Abstract
The role of π-stacking in controlling redox and ligand binding properties of porphyrins has been of interest for many years. The recent discovery of H-NOX domains has provided a model system to investigate the role of porphyrin π-stacking within a heme protein scaffold. Removal of a phenylalanine-porphyrin π-stack dramatically increased O2, NO, and CO affinities and caused changes in redox potential (~40 mV) without any structural changes. These results suggest that small changes in redox potential affect ligand affinity and that π-stacking may provide a novel route to engineer heme protein properties for new functions.
Keywords: Bioinorganic chemistry, pi interactions, porphyrins, heme proteins, H-NOX, redox chemistry
1. Introduction
Heme proteins are involved in a broad range of cellular functions, including catalysis, electron transfer, oxygen (O2) transport, and gaseous ligand sensing [1]. While gas-binding heme proteins bind all ligands via coordination to the heme iron, the binding affinities vary greatly. Hemoglobin and myoglobin, for example, are involved in reversible O2 binding and have O2 affinities in the low- to mid-micromolar range [2]. Other proteins involved in sensing and responding to low concentrations of gases, such as the nitric oxide (NO) sensor soluble guanylate cyclase (sGC), have no measureable affinity for O2 and exhibit picomolar binding constants for NO [3]. Previous work on the globins and Heme Nitric oxide and/or OXygen binding (H-NOX) domains has shown that the protein structure surrounding the heme cofactor plays a major role in this large range of ligand affinities through heme accessibility [4-6], steric bulk [5, 7, 8], planarity of the porphyrin ring [9, 10], proximal histidine hydrogen bonding and tilt [11, 12], and hydrogen bonding moieties in the heme pocket [5, 13, 14].
A significant amount of work has been done to establish critical determinants of ligand binding affinity and selectivity within the H-NOX family. However, it is possible that porphyrin π-stacking interactions are also an important factor and the O2-binding H-NOX protein from Thermoanaerobacter tengcongensis (Tt) [14-16] provides a model to test this since it contains a phenylalanine side chain (F78) positioned 3.5 Å above the heme in an off-set π-stack (Figure 1).
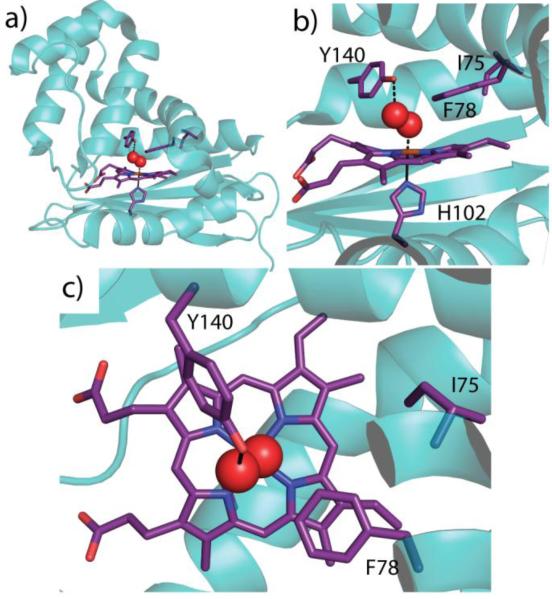
Figure 1.
Wild type Tt H-NOX (PDB ID 1U55, Reference [16]). a) Structure showing the heme and key amino acid residues shown in purple (shown in darker bond lines; amino acid residues numbered) with coordinating oxygen molecule in red spheres. b) Heme is coordinated by H102 with O2 bound above heme and stabilized by a hydrogen bond with Y140. F78 forms an off-set π-stack with the heme and I75 sits slightly further back in the heme pocket. c) Top-view of the heme pocket illustrating F78 stacked above the porphyrin
Four naturally occurring amino acids contain aromatic side chains that could π-stack with the porphyrin and alter the electronics and reactivity of the iron center [17]. There are two types of π-stacking that have been described: an off-set π-stack where the π-stacking side chain is parallel and slightly off-set relative to a pyrrole ring, and the T-stack where the aromatic side chain is perpendicular to the porphyrin ring [18]. Both types of π-stacking have been seen in model systems [17, 19-22] as well as enzymes such as nitric oxide synthase [23-25] and peroxidases [26, 27], and have been shown to influence enzymatic activity.
