Abstract
Class IV homeodomain leucine zipper (C4HDZ) genes are plant-specific transcription factors that, based on phenotypes in Arabidopsis thaliana, play an important role in epidermal development. In this study, we sampled all major extant lineages and their closest algal relatives for C4HDZ homologs and phylogenetic analyses result in a gene tree that mirrors land plant evolution with evidence for gene duplications in many lineages, but minimal evidence for gene losses. Our analysis suggests an ancestral C4HDZ gene originated in an algal ancestor of land plants and a single ancestral gene was present in the last common ancestor of land plants. Independent gene duplications are evident within several lineages including mosses, lycophytes, euphyllophytes, seed plants, and, most notably, angiosperms. In recently evolved angiosperm paralogs, we find evidence of pseudogenization via mutations in both coding and regulatory sequences. The increasing complexity of the C4HDZ gene family through the diversification of land plants correlates to increasing complexity in epidermal characters.
Keywords: gene family evolution, gene duplication, transcription factor, homeodomain leucine zipper
Introduction
Knowledge of the phylogenetic relationships of organisms allows hypotheses of ancestral states and derived conditions to be proposed. Likewise, knowledge of the phylogenetic relationships of genes within gene families provides a foundation for formulating hypotheses concerning the fates of genes following gene duplication and speciation events. Gene duplications provide the raw material for evolutionary change (Ohno 1970). Paralogs produced via gene duplication often become pseudogenes, but can undergo sub- and neofunctionalization facilitating the evolution of novel biochemistries, anatomies and morphologies (Ohno 1970; Force et al. 1999). Orthologs in diverging lineages can acquire new lineage-specific functions in addition to retaining more ancestral ones.
The evolution of an epidermis with a specialized biochemistry and cell types was instrumental to the colonization of land by plants. The epidermal cuticle and stomata are two of the key components of homoiohydry, the ability to internally regulate water content (Raven 1993, 2002), along with an internal water conducting system (Raven 1999). The algal ancestors of land plants depended on water for hydration and the earliest land plants also depended on external water and atmospheric humidity for hydration, a condition known as poikilohydry (Walter and Stadelmann 1968; Raven 2002). The free-living haploid gametophytes of the bryophytes as well as vascular plant gametophytes retain the poikilohydric condition (Raven 1999; Ligrone et al. 2012). The evolution of the homoiohydric condition freed the land plant sporophyte from size constraints of poikilohydry (Raven 1999) and was critical to the subsequent evolution of large and dominant vascular plant floras (Raven 1999; Ligrone et al. 2012). It has been suggested that investigation of genes that are orthologous to those known to regulate epidermal and cuticle development in flowering plants should provide insight into the origin and evolution of the epidermis and important functional components of homoiohydry including stomata and cuticle (Graham et al. 2000; Ligrone et al. 2012).
The class IV Homeodomain Leucine Zipper (C4HDZ) genes have been implicated as “master regulators” of epidermal development in flowering plants (Javelle, Vernoud et al. 2011). Multiple homologs in seed plants show expression patterns that are restricted to the epidermal or subepidermal layers of vegetative, floral, and root meristems and lateral organs (Lu et al. 1996; Nadeau et al. 1996; Ingram et al. 1999, 2000; Ingouff et al. 2001; Ito et al. 2002; Nakamura et al. 2006; Javelle, Klein-Cosson, et al. 2011; Nadakuduti et al. 2012; Peterson et al. 2013; Takada et al. 2013). Loss-of-function phenotypes indicate that Protodermal Factor2 (PDF2/At4g04890) and Meristem Layer1 (ML1/At4g21750) specify protoderm identity in Arabidopsis (Abe et al. 2001, 2003). Nakamura et al. (2006) showed that some Arabidopsis C4HDZs were strongly expressed in developing stomatal complexes and it was recently demonstrated that both HDG2/At1g05230 and ML1 can induce ectopic stomata when overexpressed and delayed stomatal development in loss-of-function mutants (Peterson et al. 2013) indicating a role for C4HDZ transcription factors in regulating stomatal development. C4HDZ genes have also been shown to regulate other specialized epidermal processes including cuticle deposition, trichome development, root hair development, and mucilage and oil secretion (Rerie et al. 1994; Di Cristina et al. 1996; Nakamura et al. 2006; Javelle et al. 2010; Javelle, Klein-Cosson, et al. 2011; Wu et al. 2011; Nadakuduti et al. 2012).
C4HDZ genes encode plant-specific transcription factors, and belong to a larger family of genes that encode proteins characterized by an N-terminal DNA-binding homeodomain (HD) followed by a leucine zipper (Zip) (Ruberti et al. 1991; Schena and Davis 1992). HD-Zip genes are divided into four subclasses—HD-Zip I, II, III, and IV based on their molecular characteristics (Sessa et al. 1994). All members encode HD and Zip domains, but beyond this only class III and IV genes share a putative lipid/sterol binding region called a START domain (Ponting and Aravind 1999) followed by a conserved region of unknown function referred to as the start adjacent domain (SAD) (Schrick et al. 2004; Mukherjee and Burglin 2006). C3HDZ genes possess an additional domain downstream from the SAD called the MEKHLA domain that is similar to domains that function in sensing light, oxygen, and redox activity (Mukherjee and Burglin 2006). Phylogenetic analyses of HD-Zip genes resolved C1HDZ and C2HDZ genes as a clade sister to a clade of C3HDZ and C4HDZ genes (Sessa et al. 1994; Chan et al. 1998; Schrick et al. 2004).
Investigations of the evolution of C3HDZ genes revealed that these transcription factors are ancient, with homologs present in charophyte aglae, but not in chlorophyte algae. C3HDZ genes have been identified in all land plant lineages as well as their charophycean algal relative Chara (Floyd et al. 2006; Prigge and Clark 2006). Homologs of C4HDZs have been identified in the genomes of the lycophyte Selaginella moellendorffii, the moss Physcomitrella patens (Nakamura et al. 2006; Banks et al. 2011; Javelle, Klein-Cosson, et al. 2011) and the transcriptomes of the charophycean algae Coleochaete and Spirogyra (Timme and Delwiche 2010). Thus, both classes of genes evolved in an algal ancestor prior to the origin of land plants. The sister relationship of C4HDZ and C3HDZ genes indicates a common origin, but which class is more ancient is unknown.
Two additional Arabidopsis START domain-encoding genes (At4g26920 and At5g07260) are related to the C3HDZ and C4HDZ gene families (Schrick et al. 2004). These genes lack the homeobox and leucine zipper domains found in C3HDZ and C4HDZ genes and previous phylogenetic analyses using START domain sequences placed these two genes as a clade distinct from but related to clades of C3HDZ and C4HDZ sequences (Schrick et al. 2004). The putative sister relationship of these genes with C4HDZ genes implies an origin in the charophycean algae, although neither the functions nor the phylogenetic distributions of At4g26920 and At5g07260 orthologs has been investigated.
Previous phylogenetic analyses of the plant-specific C4HDZ gene family have either focused on a single taxon (Schrick et al. 2004; Ariel et al. 2007) or, if sequences from a broad range of land plants were included, taxon sampling was sparse (Mukherjee et al. 2009; Javelle, Klein-Cosson, et al. 2011; Zhao et al. 2011; Hu et al. 2012). The published gene trees are incongruent with each other and bear little resemblance to accepted land plant phylogeny, consequently implying extensive gene losses in several linages. These inconsistencies may be due to extensive homoplasy leading to random sampling errors in phylogenetic reconstructions (Yang and Rannala 2012).
To fully address the evolutionary history of the C4HDZ transcription factors and begin to assess the possible roles of these genes in the evolution of epidermal features, we investigated the phylogenetic distribution of C4HDZ genes by sampling taxa representing every major land plant clade and three taxa of charophycean algae lineages most closely related to embryophytes. We also investigated the phylogenetic distribution of the Arabidopsis At4g26920 and At5g07260 genes and their relationship to the C4HDZ gene family. Broad phylogenetic sampling and analysis of recently derived paralogs provides insights, which may be more broadly applicable, into the evolution of the C4HDZ gene family.
Results
C4HDZ Genes Are Present in All Major Land Plant Clades and Charophycean Algae
C4HDZ gene family members were detected in all lineages of land plants, and some lineages of charophycean algae, but were not identified in the genome of any sequenced chlorophycean alga (supplementary table S1, Supplementary Material online).
Within the charophycean algae, partial sequences of C4HDZ homologs were previously identified in Spirogyra pratensis and Coleochaete orbicularis (Timme and Delwiche 2010) and we amplified a partial sequence of a single homolog in C. scutata. We failed to identify a homolog in Chara corallina. Among bryophytes, single homologs were cloned from the liverwort Marchantia polymorpha, the hornwort Phaeoceros carolinianus, and the moss Sphagnum, however, in the moss P. patens, multiple paralogs were identified. Multiple paralogs were also identified in each taxon of the major vascular plants lineages surveyed. In the lycophyte S. moellendorffii four paralogs were identified in its sequenced genome. No whole-genome sequences are available for any fern species, but multiple paralogs were identified in transcriptomes of the leptosporangiate ferns Asplenium platyneuron, and Ceratopteris richardii, the eusporangiate fern Angiopteris evecta, the horsetail Equisetum diffusum, and Psilotum nudum. Likewise, no genome sequences are available for any gymnosperm taxa, but multiple C4HDZ paralogs were identified in transcriptomes of Cycas rumphii, Ginkgo biloba, Pinus taeda, Picea alba, and Pseudotsuga menziesii. Multiple paralogs were also identified in the sequenced genomes of the flowering plants A. thaliana (a rosid), Vitis vinifera (a rosid), Solanum lycopersicum (an asterid), Oryza sativa (a monocot), and multiple transcripts in Zea mays. Single transcripts were identified for the orchids Phaelenopsis sp. and Vanilla planifolia.
Genomic Architecture of C4HDZ Genes
C4HDZ genes from Marchantia, Phaeoceros, and Selaginella have a structure of 11 exons and 10 introns within the coding regions (fig. 1). Exceptions to this basic structure are an additional intron in exon 1 of SmC4HDZ2. Strikingly, there is a complete absence of introns in all the moss genes. The four P. patens C4HDZ genes annotated in the genome lack all introns and amplification of the genomic Sphagnum SspC4HDZ1 gene sequence indicates a lack of introns also.
Fig. 1.
Intron–exon structure of C4HDZ coding regions. Exons are represented by wide bars; introns by folded black lines. Lines and bars are proportional in length and represent total sequence length. Introns within the coding region are numbered. Key refers to encoded protein domains—red, HD; blue, leucine zipper (LZ); yellow, START domain (START); green, SAD; white, 3′-UTR region; gray lines, 3′-UTR motif (UTR). Scale bar represents 500 nucleotides.
