Summary
Background
Reports of adhesions in cells growing in 3D vary widely—from non-existent to very large and elongated—and are often in apparent conflict, due largely to our minimal understanding of the underlying mechanisms that determine 3D cell phenotype. We address this problem directly by systematically identifying mechanisms that determine adhesion in 3D matrices, and from our observations, develop principles widely applicable across 2D and 3D substrates.
Results
We demonstrate that non-muscle myosin II activity guides adhesion phenotype in 3D as it does in 2D; however, in contrast to 2D, decreasing bulk matrix stiffness does not necessarily inhibit the formation of elongated adhesions. Even in soft 3D matrices, cells can form large adhesions in areas with appropriate local matrix fiber alignment. We further show that fiber orientation, apart from influencing local stiffness, modulates the available adhesive area and thereby limits or allows adhesion maturation.
Conclusion
Thus adhesion in 3D is determined by both myosin activity and the immediate microenvironment of each adhesion, as defined by the local matrix architecture. Important parameters include not only the resistance of the fiber to pulling (i.e. stiffness) but also the orientation and diameter of the fiber itself. These principles not only clarify conflicts in the literature and point to adhesion modulating factors other than stiffness; they have important implications for tissue engineering and studies of tumor cell invasion.
Keywords: 3D, extracellular matrix, adhesion, myosin II, stiffness, myosin light chain, collagen, microenvironment, matrix fibers, fiber alignment, migration, cancer invasion, tissue engineering, polarity
Introduction
Driven by a desire for cell culture models with greater physiological relevance, an increasing number of studies are examining cell behavior in three-dimensional (3D) matrices. These studies have identified numerous differences in cell phenotype between standard two-dimensional (2D) substrates and 3D matrices [1, 2]. For example, fibroblasts migrating in 2D typically assume a spread morphology with a broad leading edge, while the same cells in 3D and in vivo typically develop an elongated or dendritic morphology with narrower protrusions [3, 4]. Cells have also been reported to differ in adhesion, adhesive signaling, and overall migration mode not only between 2D and 3D, but also between different 3D matrices [3, 5–8]. Yet despite these intriguing observations, the mechanisms underlying the differences are not well understood.
Adhesion is a good example of a cell characteristic that is well characterized in 2D, but lacks a convincing mechanistic explanation for how it is determined in 3D. Adhesions in 2D range from small, dot-like structures at the cell periphery to large, elongated focal or matrix adhesions in more central regions and at the cell rear [9]. In contrast, the observed phenotypes in 3D vary widely among substrate types and even among studies using the same substrate [5]. For example, one study showed that fibroblasts in 3D cell-deposited extracellular matrix (ECM) generate longer adhesions than on a flattened (2D) version of the ECM; whereas cells in a 3D collagen matrix and on collagen-coated glass form similar adhesions [10]. In contrast, another study showed that fibroblasts in 3D collagen matrices form very long adhesions [11]; while a third study reported that multiple cell lines produce very small adhesions in 3D collagen [12], and lastly other studies report no detectable adhesions [13]. The diversity of observations is not surprising considering the variation both in matrix types and experimental conditions (e.g. presence of different growth factors [3]) among studies. In 2D, adhesion phenotypes have been mechanistically linked to specific parameters including ligand density, integrin avidity, and most importantly, cell contractility (including response to substrate stiffness) [8, 9]. However, the essential properties that determine adhesion in 3D have not been studied systematically, resulting in incomplete or unclear explanations for observed cell adhesion and behaviors in 3D and hindering the comparison data from different substrate types.
A prominent hypothesis is that matrix stiffness determines adhesion in 3D. This is intuitive, given that the stiffness of in vitro matrices and most tissues is much lower than that of glass and plastic culture dishes [14]. Furthermore, adhesion size and number in 3D can be reduced by global pharmacological inhibition of myosin contractility [15–17]. In addition, low-resolution imaging and biochemical analyses show that increasing 3D ECM density or crosslinking (both increase stiffness as well as affecting other properties) increases total adhesive area and tyrosine signaling [18–21]. In this hypothesis, matrix stiffness alters myosin-activity-regulated adhesion maturation through mechanisms that are assumed to be analogous to 2D. However, there also is evidence suggesting that bulk matrix stiffness is not the sole determinant of adhesion in 3D [1, 2]. For example, the adhesion and migration behavior between cells in 3D ECMs and on a one-dimensional (1D) line of adsorbed fibronectin are similar despite the 1D lines being on deposited on rigid glass [22, 23]. Moreover, cells in cell-derived 3D ECMs can form adhesions longer than those formed by cells on 2D glass substrates [10].
In this study, we identify basic mechanisms that determine adhesion in 3D collagen matrices. We show that non-muscle myosin II (MII) activity modulates adhesion maturation and localizes adhesions to the ends of long, thin protrusions, analogously to its function in 2D. However, we also show that adhesion in 3D does not respond predictably to alterations in bulk matrix stiffness, but is highly responsive to the local microenvironment—in particular matrix fiber architecture. We demonstrate that the orientation of fibers relative to a cell’s direction of movement, in addition to its known effect on local stiffness [2, 24], affects adhesion maturation by modifying the area available for adhesion. Our results indicate that, while MII-mediated contractility plays similar roles in 2D and 3D, the effect of microenvironment on adhesion is distinctly local and determined by fiber architecture.
Results
Adhesion maturation in 3D depends on MII activity
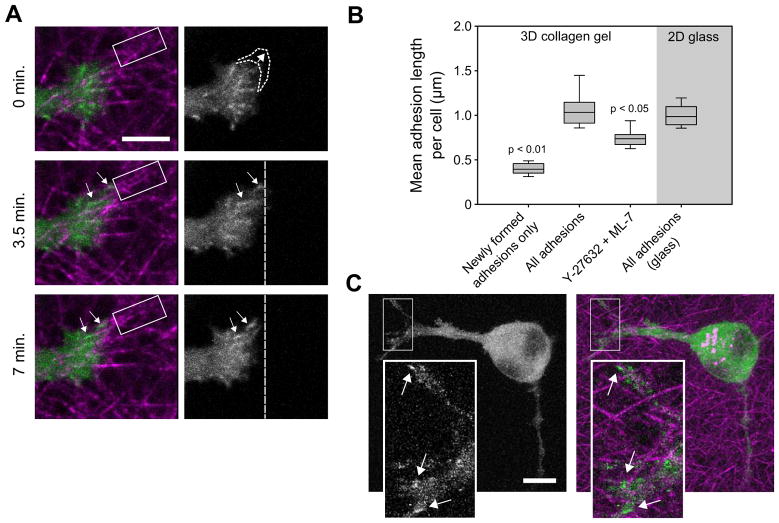
We previously used U2OS osteosarcoma and HT-1080 fibrosarcoma cells, expressing GFP-paxillin, and cultured in 3D collagen I matrices for 3–5 h to observe adhesion formation and maturation [16]. We chose to examine U2OS cells at early time points (~3 hours, when protrusions are first observed) to ensure that the cells were actively protruding and to limit cell-mediated changes to the matrix properties [2, 15, 25]. Under these conditions, the cells are usually elongated with one or more long extensions, which we will refer to as pseudopodia, extending into the matrix (Fig. 1a, Supplemental Movie 1). Protrusions deploy from the distal end of the pseudopod (Fig. 1a, 0 min.), pause, and adhesions (arrows, GFP-paxillin) form at the leading edge on collagen fibers (magenta). The adhesions typically mature while moving retrograde (compare relative to vertical, dashed lines) as the protrusion retracts, and eventually stabilize or disassemble. The retrograde adhesion movement pulls the associated matrix fibers, sometimes resulting in fiber alignment (compare fibers in the rectangles).
Figure 1. Myosin II (MII) activity guides adhesion maturation in 3D.