In myoglobin, phenylalanine 43 slightly overlaps a methine of the porphyrin and has been investigated as a π-stacking residue. However, mutation of F43 results in structural changes within the heme pocket [28]. The F43V mutant exhibits a rotation of H64 (the hydrogen bond donor) into the newly formed pocket, significantly increasing solvent accessibility and heme dissociation, as well as resulting in biphasic O2 binding kinetics and weaker O2 affinity [13]. Mutation of F43 to tryptophan also resulted in decreased O2 affinity, potentially due to structural changes or steric clashes. Mb F43W/H64D/V68I has been crystallized but exhibited a number of differences within the distal pocket, including steric interaction with water bound to the iron [29].
Additional mutations have been made in Mb that introduce aromatic residues into the heme pocket. Through photo-dissociation of Mb crystals, it was previously been found that Mb has multiple off-heme ligand binding pockets.[6] These pockets were blocked through introduction of tryptophan residues and the mutants were found to significantly alter the ligand binding kinetics. For example, the L29W mutation, which blocks the B-state off-heme binding site, resulted in greater than 30-fold decrease in O2 affinity due to extensive escape of the ligand from the protein [30].
However, the influence of π-stacking on ligand affinity in gas transport and sensing proteins without concomitant structural and stability changes has yet to be reported. Therefore, the O2-binding Tt H-NOX [15, 16] was chosen as a model system to probe π-stacking due to the phenylalanine side chain (F78) that forms an offset π-stack with the heme (Figure 1). Tt H-NOX provides an opportunity to probe the effect of a π-stacking interaction on heme ligand affinity.
Tt H-NOX binds O2 with a Kd in the nM range and a number of mutants have been designed to dissect the various factors contributing to this tight O2 binding constant [7-9, 14]. However, the effect of the off-set π-stack between the heme cofactor and phenylalanine 78 on ligand binding has not yet been studied. The phenylalanine ring is aligned parallel to the heme porphyrin and the edge of the aromatic ring of F78 lies above the center of pyrrole B (Figure 1b,c). To probe the role of π-stacking on heme electronic and ligand binding properties, the conservative F78L mutant was generated and ligand binding constants, an X-ray crystal structure, and heme redox potential were determined. In addition, another residue above the porphyrin, isoleucine 75 (Figure 1c), was mutated to a phenylalanine to evaluate the effect of a potential alternate off-set π-stack with the heme. Accordingly, the I75F/F78L mutant was also characterized. Together, our results provide insight into the role of porphyrin π-stacking interactions on heme protein chemistry.
2. Materials and Methods
2.1. Materials
Unless otherwise noted, all reagents were purchased in the highest available purity and used as received.
2.2. Protein expression and purification
The expression and purification of H-NOX proteins were performed as previously described [7]. Briefly, cultures were grown at 37 °C to an OD600 of 0.6–1 in media (45 g yeast extract, 1.6 g KH2PO4, 11.5 g K2HPO4·3H2O, 1.3% glycerol per 1 L) and cooled to 20 °C prior to induction. Isopropyl β-D-thiogalactopyranoside (Research Products International Corp.) was added to 10 μM and aminolevulinic acid (Cosmo Bio Co. Ltd.) was added to 500 μM. Cultures were grown overnight for 18-24 h and then harvested. Cells were lysed using a homogenizer in 50 mM TEA (pH 7.5), 300 mM NaCl, 10 mM imidazole (Buffer A) with Pefabloc (Centerchem Inc.) and benzamidine (Sigma Aldrich) added to 1 mM. Lysates were heat denatured at 70 °C for 40 min and then pelleted at 130,000 × g. Supernatants were applied to HisPure columns (Pierce) pre-equilibrated with Buffer A. The protein-loaded columns were then washed with 25–30 column volumes of Buffer A. H-NOX proteins were eluted in Buffer A containing 150 mM imidazole and concentrated to < 2 mL. The proteins were then desalted into 50 mM triethanolamine buffer (TEA) (pH 7.5), 20 mM NaCl, 5% glycerol (Buffer B) using PD10 columns (GE Lifesciences) and stored at −80 °C.
2.3. UV–visible (UV-Vis) spectroscopy
All spectra were recorded on either a Cary 3E spectrophotometer equipped with a Neslab RTE-100 constant temperature bath or a Cary 300Bio spectrophotometer equipped with a Peltier accessory. Preparation of complexes was carried out in Buffer B as previously described [7, 14, 15]. Representative spectra for wild type and the two mutants are shown in Supplemental Figure S1.
2.4. O2 dissociation rate
O2 dissociation rate experiments were performed as previously described [7, 14]. Protein and dithionite traps were prepared in Buffer B. Dithionite concentration and presence of CO did not affect the O2 dissociation rate. The dissociation of O2 from the heme was monitored and fit globally using SpecFit32 (HiTech Scientific) [7, 14].