Phylogenetic Analyses of C4HDZ Genes
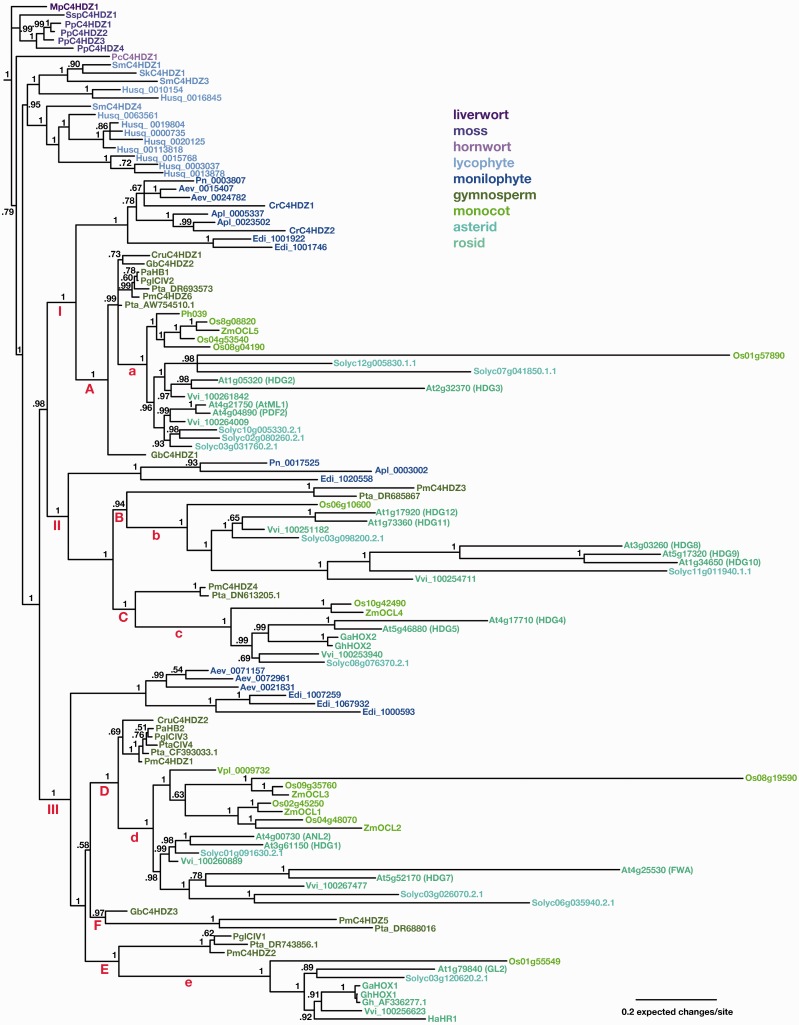
We initially conducted a Bayesian analysis of aligned amino acid sequences with representatives from all land plant lineages plus the charophycean algal sequences. We were unable to run this analysis long enough for the separate runs to converge. The resulting tree was not consistent with land plant phylogeny nor was it well resolved when rooted with either algal sequence (supplementary fig. S1, Supplementary Material online). Alternatively, when the tree was rooted with the liverwort sequence (MpC3HDZ1), the branching order of the land plant sequences was in agreement with accepted hypotheses of land plant evolution with the exception that the Coleochaete sequence was resolved in a poorly supported clade with the hornwort sequence and the Spirogyra sequence was resolved in a clade with euphyllophyte sequences. We determined that the algal sequences were too fragmentary and divergent to provide good phylogenetic signal in this analysis. An identical analysis was performed excluding the algal sequences as well as the most divergent Selaginella sequence, SmC4HDZ2. The topology of the resulting tree when rooted with the liverwort gene, MpC4HDZ1, is mostly consistent with accepted toplogy of land plant evolution (fig. 2).
Fig. 2.
Bayesian phylogram of land plant C4HDZ genes. Tree was constructed using amino acid alignment in supplementary data (Suppl.dataC4lpBAYES.txt, Supplementary Material online). Numbers at branches indicate posterior probability values. Taxa are color coded according to major land plant classification. Three clades comprised of euphyllophyte sequences (I, II, and III), six monophyletic clades of seed plants sequences (A–F) and five clades of angiosperm sequences (a–e) are indicated. Taxon abbreviations: At, Arabidopsis thaliana; Aev, Angiopteris evecta; Apl, Asplenium platyneuron; Cr, Ceratopteris richardii; Cru, Cycas rumphii; Edi, Equisetum diffusum; Ga, Gossypium arboreum; Gb, Ginkgo biloba; Gh, Gossypium hirsutum; Ha, Helianthus annuus; Mp, Marchantia polymorpha; Os, Oryza sativa; Pa, Picea abies; Pc, Phaeoceros carolinianus; Pgl, Picea glauca; Phsp, Phalaenopsis sp; Pm, Pseudotsuga menziesii; Pn, Psilotum nudum; Pp, Physcomitrella patens; Pta, Pinus taeda; Sk, Selaginella kraussiana; Sm, Selaginella moellendorffii; Solyc, Solanum lycopersicum; Ssp, Sphagnum sp; Vpl, Vanilla planifolia; Vvi, Vitis vinifera; Zm, Zea mays.
C4HDZ Gene Phylogeny—Bryophyte Sequences
By defining the liverwort gene MpC4HDZ1 as the outgroup, the remaining bryophyte genes form a grade that mirrors the grade of bryophyte taxa within land plant phylogeny. All moss genes form a well-supported clade (98%) sister to Phaeoceros PcC4HDZ1 plus vascular plants with 100% support. Within the moss clade, the Sphagnum C4HDZ sequence is sister to a clade of the four P. patens sequences with all relationships strongly supported. The hornwort representative, PcC4HDZ1, is sister to all vascular plant C4HDZ genes, although this topology is not strongly supported (78%).
C4HDZ Gene Phylogeny—Vascular Plant Sequences
All vascular plant C4HDZ sequences were resolved as a well-supported clade (99%). Initial analyses including all four S. moellendorffii sequences resulted in a topology with SmC4HDZ2 plus SmC4HDZ4 sister to euphyllophytes with weak support (65%), and SmC4HDZ1 plus SmC4HDZ3 sister to remaining vascular plants. SmC4HDZ2 is divergent relative to the other sequences and was resolved with a long branch. Subsequent analysis omitting SmC4HDZ2 resulted in a topology in which lycophyte sequences resolved as a clade with two highly-supported subclades including both Selaginella and Huperzia sequences. The lycophyte clade is sister (99% support) to a highly supported (100%) clade including all of the euphyllophyte sequences (fig. 2).
C4HDZ Gene Phylogeny—Euphyllophyte Sequences
Within the euphyllophyte clade there are three well-supported (100%) clades of sequences, each containing monilophyte, gymnosperm, and ansgiosperm taxa (clades I, II, and III). Euphyllophyte clade III is sister to clades I + II, which together form a well-supported clade (98%). Within each of these three euphyllophyte clades, a clade of monilophyte sequences is sister to a clade of seed plant sequences, mirroring the phylogenetic relationships between these taxa.
C4HDZ Gene Phylogeny—Euphyll Clade I
Within euphyll clade I, the monilophyte clade includes three paralogs from the eusporangiate fern Angiopteris, one sequence from Psilotum, two sequences from Equisetum and three from the leptosporangiate ferns Asplenium and Ceratopteris. Sister to the monilophyte sequences is seed plant clade “A” with a basal grade of gymnosperm sequences from Cycas and Ginkgo and then subclade “a” with a clade of conifer sequences including Pinus, Picea, and Pseudotsuga sister (albeit with weak support) to a clade of angiosperm sequences. The gymnosperm grade includes multiple sequences from Ginkgo and Pinus. The angiosperm sequences of euphyll clade I include a well-supported (100%) clade of monocot sequences sister to a clade including mostly eudicot sequences from Vitis, Solanum, and Arabidopsis and one Oryza sequence. Four of the sequences in this clade, including the Oryza sequence, have long branches. The remaining clade of sequences is well supported and includes eudicot sequences with asterid sequences (Solanum) sister to rosid sequences (Vitis and Arabidopsis). The Arabidopsis sequences in this clade are ML1 and PDF2, which are sister to each other.
C4HDZ Gene Phylogeny—Euphyll Clade II
Within euphyll clade II, a clade of three monilophyte sequences (one each from Equisetum, Psilotum, and Asplenium) is sister to a clade of seed plant sequences with two subclades (“B” and “C” in fig. 2) that both include gymnosperm and angiosperm sequences). The angiosperm subclades (“b” and “c”) each include single monocot sequences sister to a clade of eudicot sequences. The eudicot sequences of subclade “b” are further resolved into two clades that both include single Solanum and Vitis sequences and multiple Arabidopsis sequences. In subclades “c,” there are also single sequences from Solanum, Vitis, and Gossypium species and two sister sequences from Arabidopsis.
C4HDZ Gene Phylogeny—Euphyll Clade III
The monilophyte sequences in euphyll clade III are resolved into a clade of three Angiopteris sequences sister to a clade of three Equisetum sequences. No leptosporangiate fern sequences were resolved in this clade. The monilophyte clade is sister to a large clade of seed plant sequences, which is further resolved into two clades that include gymnosperm and angiosperm sequences (“D” and “E”) and a third that includes only gymnosperm sequences (“F”). The relationship of clades “D,” “E,” and “F” is not resolved. Within subclade “D,” there is an angiosperm clade (“d”) of monocot sequences sister to two sister eudicot clades. Within the monocot subclade, a single orchid sequence is sister to a grass clade that includes Oryza and Zea sequences. The two eudicot clades include sequences of Vitis and Solanum and a clade of two Arabidopsis sequences, one including ANL2 + At3g61150/HDG1 and the other including FWA + At5g52170/HDG7. Subclade E includes a clade of conifer sequences sister to a single clade of angiosperm sequences (e) each flowering plant species is represented by a single sequence, including Arabidopsis GL2. Seed plant subclade F includes only gymnosperm sequences, one each from Ginkgo, Pseudotsuga, and Pinus.
C4HDZ Gene Phylogeny—Arabidopsis
Sixteen C4HDZ genes are encoded in the Arabidopsis genome, with most represented as sister pairs of genes often with one exhibiting a much longer branch as compared with its paralog. In general, the paralog with the long branch is characterized by a limited expression pattern. Based on syntenic relationships, these duplicate paralogs likely date to the latest whole-genome duplication in the lineage leading to Arabidopsis (Blanc et al. 2003; Jiao et al. 2012).
BUZIGBUZIG Genes Nest within the C4HDZ Phylogeny
We have named the two Arabidopsis HDZ-related START domain-encoding genes lacking the homeobox and leucine zipper domains genes BUZIGBUZIG (BZG), the Meryam Mir (a native language of the eastern Torres Strait Islands, Queensland, Australia) word for decaying or rotten (BZG1/At4g26920 and BZG2/At5g07260). We searched throughout land plant genomes and surprisingly found homologs of BZG genes only in A. lyrata, Capsella rubella, and Eutrema parvulum, all species within the core Brassicaceae (Couvreur et al. 2010). We did not find similar sequences in sequenced genomes of other related rosids, for example, Populus trichocarpa and Carica papaya, suggesting a recent origin for these genes.
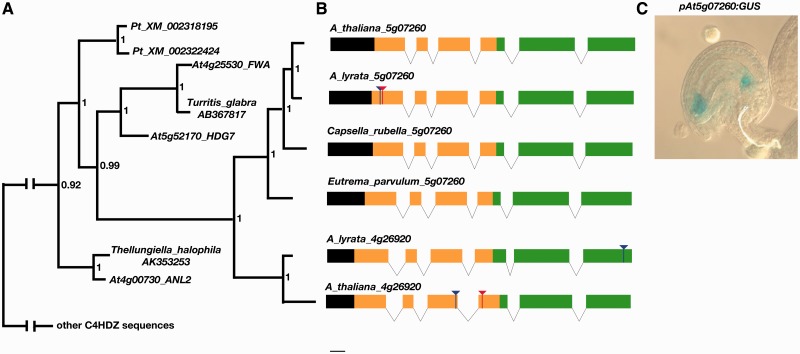
Manual alignment of predicted coding sequences of BZG genes with C4HDZ sequences revealed similarity not only in the START domain but also throughout the SAD domain despite some regions of the predicted proteins being divergent (supplementary fig. S2, Supplementary Material online). The predicted genomic architecture of the genes is also similar to those of C4HDZ genes, with intron positions within the START and SAD domains being conserved in BZG genes with the exception of the absence of the middle intron of the SAD domain of C4HDZ genes (fig. 3). Using an alignment of homologous amino acids, a Bayesian analysis of the corresponding nucleotide alignment produced a gene tree that, when rooted with the MpC4HDZ1 sequence, places the BZG genes in a clade nested within the angiosperm subclade “d,” sister to a clade of genes containing At4g25530/FWA and At5g52170/HDG7 (fig. 3; supplementary fig. S3, Supplementary Material online). The BZG genes are more closely related to FWA and At5g52170/HDG7, than the Populus orthologs of these genes.