(a) Adhesion and protrusion dynamics at the distal end of a pseudopod (images from Supplemental Movie 1). U2OS cells were transfected with GFP-paxillin (green) and cultured for 3 h in 2 mg/ml bovine collagen I matrices (magenta). A protrusion occurs after the first frame (indicated by dotted curves), and then new adhesions form (arrows), move rearward and grow. The vertical dashed lines provide fiduciary marks to judge adhesion movement and growth. Note the progressive alignment of the collagen fibers (magenta) highlighted by the rectangles, which is caused by force exerted through the rearward movement of the adhesions (arrows). Bar, 5 μm. (b) Box plots show the distribution of average (geometric mean) adhesion lengths per cell for each treatment group. Cells in 3D collagen gels were cultured for 3–5 h and, where indicated, treated with ~7 μM Y-27632 and ~3.5 μM ML-7. The “newly formed adhesions” are from cells in non-inhibited matrices (timelapse movies were used to identify and measure adhesions at the moment they were first detected). Bars show the 10th and 90th percentiles; the bottom and top box edges show the 25th and 75th percentiles; and the central line shows the median. Adhesions elongated significantly after initial formation, but length decreased upon ROCK/MLCK inhibition (Kruskal-Wallis test; from left to right, n = 20, 25, 22, and 14 cells). (c) Image of adhesions (green, GFP-paxillin) in a U2OS cell treated with Y-27632/ML-7 in a collagen matrix (magenta). Bar, 10 μm (insets, 3 μm).
Superficially, this adhesion behavior appears similar to that in 2D [26]. Since adhesion maturation in 2D is mediated in large part by cell contractility [9], we hypothesized that the same dependency operates in 3D. We quantified adhesion length as a measure of adhesion maturity in U2OS cells expressing GFP-paxillin in 3D collagen matrices (Fig. 1b–c). New adhesions, when first observable, were approximately 0.4 μm in diameter, putting them in the range of small focal complexes in 2D, while the average adhesion length per cell in short-term (3–5 h) cultures was ~1 μm [16]. Interestingly, U2OS cells on 2D glass produced a similar range of adhesion sizes, despite the large difference in substrate stiffness [18, 27], but in support of some previous studies [10, 11]. Treating cells with the Rho kinase (ROCK) inhibitor Y-27632 and the myosin light chain kinase (MLCK) inhibitor ML-7 produced a significant reduction in average adhesion length. This confirms previous qualitative observations implicating MII contractility in adhesion maturation in 3D [11, 15, 16]. We used this combination of agents to inhibit two major activators of myosin and approximate the effect of blebbistatin, which is incompatible with GFP excitation wavelengths. Individually, they give intermediate phenotypes (not shown), suggesting an additive response. To directly assess the role of MII activation in 3D, we evaluated specific myosin regulatory light chain (RLC) activation states.
The control of adhesion maturation in 2D by MII activity has recently been tied to the specific phosphorylation states of Thr18 and Ser19 on RLC [28]. Although nascent adhesions can form independently of MII contractility [29], their subsequent maturation into focal adhesions is promoted by phosphorylation of Ser19 [28]. Phosphorylation of both Thr18 and Ser19 produces large, stable MII bundles that terminate in large stable, adhesions; these highly stable complexes of MII and adhesions localize to and define the non-protruding “sides” and “rear” of a cell in 2D [28, 30].
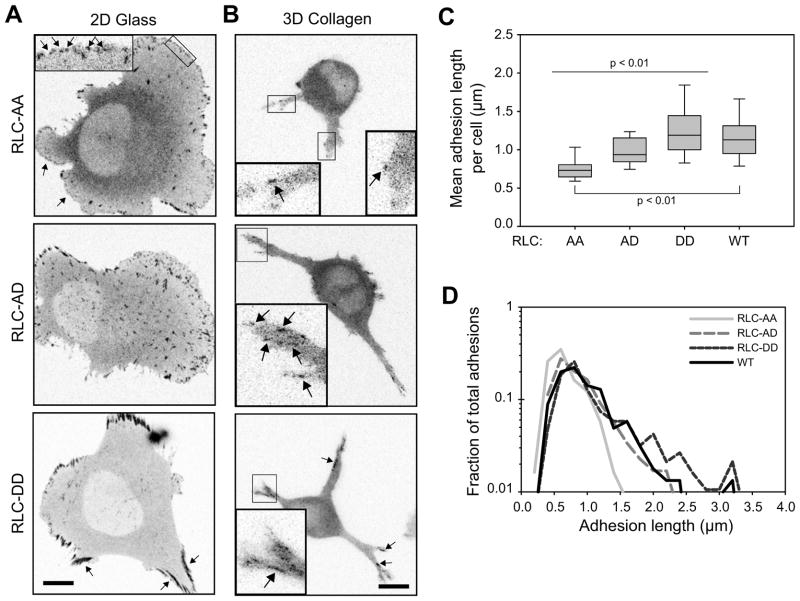
To determine if RLC phosphorylation state has similar effects on adhesions in 3D, we quantified adhesion length in U2OS cells over-expressing different RLC mutants: di-phosphomimetic RLC-T18D/S19D (“RLC-DD”), mono-phosphomimetic RLC-T18A/S19D (“RLC-AD”), and non-phosphorylatable RLC-T18A/S19A (“RLC-AA”) [28]. Mutating to Asp (D) is phospho-mimetic, while mutating to Ala (A) renders the site non-posphorylatable. On 2D substrates, U2OS cells expressing RLC-AD had mature focal adhesions, while those expressing RLC-DD developed large, robust adhesions along non-protruding areas of the cell perimeter (Fig. 2a, quantification in Supplemental Fig. S1a–b), similar to previous studies using CHO.K1 cells [28]. RLC-AA over-expression limited adhesion maturation and increased the population of small, early adhesions (Fig. 2a). Cells expressing RLC-AA still contained some maturing adhesions, likely due to the lower affinity of MII and non-phosphorylated RLC [28, 31], which decreases the ability of RLC-AA to out-compete the endogenous RLC (particularly in highly contractile cells such as fibroblasts).
Figure 2. Myosin regulatory light chain (RLC) phosphorylation state directs adhesion maturation in 3D.
U2OS cells were co-transfected to express GFP-paxillin and over-express RLC phospho-mutants (-AA, -AD, and -DD). (a) Paxillin images of cells cultured on fibronectin-adsorbed glass for 2 h. RLC-AA transfected cells had numerous protrusions with lammellipodia characterized by nascent adhesions and focal complexes (arrows, see inset). RLC-AD transfected cells had more mature focal adhesions and fewer protrusive zones. RLC-DD transfected cells had large, robust adhesions bordering non-protrusive areas of the cell body. (b) Paxillin in cells cultured in 2 mg/ml bovine collagen matrices. RLC-AA: cells had few visible adhesions; those detected were very small. RLC-AD: cells had overall larger adhesions and a greater range of adhesion sizes. RLC-DD: cells had a population of very long adhesions that localized to the lateral edges of the pseudopodia and more proximal to the cell body. (c) Quantification of mean adhesion length for cells cultured in 3D (2D quantification in Fig. S1a, b). Data from cells expressing wild-type RLC (RLC-WT) are shown for comparison. All populations are significantly different except AD-WT and DD-WT (Rank-sum test; from left to right, n = 18, 20, 21 and 26 cells). (d) Histogram of adhesion lengths from cells in panel c. The histograms are normalized based on area under the curve. Overexpression of the RLC mutants results primarily in changes in the population of the longest adhesions. Bar, 10 μm (for insets, 3 μm).
Similar to 2D, cells over-expressing RLC-DD developed relatively long adhesions that localized to the non-protruding edges of the pseudopodia (Fig. 2b). Quantification showed that cells expressing RLC-DD had a higher mean adhesion length (Fig. 2c), due primarily to an increase in the population of very long adhesions (Fig. 2d). In contrast, over-expression of RLC-AA prevented cells from developing large adhesions (even over 24 h culture; not shown), while RLC-AD resulted in cells with fewer small, but more large, adhesions (Fig. 2b–d). Wild-type (WT) cells had adhesion sizes intermediate between the RLC-AD and RLC-DD cells. Similar results were obtained if adhesion area was used instead of adhesion length (Fig. S1c). Therefore, adhesion maturation in 3D is controlled by MII activity, which is regulated by the RLC phosphorylation state similar to in 2D.
Adhesion localization depends on MII organization and RLC activation
While the above results suggest that adhesion maturation in 3D is, at least in part, due to differences in MII activity, we also asked whether MII also dictates where adhesions form in 3D as it does in 2D. In an earlier study [16] and in the experiments presented in the previous section, we noted that adhesions form only near the leading edge of actively protruding/retracting areas of the cell in 3D (Figs. 1a, 2b). In contrast, adhesions do not form along the non-protruding sides of elongated pseudopodia. Adhesions in these areas are usually long, oriented parallel to the sides, and derive from older adhesions that formed at the tip of the pseudopod. This polarity is reminiscent of polarized cells on 2D surfaces, which possess protrusive leading edges with new adhesions, and stable, non-protrusive regions that comprise the sides and rear. Large, contractile actomyosin bundles containing non-muscle myosin IIB (MIIB), di-phospho-RLC, and large, anchoring adhesions line the edges of the cell rear in 2D and inhibit protrusive Rac signaling and new adhesion formation [28, 30].