2.5. O2 association rate
O2 association rates were observed using flash photolysis and transient absorption spectroscopy as previously described [7]. Briefly, the FeII–CO complex was generated in an anaerobic chamber, as described in an earlier publication [14], and diluted to ~8 μM with anaerobic buffer in a 1 cm pathlength quartz cuvette. The Fe–CO bond was photolyzed by excitation with 8 ns pulses of 532 nm from a doubled Nd:YAG laser, using an apparatus described elsewhere [31]. The cuvette was opened to air and stirred with a steady stream of air blown into the mouth of the cuvette to fully saturate the solution for measurement of the O2 association rates. Spectra were taken at ~20 °C. Data points were collected at a rate of 1× 109 s–1 using a LeCroy digital oscilloscope.
2.6. NO dissociation rate
NO dissociation rates were performed as previously described [7, 14]. Briefly, FeII-NO complexes were rapidly mixed with equivolume of buffer B containing saturated CO and 30 mM dithionite (final concentration) as an NO trap. Data were acquired on either a Cary 3E spectrophotometer equipped with a Neslab RTE-100 constant temperature bath or a Cary 300Bio with Peltier accessory with the temperature bath set to 20 °C. The NO dissociation rate was determined from the increase in the maximum of the Fe(II)-CO spectra over time.
2.7. CO dissociation rate
CO dissociation rates were performed as previously described [4, 32, 33]. Briefly, FeII-CO protein was rapidly mixed with a NO (99.5%; Praxair, Inc.) trap using a stopped flow spectrometer (HiTech Scientific). Rates were found to be independent of trap concentration. UV–visible spectra were recorded from 350-700 nm and the difference in absorbance (424 and 419 nm) versus time was fit to a single exponential equation using Igor Pro.
2.8. Purification of untagged F78L Tt H-NOX for crystallization
Untagged F78L Tt H-NOX was expressed and purified as previously described [8].
2.9. Crystallization, data collection, data processing, and structure refinement of F78L
The non-His tagged F78L Tt H-NOX protein was thawed and a UV-Vis spectrum was taken to ensure the protein was in the ferrous-O2 state. Next, the F78L protein was then exchanged into 20 mM TEA (pH 7.5) using a PD-10 column and then concentrated to 25 mg/mL. The protein was crystallized via sitting drop vapor diffusion by mixing 1 μL of protein with 1 μL of well solution (0.2 M sodium citrate, pH 4.4, 25% (w/v) PEG 3350) and equilibrating the drop against a 400 μL well volume. Crystals formed within two days. These crystals were cryoprotected by a brief soak in well solution containing 25 % (v/v) ethylene glycol, and then rapidly frozen in liquid nitrogen.
X-ray diffraction data were collected at the Advanced Light Source (ALS) at Lawrence Berkeley National Laboratory (Berkeley, CA) on beamline 8.3.1 (λ = 1.127144 Å). Crystals diffracted to 2.1 Å and data were processed in HKL2000 with space group P21212.
The protein crystallized with two protein monomers in the asymmetric unit. The protein component (heme and water molecules removed) of the wild type (WT) structure (PDB ID 1U55) with phenylalanine 78 truncated to an alanine and all B-factors reset to 25 Å2 was used for molecular replacement in PHASER. After an initial round of rigid body refinement in Phenix the 2Fo-Fc and Fo-Fc electron density maps clearly showed density for the heme (Figure S2) and heme molecules were modeled in at this point and maintained for subsequent rounds of refinement. Following addition of the hemes, a single round of positional refinement indicated clear electron density for a leucine residue at position 78 (Figure S3). Thus L78 was modeled and the resulting structure was refined in Phenix against 2.10 Å data with alternate cycles of positional and ADP refinement with manual refitting in Coot. A TLS model was incorporated towards the end of refinement. The final model contains 2 chains with 188 residues in both chains A and B, 280 water molecules, two heme molecules, and two O2 molecules to produce the final refinement parameters of Rwork of 17.0 % and Rfree of 23.0 %.
2.10. Measurement of reduction potentials
Reduction potentials of the proteins were measured as previously described,[9, 34] with the following changes. The following mediators were used in for the measurements: methyl viologen (15 μM), anthraquinone-2-sulfonic acid (20 μM), 2-hydroxy-1,4-napthoquinone (20 μM), Ru(NH3)6Cl3 (33 μM), toluylene blue (10 μM), 3’-chloroindophenol (20 μM), and p-benzoquinone (33 μM). ODQ (Cayman Chemical Company) was used as the oxidant during oxidative titrations.