Fig. 3.
START domain encoding genes that lack an HD are Brassicaceae-specific genes. (A) Phylogenetic relationship of BZG genes within the C4HDZ gene family. (B) Genomic architecture of BZG genes with six intron positions conserved with C4HDZ genes. Blue lines indicate frameshifts in coding regions and red lines indicate stop codons within exons; yellow, START domain; green, SAD domain. (C) Reporter gene activity when GUS was transcriptionally fused with genomic DNA 5′ of the coding regions of BZG1. Expression of BZG1 is from the maternal allele only.
Previous analyses indicate that expression of BZG2 is limited to early seed development (Schmid et al. 2005). A transcriptional fusion of 2 kb of sequence 5′ to the BZG2 coding sequence with β-glucuronidase (GUS) results in GUS activity in the embryo sac and the chalazal and micropylar developing endosperm (fig. 3). Reciprocal crosses revealed this expression results from only maternally derived transgenes. The orthologous sequence in A. lyrata is not annotated as a gene due to a one-basepair deletion resulting in a frameshift and downstream stop codon in the first coding exon of the predicted gene. Thus, this allele of A. lyrata 5g07260 is a pseudogene.
There is little evidence that BZG1 is expressed (Yamada et al. 2003; Schmid et al. 2005). We were unable to detect transcripts by reverse transcriptase-polymerase chain reaction (RT-PCR) using mRNA isolated from developing flowers. The annotated version of BZG1 (www.arabidopsis.org, last accessed August 9, 2013) skips the fourth potential coding exon due to the presence of an in-frame stop codon in this exon. The presence of this stop codon was not polymorphic among 20 accessions of A. thaliana (Clark et al. 2007). The stability of any transcript including this exon would be reduced due to nonsense-mediated decay and might account for the lack of detectable transcript. In a situation converse to BZG2 orthologs, the sequence orthologous to BZG1 in A. lyrata has the potential to encode a “full-length” protein; however, there is a two basepair deletion in the last coding exon leading to a frameshift and an altered carboxyl terminal sequence relative to other C4HDZ genes.
Expansion of C4HDZ Sequences in Sol. lycopersicum
To examine whether the Arabidopsis C4HDZ gene family is unique, we surveyed the C4HDZ family in Sol. lycopersicum where 39 predicted coding sequences are annotated as homologous to C4HDZ genes (Sato et al. 2012). However, only 12 of the 39 potentially encode full-length C4HDZ proteins are detectably expressed (http://solgenomics.net/organism/Solanum_lycopersicum/genome, last accessed August 9, 2013). The remainder is not detectably expressed and most of this category is predicted to encode truncated proteins, with many lacking HD-encoding sequences. We resolved the phylogenetic relationships of 13 of the nonexpressed sequences relative to other seed plant sequences (supplementary fig. S4, Supplementary Material online). In contrast to the situation in Arabidopsis, many of the nonexpressed sequences in Sol. lycopersicum are closely related and are derived from a single ancestral gene rather than the result of whole-genome duplications (supplementary fig. S4, Supplementary Material online). The closely related paralogs are a combination of physically linked tandem genes on chromosome 9 and genes widely scattered throughout the genome. As all of the genes possess introns, their mobility is unlikely through an RNA intermediate.
Most C4HDZ Genes Contain Conserved 3′-UTR Sequence Motifs
It has been noted that many C4HDZ genes, from Physcomitrella, Selaginella, gymnosperms and angiosperms, possess two conserved sequence motifs in their 3′-UTRs (Ingouff et al. 2003; Nakamura et al. 2006; Javelle, Klein-Cosson, et al. 2011). We found that the conserved sequences are present in the 3′-UTRs of liverwort (MpC4HDZ1) and hornwort (PcC4HDZ1) genes (supplementary fig. S5, Supplementary Material online), consistent with it being present in the ancestral land plant C4HDZ gene (Javelle, Klein-Cosson, et al. 2011). To investigate the evolution of these sequences further, we determined consensus sequences of 17 and 18 nucleotides using aligned conserved sequences from selected land plant C4HDZ sequences with the exclusion of angiosperms (fig. 4). When aligned to the consensus, a potentially homologous sequence in the 3′-UTR of the C. orbicularis C4HDZ gene was identified.
Fig. 4.
Analysis of the conserved 3′-UTR sequence motifs in C4HDZ genes. (A) A consensus sequence was computed using nonangiosperm land plant C4HDZ sequences and is displayed as a sequence logo (Schneider and Stephens 1990; Crooks et al. 2004). A potentially homologous sequence in the 3′-UTR of the Coleochaete orbicularis C4HDZ gene is shown below the sequence logo. The corresponding sequences, where present, of the sixteen Arabidopsis genes are shown above the sequence logo; differences from the consensus are in red. Those genes with limited expression patterns and long branches are noted. (B) Many C4HDZ 3′-UTRs have the potential to form secondary structures similar to that shown for the MpC4HDZ1 gene. The conserved 17 and 18 basepair sequences are highlighted in green and yellow, respectively, and the short motifs conserved in many nonangiosperm 3′-UTRs are highlighted in turquoise.
The 3′-UTRs of angiosperm C4HDZ genes possessing both motifs have the potential to fold into a stem-loop structure, with the conserved motifs being partially complementary (Javelle, Klein-Cosson, et al. 2011). Using secondary structure prediction software (Zuker 2003), the 3′-UTRs of other land plant C4HDZ genes also have the potential to fold into a stem-loop structure (supplemenatry fig. S6, Supplementary Material online). The predicted secondary structures of 3′-UTR sequences from genes spanning the diversity of land plants exhibit a stereotypical topology, with the two conserved sequences pairing near the bases of stem-loops of variable length (fig. 4; supplementary fig. S5, Supplementary Material online). While most of the sequence in the stem-loop outside the two previously identified motifs is not conserved among the C4HDZ genes from divergent lineages of land plants, we identified four additional motifs, five nucleotides in length, that potentially basepair in the stem-loop in most nonangiosperm C4HDZ genes (fig. 4). The 3′-UTR sequence of the C. orbicularis C4HDZ gene is not predicted to form a stem-loop structure.
In contrast, while similar stem-loop structures are predicted to form in the 3′-UTR of some A. thaliana C4HDZ genes, the four additional short motifs are not identifiable in the predicted structures. In eight of the 16 Arabidopsis C4HDZ genes, the 17- and 18-nucleotide conserved 3′-UTR motifs are present with only 1–3 differences (in total) from the consensus based on nonangiosperm land plants (fig. 4). In two additional genes, the sequences have a total of six differences each from the consensus sequence, in another two genes a potentially homologous sequence could be identified, and in four other genes neither motif was identified. None of the 3′-UTRs with significant deviations from the consensus sequences are predicted to form secondary structures with a topology similar to that formed by those with conserved 3′-UTR sequences. Of the six Arabidopsis genes characterized by limited expression patterns and long branches in phylogenetic analyses (fig. 2), five are characterized by complete loss or significant changes in the 3′-UTR motifs. Only two genes, At5g52170/HDG7 and At1g05230/HDG2, with broad expression patterns have degenerate 3′-UTR motifs. In addition, of six gymnosperm C4HDZs for which we have the 3′-UTR sequences, one exhibits significant deviation from the conserved sequences, PmC4HDZ2 (in subclade “E” in fig. 2). All monilophyte, lycophyte, and bryophyte C4HDZs for which we have the 3′-UTR sequences include the conserved motif.
Loss of the 3′-UTR Motif Has Occurred Multiple Times
Despite the broad conservation in land plants, there are several Arabidopsis sequences that lack the C4HDZ 3′-UTR motif or have significant deviations from the consensus (fig. 4). These sequences are resolved in three different seed plant clades, “B” (euphyll clade II), “D,” and “E” (euphyll clade III; fig. 2). Within clade “B,” only the closely related paralogs At3g03260/HDG8, At5g17320/HDG9, and At1g34650/HDG10 are lacking the conserved motif. These are all genes with limited expression and no known function and interestingly they are also all truncated at the 5′-end, having lost ancestral exon and intron 1. At3g03260/HDG8 + At5g17320/HDG9 + At1g34650/HDG10 were resolved in a eudicot subclade with 12 Solanum genes (supplementary fig. S4, Supplementary Material online) and a Vitis gene (Vvi_100254711), none of which have the conserved 3′-UTR motif. RNA seq data suggest that none of the 12 Solanum genes in clade “b” are expressed (http://solgenomics.net/organism/Solanum_lycopersicum/genome, last accessed August 9, 2013). The other eudicot sequences, monocot and gymnosperm sequences in subclade “b” have the conserved motif. This suggests a loss of the 3′-UTR motif in a eudicot-specific paralog prior to the divergence of core eudicots.
Expression of C4HDZ Genes in Nonflowering Plants
Charophycean algae have a haplontic life cycle, whereas land plants undergo an alternation of generations with multicellular development in both haploid and diploid phases of their life cycle. The C. scutata C4HDZ sequence was cloned from complementary DNA (cDNA) produced from mRNA isolated from the haploid stage of the life cycle, similar to the previously isolated Spirogyra C4HDZ gene (Timme and Delwiche 2010), indicating C4HDZ genes are expressed in the haploid stage of the life cycle in charophycean algae.
In each of the bryophyte lineages, C3HDZ gene expression is detectable in both the haploid and diploid stages of the life cycle by RT-PCR (data not shown). We analyzed C4HDZ gene expression patterns in P. patens in more detail. Using mRNA isolated from protonemata, gametophores, and sporophytes, PpC4HDZ expression was detected in haploid gametophores and diploid sporophytes but not in haploid protonemata (supplementary fig. S7, Supplementary Material online). The spatial and temporal expression pattern of PpC4HDZ2 was determined by constructing a translational fusion of the GUS reporter gene with the PpC4HDZ2 gene via homologous recombination (supplementary fig. S8, Supplementary Material online). During the haploid generation, GUS staining for PpC4HDZ2 was not detected during protonemal growth (fig. 5A), with the earliest signal detected in the shoot initial of the gametophore during its formation from caulonemal cells of the protonemata (fig. 5B). GUS activity is detectable in the apical cell, apical cell derivatives, and cells immediately below the shoot apex of the gametophores (fig. 5C). In developing sporophytes, GUS staining is evident throughout the apical region and later in the region of the sporophyte that will give to the capsule (fig. 5D). C4HDZ sequences have been identified in expressed sequence tags (ESTs) from the sporophyte generation of ferns and other monilophyte species, and one C4HDZ sequence was identified from gametophyte of the fern Adiantum capillus-venerus from the GenBank EST database (BP917671.1). All the gymnosperm C4HDZ ESTs were identified from sporophyte tissue.
Fig. 5.
Expression patterns of Physcomitrella patens C4HDZ genes inferred from a GUS fusion line (PpC4HDZ2-GUS). GUS activity was not detected in the protenemata (A) but was detected in apical cells and meristematic regions of bud initials (B) and in developing gametophores (C). Expression is detected in the apical cell and its immediate derivatives and in all cells of initiating leaves. During sporophyte development GUS activity was at first broadly detected throughout the embryo before becoming restricted to the apical regions and later the capsule (D). Scale bars = 50 µm except rightmost sporophyte (bar = 100 µm).