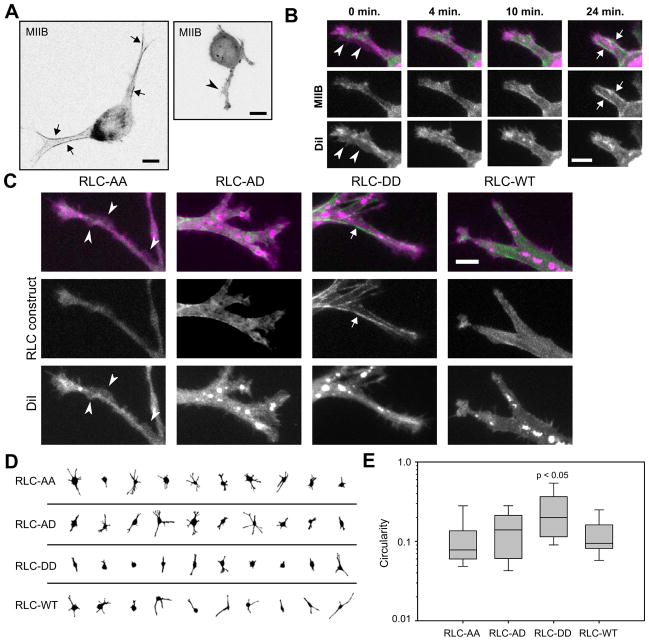
Because of these similarities, we asked whether MIIB similarly blocked protrusion, and therefore adhesion formation along the edges of pseudopodia in 3D. To examine MII organization in 3D, we transfected U2OS cells with GFP-RLC-WT or GFP-MIIB and cultured the cells in 3D collagen matrices. Both RLC (Supplemental Fig. S2a) and MIIB (Fig. 3a) localized to the sides of well-defined pseudopodia, but were more diffuse in pseudopodia that had multiple actin-containing protrusions (Fig. S2b) or ruffles (non-productive protrusions) along their length. Figure 3b shows time-lapse images (see Supplemental Movie 2) of a cell expressing GFP-MIIB and stained with DiI (to visualize membrane). At the beginning of the sequence, the pseudopod does not have any visible MIIB structure and is ruffling over its entire surface. Over the course of the movie, the pseudopod pulls on the collagen matrix as the cell body advances. As the cell pulls, MIIB accumulates in distinct structures along the edges of the pseudopod, and concomitantly, ruffling along the edges ceases. Similar results were obtained with wild-type RLC (Supplemental Movie 3). This is analogous to the behavior of MII in endothelial branching [32].
Figure 3. RLC phosphorylation state localizes protrusion in 3D.
(a) U2OS cells expressing GFP-MIIB in 2 mg/ml bovine collagen matrices. Left image: MIIB lining the non-protrusive edges of the pseudopodia (arrows). Right image: cell lacks organized MIIB and is instead protruding along its pseudopodia edges (arrowhead). Bar, 10 μm. (b) Frames from Supplemental Movie 2 of a cell in a 2 mg/ml bovine collagen matrix (green, GFP-MIIB; magenta, DiI). Initially the pseudopod is ruffling along most of its surface (arrowheads). Over time the pseudopod contracts, applying tensile force to the matrix, as visualized by forward cell body movement. Concurrently, MIIB becomes enhanced along the lateral edges of the pseudopod (arrows) and ruffling in those regions diminishes. Bar, 5 μM. (c–e) U2OS cells expressing GFP-RLC-WT or over-expressing GFP-RLC-AA, -AD, or -DD (green) and labeled with DiI (magenta) were cultured in 2 mg/ml bovine collagen matrices. (c) Cells expressing RLC-AA show ruffling/protrusions along the edges of their pseudopodia (arrowheads) while cells expressing RLC-DD show robust localization to fibers along pseudopodia edges and locally reduced protrusion (arrow). Bar, 5 μm. (d–e) Quantification of cell protrusiveness using the circularity parameter. A circularity of 1 indicates a perfect circle; increasing deviations in shape due to protrusions result in decreasing circularity. Cells expressing RLC-DD were significantly less protrusive (higher circularity) than the other groups (Kruskal-Wallis test; from left to right, n = 35, 36, 37, and 42 cells).
These observations support the hypothesis that MIIB contractile activity along the edges of a pseudopod prevents local protrusion. We therefore tested the effect of RLC di-phosphorylation on protrusion using phosphomimetic forms of RLC (Fig. 3c–e). U2OS cells over-expressing RLC-DD had fewer pseudopodia (Fig. 3d), protrusions and ruffles only appeared at their tips, and robust myosin fibers localized to their sides (Fig. 3c). Cells over-expressing RLC-AA were more protrusive (Fig. 3d), contained little visible myosin organization, and had small protrusions or ruffles along the edges of their pseudopodia (Fig. 3e). We quantified this using a circularity parameter (Fig. 3e), which describes the deviation of the cell shape from a perfect circle (circularity = 1) and correlates with the abundance of protrusions (see Supplemental Experimental Procedures and Fig. S2c–d). These results were recapitulated using blebbistatin, an inhibitor of MII activity, and calyculin A, an inhibitor of myosin regulatory light chain phosphatase (Fig. S2e–f) and were also seen with primary MEFs (Fig. S2g–h). Thus, MII contractile activity through di-phosphorylated RLC aligns along the edges of pseudopodia, locally prevents protrusive activity, and relegates new protrusions and therefore new adhesion formation to the distal end (leading edge). Taken with our observations in the previous section, these results indicate that MII activity directs adhesion maturation, stabilization and location of adhesion formation in a manner similar to 2D, albeit within the context of the very different (elongated) cell morphology.
Local fiber orientation directs adhesion maturation
Having determined that MII activity plays an important role in 3D adhesion maturation and localization, we next asked if changes in bulk 3D matrix stiffness modulate adhesion size. Previous studies of cell responses to changes in 3D matrix stiffness relied primarily on qualitative or ensemble measurements, e.g. changes in the overall intensity of immunostained focal adhesion proteins in a group of cells [18–21]. Here we quantified the effect of changing 3D matrix stiffness at the level of individual cells and adhesions.
U2OS cells in 2 mg/ml matrices had significantly elongated their adhesions, relative to newly formed or contractility-inhibited adhesions (Fig. 1b, Fig 2c). We therefore reduced matrix stiffness by lowering the collagen concentration [18, 27, 33], hypothesizing that this would result in smaller adhesions. We first verified that lower-concentration matrices provided decreasing stiffness over a range relevant to U2OS cells by measuring the ability of the cells to contract the matrices macroscopically. Softer matrices should offer less overall resistance (stiffness) to cell-generated forces and, as a result, contract over time [15, 18, 33]. Indeed, cells contracted 0.3, 0.6, 1.2, and 1.8 mg/ml matrices in a concentration-dependent manner; the lowest-concentration matrices contracted the most (Supplemental Fig. S3a–b).