3. Results and Discussion
3.1. The F78L mutation alters ligand binding
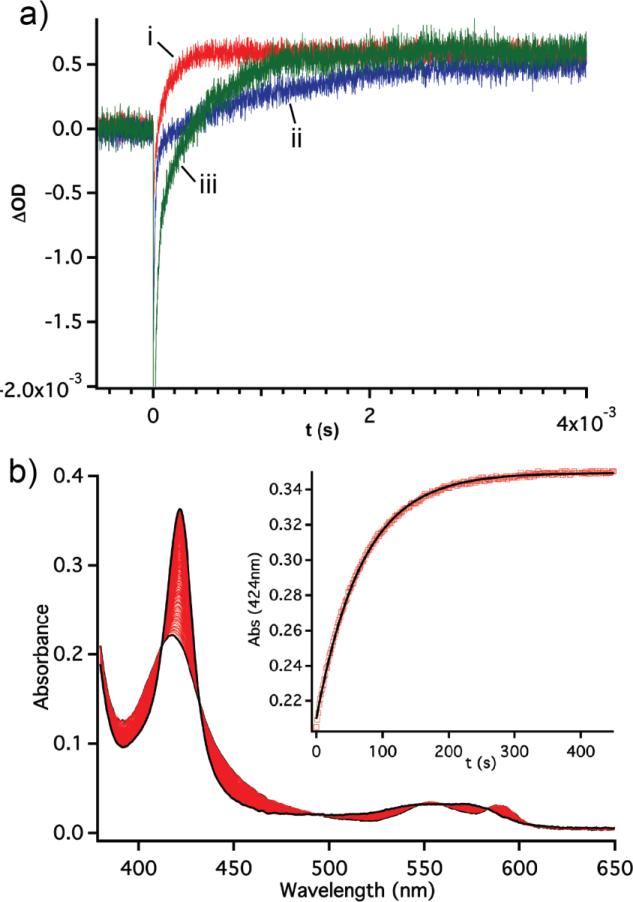
The Tt H-NOX F78L and I75F/F78L mutants were purified and characterized by steady-state and time-resolved absorbance measurements. Changes were observed in the Q-band region (~500-640 nm) of the UV/visible spectra for the ferrous oxidation state of the mutants (Figure S1). Whereas WT ferrous unligated Tt H-NOX does not exhibit splitting in the Q-band, the Q-bands in the F78L and I75F/F78L mutants have a distinct second peak, suggesting that removing the off-set π-stack results in changes to the heme electronic properties (Figure S1, black traces). To investigate the effect of F78 on O2 affinity, the rates of O2 association and dissociation were measured using laser flash photolysis and UV/visible stopped-flow spectroscopy, respectively (Figure 2). The F78L mutation resulted in a two order of magnitude decrease in the O2 dissociation rate (1.20 s−1 vs. 0.015 s−1 for WT and F78L, respectively, Table 1) and a 9-fold decrease in O2 association rate (25 μM−1 s−1 vs. 2.8 μM−1 s−1 for WT and F78L, respectively, Table 1). These changes in association and dissociation rates resulted in an overall increase in affinity of O2 for the F78L mutant versus WT (WT Kd = 48 nM, F78L Kd = 5.4 nM, Table 1). When a phenylalanine was reintroduced into the distal pocket at position 75 (Figure 1), both the O2 dissociation and association rates decreased in comparison to WT (I75F/F78L koff = 0.410 s−1, kon = 8.1 μM−1s−1; WT koff = 1.20 s−1, kon = 25 μM−1s−1), but to a lesser extent than the F78L mutant. This resulted in a similar Kd for O2 to WT Tt H-NOX (50.6 nM for F78L/I75F and 48 nM for WT, Table 1), although the individual kinetic parameters had changed. The introduction of a phenylalanine at position 75 may have generated a weaker π-stacking interaction than in WT, resulting in the intermediate kinetic behavior.
Figure 2.
(a) Laser photolysis traces of the photolysis of the FeII-CO complex followed by binding of O2; WT in red (i); F78L in blue (ii), I75F/F78L in green (iii). (b) Representative stopped flow kinetic data for F78L O2 dissociation rates.
Table 1.