Discussion
Elucidation of the evolutionary history of a gene family requires broad phylogenetic sampling to identify gene duplications as well as gene losses. We provide a comprehensive view of the C4HDZ gene family, from its charophycean algal origin through the evolution of land plants and finally to the fates of C4HDZ genes that have evolved recently in extant angiosperms.
The Deep Charophycean Roots of “Land Plant” Genes
It is becoming increasingly apparent that gene families present in embryophytes (land plants) and absent from sequenced chlorophyte genomes have their origins in the grade of charophyte algae from which the ancestral land plant evolved. Both C3HDZ and C4HDZ gene families originated during the evolution of the charophyte algal grade sister to land plants. Based on the derived HD of C3HDZ proteins, which has an addition of four amino acids relative to those encoded by C1HDZ, C2HDZ, and C4HDZ genes, Mukherjee and Burglin (2006) hypothesized that C3HDZ genes were derived from an addition of a MEKHLA encoding sequence to a C4HDZ gene. This implies that a C4HDZ was the ancestral gene that was then duplicated and modified to produce a C3HDZ gene. Among the charophyte algae, C4HDZ genes have been detected in Coleochaete and Spirogyra, but not Chara. C3HDZ genes have been previously found in Chara (Floyd et al. 2006) and we detected partial EST sequences of MEKHLA domains similar to those of C3HDZ genes in Coleochaete and Spirogyra transcriptome databases (Timme and Delwiche 2010). Further work is required to determine whether these sequences do indeed represent full C3HDZ coding sequences.
Although it is accepted that the monophyletic clade of land plants is nested within a grade of charophycean algae, the precise identity of the sister lineage of land plants is enigmatic. Different molecular data sets have resolved members of the Charales (Karol et al. 2001), the Coleochaetales (Finet et al. 2010), the Zygnematales (Wodniok et al. 2011; Timme et al. 2012), or a clade comprising the Coleochaetales and the Zygnematales (Finet et al. 2012; Laurin-Lemay et al. 2012) as the sister taxa of land plants, the latter topology being favored by analyses based on broad transcriptome sampling. Mapping the presence of C3HDZ and C4HDZ genes onto the latter topology would imply that a C3HDZ gene originated first and that the ancestral C4HDZ gene was derived from a duplication of the ancestral C3HDZ, in contrast to the hypothesized derivation of C3HDZ genes from C4HDZ genes based on HD sequences (Mukherjee and Burglin 2006). However, given the uncertainty of charophyte phylogeny and the possibility of gene loss within lineages, deeper sampling in the charophytes will be required to determine at what point the C3HDZ/C4HDZ ancestral gene evolved and the timing of duplication and divergence during streptophyte evolution.
C4HDZ genes are expressed in the haploid phase of the life cycle in charophycean algae, but no data are available on expression in the diploid zygotes of these taxa. In both bryophytes and ferns, C4HDZ genes are expressed in both the haploid gametophyte and diploid sporophyte generations, indicating this was likely also the case for the common ancestor of extant land plants and that expression in both generations was retained during the evolution of euphyllophytes. In surveys of gene expression in Arabidopsis, C4HDZ gene expression is also detected in both gametophytic and sporophytic generations (Nakamura et al. 2006). However, if the hypothesis that gametophytic C4HDZ gene expression is a consequence of general derepression of gene expression in certain cell types during this generation, C4HDZ gene expression in this generation might be considered a secondary gain after a loss in the lineage leading to the flowering plants. Thus, across streptophytes C4HDZ expression is detectable in those generations in which a complex multicellular body develops.
Evolution of the C4HDZ Gene Family in Land Plants
C4HDZ Sequences in Bryophytes
Rooting the land plant C4HDZ gene tree (fig. 1) with the M. polymorpha gene, MpC4HDZ1, causes the other bryophyte genes to fall into a grade that reflects their accepted phylogenetic relationships (Qiu et al. 2006). As all moss sequences form a monophyletic clade and single sequences were present in the liverwort and hornwort taxa, the tree topology implies that the ancestral land plant genome encoded a single C4HDZ gene (fig. 6). Furthermore, the common ancestors of moss and hornworts + vascular plants, and of hornworts and vascular plants are also predicted to have had a single C4HDZ gene.
Fig. 6.
Hypothesis of the evolution of C4HDZ gene family in Streptophytes. Charophycean algae are in blue, bryophytes in light green and vascular plants in dark green. Predominant species used in this study are listed at the right. Numbers represent an estimate of C4HDZ gene number in the common ancestor at that node. Red circles represent gene duplication events deduced from the phylogenetic relationships of genes in extant species. The purple pentagon represents a loss of introns in the lineage leading to the extant mosses, Sphagnum and Physcomitrella.
As in the case of the C3HDZ gene tree (Floyd et al. 2006; Prigge and Clark 2006), multiple moss paralogs are the result of gene duplications within the moss lineage, consistent with the predicted whole-genome duplication of the P. patens genome within the moss lineage (Rensing et al. 2008). In contrast, in both the C3HDZ and C4HDZ gene trees, single homologs are found in the liverwort (M. polymorpha) and hornwort (Pha. carolinianus) sampled. Whether these conditions represent a general phenomenon is not yet clear, but the observation is consistent with a paucity of evidence for whole-genome duplications in liverworts (Berrie 1963) and with the topology in figure 6.
C4HDZ Sequences in Vascular Plants—Lycophytes
The monophyly of all vascular plant C4HDZ sequences and monophyly of all lycophyte sequences together imply that the genome of the common ancestor of vascular plants contained a single C4HDZ gene (fig. 6). Subsequent to their divergence from other vascular plants, a duplication must have occurred in a common ancestor of Selaginella and Huperzia giving rise to two paralogs that have undergone lineage-specific duplications to produce multiple Selaginella and Huperzia parlogs. According to the phylogenetic analysis of Kenrick and Crane (Kenrick and Crane 1997), the last common ancestor of Huperzia and Selaginella included all lycopsids except the earliest diverging lineages that are known as fossils from the Devonian (∼370 Ma). This would place the lycophyte C4HDZ duplication very early in the history of the lineage, or in the common ancestor of the lycophytes.
C4HDZ Sequences in Euphyllophytes—Monilophytes
The euphyllophyte sequences fall into three well-supported lineages (I, II, and III in fig. 2), with clades of monilophyte sequences sister to clades of seed plant sequences in each case, implying that the ancestral euphyllophyte genome encoded three C4HDZ genes (fig. 6), which must have resulted from two duplications of the single ancestral C4HDZ in the vascular plant ancestor. A similar scenario was postulated for C3HDZ genes, with the common ancestor of vascular plants having a single gene, and the common ancestor of euphyllophytes having two genes (Floyd et al. 2006; Prigge and Clark 2006). Comprehensive analyses of additional gene families are required to determine whether this is a general pattern, which could be indicative of one or more whole-genome duplications in the lineage leading to the extant euphyllophytes following divergence from the lycophyte lineage. As in the lycophytes, there is evidence for independent gene duplications in both “basal” monilophytes (Equisetum and Angiopteris) and leptosporangiate ferns (Ceratopteris and Asplenium) (Pryer et al. 2004). In most cases, the gene duplications are species specific with respect to our taxon sampling, implying duplication events in the lineages leading to the extant species since their divergence from one another. The monophylly of three clades of monilophyte sequences is consistent with other molecular evidence for monophylly of these diverse eusporangiate taxa and leptosporangiate ferns (Pryer et al. 2004).
C4HDZ Sequences in Seed Plants
Seed plant sequences are represented in six clades (A–F in fig. 2). In four of the clades (B–E), gymnosperm sequences reside in monophyletic clades sister to clades of angiosperm sequences, and in another case (A) gymnosperm sequences form a grade, which is not well supported, sister to a monophyletic angiosperm clade. The final seed plant clade (F) contains only gymnosperm genes, suggesting a gene loss from this lineage in the angiosperm ancestor.
Based on phylogenomic analyses of sequenced seed plant genomes and transcriptomes, a whole-genome duplication in the common ancestor of seed plants and another in the common ancestor of flowering plants were proposed (De Bodt et al. 2005; Jiao et al. 2011). The topology of the C4HDZ gene tree is consistent with a whole-genome duplication in the ancestor of seed plants with two (or more) clades of seed plant sequences in two (II and III) of the three euphyllophyte clades. This could also be the case in the other euphyllophyte clade (I) if the gymnosperm sequences reside in a grade, but additional sampling of gymnosperm sequences is required to resolve ambiguity within seed plant clade A. In contrast, the C4HDZ gene phylogeny does not provide any support for a whole-genome duplication at the base of the flowering plants (De Bodt et al. 2005; Jiao et al. 2011). For each clade of gymnosperm sequences, the angiosperm sister clade, if one exists, consists of a single clade of monocot sequences sister to a single clade of eudicot sequences. Thus, if a whole-genome duplication occurred in the common ancestor of angiosperms, one of each of the duplicate paralogs was lost prior to the divergence of monocots and eudicots. Although we did not survey extensively within the angiosperms, the patterns of paralogous monocot and eudicot sequences are consistent with postulated whole-genome duplications with the eudicot and grass lineages (Vision et al. 2000; Tuskan et al. 2006; Tang et al. 2010; Jiao et al. 2012).
Evolution of C4HDZ Genomic Architechture in Land Plants
C4HDZ genes from the liverwort (MpC4HDZ1), hornwort (PcC4HDZ1), and all four S. moellendorffii genes (SmC4HDZ1, 2 and 4) include 10 introns with conserved splice sites within the coding region (fig. 1; supplementary table S2, Supplementary Material online). The same structure has been shown to occur in at least one gymnosperm C4HDZ gene (PaHB1) (Ingouff et al. 2003) as well as in several eudicot and monocot C4HDZ genes, although several angiosperm genes have fewer than 10 introns (Nakamura et al. 2006; Javelle, Klein-Cosson, et al. 2011). Previous analysis, based on flowering plant genes, concluded that a gene structure comprising seven introns and eight exons was ancestral (Javelle, Klein-Cosson, et al. 2011). However, comparison of the C4HDZ genomic architecture including earlier diverging land plants clearly suggests an ancestral structure with 10 introns, with conserved splice site positions within the coding region. C4HDZ genes in flowering plants that differ from the ancestral structure can most parsimoniously be explained by proposing intron losses. In some cases, these appear to be lineage specific. For example, flowering plant genes in angiosperm subclade “d” (fig. 2) all lack ancestral introns 4 and 9.
Our comparative analysis incuding C4HDZ sequences from earlier-diverging plants indicate that the first intron was originally within the coding region. Some flowering plant genes, including ANL2, its close paralog At3g61150/HDG1, and related monocot genes (all in angiosperm subclade “d”) share this structure, with the start codon upstream of intron 1, and align in this region with Marchantia and Selaginella (data not shown). Our data suggest that these flowering plant genes retain the ancestral structure of both gene and transcript. In contrast, eudicot and monocot genes in angiosperm subclade “b” lack intron 1 and do not encode start codons upstream of those annotated. Our analysis would suggest that this represents an evolutionary truncation of the transcript, including the loss of an exon and intron 1 that were originally part of the coding sequence. Five Arabidopsis C4HDZ genes, ML1, PDF2, AT1g05230/HDG2, AT2g32370/HDG3 (all in subclade “a” [fig. 2]), and FWA (in sublade “d” [fig. 2]) encode an intron upstream of the start codon. However, most of the monocot genes in subclades “a” and “d” encode intron 1 downstream of the start codon. These data indicate that mutations have occurred within the eudicot lineage causing the loss of the original start codon but retaining ancestral intron 1.