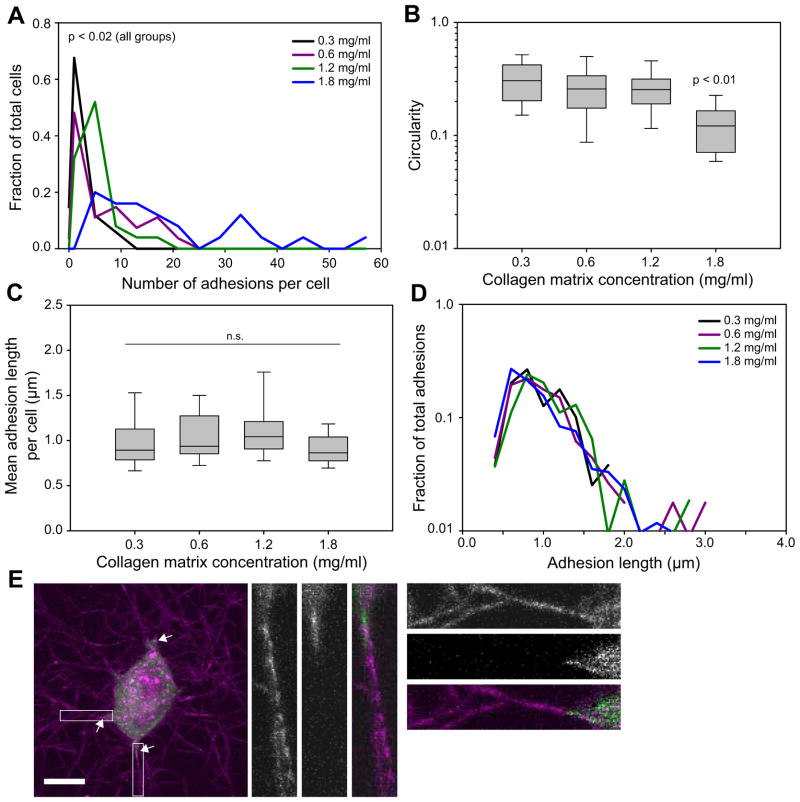
We next examined cell morphology and adhesion in this range of matrix concentrations. In agreement with earlier studies, cells in soft matrices were less spread under serum stimulation (Fig. 4b) and had fewer visible adhesions (Fig. 4a) [15, 33]. Indeed, 15% of cells in 0.3 mg/ml and 4% of cells in 0.6 mg/ml matrices had no clearly visible adhesions. Surprisingly, however, those cells in softer matrices that did exhibit adhesions showed no significant reduction in adhesion length (Fig. 4c). The softest matrices still enabled adhesion elongation at levels similar to globally stiffer matrices, and unlike the effects of varying MII activity (Fig. 2d), there was relatively little effect on the overall distribution of adhesion sizes (Fig. 4d). Because some cancer cells can be less sensitive to microenvironment stiffness, we verified that our U2OS cells do indeed alter adhesion size in response to changes in 2D substrate stiffness (Fig. S3c) and recapitulated our results with primary MEFs (Fig. S3d). Finally, similar results were obtained from overnight cultures (Fig. S3d).
Figure 4. Effect of bulk substrate stiffness on adhesion in 2D and 3D.
U2OS cells expressing GFP-paxillin were cultured in 3D rat-tail collagen matrices of varying stiffness. (a) Histogram of number of adhesions per cell. Cells in denser/stiffer matrices had higher numbers of adhesions (p<0.02, Kruskal-Wallis test; from low to high concentration, n = 29, 26, 25, and 25 cells). (b) Cells were more spread (lower circularity) in stiffer matrices with cells the 1.8 mg/ml matrix having significantly more protrusions (p<0.01, Kruskal-Wallis test; same cells as in (a)). (c–d) Reducing bulk matrix stiffness did not significantly alter the distribution of adhesion lengths (Rank-Sum test; same cells as (a)). Note that, unlike Fig. 2d, there is relatively little change in the overall distribution of adhesion lengths. (e) Cells expressing GFP-paxillin (green) were cultured in 0.3 mg/ml matrices (magenta). Adhesions (arrows) are localized to areas of elevated matrix alignment and density. Bar, 10 μm (for insets, 2.5 μm).
Interestingly, upon closer inspection of the U2OS (Fig. 4e) and MEF (not shown) cells in 0.3 mg/ml matrices, we observed that adhesions were typically localized to areas with increased fiber alignment. It has long been known that cells cultured in fibrillar ECMs reorganize their surrounding fibers, and over the course of many hours cause widespread fiber alignment in the direction of pulling and/or migration [3]. Indeed, cells cultured overnight in 0.3 mg/ml matrices showed even higher fiber alignment (Fig. S3e), concurrent with a large increase in the number of visible adhesions per cell (Fig. S3g). The increases were even more dramatic for the highly contractile MEFs (Fig. S3f, h). In contrast, U2OS cells over-expressing RLC-AA did not show substantial matrix alignment and had no significant increase in adhesion number over 24 h (not shown). The correlation between matrix fiber alignment and adhesion led us to hypothesize that the large adhesions only occur in locally favorable areas, e.g. along aligned fibers.
Localized inhomogeneities in matrix architecture due to variance in fiber polymerization or to cell-generated forces could alter local matrix properties. Indeed, there was greater variation in adhesion size within the 3D matrices versus 2D (e.g. Fig. 1b; compare 4c and S3c; compare 2d and S1b). Previous studies have shown that the mechanical properties of a collagen matrix, on a sub-cellular scale, can vary widely between different locations and therefore not reflect macroscopic measurements [2, 24, 34], which are averages over the entire matrix. Moreover, cell-generated forces have been shown to alter their microenvironment, e.g. by increasing matrix density and fiber alignment [15, 25, 35, 36].
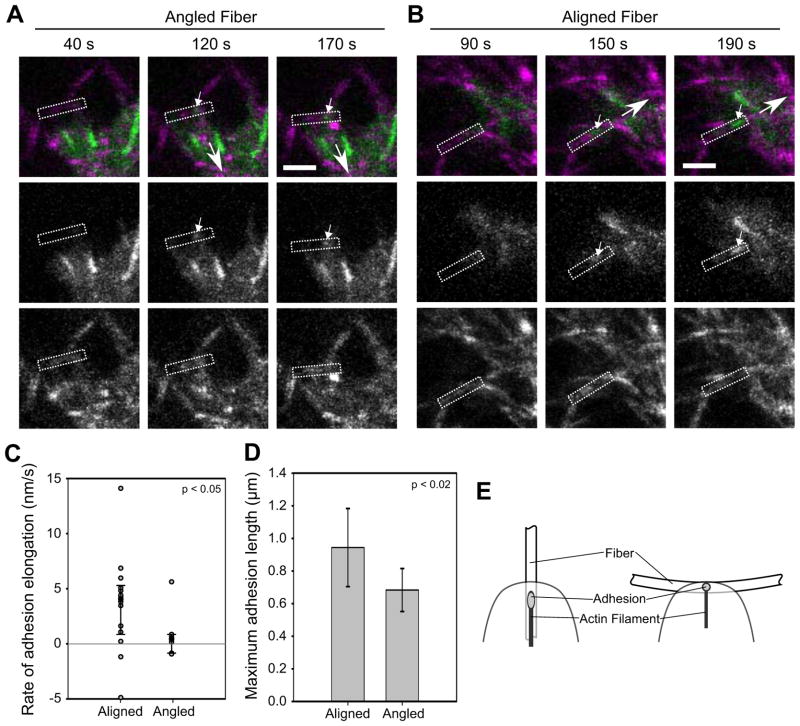
Fiber alignment, in particular, could modulate the stiffness experienced by the cell. In a relatively loose network of semi-flexible fibers, such as a collagen matrix, elongated, aligned fibers generally offer greater resistance to cell pulling than curved, non-aligned fibers [2, 24] (Fig. 5e). For example, forces applied to angled fibers can be dissipated through bending or straightening and aligning them along the force vector, whereas a force applied along the long-axis can experience the full stiffness of the collagen fiber (up to 7.5 GPa [37]). Moreover, repeated pulling and progressive alignment of fibers by a cell will cause an increase in local stiffness (strain-stiffening) [2, 24]. To determine if matrix fiber orientation affects adhesion maturation, we acquired time-lapse images of adhesions forming on fibers in 3D collagen matrices. We identified adhesions that formed on fibers aligned with (Fig. 5b) or at an angle greater than 30° to (Fig. 5a) the direction of the adhesion axis and monitored their size over time. Adhesions forming on aligned fibers primarily grew and achieved an average length of approximately 1 μm (Fig. 5c–d). In contrast, most adhesions that formed on angled fibers translocated rearward and caused fiber movement, but remained small. Interestingly, the one adhesion on an angled fiber that showed consistent growth (Fig. 5c) was on a fiber that gradually bent in response to the applied force, thereby reducing the angle between the fiber and the path of adhesion movement. These results demonstrate a relation between fiber orientation and adhesion maturation.
Figure 5. Local fiber orientation guides adhesion growth.