Measured binding affinities and rate constants for gas association/dissociation in WT Tt H-NOX and mutants.
| Tt H-NOX Protein | Kd O2 (nM) | koff O2 (s−1) | kon O2 (μM−1s−1) | koff NO (×10−4, s−1)a | koff CO (s−1) |
|---|---|---|---|---|---|
| WT | 48 ± 6b | 1.20 ± 0.02b | 25 ± 3b | 6.29b | 3.56 ± 0.42c |
| F78L | 5.4 ± 0.08 | 0.015 ± 0.001 | 2.8 ± 0.4 | 0.333 | 0.83 ± 0.02 |
| I75F/F78L | 50.6 ± 9.8 | 0.410 ± 0.024 | 8.1 ± 1.5 | 1.3 | 1.36 ± 0.04 |
| P115A | 21.1 ± 2.1d | 0.022 ± 0.01d | 10.4 ± 1.1d | n/d | n/d |
We next investigated NO and CO dissociation rates. A significant decrease in dissociation rate was observed for the F78L mutant (Table 1). As was observed with O2, introduction of the second mutation in the I75F/F78L mutant also resulted in decreased dissociation rate for NO and CO relative to WT, though these rates were greater than those measured for the F78L mutant. For the F78L mutant, the NO dissociation rate decreased to 0.333 × 10−4 s−1, which corresponds to a 19-fold decrease compared to the NO dissociation rate for WT [8]. I75F/F78L shows a moderate decrease in NO dissociation rate, resulting in a nearly 5-fold decrease relative to the rate of WT Tt HNOX (Table 1).
The CO dissociation rates undergo a small change upon introduction of the mutations. The F78L mutant resulted in an ~4-fold decrease in CO dissociation rate while the I75F/F78L double mutant resulted in ~2.5-fold decrease in rate as compared to WT [4]. The modest changes observed for CO are consistent with previous work done with myoglobin where substitution of the proximal ligand, and concomitant alteration of the heme electronics, resulted in moderate changes in CO association and dissociation rates [12]. The finding that CO is least affected by subtle electronic changes of the heme is consistent with it being an innocent heme-binding ligand, while rates of the non-innocent ligands NO and O2 (ligands which can undergo electron transfer when binding the iron) are more greatly affected by the subtle electronic changes [35].
3.2. Removal of the π-stack does not affect protein structure
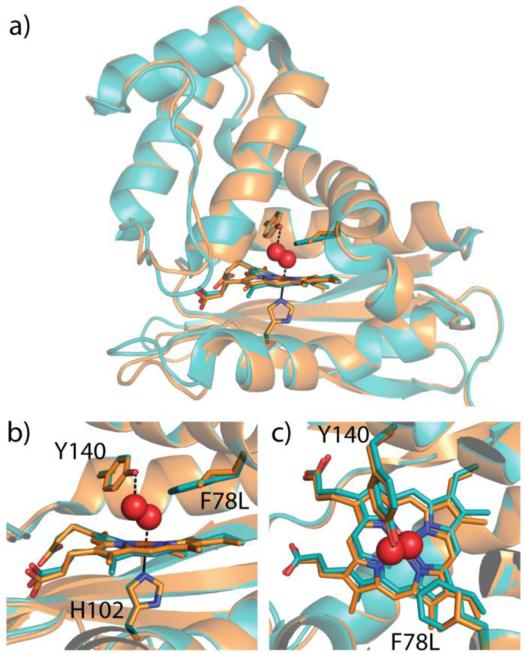
To ensure that the observed changes in ligand binding of the F78L mutant are due to removal of the π-stack and not to a structural change inside the heme pocket, the crystal structure of the F78L mutant was solved (Figure 3, Table S1). The X-ray structure of the F78L mutant is nearly identical to WT Tt H-NOX with an all-atom RMSD of 0.40 Å (Figure 3). In addition, the heme remained distorted from planarity, with no observable differences between F78L and WT. Although the F78L mutant may alter protein dynamics, it does not result in any structural changes, suggesting that the changes in ligand binding kinetics are likely due to removal of the off-set π-stack and the resultant changes in heme electronics.
Figure 3.
Structural alignment of wild type Tt H-NOX (teal, PDB ID 1U55) and F78L Tt H-NOX (orange). Heme, H102, Y140, and F78 or L78 are shown in stick representation. O2 modeled in spheres from wild type structure. a) Overall structural alignment and heme pocket overlay b) side view and c) top view.
