUNDING SOURCES
This study was supported by National Institutes of Health grants NIH U01 GM074518, U01 HL105198, U01 HL084756, U01 GM074492, R01074730, and K23 GM102678, GM074518-05S1, the Mid-Atlantic Nutrition and Obesity Center (P30 DK072488), the University of Maryland General Clinical Research Center (M01 RR16500), and the Baltimore Veterans Administration Geriatric Research and Education Clinical Center.
CONFLICTS OF INTEREST
Dr. Shuldiner receives grant support from NIH to study the pharmacogenomics of anti-platelet therapy. He is also a consultant for United States Diagnostic Standards, Inc.
Disclaimer:
The manuscript is a result of independent research and no official support or endorsement of this manuscript by the Food and Drug Administration is intended or should be inferred.
Footnotes
ADDENDUM
JP Lewis* and SH Stephens* conceived and designed the research, analyzed and interpreted the data, performed statistical analyses, and drafted the manuscript. RB Horenstein*, CJ Peer†, WD. Figg†, SD Spencer‡, and MA Pacanowski§ conceived, designed, and performed the clopidogrel metabolite quantification assay. JR O'Connell*, K Ryan*, and BD Mitchell* performed statistical analyses and interpreted data. AR Shuldiner*, ¶ conceived and designed the research, acquired the data, and interpreted the data. All authors reviewed and made critical revisions to the manuscript.
*Division of Endocrinology, Diabetes and Nutrition and Program in Personalized and Genomic Medicine, University of Maryland School of Medicine, Baltimore, Maryland, USA. †Clinical Pharmacology Program, National Cancer Institute, Bethesda, Maryland, USA. ‡Applied and Developmental Research, SAIC-Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. §U.S. Food and Drug Administration, Center for Drug Evaluation and Research, Office of Clinical Pharmacology, Silver Spring, Maryland, USA. ¶Geriatric Research and Education Clinical Center, Veterans Administration Medical Center, Baltimore, Maryland, USA.
REFERENCES
- 1.Kushner FG, Hand M, Smith SC, Jr., King SB, 3rd, Anderson JL, Antman EM, Bailey SR, Bates ER, Blankenship JC, Casey DE, Jr., Green LA, Hochman JS, Jacobs AK, Krumholz HM, Morrison DA, Ornato JP, Pearle DL, Peterson ED, Sloan MA, Whitlow PL, et al. 2009 focused updates: ACC/AHA guidelines for the management of patients with ST-elevation myocardial infarction (updating the 2004 guideline and 2007 focused update) and ACC/AHA/SCAI guidelines on percutaneous coronary intervention (updating the 2005 guideline and 2007 focused update) a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2009;54:2205–41. doi: 10.1016/j.jacc.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 2.King SB, 3rd, Smith SC, Jr., Hirshfeld JW, Jr., Jacobs AK, Morrison DA, Williams DO, Feldman TE, Kern MJ, O'Neill WW, Schaff HV, Whitlow PL, Adams CD, Anderson JL, Buller CE, Creager MA, Ettinger SM, Halperin JL, Hunt SA, Krumholz HM, Kushner FG, et al. 2007 focused update of the ACC/AHA/SCAI 2005 guideline update for percutaneous coronary intervention: a report of the American College of Cardiology/American Heart Association Task Force on Practice guidelines. J Am Coll Cardiol. 2008;51:172–209. doi: 10.1016/j.jacc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, Damcott CM, Pakyz R, Tantry US, Gibson Q, Pollin TI, Post W, Parsa A, Mitchell BD, Faraday N, Herzog W, Gurbel PA. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–62. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 5.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Karolinska Institute [September 23, 2012];Human Cytochrome P450 (CYP) Allele Nomenclature Committee home page. ( http://www.cypalleles.ki.se.)
- 7. [September 23, 2012]; Available from http://products.sanofi.us/PLAVIX/PLAVIX.html.
- 8.Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–13. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Peer CJ, Spencer SD, VanDenBerg DA, Pacanowski MA, Horenstein RB, Figg WD. A sensitive and rapid ultra HPLC-MS/MS method for the simultaneous detection of clopidogrel and its derivatized active thiol metabolite in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;880:132–9. doi: 10.1016/j.jchromb.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis JP, Horenstein RB, Ryan K, O'Connell JR, Gibson Q, Mitchell BD, Tanner K, Chai S, Bliden KP, Tantry US, Peer CJ, Figg WD, Spencer SD, Pacanowski MA, Gurbel PA, Shuldiner AR. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet Genomics. 2012 doi: 10.1097/FPC.0b013e32835aa8a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 12.Dansette PM, Rosi J, Debernardi J, Bertho G, Mansuy D. Metabolic activation of prasugrel: nature of the two competitive pathways resulting in the opening of its thiophene ring. Chem Res Toxicol. 2012;25:1058–65. doi: 10.1021/tx3000279. [DOI] [PubMed] [Google Scholar]
- 13.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–84. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 15.Tiroch KA, Sibbing D, Koch W, Roosen-Runge T, Mehilli J, Schomig A, Kastrati A. Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J. 2010;160:506–12. doi: 10.1016/j.ahj.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 16.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, Steg PG, Ferrieres J, Danchin N, Becquemont L. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–75. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 17.Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, Morath T, Schomig A, von Beckerath N, Kastrati A. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–8. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 18.Gurbel PA, Tantry US, Shuldiner AR. Letter by Gurbel et al regarding article, “Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement”. Circulation. 2010;122:e478. doi: 10.1161/CIRCULATIONAHA.110.943548. author reply e9. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Tang HL, Hu YF, Xie HG. The gain-of-function variant allele CYP2C19*17: a double-edged sword between thrombosis and bleeding in clopidogrel-treated patients. J Thromb Haemost. 2012;10:199–206. doi: 10.1111/j.1538-7836.2011.04570.x. [DOI] [PubMed] [Google Scholar]
- 20.Zabalza M, Subirana I, Sala J, Lluis-Ganella C, Lucas G, Tomas M, Masia R, Marrugat J, Brugada R, Elosua R. Meta-analyses of the association between cytochrome CYP2C19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart. 2012;98:100–8. doi: 10.1136/hrt.2011.227652. [DOI] [PubMed] [Google Scholar]
- 21.Bauer T, Bouman HJ, van Werkum JW, Ford NF, ten Berg JM, Taubert D. Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. BMJ. 2011;343:d4588. doi: 10.1136/bmj.d4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pendergrass SA, Dudek SM, Crawford DC, Ritchie MD. Synthesis-View: visualization and interpretation of SNP association results for multi-cohort, multi-phenotype data and meta-analysis. BioData Min. 2010;3:10. doi: 10.1186/1756-0381-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, Froeschl M, Dick A, Marquis JF, O'Brien E, Goncalves S, Druce I, Stewart A, Gollob MH, So DY. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012;379:1705–11. doi: 10.1016/S0140-6736(12)60161-5. [DOI] [PubMed] [Google Scholar]
- 24.Shuldiner MVR Alan R., Peterson Josh F., Hicks Kevin, Freimuth Robert R., Sadee Wolfgang, Pereira Naveen L., Roden Dan M., Johnson Julie A., Klein Teri E., for the Pharmacogenomics Research Network Translational Pharmacogenetics Program Group The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Overcoming Challenges of Real-World Implementation. Clin Pharmacol Ther. doi: 10.1038/clpt.2013.59. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA, Roden DM, Klein TE, Shuldiner AR. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90:328–32. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]