U2OS cells expressing GFP-paxillin (green) were cultured in 2 mg/ml bovine collagen matrices (magenta). (a–b) Representative image series: (a) Formation of an adhesion (small arrow) on a fiber (rectangle) at an angle to the direction of translocation (large arrow). The adhesion forms but does not increase in size (Supplemental Movie 4). Bar, 5 μm. (b) Analogous image series for an adhesion forming on a fiber parallel to the direction of translocation. The adhesion elongates (Supplemental Movie 5). (c) Dot plot of slopes from plots of adhesion length over time. Adhesions on aligned fibers predominately elongated, while those on angled fibers did not (rate ~0). The distributions are significantly different (p < 0.05, Kolmogorov-Smirnov test; n = 14 and 7 adhesions for aligned and angled, respectively). (d) The maximum length of adhesions on aligned fibers was greater than on angled fibers (p < 0.02, t-test; errorbars, s.d.; same cells as (c)). (e) Diagram of how fiber orientation might affect adhesion growth. A fiber oblique to the direction of translocation could limit adhesion growth by offering less resistance (e.g. by bending in response to pulling) and/or by presenting less adhesive area. In contrast, a parallel fiber offers more contiguous area and potentially greater stiffness.
Fiber orientation restricts adhesion elongation by limiting adhesive area
The effect of fiber alignment on adhesion maturation could be due to orientation-dependent differences in resistance to cell pulling (stiffness); however the limited area for adhesion growth could also contribute. In general, adhesions mature and elongate along actin filaments oriented perpendicular to the leading edge [38]. In contrast to 2D, where the ECM is isotropic, fibrillar matrices have a discontinuous adhesive area, i.e., the fibers are only continuous in one dimension and are separated by pores. Given that in vitro preparations of collagen matrices have fibers with a diameter of 30–800 nm [24], matrix fiber orientation could pose a significant restriction to adhesion size or length. Indeed significantly limiting adhesive area could ultimately inhibit adhesion formation or force transmission to the substrate [39, 40].
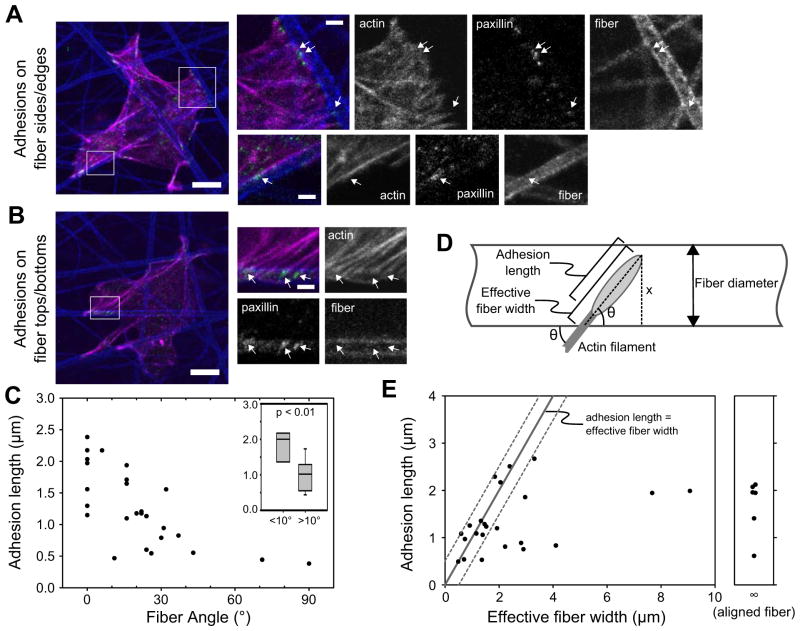
To determine whether fiber orientation affects adhesion size by modulating adhesive area, we cultured U2OS cells on rigid, fibrillar, electrospun polycaprolactone (PCL) scaffolds [41]. Unlike our collagen matrices, these scaffolds were sufficiently stiff (7 MPa vs. 2–10 kPa [41]) that the cells did not move the fibers, indicating that the cells sensed a similar stiffness regardless of fiber orientation. Monomeric collagen I was adsorbed to the scaffold fibers, the cells were seeded and cultured for 2 h, and then the samples were immunostained for paxillin, actin (phalloidin), and collagen (to visualize the fibers). The cells attached primarily to the surface of the scaffolds, but also penetrated under the first layer of fibers. Because the PCL fiber diameters were larger than those of the collagen matrices, we were able to identify two types of adhesion: adhesions to the lateral side or edge of a fiber (Fig. 6a) and adhesions on top or bottom of a fiber (Fig. 6b).
Figure 6. Fiber orientation limits adhesion growth by limiting adhesive area.
U2OS cells were cultured on stiff, electrospun PCL fibers that were adsorbed with collagen. (a–b) The samples were stained for paxillin (green), actin (phalloidin, magenta), and collagen to visualize the fibers (blue). Adhesions (arrows) either attached to the sides/edges (a) or on the tops/bottoms (b) of the PCL fibers. Bars, 10 μm (insets, 2 μm). (c) For adhesions on the sides/edges of PCL fibers, length was plotted as a function of the angle between the associated actin fiber and PCL fiber (dots show individual adhesions). Adhesion length decreases as fiber angle increases. Inset: Difference between adhesions at angles < 10° vs. > 10° is significant, p < 0.01 (rank sum test; from left to right, n = 9 and 18 adhesions). (d) Diagram of how the “effective fiber width” (an estimate of the area available for the adhesion to elongate) was determined for adhesions on top/bottom of PCL fibers (see the Supplemental Experimental Procedures). (e) Plot of adhesion length as a function of effective fiber width. The line indicates where adhesion length and effective fiber width are equal; dotted lines show 0.5 μm error range. All points are within or below this range, indicating that adhesions do not exceed the effective fiber width.
The first type of adhesion would be limited in its adhesive area if it were to elongate along an actin filament perpendicular to the PCL fiber (Fig. 6a). We measured the lengths of this type of adhesion and the angle between its associated actin filament and the PCL fiber. Adhesions elongating along PCL fibers (angle = 0°) varied in length, likely due to differences in age and/or the level of applied force (Fig. 6c). However, as the angle increased, adhesions were significantly smaller (Fig. 6c inset), suggesting that adhesive area is limiting adhesion elongation. These smaller adhesions still exhibited variation in length due to the aforementioned reasons and also likely because of variation in the actual amount of available adhesive area, which could not be accurately determined.
For the second type of adhesions—those on top or on bottom of a fiber—it was, however, possible to estimate the available adhesive area. We calculated an “effective fiber width” parameter that quantified the maximum possible length of the adhesive area, from the distal end of the adhesion to the edge of the PCL fiber, at the angle specified by the associated actin filament (Fig. 6d). We note, however, that the actual adhesive area is determined by the curvature of the fiber, which is difficult to characterize, and is not included in the calculation of the effective fiber width (see Supplemental Experimental Procedures for further discussion). Because accounting for curvature would increase the actual adhesive area measurement, the effective fiber width parameter is an underestimate. Plotting effective fiber width against adhesion length demonstrated that, although there was variability in adhesion length, there were no adhesions that exceeded the effective fiber width (i.e. above the line shown in Fig. 6e) by more than 0.5 μm, which is well within our measurement error. Because most of the fibers were relatively thick and even adhesions at high angles could elongate considerably, there was no obvious correlation with fiber angle discernable above the natural variation in adhesion length. In this regard, the adhesions on the sides or edges of fibers can be viewed as a limiting case in which the adhesive area is greatly limited by angle (Fig. 6c). Similar observations were made using MEFs (Fig. S4). Taken together, these data with PCL fibers and collagen fibers (Fig. 5) demonstrate that fiber width and orientation limit the available adhesive area and thereby can restrict adhesion size.