3.3. Reduction potentials of the F78L and I75F/F78L mutants
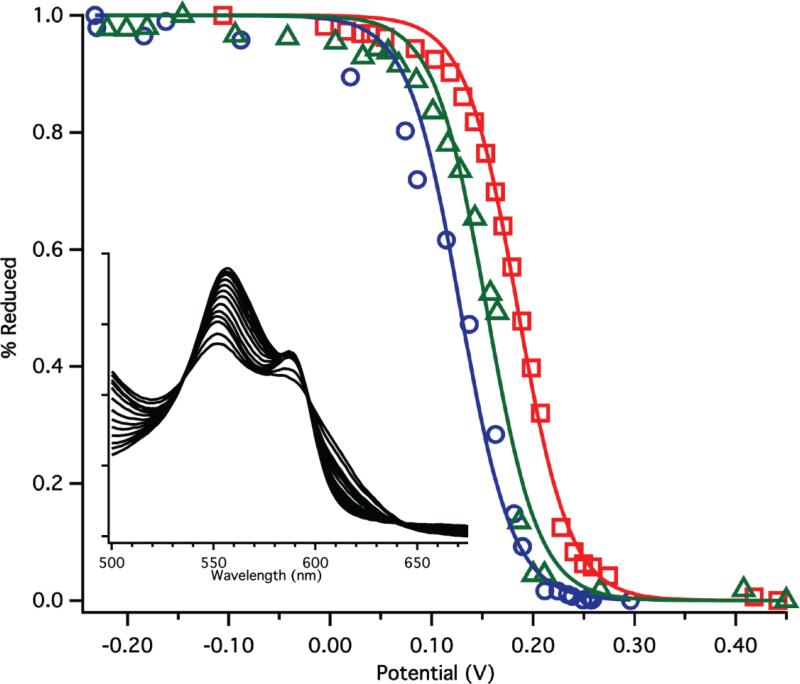
To evaluate the effects of the mutations on heme electronic properties in more detail, the redox potentials of the mutants were measured [34]. The redox potential of WT Tt H-NOX was previously reported to be 167.0 mV (Table 2) [9], which is significantly higher than that of Mb (59 mV) [36]. Mutation of F78 to leucine resulted in a 45 mV decrease in redox potential to 123.8 mV (Figure 4, Table 2). This demonstrates increased electron density at the heme iron, which has been shown to result in stronger binding of non-innocent ligands such as O2 and NO [37]. This result is consistent with the removal of the F78 off-set π-stack in WT Tt H-NOX. Indeed, a similar decrease in redox potential associated with the removal of an off-set π-stack also has been observed for the flavin cofactor of short-chain acyl-coenzyme A dehydrogenase [38]. This may be due to loss of both attractive and repulsive interactions between F78 and the porphyrin, as described by Hunter and Sanders [18], and suggests that overall interactions between the side chain and the heme stabilize the ferrous state. However, theoretical calculations will be needed to further investigate this possibility. The midpoint potential of the I75F/F78L mutant was intermediate between that of WT and F78L (150.6 mV). This supports the hypothesis that introduction of a phenylalanine at position 75 forms a weaker π-stack with the heme than F78, resulting in an intermediate amount of electron density at the heme iron.
Table 2.
Reduction potentials determined for WT Tt H-NOX and mutants from spectroelectrochemical titrations.
| Protein | Reduction Potential (vs SHE, mV) |
|---|---|
| Tt WT | 167.0 ± 6.7a |
| Tt P115A | −3.8 ± 10.2a |
| Tt F78L | 123.8 ± 8.2 |
| Tt I75F/F78L | 150.6 ± 5.5 |
Reference [9]
Figure 4.
Representative redox titration curves for WT Tt H-NOX (red squares), F78L (blue circles), and I75F/F78L (green triangles). Insert shows the typical change in the UV-vis α/β region for F78L during the titration.
Redox potential changes affecting O2 binding in Tt H-NOX have been examined previously for the P115A mutant. Proline 115 is in Van der Waals contact with the D pyrrole of the porphyrin and causes significant distortion of the heme from planarity. This mutant was found to have a significantly lower redox potential (−3.8 mV vs. 167 mV) and tighter O2 Kd than WT (21 nM vs. 48 nM).[9] Although the P115A mutant has a lower midpoint potential than F78L, the O2 affinity of the P115A mutant is unexpectedly weaker than that of the F78L mutant. However, the P115A mutation has a profound impact on the structure of the protein, especially in the heme pocket. Introduction of the P115A mutation decreases the degree of heme distortion, alters the histidine-Fe tilt, and induces a significant conformational change to the protein. Unlike in the previous study, the F78L mutant appears to afford an opportunity to carefully dissect the impact of altered heme redox potential on O2 affinity.