The lack of introns in the moss C4HDZ genes can also be considered a derived condition. The lack of introns in the Sphagnum gene, SspC4HDZ1, indicates that the intron loss occurred early in the evolution of extant mosses, prior to the divergence of the ancestors of Sphagnum and Physcomitrella, (Shaw et al. 2011). Further sampling of Sphagnum and Takakia, representing the basal lineags of mosses, is required to determine the precise evolutionary timing of the loss of introns. One mechanism to account for the loss of all introns from the ancestral P. patens C4HDZ gene is via reverse transcription of an intronless mRNA followed by homologous recombination between the resulting intronless cDNA and the original genomic locus (Baltimore 1985; Fink 1987). Reverse transcriptase mediated gene conversion has been demonstrated to occur in vivo (Derr and Strathern 1993) and could explain a bias of 3′ intron loss due to insufficient processivity of reverse transcriptase (Mourier and Jeffares 2003). Given the propensity of exogenous DNA to be incorporated into the P. patens genome via homologous recombination (Schaefer and Zryd 1997) and the presence of possible sources of reverse transcriptase (Rensing et al. 2008), such a mechanism appears to be a plausible explanation for intron loss in an ancestral moss C4HDZ gene.
The Brassicaceae-Specific BZG Genes Are Young and Dying
The BZG genes can be considered to be lineage-specific genes (Fischer and Eisenberg 1999; Schmid and Aquadro 2001; Wilson et al. 2005; Khalturin et al. 2009). Although one previous study identified BZG1 as Brassicaceae specific (Donoghue et al. 2011), none found BZG2 to be so (Yang et al. 2009; Lin et al. 2010; Donoghue et al. 2011). Our phylogenetic analysis clearly places the BZG genes in a Brassicaceae-specific clade of genes, more closely related to Brassicaceae genes (At5g52170/HDG7 + At4g25530/FWA) than the Populus orthologs of these genes (fig. 4). Lineage-specific genes are often characterized by a short length, fewer introns, reduced expression patterns, and increased evolutionary rates, attributes evident in the BZG genes. The BZG genes lack the HD and leucine zipper domains and are also missing an intron in the SAD domain that is found in all other land plant C4HDZ genes (except those of mosses). The BZG genes have a restricted expression pattern and an increased rate of molecular evolution based on their long branches in the phylogram (fig. 2).
BZG orthologs are found in both A. thaliana and A. lyrata. The presence of a stop codon in a predicted exon and the lack of detectable transcript strongly suggest that BZG1 is a pseudogene in A. thaliana. The orthologous gene in A. lyrata still has the potential to encode a “full length” protein although a frameshift in the last exon results in an altered carboxyl terminus. Conversely, BZG2 is an expressed gene in A. thaliana while the orthologous sequence in A. lyrata is likely a pseudogene. Based on our reporter lines (fig. 4), BZG2 expression is only in the embryo sac and endosperm and only from the maternally contributed allele, and we find no evidence of expression of BZG1. Taken together, we speculate that these two lineage-specific genes are reaching the ends of their existence and will continue their evolution into pseudogenes.
Four other A. thaliana C4HDZ genes (At4g25530/FWA, At2g32370/HDG3, At3g03260/HDG8, and At5g17320/HDG9) are imprinted, with expression of only either the maternal or the paternal allele in the endosperm (Kinoshita et al. 2004; Gehring et al. 2009). Based on microarray data and reporter lines, the expression of these genes is confined to pollen or early seed (e.g., endosperm) development, except At5g17320/HDG9, which also exhibited expression in the tapetum (Schmid et al. 2005; Nakamura et al. 2006). The four imprinted C4HDZ genes are widely dispersed within the C4HDZ phylogeny, but are united in that each is a long branch relative to closely related paralogs, indicative of an accelerated rate of evolution. In each case, one (or more) closely related paralogs with a shorter branch length is broadly expressed, primarily in the epidermal cell layers of the sporophyte (Nakamura et al. 2006). In the phylogram (fig. 2), two other A. thaliana genes represent long branches relative to related C4HDZ paralogs. One of these, At1g34650/HDG10, forms a clade with two of the imprinted genes, and its expression is also limited to the tapetum and pollen (Schmid et al. 2005; Nakamura et al. 2006). The other, At4g17710/HDG4, is a paralog of broadly expressed At5g46880/HDG5, but expression of the former is restricted to pollen (Schmid et al. 2005).
A general theme emerges from the phylogenetic and expression analyses of the C4HDZ and BZG genes in A. thaliana. Following gene duplication, often the result of the last whole-genome duplication in the lineage leading to Arabidopsis (Vision et al. 2000; Blanc et al. 2003), the two paralogs diverged, with one undergoing a slow rate of molecular evolution and retaining the presumed ancestral epidermal expression pattern, and the other paralog undergoing a more rapid rate of molecular evolution and expression becoming limited to the endosperm, pollen and/or tapetum. In the cases where loss-of-function alleles in the latter genes have been examined, aberrant phenotypes have been observed only for At2g32370/HDG3 when in combination with pdf2 or ml1 mutations (Nakamura et al. 2006). In addition, these genes also either lost or possess degenerating 3′-UTR motifs (fig. 4). One hypothesis consistent with these observations is that all of these genes are on the path to pseudogenization, having already lost ancestral regulatory sequences (Yang et al. 2011) and accumulating mutations in the coding regions more rapidly than their paralogs.
The residual expression of these genes in the endosperm, pollen, and tapetum may be a consequence of a relaxation of constraints on gene expression in these tissues. In both the endosperm and the vegetative cell of pollen, a general DNA demethylation leads to activation of expression of repetitive elements in these cells (Gehring et al. 2009; Hsieh et al. 2009; Slotkin et al. 2009). The activation of transposable element expression in the endosperm and the vegetative cell of the pollen is proposed to reinforce their silenced state in the pollen germ cells and embryo, likely mediated by small RNAs (Gehring et al. 2009; Hsieh et al. 2009; Slotkin et al. 2009). Based on our results, we postulate cells of the tapetum may also experience a relaxation of gene expression constraints and could also act as a source of small RNAs. The expression of the BZG and C4HDZ genes in these tissues could be interpreted as a consequence of being located nearby a transposon or other repetitive DNA (Gehring et al. 2009; Kohler and Weinhofer-Molisch 2010). In this scenario, expression of these genes, including imprinting, is merely a byproduct of the general relaxation on gene expression constraints in these tissues.
It is unlikely that the Arabidopsis C4HDZ gene family is unique, but rather, close inspection of many gene families, especially in organisms that frequently undergo whole-genome duplications, might reveal a plethora of such genes, especially among lineage-specific genes or paralogs. Indeed, when the C4HDZ family is examined in Sol. lycopersicum (Sato et al. 2012), a similar story emerges, with 39 predicted coding sequences annotated as homologous to C4HDZ genes, but only 12 of these are full-length and detectably expressed. Thus, gene number estimates based on present annotations are likely an overestimate of functional gene number in species that have recently experienced a whole-genome duplication. Although there is little evidence that the BZG or C4HDZ “pseudogenes” have any function, because most have the potential to encode proteins, they embody evolutionary potential on which selection could act following changing environmental conditions. In this sense, the genes exemplify the “waiting” model of evolution, whereby new genes initially experience increased rates of evolution due to lack of functionality, with eventual fates either being pseudogenization or occasionally, preservation due to selection (Long et al. 2003).
A 3′-UTR Motif Is Conserved in Land Plant C4HDZ Genes
Sequences within 3′-UTRs are well documented in playing an important role in posttranscriptional regulation of gene expression via the binding of regulatory proteins mediating translational repression, mRNA stability, and mRNA localization (Grzybowska et al. 2001; de Moor et al. 2005; Kong and Lasko 2012). The two 3′-UTR 17–18 nucleotide motifs initially identified in angiosperm and gymnosperm C4HDZ sequences are conserved throughout land plants (Ingouff et al. 2003; Nakamura et al. 2006; Javelle, Klein-Cosson, et al. 2011). As noted previously, the C4HDZ genes in flowering plants either have both or neither of these conserved sequences, and their ability to complementary basepair suggests that the formation of a stem-loop is required for function (Javelle, Klein-Cosson, et al. 2011). We have identified four additional 5-nucleotide sequences, also with the potential to complementary basepair in the predicted stem loop structure, in land plant C4HDZ 3′-UTRs, strengthening the hypothesis that ability to form a stem-loop is a conserved feature (fig. 4). However, these latter sequences were not conserved in any Arabidopsis C4HDZ gene. Three possible functions of this conserved sequence are the generation of regulatory small RNAs, a riboswitch, or a protein binding sequence.
Because of the ability to fold into a stem-loop structure, the conserved sequences in the 3′-UTRs of C4HDZ genes were predicted to encode potential microRNAs (Adai et al. 2005). However, multiple lines of evidence argue against the production of regulatory small RNAs from the 3′-UTRs of C4HDZ genes. For example, the most variable sites are those near the middle of the complementary sequences, whose identity is usually critical in functionaing microRNAs. And secondly, there is no evidence of small RNAs produced from any of the Arabidopsis loci harboring the conserved 3′-UTR sequences (Gustafson et al. 2005).
The other two hypotheses, that the conserved sequences act as a riboswitch or a binding site for regulatory proteins suggest that the stem-loop structure could act in translational regulation or stability of C4HDZ mRNAs. In either case, the conserved secondary structure would act as a binding site for a trans-acting molecule, either a metabolite (e.g., auxin) or protein(s). Riboswitches, originally discovered in bacteria, are regulatory elements located in noncoding sequences of mRNAs that often regulate metabolic processes in many organisms by altering the mRNA transcript structure upon binding a ligand, usually a metabolite (Sudarsan et al. 2003; Mandal and Breaker 2004; Wachter et al. 2007). The lack of conservation of stem-loop length, and sequence motifs lacking in Arabidopsis 3′-UTR sequences argues against the sequence acting as a riboswitch. That the 5′ genomic sequences of the Arabidopsis genes PDF2 and ML1 are sufficient to drive epidermal-specific expression of reporter genes (Sessions et al. 1999; Abe et al. 2001; Takada and Jurgens 2007) suggests that the putative regulatory element in the 3′-UTR does not function to restrict C4HDZ activity spatially, at least with respect to the L1. Nonetheless, conservation of both the primary sequence and secondary structure for the entirety of land plant evolution implicates a role in gene regulation, which require functional studies to reveal.
C4HDZ Genes and Epidermal Evolution
C4HDZ genes are considered master regulators of epidermal development in flowering plants (Javelle, Vernoud, et al. 2011; Nadakuduti 2012) and it has been postulated that C4HDZ homologs may have been instrumental in the evolution of epidermal adaptations required for life on land (Graham et al. 2000; Ligrone et al. 2012). With a robust C4HDZ gene tree (fig. 2), we can ask whether the diversification of the gene family correlates with increasing epidermal elaboration in the land plant lineage. The C4HDZ gene tree displays a pattern of increasing complexity during land plant evolution (figs. 2 and 6). Single homologs are predicted in the common ancestors at each of the nodes of land plant evolution from their origin through to the evolution of vascular plants. Following the divergence of lycophyte and euphyllophyte lineages, there was a triplication in the euplyllophyte lineage. Additional duplications occurred in at least two of the ancestral euphyllophyte paralog (clades II and III) in the seed plant ancestor (six C4HDZ genes), and independently within the lycophyte and moss lineages. The phylogeny suggests a single loss in the angiosperm ancestor (five C4HDZ genes). Thus, the pattern is one of increasing complexity (in terms of numbers of C4HDZ gene family members) during early vascular plant evolution, and particularly at the origin of the euphyllophytes, the seed plants, and the eudicots (fig. 6). Lacking knowledge of expression patterns or function of C4HDZ genes throughout streptophytes, it is difficult to state whether C4HDZ epidermal functions are conserved. However, we can examine the evidence for epidermal complexity (focusing on the cuticle, stomata, and trichomes) in the history of land plants to see how this correlates with gene evolution to make some predictions of ancestral and derived functions.