Adhesion composition correlates with size in 3D
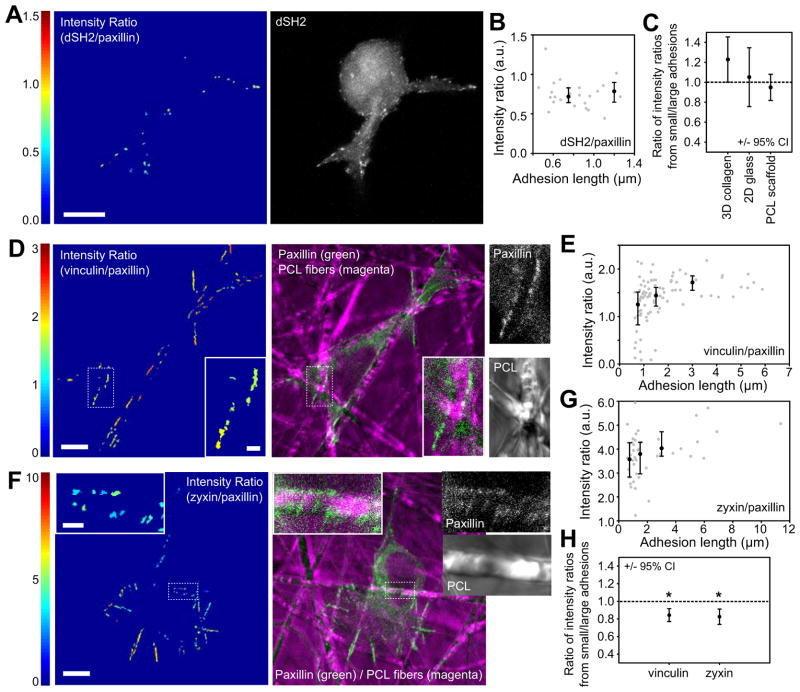
To identify possible functional differences among adhesions in 3D, we investigated tyrosine phosphorylation levels (a marker for adhesive signaling) using the GFP-dSH2 probe driven by a reduced expression system [30, 42]. U2OS cells were co-transfected with this probe and DsRed-paxillin, which localizes more or less uniformly to all adhesions [29, 43], and then cultured in 3D collagen matrices and imaged. The levels of GFP-dSH2 fluorescence were normalized to that of DsRed-paxillin using an intensity ratio [43], which allowed a relative comparison among adhesions within each cell. A representative heat-map image and plot of intensity ratio vs. adhesion length is shown in Fig. 7a–b (plotting vs. adhesion area yields similar trends; not shown). Because intensity ratios could not be directly compared between cells, results were quantified across multiple cells by dividing the median intensity ratio for adhesions < 1 μm by that for adhesions > 1 μm (Fig. 7c). While adhesions exhibited a range of intensity ratios, there was no significant correlation with adhesion size. Similar results were seen for cells plated on glass (Fig. 7c and S5c). This agrees with previous studies showing that most adhesions exhibit some tyrosine signaling and elevated levels only occur in very new nascent adhesions [42, 43].
Figure 7. Adhesion composition correlates with size in 3D.
U2OS cells were co-transfected with DsRed-paxillin and with GFP-dSH2, GFP-vinculin, or GFP-zyxin. (a–b) Cells were cultured in 2 mg/ml bovine collagen gels. The ratio of dSH2 to paxillin intensities for each adhesion was determined and presented as a heatmap image (a) and plotted as a function of adhesion length (b, gray dots). Black dots show the median values for adhesions < 1 μm, 1–2 μm, and > 2 μm (error bars are 25th and 75th percentiles). There is no trend of dSH2 levels with adhesion size of cells in collagen matrices. (c) Because intensity ratio values can only be compared within a single cell, data from multiple cells were summarized by dividing the median intensity ratio values for adhesions < 1 μm by those of adhesions > 1 μm. Error bars are the 95% confidence intervals. Cells cultured in collagen matrices, on glass, or on PCL scaffolds did not show a significant difference in dSH2 levels between small and large adhesions (the 95% confidence intervals include 1 (identical intensity ratios); from left to right, n = 6, 3 and 9 cells). See Fig. S5c–d for glass and PCL scaffold images. (d–h) A similar analysis is presented for cells expressing vinculin/paxillin (d–e) or zyxin/paxillin (f–g) and cultured on PCL scaffolds. Both vinculin and zyxin show increased levels (relative to paxillin) with increasing adhesion size. Insets show small adhesions in locations with limited adhesive area. (h) Quantification across multiple cells by dividing intensity ratios of adhesions < 1 μm by those of adhesions > 2 μm. Both vinculin and zyxin levels are significantly higher in larger adhesions (n = 9 cells for each group). Bars, 10 μm (for insets, 2 μm).
We next investigated compositional differences among adhesions using three markers: paxillin, and the putative mechanosensitive proteins vinculin and zyxin [29, 42–44]. Paxillin is generally present in all adhesions, vinculin levels increase as nascent adhesions mature, and zyxin is primarily in large focal adhesions [29, 43, 44]. While vinculin and zyxin were generally present in all adhesions in cells in 3D collagen, qualitatively, the levels tended to vary with regard to paxillin (Fig S5a–b). Quantitative analyses of vinculin and zyxin fluorescence relative to paxillin suggested a trend opposite that of dSH2: levels increased with increasing adhesion size (data not shown). However, due to poor signal-to-noise, we were unable to obtain sufficient data to be statistically certain of this trend. We therefore turned again to the electrospun PCL scaffolds, which both allowed better image quality and held substrate stiffness constant. Since stiffness is known to affect adhesion composition, this allowed us to extend our earlier findings (Fig. 6) to ask if fibrillar environments that restrict adhesion size by limiting adhesive area independent of stiffness also alter adhesion composition.
U2OS cells were co-transfected with DsRed-paxillin and GFP-dSH2, GFP-vinculin or GFP-zyxin, cultured on PCL scaffolds and imaged as before. Representative intensity ratio and fluorescence images are shown for each of the constructs (Fig. 7d–g and S5d). Adhesions that formed in locations with severely limited adhesive area were both small and contained relatively low levels of zyxin and vinculin (insets in Fig. 7d, f). Plots of intensity ratio vs. adhesion length confirmed this trend. In contrast, dSH2 levels were relatively constant (Fig. S5d), as in 2D and collagen matrices (Fig. 7a–c). The data were quantified across multiple cells by dividing the median intensity ratio for adhesions < 1 μm by that for adhesions > 2 μm (Fig. 7h; similar results were seen when based on adhesion area, Fig. S5h). The aggregated data confirm that vinculin and zyxin levels were lower in smaller adhesions than in larger adhesions (i.e. values significantly less than 1 in Fig. 7h); while dSH2 showed no significant difference (Fig. 7c), suggesting that most of the adhesions were not nascent. These results are similar to those from 2D cultures (Fig. S5), which agree with previously published observations [29, 30, 42, 43]. Therefore, the size restriction induced by the discontinuous adhesive area of fibers can affect the presence of adhesion components.
Discussion
The study of cells in 3D environments, and therefore in many in vivo situations, is hindered by our limited understanding of how the properties of 3D substrates affect cell phenotype. This inhibits attempts to compare results obtained in different 3D models and compromises our ability to apply principles learned from 2D studies to cells in 3D. In particular, observations of cell adhesive behavior in 3D vary widely across the literature, and there is little consensus about how adhesion phenotype is determined in 3D except, perhaps, that stiffness is somehow involved. In this study we identify basic principles that guide adhesion in 3D environments. These principles provide a basis for understanding the disparity in adhesion phenotypes among cells and matrices in 3D. We show that MII activity, and in particular RLC activation state, modulates adhesion localization and maturation in 3D as it does in 2D, but adapted to a different cell morphology: the sides of an elongated pseudopodium in 3D are analogous to the non-protrusive sides and rear of a mesenchymal cell migrating in 2D. However, while the control of adhesion by MII activity is conserved in 3D, our results further show a significant role for local matrix fiber size and alignment. While it is known that the size and orientation of a fiber, relative to an applied tensile force, affect its stiffness, we now demonstrate that these parameters also modulate adhesive area and thereby exert an additional influence on adhesion maturation that is independent of stiffness. It is well known from 2D studies that substrate stiffness can limit adhesion maturation—even when provided with an essentially infinite area over which to elongate [14]. However, we show here that, given a sufficiently stiff substrate, adhesion growth can still be restricted by adhesive area (Fig. 6). Therefore, both stiffness and adhesive area are necessary for adhesion in 3D and each can limit the other.
The presence of fibers is a key difference between many commonly used 2D and 3D substrates. Although fiber architecture has been a focus of recent 3D directional migration studies (e.g. [33, 45–47]), its influence on adhesion in 3D is largely unstudied. Many 3D substrates are composed of fibers, each with a distinct diameter and orientation, and separated by non-adhesive space (pores). Because the fibers are generally thinner than the cell and each cell can interact with many fibers, this creates a situation in which local stiffness and adhesive area are not uniform and isotropic. Except in very stiff or highly cross-linked matrices, some fibers will offer more or less resistance and adhesive area due to their orientation and size [2, 24]. Moreover, in most biologically-derived 3D matrices, the fibers can move, allowing cells to modify their local stiffness and adhesive area. While microfabricated 2D substrates are beginning to allow the recapitulation of certain aspects of the fibrillar environment [1], most 2D studies of adhesion still rely on substrates with a ligand density and stiffness that are uniform and isotropic over the scale of a cell. Our study demonstrates that the fibrillar nature of matrix is important because it defines the properties seen on the scale of a single adhesion. These properties will determine not only local mechanical properties and adhesive area, but also ECM ligand type and substrate curvature (see Supplemental Table S1 for further discussion). For example, although most studies of curvature have focused on cell-level responses, it would be interesting to know how curvature affects the maturation of individual adhesions, particularly in light of the recent finding that adhesions apply rotational torque [48]. While a matrix of random collagen fibers may appear uniform and isotropic on a macroscopic scale, the microenvironment of an adhesion may be defined by a single fiber, which is a distinctly anisotropic structure.