3.4. Potential effects of π-stack removal
The mutation of F78 to leucine slightly increases the volume within the heme pocket, so it is possible that changes in O2 affinity and redox potential are due to a combination of removal of the π-stack and increased solvent accessibility. Previous work on the globins found numerous off-heme binding pockets that allow other gases or solvent to reside within the heme pocket without directly interacting with the iron. However, a recent study on H-NOX domains did not find any off-heme binding sites within the Tt H-NOX heme pocket [4]. In addition, water has not been observed close to the pyrroles groups within the crystal structures of any ferrous Tt H-NOX mutants [7-9, 16]. Water molecules have been observed within hydrogen bonding distance of the propionates in some of the Tt H-NOX mutant structures [7, 8], which is in keeping with the propionates being partially solvent exposed and within a flexible region of the protein as determined by B-factors, MD simulations (unpublished data), and an NMR structure of a homologous H-NOX domain [39]. The only water that has been found within contact of the porphyrin ring was bound to the iron in the Tt I5F H-NOX FeIII-H2O structure [8]. Therefore, it seems unlikely that Tt F78L, which does not appear to have increased backbone or heme flexibility, has significantly more solvent accessibility than WT Tt H-NOX, suggesting that the change in reduction potential is most likely caused by the removal of the π-stack. Additional testing of the π-stacking-redox potential hypothesis through systematic substitutions at position 78 in the future will further delineate the roles of sterics, solvent accessibility, and π-stacking.
4. Conclusions
To our knowledge, this is the first example of removal of a π-stack above a heme without a change in protein structure. It has allowed for direct investigation into the effect of π-stacking on heme electronics and reversible ligand affinity. The off-set π-stack within the distal pocket of Tt H-NOX can tune both the redox potential and ligand affinity of the protein. Small changes in redox potential (~40 mV) influence ligand affinities, suggesting an additional method by which proteins can tune ligand-binding properties. In addition, introduction of π-stacking interactions may be a new strategy for rationally designing novel heme proteins with tailored properties for a variety of biomedical and biotechnological applications, such as novel gas sensors or therapeutic gas delivery agents.
Supplementary Material
Highlights.
We examine the role of a heme-phenylalanine π-stack in a heme protein scaffold.
Removal of the phenylalanine-heme π-stack did not cause any structural changes.
Modest changes in redox potential were observed for the mutant.
Significant changes in ligand binding kinetics are observed upon removal of the π-stack.
Acknowledgements
Funding for this research was provided by the National Institutes of Health National Heart, Lung, and Blood Institute Award F32L090174 (E.E.W.), NIH grant GM 070671 (M.A.M.), and a grant from the Rogers Family Foundation (M.A.M.). We are grateful to Dr. Charlotte Whited, Professor Harry Gray, Dr. Jay Winkler and the Beckman Institute Laser Resource Center at the California Institute of Technology for assistance with on-rate measurements, Professor John Kuriyan for use of crystallography equipment, Dr. Michael Winter for helpful discussions and members of the Marletta laboratory for discussions and critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mayfield JA, Dehner CA, DuBois JL. Curr. Opin. Chem. Biol. 2011;15:260–266. doi: 10.1016/j.cbpa.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonini E, Brunori Journal M. Hemoglobin and myoglobin in their reactions with ligands. North-Holland Pub. Co; Amsterdam: 1971. [Google Scholar]
- 3.Derbyshire ER, Marletta MA. Annu. Rev. Biochem. 2012;81:533–559. doi: 10.1146/annurev-biochem-050410-100030. [DOI] [PubMed] [Google Scholar]
- 4.Winter MB, Herzik MA, Jr., Kuriyan J, Marletta MA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E881–889. doi: 10.1073/pnas.1114038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dou Y, Maillett DH, Eich RF, Olson JS. Biophys. Chem. 2002;98:127–148. doi: 10.1016/s0301-4622(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 6.Olson JS, Soman J, Phillips GN., Jr. IUBMB Life. 2007;59:552–562. doi: 10.1080/15216540701230495. [DOI] [PubMed] [Google Scholar]