Cuticle
External cuticle-like layers in some of the charophycean relatives of land plants suggest the cuticle had its origin in an algal ancestor of land plants (Cook and Graham 1998). Thus, a role in cuticle synthesis deposition may represent an ancient function for C4HDZ genes, one that predates the origin of the land plant epidermis. Specific functions of C4HDZ transcription factors in flowering plants that have been identified include upregulating genes involved in lipid metabolism and transport (Javelle et al. 2010; Nadakuduti et al. 2012). Disruption of cuticle-related lipid synthesis and transport leads to a variety of epidermal defects that are also associated with loss of C4HDZ function in flowering plants. This is consistent with the emerging understanding of the important role of cuticular lipid signaling in plant development (Javelle, Vernoud, et al. 2011). The association of C4HDZ genes with lipid signaling and transport could then be more ancient than the cuticle itself, and could represent an ancestral role for the C4HDZ transcription factors. Assessing this hypothesis will require complete knowledge of the distribution of the C4HDZ genes in charophytes and the ability to assess function of these genes in algae and bryophytes.
Stomata
Stomata evolved after the divergence of land plants, in the common ancestor of hornworts, mosses, and vascular plants (Kenrick and Crane 1997; Bateman et al. 1998; Raven 2002). Stomata of mosses and hornworts develop on the sporophyte capsule and are described as being relatively simple in structure, with a simple ontogeny in which an epidermal cell undergoes a single symmetric cell division to form a guard mother cell (GMC) that divides equally to form the two guard cells (Payne 1979; Sack and Paollilo 1983; Ziegler 1987). The earliest sporophyte-dominant land plants developed stomata on leafless axes and sporangia of the sporophyte and in some cases the gametophyte (Kenrick and Crane 1997; Edwards and Haas 1998). These plants are known only as fossils, so stomatal ontogeny cannot be known for sure, however the lack of distinct subsidiary cells suggests that stomatal ontogeny was similar to that of the bryophytes (Edwards and Haas 1998; Ziegler 1987). This simple, ancestral pattern of stomatal development was retained in the lycophyte lineage (including extant species) and also characterizes the basal lineages of monilophtyes and seed plants (Payne 1979; Ziegler 1987). More complex stomatal ontogenies (involving one or more distinctly asymmetrical divisions prior to determination of the GMC), like the well-characterized ontogeny of Arabidopsis (Pillitteri et al. 2007; Vaten and Bergmann 2012) or the leptosporangiate ferns (Ziegler 1987; Sen and De 1992) evolved much later within the fern and angiosperm lineages. The greatest diversity of stomatal patterns occurs in the eudicot angiosperms (Ziegler 1987).
Arabidopsis C4HDZ genes in two different euphyll clades (I and II) are expressed at high levels in stomatal complexes. AtML1 and At1g05230/HDG2 (euphyll clade I) have been shown to regulate stomatal development (Peterson et al. 2013) and At5g46880/HDG5 (euphyll clade II) is expressed at high levels in developing stomatal complexes (Nakamura et al. 2006) and awaits functional characterization. This distribution of stomata-related C4HDZs is consistant with a role in stomatal development being an ancient function, mapping potentially to the euphyllophyte ancestral C4HDZ gene. Because a single C4HDZ gene was present in the plant genome when stomata evolved, it is possible that the ancestral C4HDZ gene played a role in the origin of stomata. If C4HDZ genes are shown to regulate stomatal development in mosses and hornworts this would be strong evidence in favor of an ancient role in C4HDZ genes in the origin of stomata. The number of C4HDZ genes increased at each major node of the euphyllophyte tree, with the eudicot ancestor having the greatest number. Thus, the early simplicity of stomatal ontogeny and increases in the complexity of stomatal development correlate with an early simplicity, followed by an increase in complexity in the C4HDZ gene family.
On the other hand, if the ancestral C4HDZ was important for the origin of stomata, it would represent an increase in epidermal complexity (neofunctionaliztion) in the absence of C4HDZ gene family complexity. This could have occurred via the recruitment of novel guard-cell promoting targets for the ancestral C4HDZ transcription factor. Three closely related genes in the bHLH transcription factor family (FAMA, MUTE, and SPCH) and a more distant bHLH (SCRM) have been shown to specify specific stages of stomatal patterning in Arabidopsis (Pillitteri et al. 2007; Vaten and Bergmann 2012). Recent evidence suggests that the stomata bHLH genes may be targets of C4HDZ transcription factors (Peterson et al. 2013) and it has been suggested that diversification of the stomata bHLH gene family may have played a role in the evolution of stomatal complexity (Pillitteri et al. 2007). Cross species analysis has shown that a stomata bHLH homolog from the moss, Physcomitrella, can partially complement A. mute and fama mutants (MacAlister and Bergmann 2011). However, it remains to be determined if the same genes promote stomatal development in the moss. Phylogenetic analyses of stomata bHLH genes and homologs suggest a pattern of increasing gene family complexity in land plants, however published analyses to date (Peterson et al. 2010; MacAlister and Bergmann 2011) have failed to include all the related paralogs in flowering plants, have not sampled any monilophyte species, and have not determined whether the stomata bHLH genes occur in the liverworts, the only land plant group lacking stomata. Further work is needed to clarify the phylogenetic distribution, relationships, and functions of the stomata bHLH genes in land plants in order to assess their possible role as targets of C4HDZ genes in stomatal evolution.
Trichomes
Trichomes are hair-like epidermal outgrowths in plants. Structures called trichomes are said to occur in all land plants, but are most common and diverse in the eudicot angiosperms (Johnson 1975). In bryophytes, the trichome-like structures are rhizoids, axillary hairs, and awns on the leaves, all in the gametophyte. Trichomes are absent from the sporophytes of bryophytes. The earliest sporophyte-dominant land plants also lacked sporophytic trichomes (Kenrick and Crane 1997). The ancestors and early relatives of the lycophyte lineage are known as fossils, many of which showed a diversity of spiny outgrowths or “enations” on their axes (Gensel et al. 1975, 2001; Hao and Gensel 2001). Although some of the enations appear to have been unicellular (like a trichome), most were multicellular and described as cortical outgrowths (Gensel et al. 2001). Trichomes are uncommon in the lycopsids, being absent in the Lycopodiales and occurring uncommonly in living and extinct ligulate taxa (Graham 1935; Uphof 1962). The earliest fossil plants in the euphyllophyte lineage clade were also variously spiny, although some were “naked.” Trichomes appear to become increasingly common in later in euphyllophyte evolution. There are trichomes on Pertica, a fossil euphyllophyte that evolved prior to the divergence of monilophyte and seed plant lineages (Hotton et al. 2001). Trichomes are common in extant leptosporangiate ferns and the Marratialles, and have also been described for various fossils that were part of the monilophyte lineage. Trichomes are uncommon in extant gymnosperms except in cycads, but do occur, and have been described from extinct Paleozoic and Mesozoic seed plants (Townrow 1960; Krings et al. 2003) and are nearly ubiquitous in the flowering plants.
Trichomes were clearly not present in the sporophytes of the earliest vascular plants, which had a single C4HDZ gene. The early ancestors and relatives of the lycophyte and euphyllophyte lineages ranged from smooth to variously spiny, suggesting a plasticity in epidermal development that is not present in extant land plants. It is not clear that true trichomes were part of the vascular plant developmental program at the time lycophyte and euphyllophyte lineages diverged. The trichomes described for some ligulate lyocpsids may represent an independent innovation from the trichomes that likely evolved in an ancestor of the euphyllophyte clade. This period of epidermal plasticity and the origin of trichomes correlates with the timing of the first C4HDZ gene duplications. Trichomes became much more common and diverse later in the leptosporangiate ferns and eudicot angiosperms, also to some extent mirroring the increasing numbers of C4HDZ genes in plant genomes (fig. 6).
Conclusions
As “nothing in biology makes sense except in the light of evolution” (Dobzhansky 1973), analyses of gene families are often placed in an evolutionary context with available sequence data. However, as amino acid and nucleotide sequences have a limited number of character states, homoplasy can be a frequent occurrence when sequences are derived from phylogenetically distant organisms. To compound this problem in the land plant clade, those taxa for which genome sequence is available often exhibit long branches, indicative of an accelerated rate of molecular evolution, compared with related taxa. The result is that most land plant gene trees constructed with limited data contain random sampling errors (Yang and Rannala 2012), and if interpreted literally, imply significant gene losses in multiple lineages. We have demonstrated that comprehensive sampling across all extant lineages of land plants results in a C4HDZ gene tree that mirrors land plant evolution with evidence for gene duplications in many lineages, but minimal evidence for gene losses. Our results suggest caution when interpreting gene trees constructed with minimal taxon coverage across land plants.
Paralogs produced via gene duplication events most often evolve into pseudogenes. Whole-genome duplications simultaneously produce many duplicate paralogs whose fates can be compared across gene families. Based on long phylognetic branches and limited expression patterns, we propose that several Arabidopsis C4HDZ paralogs produced in the latest whole-genome duplication of this lineage are undergoing pseudogenization. Given the propensity of flowering plant genomes to duplicate (Jiao et al. 2012), this may be a widespread phenomenon, and differential branch lengths can be used to identify pseudogene candidates, as well as representing a better estimate of functional gene numbers in species that have recently experienced a whole-genome duplication. Genes experiencing a fast rate of molecular evolution may also distort gene trees, as was the case of the BZG genes in previous phylogenetic analyses (Schrick et al. 2004). Finally, some genes (e.g., BZG) we identify as lineage specific were not identified as such in recent genome-wide analyses in Arabidopsis (Yang et al. 2009; Lin et al. 2010; Rutter et al. 2012), suggesting that careful phylogenetic analysis of individual gene families may often reveal insights missed by genome-wide studies.
Finally, C4HDZ genes evolved prior to most of the epidermal features with which these transcription factors are associated in flowering plants. Of all of the known functions of these genes, a role in lipid synthesis and transport is a candidate for an ancestral function and it is possible that the C4HDZ in charophycean algae played a role in the origin of the cuticle. The C4HDZ family did not diversify during the earliest radiation of land plants, but duplications occurred early after the two major vascular plant lineages diverged, increasing the number of gene family members at key nodes in the land plant tree. Increasing C4HDZ family complexity correlates roughly with increasing complexity of some of the important epidermal features these genes are known to regulate including stomatal development and trichomes. This suggests that the C4HDZ transcription factors could have played a role in the evolution of some of the key innovations of the embryophytes. Further work to characterize function and expression of C4HDZ genes in bryophytes, lycophytes, monilophytes, and eventually charophycean algae promise to reveal further insight into the functional evolution of these important transcription factors and their role in the evolution of the land plant epidermis.
Materials and Methods
Selection of Taxa and Sequence Aquisition
We attempted to identify C4HDZ gene sequences in taxa representing all major land plant clades as well as charophycean algae. Sequences were obtained from publicly available databases when possible (details discussed later and supplementary table S1, Supplementary Material online) and by cloning using degenerate primers. RNA extraction, cDNA synthesis, degenerate primer design, and other methods for cloning new C4HDZ sequences were as described in Floyd et al. (2006) except that some of the sequencing was performed by Micromon DNA sequencing Facility, Monash University, Clayton, Australia, using an Applied Biosystems 3730S Genetic Analyzer. Degenerate primer sequences are provided in supplementary table S3, Supplementary Material online. All nucleotide sequences are provided as a FASTA file available as supplementary data, Supplementary Material online.