The implications of the fibrillar nature of ECM could be very important for interpreting observed 3D phenotypes. Fiber alignment is implicated in facilitating cancer cell invasion from the tumor into the surrounding tissue [49]. Invading cancer cells and cancer-associated fibroblasts can establish “tracks” through the proteolytic cleavage and force-mediated alignment of fibers [45, 50, 51], enabling other cells to follow. The aligned fibers are stiffer and implicate durotaxis; but they also provide a defined axis for adhesion elongation and can increase migration persistence. Moreover, the added finding that fiber orientation dictates adhesion size and composition suggests it may also determine adhesion-generated cellular phenotypes. Finally, cancer cells may encounter very different matrix structures at different points in tumor progression, for example from basement membrane with thin, dense fibers to connective tissue with thick, sparse fibers. How these different microenvironments limit or enable different adhesion behaviors and thereby influence cell phenotype remains to be answered.
Fiber architecture also helps to explain diverging observations in the literature. For example, the long adhesions observed in 3D cell-derived ECMs and the shorter adhesions observed in 3D collagen gels may be due to the former having more aligned fibers while the latter have predominantly randomly oriented fibers [10, 12, 20]. Fiber architecture may also explain why stiff “1D” substrates replicate many aspects of cell phenotype in soft 3D cell-derived ECMs with aligned fibers [23]. Therefore fiber architecture is an important consideration when selecting in vitro models. For example, collagen matrices, collagen/Matrigel composites, and poly-ethylene glycol (PEG) gels can be made to have a similar macroscopic stiffness; but their fiber structures are very different [27, 52, 53].
Finally, it is notable that the roles of MII activity and fibers identified in this study are independent of dimensionality. For example, cells in 3D may distribute forces in any direction within the matrix, rather than over a 2D plane; however this will simply result in reduced MII contractile force being applied in a given direction. The lower force may result in smaller adhesions in 3D; but still arises from the relationship between MII activity and adhesion maturation. In addition, although commonly used 2D substrates are typically isotropic, a 2D mat of fibers will impose the same restrictions on adhesion formation as in 3D.
Conclusion
In this study, we have identified mechanisms that determine adhesion in 3D matrices. Our observations reveal principles that determine adhesion; they include MII activity, local stiffness, and restricted adhesive area. It is likely that these can be applied to understand adhesion on essentially any substrate. As the use of 3D culture increases throughout the biomedical sciences, selecting and comparing results between different 3D models will become increasingly important. Our results not only help to define these differences, they also enhance our understanding of how cells respond to ECM fiber structure. This should facilitate medical applications such as tissue engineering and inhibiting tumor cell invasion.
Experimental Procedures
Plasmids, antibodies, fluorescence reagents, and inhibitors
See the Supplemental Experimental Procedures for a detailed list of the plasmids. Antibodies/stains: DiI (Molecular Probes, Invitrogen), paxillin (mouse pAb, BD Biosciences), collagen I (rabbit pAb, Abcam), rhodamine-phalloidin (Cytoskeleton), goat-anti-mouse Alexa488 and goat-anti-rabbit Alexa647 (Invitrogen). Inhibitors: Y-27632 (~7 μM, in deionized water), ML-7 (~4 μM, in DMSO), +/−blebbistatin (50 and 100 μM, in DMSO), and calyculin A (1 and 2 nM, in DMSO). All inhibitors were from Calbiochem (EMD Millipore). See the Supplemental Experimental Procedures for more information about inhibitor dosing.
Cell culture and transfection
U2OS cells were cultured in McCoy’s 5A medium + 10% FBS and were transfected withLipofectamine 2000 (Invitrogen), TransIT-2020 (Mirus Bio; Madison, WI) or by electroporation (Amaxa Nucleofector II). MEFs were cultured in high-glucose DMEM + 10% FBS + non-essential amino-acids and transfected by electroporation. During experiments, the cells were cultured in CCM1 (a CO2-independent medium; Hyclone, Thermo Fisher Scientific) supplemented with 10% FBS except in the RLC perturbation experiments (to eliminate confounding effects of serum stimulation; Figs. 2, 3, S1, and S2) and in Fig. 5 (to enhance cell protrusiveness). All cell-culture reagents were from Invitrogen unless otherwise indicated. Expression levels for adhesion markers were kept low either by co-expression with other constructs or by a low-expression promoter to allow their visualization in 3D [16]. Over-expression of RLC-AA, -AD, and –DD to generate phenotypes [28, 30] was optimized for each cell type. At the optimized transfection levels, there was still a large range in expression (as assessed by the fluorescent signal of the construct), yet these differences did not correlate with differences in phenotype, suggesting that we were using levels well above saturation (data not shown).
Cell seeding in 3D collagen and on glass
We followed our previously published protocol [16], with the following changes: cells were seeded at 30–60 × 103 cells per matrix; gel volume was either 200 or 300 μl; and collagen concentrations were varied from 0.3–2 mg/ml as indicated. For MEFs, collagen matrices were supplemented with 10 μg/ml fibronectin. One of two collagen type I sources was used, depending on the experiment type: Nutragen (Advanced BioMatrix; San Diego, CA) pepsin-extracted bovine collagen; or rat-tail collagen (Invitrogren). Unless otherwise indicated, cells were imaged beginning 3 h after seeding, which is when the cells begin actively protruding. For 2D experiments on glass, cells were seeded in CCM1 on glass pre-adsorbed with 2 μg/ml fibronectin (Invitrogen), and cultured for 1.5–2 h.
Basic imaging protocol and analyses
Except where indicated, samples were imaged on an Olympus Fluoview 1000 laser scanning confocal microscope with a UPlanSApo 60x (1.20NA) water-immersion objective equipped with a stage heater. Samples were excited with the appropriate laser lines: 488 nm Ar-ion laser (for GFP); 543 nm HeNe laser (for rhodamine, DiI, and mCherry); 635 diode laser (for Alexa647). Collagen fibers were imaged simultaneously by confocal reflectance microscopy. Settings were adjusted to minimize photodamage. Timelapse images were acquired as previously described [16]. Images for experiments in Figs. 7 and S5 were acquired on a Zeiss LSM 700 confocal microscope with a 63x (1.4NA) oil-immersion objective. GFP was excited by the 488 nm line of an Ar-ion laser and DsRed was excited by a 555 nm diode laser. Images and videos were produced and analyzed with MATLAB (MathWorks) and ImageJ (http://rsb.info.nih.gov/ij/). Intensities were not modified except for linear adjustments to the display range. Unless otherwise indicated, micrographs are maximum intensity projections of multiple z-slices.
Adhesion length on electrospun polycaprolactone (PCL) experiments
Randomly oriented electrospun PCL fibers on 15 mm diameter plastic coverslips (NanoECM 24-well plate inserts, Nanofiber Solutions) were placed in 12-well plates and adsorbed with 50 μg/ml bovine dermis collagen (Nutragen, Advanced Biomatrix) in 20 mM acetic acid (for U2OS) or 10 μg/ml fibronectin in PBS (for MEFs) overnight at 5°C. Cells were seeded on the scaffolds and cultured for 2 h. Cells were then fixed and either processed for immunostaining or, if transfected for ratio imaging, immediately mounted. See Supplemental Experimental Procedures for more details.
Data presentation and statistics
Non-normally distributed data is displayed using box-and-whisker plots: error bars show the 10th and 90th percentiles; lower and upper sides of the box show the 25th and 75th percentiles, respectively; the line within the box shows the median. Statistical tests are indicated in the figure legends. Unplanned comparisons were performed with the Tukey-Kramer test. Planned, nonorthogonal comparisons were corrected using the sequential Dunn-Sidak method. Because adhesions lengths were not normally distributed, the mean values in Figs. 1, 2, 4, S1, and S3 are geometric rather than arithmetic means. Except in Figs. 5, 6, and S4, adhesion statistics were based on cells, not individual adhesions. Statistics were computed with MATLAB. Graphs were made in SigmaPlot.