- 7.Weinert EE, Plate L, Whited CA, Olea C, Jr., Marletta MA. Angew. Chem. Int. Ed. Engl. 2010;49:720–723. doi: 10.1002/anie.200904799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinert EE, Phillips-Piro CM, Tran R, Mathies RA, Marletta MA. Biochemistry. 2011;50:6832–6840. doi: 10.1021/bi200788x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. ACS Chem. Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bikiel DE, Forti F, Boechi L, Nardini M, Luque FJ, Marti MA, Estrin DA. J. Phys. Chem. B. 2010;114:8536–8543. doi: 10.1021/jp102135p. [DOI] [PubMed] [Google Scholar]
- 11.Capece L, Marti MA, Crespo A, Doctorovich F, Estrin DA. J. Am. Chem. Soc. 2006;128:12455–12461. doi: 10.1021/ja0620033. [DOI] [PubMed] [Google Scholar]
- 12.Decatur SM, DePilli GS, Boxer SR. Biochemistry. 1996;35:3925–3932. doi: 10.1021/bi952625t. [DOI] [PubMed] [Google Scholar]
- 13.Springer BA, Sligar SG, Olson JS, Phillips GN. Chem. Rev. 1994;94:699–714. [Google Scholar]
- 14.Boon EM, Huang SH, Marletta MA. Nature Chem. Biol. 2005;1:53–59. doi: 10.1038/nchembio704. [DOI] [PubMed] [Google Scholar]
- 15.Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Biochemistry. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 16.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D, Williamson DA, Kennedy ML, Williams TD, Morton MM, Benson DR. J. Am. Chem. Soc. 1999;121:11798–11812. [Google Scholar]
- 18.Hunter CA, Sanders JKM. J. Am. Chem. Soc. 1990;112:5525–5534. [Google Scholar]
- 19.Juan HY, Lin WC, Wu YL, Perng JH. Polyhedron. 2008;27:3377–3382. [Google Scholar]
- 20.Zheng J, Kang YK, Therien MJ, Beratan DN. J. Am. Chem. Soc. 2005;127:11303–11310. doi: 10.1021/ja050984y. [DOI] [PubMed] [Google Scholar]
- 21.Janiak C. J. Chem. Soc., Dalton Trans. 2000:3885–3896. [Google Scholar]
- 22.C. V, Tkachenko NV, Efimov A, Guldi DM, Hirsch A, Scheloske M, Lemmetyinen H. J. Phys. Chem. B. 2004;108:16377–16385. [Google Scholar]
- 23.Adak S, Stuehr DJ. J. Inorg. Biochem. 2001;83:301–308. doi: 10.1016/s0162-0134(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 24.Wilson DJ, Rafferty SP. Biochem. Biophys. Res. Commun. 2001;287:126–129. doi: 10.1006/bbrc.2001.5559. [DOI] [PubMed] [Google Scholar]
- 25.Aoyagi M, Arvai AS, Ghosh S, Stuehr DJ, Tainer JA, Getzoff ED. Biochemistry. 2001;40:12826–12832. doi: 10.1021/bi011183k. [DOI] [PubMed] [Google Scholar]
- 26.Goodin DB, Davidson MG, Roe JA, Mauk AG, Smith M. Biochemistry. 1991;30:4953–4962. doi: 10.1021/bi00234a017. [DOI] [PubMed] [Google Scholar]
- 27.Ishimaru A. Bioorg. Chem. 1980;9:472–481. [Google Scholar]
- 28.Hargrove MS, Wilkinson AJ, Olson JS. Biochemistry. 1996;35:11300–11309. doi: 10.1021/bi960372d. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe Y, Nakajima H, Ueno T. Acc. Chem. Res. 2007;40:554–562. doi: 10.1021/ar600046a. [DOI] [PubMed] [Google Scholar]
- 30.Scott EE, Gibson QH, Olson JS. J. Biol. Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 31.Dmochowski IJ, Winkler JR, Gray HB. J. Inorg. Biochem. 2000;81:221–228. doi: 10.1016/s0162-0134(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 32.Rohlfs RJ, Mathews AJ, Carver TE, Olson JS, Springer BA, Egeberg KD, Sligar SG. J. Biol. Chem. 1990;265:3168–3176. [PubMed] [Google Scholar]
- 33.Tsai AL, Berka V, Martin F, Ma X, van den Akker F, Fabian M, Olson JS. Biochemistry. 2010;49:6587–6599. doi: 10.1021/bi1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutton PL. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 35.Kaim W, Schwederski B. Coord. Chem. Rev. 2010;254:1580–1588. [Google Scholar]
- 36.Varadarajan R, Zewert TE, Gray HB, Boxer SG. Science. 1989;243:69–72. doi: 10.1126/science.2563171. [DOI] [PubMed] [Google Scholar]
- 37.Kaim W, Schwederski B. Coordination Chem Rev. 2010;254:1580–1588. [Google Scholar]
- 38.Pellett JD, Becker DF, Saenger AK, Fuchs JA, Stankovich MT. Biochemistry. 2001;40:7720–7728. doi: 10.1021/bi010206s. [DOI] [PubMed] [Google Scholar]
- 39.Erbil WK, Price MS, Wemmer DE, Marletta MA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19753–19760. doi: 10.1073/pnas.0911645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.