Flowering plants
All 16 Arabidopsis C4HDZ gene coding sequences were obtained from the Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/, last accessed August 9, 2013). Full C4HDZ gene coding sequences for additional flowering plant species were obtained by conducting BLAST similarity searches in publicly available databases using Arabidopsis At4g04890/AtPDF2 and At1g79840/GL2 as query sequences (see supplementary table S1 [Supplementary Material online] for sequence source information). Completely sequenced genomes of the eudicot, Sol. lycopersicum (http://solgenomics.net/, last accessed August 9, 2013) and the monocot O. sativa (MSU Rice Genome Annotation Project) were searched. All Z. mays C4HDZ gene sequences previously identified (Ingram et al. 1999, 2000; Javelle, Klein-Cosson, et al. 2011) were downloaded from GenBank as were those for the eudicot V. vinifera. We also obtained the single C4HDZ gene previously identified from the orchid Phaelenopsis sp. (Nadeau et al. 1996) from GenBank and searched for additional orchid sequences in the 1KP project transcript database (http://www.onekp.com/, last accessed August 9, 2013).
Gymnosperms
Pinus taeda and Pic. abies ESTs were obtained by performing BLAST similarity searches in the EST database in GenBank. C4HDZ4 gene sequences for Cyc. rumphii and G. biloba were cloned using degenerate PCR as described earlier. Five Pse. menziesii C4HDZ cDNA orthologs were assembled from individual sequence reads from the GenBank Sequence Read Archive (SRA) by initially performing a BLAST search of each Pin. taeda EST identified and then reiteratively searching the SRA. A sixth partial Pse. menziesii EST was discovered in a BLAST search in the 1KP project transcript database (http://www.onekp.com/, last accessed August 9, 2013).
Monilophytes
Degenerate PCR was used to identify sequences from the leptosporangiate fern Cer. richardii. All other monilophyte sequences were obtained from the 1 KP plant transcriptome project website using BLAST similarity searches of known C4HDZ gene sequences. Additional monilophyte sequences include the leptosporangiate fern Asp. platyneuron, eusporangiate fern Ang. evecta, horsetail E. diffusum, and whisk fern Psi. nudum. Sequences obtained from the 1KP database were assembled scaffolds from ESTs (see supplementary table S1 [Supplementary Material online] for details).
Lycophytes
Degenerate PCR was used to clone a single C4HDZ gene sequence from the lycophyte S. kraussiana. BLAST similarity searches in the S. moelldendorffii genome portal (http://genome.jgi-psf.org/Selmo1/Selmo1.home.html, last accessed August 9, 2013) were used to identify all C4HDZ gene models in S. moellendorffii. EST contigs for Huperzia sqarrosa were identified in BLAST similarity searches of the 1KP project transcript database (http://www.onekp.com/, last accessed August 9, 2013).
Bryophytes
Degenerate PCR was used to clone C4HDZ gene sequences from the hornwort Pha. carolinianus, the liverwort M. polymorpha, and the moss Sphagnum. All C4HDZ genes from the moss P. patens were identified from BLAST similarity searches in the Physcomitrella genome portal (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html, last accessed August 9, 2013).
Algae
We used degenerate PCR to search for C4HDZ gene sequences from the charophyte algae Col. scutata and Cha. carollina. Partial cDNA sequences for C4HDZs from Spi. pratensis and Coloechaete orbicularis were obtained from Ruth Timme (personal communication). We searched the sequenced genomes of the chlorophyte algal species Chlamydomonas reinhardtii, Ostreococcus tauri, and Volvox carteri.
Sequence Analysis, Alignment, and Phylogenetic Analysis
Sequence fragments cloned for this analysis were assembled using Sequencher 4.10 for Macintosh (Gene Codes). Complete or partial coding nucleotide sequences were manually aligned as amino acid translations using Se-Al v2.0a11 for Macintosh (Rambaut 1996). We excluded ambiguously aligned sequence to produce an alignment of 671 amino acid characters in 113 C4HDZ sequences for subsequent Bayesian analysis. Bayesian phylogenetic analysis was performed using Mr. Bayes 3.2.1, run on multiple parallel processors (Huelsenbeck and Ronquist 2001; Huelsenbeck et al. 2001). Three separate analyses were performed. The first included two algal land plant C4HDZ sequences excluding the nonexpressed predicted C4HDZ genes in the Sol. lycopersicum genome. The second included only land plant C4HDZ sequences, excluding the nonexpressed Sol. lycopersicum genes and one Selaginella gene, SmC4HDZ2. The third included only seed plant genes and included all predicted Sol. lycopersicum C4HDZ genes. The fixed rate model option JTT + I was used based on analysis of the alignments with ProTest 2.4 (Abascal et al. 2005). The Bayesian analyses for the larger data sets were run for 2,500,000 generations, which was sufficient for convergence of the two simultaneous runs. To allow for the burn-in phase, 50% of the total number of saved trees was discarded in the first and second analyses, and 25% were discarded for the third analysis. Sequence alignments and command files used to run the Bayesian phylogenetic analyses are provided as supplementary data, Supplementary Material online.
Physcomitrella Plant Materials and Culture Conditions
Plant materials and culture conditions are similar to those outlined in Nishiyama et al. (2000) and Sakakibara et al. (2008). Physcomitrella patens strain David-NIBB as grown on BCDATG medium or BCD medium at 25 °C under continuous light for protonemata and gametophore growth. For vegetative propagation, the protonemata were collected every 5–7 days, and ground with a Polytron homogenizer (Kinematica, Littau, Switzerland) or a mortar and pestle. For the growth of gametangia and sporophytes, protonemata were transplanted onto sterile peat pellets (Jiffy-7; Jiffy Products International AS, Kristanansand, Norway) cultured for 1 month at 25 °C under continuous light. We then induced gametangia and sporophytes by moving cultures under 8 h light and 16 h dark conditions at 15 °C.
GUS Expression
Arabidopsis
The 2,052-bp sequence upstream of the translational start site was amplified from the Ler genomic DNA using the following primer pair, pBZG2-PstI-F (5′-ccgctgcagTTGGATTGAAGGCGGTAAG-3′) and pBZG2-NcoI-R (5′-ggccatggATCTGACCTTTTCATGTG-3′), and subsequently inserted into PstI and NcoI sites upstream of the GUS gene in the vector pRITA. The promoter:GUS cassette was then subcloned into the binary plasmid pMLBART as a NotI fragment in the same orientation as the BASTA resistance gene. Transformation was performed into wild-type Landsberg erecta using Agrobacterium tumefaciens strain GV3001.
Physcomitrella
We created in-frame PpC4HDZ-GUS plasmids using pTN85 (AB267707). 5′ and 3′ genomic fragments from PpC4HDZ1-4 were obtained using PCR. For the 5′-end, we amplified a genomic fragment—1 kb from the stop codon of the target gene, removing the stop codon in the process, and inserted this fragment, in frame, 5′ to the coding region of the GUS gene in the pTN85 vector (AB267707)—creating an in-frame fusion of GUS with the targeted class IV HD-Zip. For the 3′ fragment, we amplified a 1 kb genomic fragment starting from the codon immediately proceeding the stop codon. This fragment was inserted into the 3′ region of the pTN85 vector directly subsequent to the NPTII expression cassette of pTN85. Polyethylene glycol-mediated transformation was performed as described previously using 10–15 µg of a linearized plasmid. Stable transformants were screened by PCR for those having the construct integrated with homologous recombination in both 5′- and 3′-ends (Nishiyama et al. 2000). Candidates were further analyzed using Southern hybridization to exclude transformants with nonhomologous or tandem-integrated transgenes using a probe on the homologous region of the transgene.
Histology
The histochemical detection of GUS activity was performed as described previously (Nishiyama et al. 2000). Physcomitrella gametophytes were fixed in a series of 5% formaldehyde the 5% acetic acid after GUS staining for 10 min. We used a Zeiss steREO Lumar.V12 stereoscope with a Zeiss Axiocam digital camera for the observation and imaging of stained gametophytes.
For plastic sectioning, specimens were then dehydrated through an ethanol series to 100% ethanol prior to infiltration with catalyzed monomer A of the JB-4 embedding kit (Polysciences, Warrington, PA) and embedded in an oxygen-free environment following the basic protocol provided with the kit. Blocks were serially sectioned at 4 µm on a Jung 2065 Supercut rotary microtome (Leica, Heidleberg, Germany) using glass knives. Slides were observed and photographed on a Zeiss Axioskop microscope equipped with a Zeiss Axiocam digital camera using bright-field microscopy.
Expression Analysis of PpC4HDZ01-04 by Semiquantitative RT-PCR
Spores were grown on solidified BCDAT medium for two weeks. Between two and five plates were collected every day for 14 days and contents were flash frozen and stored at −80 °C. Gametophores were grown on solidified BCD for 4 weeks (Sakakibara et al. 2001). Total RNA was extracted using the RNeasy Plant Mini kit (Qiagen). cDNA was synthesized from 100 ng of total RNA with Primescript reverse transcriptase (Takara Bio) using the SMART RACE kit (Clonetech).