Additional information in the Supplemental Experimental Procedures
The following experimental and analysis protocols are described in detail in the Supplemental Experimental Procedures: adhesion size measurements; collagen matrix contraction assays; polyacrylamide gel experiments; adhesion dynamics and fiber orientation experiments; and ratiometric imaging of adhesion components.
Supplementary Material
Highlights.
Myosin II activity regulates adhesion and localization in 3D as in 2D
Each adhesion is determined by its specific microenvironment, not bulk properties
Adhesion microenvironment is defined by matrix fiber stiffness, orientation, and size
Fiber architecture determines not only stiffness but also available adhesive area
Acknowledgments
We thank H. Asmussen for preparation of the polyacrylamide gels, B. Hoffman (Duke University) for assistance with the Water algorithm, and the University of Virginia Advanced Microscopy Facility for use of their Zeiss confocal microscope. This research was supported by GM138330 and the Cell Migration Consortium (U54 GM064346). KEK is supported by a Postdoctoral Fellowship (PF-12-136-01-CSM) from the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baker BM, Chen CS. Deconstructing the third dimension - how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012:125. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol (Camb) 2011;3:267–278. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 3.Grinnell F, Petroll WM. Cell motility and mechanics in three-dimensional collagen matrices. Annu Rev Cell Dev Biol. 2010;26:335–361. doi: 10.1146/annurev.cellbio.042308.113318. [DOI] [PubMed] [Google Scholar]
- 4.Knight B, Laukaitis C, Akhtar N, Hotchin NA, Edlund M, Horwitz AR. Visualizing muscle cell migration in situ. Curr Biol. 2000;10:576–585. doi: 10.1016/s0960-9822(00)00486-3. [DOI] [PubMed] [Google Scholar]
- 5.Harunaga JS, Yamada KM. Cell-matrix adhesions in 3D. Matrix Biol. 2011;30:363–368. doi: 10.1016/j.matbio.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130:601. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. BBA-Mol Cell Res. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 10.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct Comparisons of the Morphology, Migration, Cell Adhesions, and Actin Cytoskeleton of Fibroblasts in Four Different Three-dimensional Extracellular Matrices. Tissue Eng Part A. 2011;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petroll MW, Ma L. Direct, dynamic assessment of cell-matrix interactions inside fibrillar collagen lattices. Cell Motil Cytoskeleton. 2003;55:254–264. doi: 10.1002/cm.10126. [DOI] [PubMed] [Google Scholar]
- 12.Geraldo S, Simon A, Elkhatib N, Louvard D, Fetler L, Vignjevic DM. Do cancer cells have distinct adhesions in 3D collagen matrices and in vivo? Eur J Cell Biol. 2012;91:930–937. doi: 10.1016/j.ejcb.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, Longmore GD, Wirtz D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Discher D, Janmey PA, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 15.Tamariz E, Grinnell F. Modulation of Fibroblast Morphology and Adhesion during Collagen Matrix Remodeling. Mol Biol Cell. 2002;13:3915–3929. doi: 10.1091/mbc.E02-05-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubow KE, Horwitz AR. Reducing background fluorescence reveals adhesions in 3D matrices. Nat Cell Biol. 2011;13:3–5. doi: 10.1038/ncb0111-3. author reply 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhee S, Jiang H, Ho CH, Grinnell F. Microtubule function in fibroblast spreading is modulated according to the tension state of cell-matrix interactions. Proc Natl Acad Sci U S A. 2007;104:5425–5430. doi: 10.1073/pnas.0608030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking Cell-Matrix Adhesions to the Third Dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 21.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Micro-environmental control of cell migration - myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J Cell Sci. 2012;125:2244–2256. doi: 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen JA, Swartz MA. Mechanobiology in the third dimension. Ann Biomed Eng. 2005;33:1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 25.Wakatsuki T, Elson EL. Reciprocal interactions between cells and extracellular matrix during remodeling of tissue constructs. Biophys Chem. 2003;100:593–605. doi: 10.1016/s0301-4622(02)00308-3. [DOI] [PubMed] [Google Scholar]
- 26.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 27.Roeder BA, Kokini K, Sturgis JE, Robinson JP, Voytik-Harbin SL. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J Biomech Eng -T ASME. 2002;124:214–222. doi: 10.1115/1.1449904. [DOI] [PubMed] [Google Scholar]
- 28.Vicente-Manzanares M, Horwitz AR. Myosin light chain mono- and di-phosphorylation differentially regulate adhesion and polarity in migrating cells. Biochem Biophys Res Commun. 2010;402:537–542. doi: 10.1016/j.bbrc.2010.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vicente-Manzanares M, Newell-Litwa K, Bachir AI, Whitmore L, Horwitz AR. Myosin IIA/IIB restrict adhesive and protrusive signaling and generate front-back polarity in migrating cells. J Cell Biol. 2011;193:381–396. doi: 10.1083/jcb.201012159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19:260–265. doi: 10.1016/j.cub.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miron-Mendoza M, Seemann J, Grinnell F. The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials. 2010;31:6425–6435. doi: 10.1016/j.biomaterials.2010.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velegol D, Lanni F. Cell Traction Forces on Soft Biomaterials. I. Microrheology of Type I Collagen Gels. Biophys J. 2001;81:1786–1792. doi: 10.1016/S0006-3495(01)75829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kniazeva E, Weidling JW, Singh R, Botvinick EL, Digman MA, Gratton E, Putnam AJ. Quantification of local matrix deformations and mechanical properties during capillary morphogenesis in 3D. Integr Biol (Camb) 2012;4:431–439. doi: 10.1039/c2ib00120a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goetz JG, Minguet S, Navarro-Lerida I, Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibanez T, Pellinen T, Echarri A, Cerezo A, Klein-Szanto AJ, Garcia R, Keely PJ, Sanchez-Mateos P, Cukierman E, Del Pozo MA. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–163. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guthold M, Liu W, Sparks EA, Jawerth LM, Peng L, Falvo M, Superfine R, Hantgan RR, Lord ST. A comparison of the mechanical and structural properties of fibrin fibers with other protein fibers. Cell Biochem Biophys. 2007;49:165–181. doi: 10.1007/s12013-007-9001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vicente-Manzanares M, Choi CK, Horwitz AR. Integrins in cell migration--the actin connection. J Cell Sci. 2009;122:199–206. doi: 10.1242/jcs.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyer SR, Singh A, Dumbauld DW, Calderwood DA, Craig SW, Delamarche E, Garcia AJ. Nanopatterning reveals an ECM area threshold for focal adhesion assembly and force transmission that is regulated by integrin activation and cytoskeleton tension. J Cell Sci. 2012;125:5110–5123. doi: 10.1242/jcs.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam J, Johnson J, Lannutti JJ, Agarwal S. Modulation of embryonic mesenchymal progenitor cell differentiation via control over pure mechanical modulus in electrospun nanofibers. Acta Biomater. 2011;7:1516–1524. doi: 10.1016/j.actbio.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirchner J, Kam Z, Tzur G, Bershadsky AD, Geiger B. Live-cell monitoringof tyrosine phosphorylation in focal adhesions following microtubule disruption. J Cell Sci. 2003;116:975–986. doi: 10.1242/jcs.00284. [DOI] [PubMed] [Google Scholar]
- 43.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 44.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Provenzano PP, Inman DR, Eliceiri KW, Trier SM, Keely PJ. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulrich TA, Jain A, Tanner K, MacKay JL, Kumar S. Probing cellular mechanobiology in three-dimensional culture with collagen-agarose matrices. Biomaterials. 2010;31:1875–1884. doi: 10.1016/j.biomaterials.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 47.Yang YL, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010;31:5678–5688. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 48.Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc Natl Acad Sci U S A. 2013;110:881–886. doi: 10.1073/pnas.1207997110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schedin P, Keely PJ. Mammary Gland ECM Remodeling, Stiffness, and Mechanosignaling in Normal Development and Tumor Progression. Cold Spring Harb Perspect Biol. 2010;3:a003228. doi: 10.1101/cshperspect.a003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 51.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 52.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: A novel model system for proteolytically mediated cell migration. Biophys J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.