Supplementary Material
Supplementary figures S1–S8 and tables S1–S3 are available at Molecular Biology and Evolution online (http://http://mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Pat Gensel for valuable communications regarding early fossil tracheophytes. This work was supported by the U.S. National Science Foundation grant IOS0515435 to J.L.B. and S.K.F., and the Australian Research Council grant FF0561326 to J.L.B.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development. 2003;130:635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- Abe M, Takahashi T, Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001;26:487–494. doi: 10.1046/j.1365-313x.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- Adai A, Johnson C, Mlotshwa S, Archer-Evans S, Manocha V, Vance V, Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Retroviruses and retrotransposons:the role of revserse transcription in shaping the eukaryotic genome. Cell. 1985;40:481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, et al. (103 co-authors) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RM, Crane PR, DiMichele WA, Kenrick PR, Rowe NP, Speck T, Stein WE. Early evolution of land plants: phylogeny, physiology, and ecology of the primary terrestrial radiation. Annu Rev Ecol Syst. 1998;29:263–292. [Google Scholar]
- Berrie GK. Cytology and phylogeny of liverworts. Evolution. 1963;17:347–357. [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH. Homeoboxes in plant development. Biochim Biophys Acta. 1998;1442:1–19. doi: 10.1016/s0167-4781(98)00119-5. [DOI] [PubMed] [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, et al. (19 co-authors) Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Cook ME, Graham LE. Structural similarities between surface layers of selected charophycean algae and bryophytes and the cuticles of vascular plants. Int J Plant Sci. 1998;159:780–787. [Google Scholar]
- Couvreur TLP, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae) Mol Biol Evol. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Derr LK, Strathern JN. A role for reverse transcripts in gene conversion. Nature. 1993;361:170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Nothing in biology makes sense except in light of evolution. Am Biol Teacher. 1973;35:125–129. [Google Scholar]
- Donoghue MTA, Keshavaiah C, Swamidatta SH, Spillane C. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol. 2011;11:47. doi: 10.1186/1471-2148-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D, Kerp H, Hass H. Stomata in early land plants: an anatomical and ecophysiological approach. J Exp Bot. 1998;49:255–278. [Google Scholar]
- Finet C, Timme RE, Delwiche CF, Marleta F. Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr Biol. 2010;20:2217–2222. doi: 10.1016/j.cub.2010.11.035. [DOI] [PubMed] [Google Scholar]
- Finet C, Timme RE, Delwiche CF, Marlétaz F. Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr Biol. 2012;22:1456–1457. doi: 10.1016/j.cub.2012.07.021. [DOI] [PubMed] [Google Scholar]
- Fink GR. Pseudogenes in yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Fischer D, Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL. Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics. 2006;173:373–388. doi: 10.1534/genetics.105.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensel PG, Andrews HN, Forbes WH. New species of Sawdonia with notes on origin of microphylls and lateral sporangia. Bot Gaz. 1975;136:50–62. [Google Scholar]
- Gensel PG, Kotyk ME, Basinger JF. Morphology of above- and below-ground structures in Early Devonian (Pragian-Emsian) plants. In: Gensel PG, Edwards D, editors. Plants invade the land. New York: Columbia University Press; 2001. [Google Scholar]
- Graham R. An anatomical study of the leaves of the Carboniferous arborescent Lycopods. Ann Bot. 1935;49:587–608. [Google Scholar]
- Graham LE, Cook ME, Busse JS. The origin of plants: body plan changes contributing to a major evolutionary radiation. Proc Natl Acad Sci U S A. 2000;97:4535–4540. doi: 10.1073/pnas.97.9.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowska EA, Wilczynska A, Siedlecki JA. Regulatory functions of 3′ UTRs. Biochem Biophys Res Commun. 2001;288:291–295. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
- Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: the Arabidopsis small RNA project database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S-G, Gensel PG. The Posongchong floral assemblages of Southeastern Yunnan, China—diversity and disparity in early devonian plant assemblages. In: Gensel PG, Edwards D, editors. Plants invade the land. New York: Columbia University Press; 2001. [Google Scholar]
- Hotton CL, Hueber FM, Griffing DH, Bridge JS. Early terrestrial plant environments: an example from the Emsian of Gaspe, Canada. In: Gensel PG, Edwards D, editors. Plants invade the land. New York: Columbia University Press; 2001. [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu RB, Chi XY, Chai GH, Kong YZ, He G, Wang XY, Shi DC, Zhang DY, Zhou GK. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa) PLoS One. 2012;7:e31149. doi: 10.1371/journal.pone.0031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Evolution—Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Farbos I, Lagercrantz U, von Arnold S. PaHB1 is an evolutionary conserved HD-GL2 homeobox gene expressed in the protoderm during Norway spruce embryo development. Genesis. 2001;30:220–230. doi: 10.1002/gene.1068. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Farbos I, Wiweger M, von Arnold S. The molecular characterization of PaHB2, a homeobox gene of the HD-GL2 family expressed during embryo development in Norway spruce. J Exp Bot. 2003;54:1343–1350. doi: 10.1093/jxb/erg145. [DOI] [PubMed] [Google Scholar]
- Ingram GC, Boisnard-Lorig C, Dumas C, Rogowsky PM. Expression patterns of genes encoding HD-ZipIV homeo domain proteins define specific domains in maize embryos and meristems. Plant J. 2000;22:401–414. doi: 10.1046/j.1365-313x.2000.00755.x. [DOI] [PubMed] [Google Scholar]
- Ingram GC, Magnard JL, Vergne P, Dumas C, Rogowsky PM. ZmOCL1, an HDGL2 family homeobox gene, is expressed in the outer cell layer throughout maize development. Plant Mol Biol. 1999;40:343–354. doi: 10.1023/a:1006271332400. [DOI] [PubMed] [Google Scholar]
- Ito M, Sentoku N, Nishimura A, Hong SK, Sato Y, Matsuoka M. Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J. 2002;29:497–507. doi: 10.1046/j.1365-313x.2002.01234.x. [DOI] [PubMed] [Google Scholar]
- Javelle M, Klein-Cosson C, Vernoud V, Boltz V, Maher C, Timmermans M, Depege-Fargeix N, Rogowsky PM. Genome-wide characterization of the HD-ZIP IV transcription factor family in maize: preferential expression in the epidermis. Plant Physiol. 2011;157:790–803. doi: 10.1104/pp.111.182147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Depege-Fargeix N, Arnould C, Oursel D, Domergue F, Sarda X, Rogowsky PM. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor outer cell layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 2010;154:273–286. doi: 10.1104/pp.109.150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC. Epidermis: the formation and functions of a fundamental plant tissue. New Phytol. 2011;189:17–39. doi: 10.1111/j.1469-8137.2010.03514.x. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Leebens-Mack J, Ayyampalayam S, et al. (26 co-authors) A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13:R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YN, Wickett NJ, Ayyampalayam S, et al. (17 co-authors) Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Johnson HB. Plant pubescence - ecological perspective. Bot Rev. 1975;41:233–258. [Google Scholar]
- Karol KG, McCourt RM, Cimino MT, Delwiche CF. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. The origin and early evolution of land plants: a cladistic study. Washington (DC): Smithsonian Institution Press; 1997. [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi YH, Kinoshita Y, Cao XF, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- Kohler C, Weinhofer-Molisch I. Mechanisms and evolution of genomic imprinting in plants. Heredity. 2010;105:57–63. doi: 10.1038/hdy.2009.176. [DOI] [PubMed] [Google Scholar]
- Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13:383–394. doi: 10.1038/nrg3184. [DOI] [PubMed] [Google Scholar]
- Krings M, Kellogg DW, Kerp H, Taylor TN. Trichomes of the seed fern Blanzyopteris praedentata: implications for plant-insect interactions in the Late Carboniferous. Bot J Linn Soc. 2003;141:133–149. [Google Scholar]
- Laurin-Lemay S, Brinkmann H, Philippe H. Origin of land plants revisited in the light of sequence contamination and missing data. Curr Biol. 2012;22:R593–R594. doi: 10.1016/j.cub.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG, Renzaglia KS. Major transitions in the evolution of early land plants: a bryological perspective. Ann Bot. 2012;109:851–871. doi: 10.1093/aob/mcs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HN, Moghe G, Ouyang S, Iezzoni A, Shiu SH, Gu X, Buell CR. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol Biol. 2010;10:41. doi: 10.1186/1471-2148-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. The origin of new genes: glismpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Lu PZ, Porat R, Nadeau JA, ONeill SD. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell. 1996;8:2155–2168. doi: 10.1105/tpc.8.12.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Bergmann DC. Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol Dev. 2011;13:182–192. doi: 10.1111/j.1525-142X.2011.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mourier T, Jeffares DC. Eukaryotic intron loss. Science. 2003;300:1393–1393. doi: 10.1126/science.1080559. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Brocchieri L, Burglin TR. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol. 2009;26:2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Burglin TR. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 2006;140:1142–1150. doi: 10.1104/pp.105.073833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti SS, Pollard M, Kosma DK, Allen C, Ohlrogge JB, Barry CS. Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol. 2012;159:945–960. doi: 10.1104/pp.112.198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Zhang XS, Li J, O’Neill SD. Ovule development: identification of stage-specific and tissue-specific cDNAs. Plant Cell. 1996;8:213–239. doi: 10.1105/tpc.8.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara K, Kato M, Hasebe M. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens, by shuttle mutagenesis. DNA Res. 2000;7:9–17. doi: 10.1093/dnares/7.1.9. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Heidelberg (Germany): Springer-Verlag; 1970. [Google Scholar]
- Payne WW. Stomatal patterns in embryophytes - their evolution, ontogeny and interpretation. Taxon. 1979;28:117–132. [Google Scholar]
- Peterson KM, Shyu C, Burr CA, Horst RJ, Kanaoka MM, Omae M, Sato Y, Torii KU. Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development. 2013;140:1924–1935. doi: 10.1242/dev.090209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L. START: a lipid binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Clark SE. Evolution of the class III HD-Zip gene family in land plants. Evol Dev. 2006;8:350–361. doi: 10.1111/j.1525-142X.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot. 2004;91:1582–1598. doi: 10.3732/ajb.91.10.1582. [DOI] [PubMed] [Google Scholar]
- Qiu YL, Li LB, Wang B, et al. (21 co-authors) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci U S A. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. Se-Al: sequence alignment editor. 1996. Available from: http://tree.bio.ed.ac.uk/software/seal/, last accessed August 9, 2013.
- Raven JA. The evolution of vascular plants in relation to quantitative functionaing of dead ater-conducting cells and stomata. Biol Rev. 1993;68:337–363. [Google Scholar]
- Raven JA. The size of cells and organisms in relation to the evolution of embryophytes. Plant Biol. 1999;1:2–12. [Google Scholar]
- Raven JA. Selection pressures on stomatal evolution. New Phytol. 2002;153:371–386. doi: 10.1046/j.0028-646X.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. (73 co-authors) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. The Glabra2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- Ruberti I, Sessa G, Lucchetti S, Morelli G. A novel class of plant-proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 1991;10:1787–1791. doi: 10.1002/j.1460-2075.1991.tb07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter MT, Cross KV, Van Woert PA. Birth, death and subfunctionalization in the Arabidopsis genome. Trends Plant Sci. 2012;17:204–212. doi: 10.1016/j.tplants.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Sack F, Paollilo DJ. Structure and development of walls in Funaria stomata. Am J Bot. 1983;70:1019–1030. [Google Scholar]
- Sakakibara K, Nishiyama T, Deguchi H, Hasebe M. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol Dev. 2008;10:555–566. doi: 10.1111/j.1525-142X.2008.00271.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Nishiyama T, Kato M, Hasebe M. Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol Biol Evol. 2001;18:491–502. doi: 10.1093/oxfordjournals.molbev.a003828. [DOI] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, et al. (317 co-authors) The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer DG, Zryd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- Schena M, Davis RW. Hd-Zip proteins—members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci U S A. 1992;89:3894–3898. doi: 10.1073/pnas.89.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid KJ, Aquadro CF. The evolutionary analysis of “orphans” from the Drosophila genome identifies rapidly diverging and incorrectly annotated genes. Genetics. 2001;159:589–598. doi: 10.1093/genetics/159.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence Logos—a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KFX. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen U, De B. Structure and ontogeny of stomata in ferns. Blumea. 1992;37:239–261. [Google Scholar]
- Sessa G, Carabelli M, Ruberti I, Lucchetti S, Baima S, Morelli G. Identification of distinct families of HD-ZIP proteins in Arabidopsis thaliana. In: Puigdomenech P, Coruzzi G, editors. Molecular-genetic analysis of plant development and metabolism. Berlin (Germany): Springer; 1994. [Google Scholar]
- Sessions A, Weigel D, Yanofsky MF. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 1999;20:259–263. doi: 10.1046/j.1365-313x.1999.00594.x. [DOI] [PubMed] [Google Scholar]
- Shaw AJ, Szovenyi P, Shaw B. Bryophyte diversity and evolution: windows into the early evolution of land plants. Am J Bot. 2011;98:352–369. doi: 10.3732/ajb.1000316. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Jurgens G. Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development. 2007;134:1141–1150. doi: 10.1242/dev.02803. [DOI] [PubMed] [Google Scholar]
- Takada S, Tabata N, Yoshida A. ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development. 2013;140:1919–1923. doi: 10.1242/dev.094417. [DOI] [PubMed] [Google Scholar]
- Tang HB, Bowers JE, Wang XY, Paterson AH. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci U S A. 2010;107:472–477. doi: 10.1073/pnas.0908007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. Broad phylogenomic sampling and the sister lineage of land plants. PLoS One. 2012;7:e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Delwiche CF. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010;10:96. doi: 10.1186/1471-2229-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townrow JA. The Peltaspermaceae, a pteridosperm family of Permian and Trassic age. Palaeontology. 1960;3:333–361. [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. (108 co-authors) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Uphof JCT. Plant hairs. In: Zimmerman W, Ozenda PG, editors. Encyclopedia of plant anatomy. Vol. 4. Berlin (Germany): Gebruder Borntrager; 1962. [Google Scholar]
- Vaten A, Bergmann DC. Mechanisms of stomatal development: an evolutionary view. Evodevo. 2012;3:11. doi: 10.1186/2041-9139-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter H, Stadelmann E. Physiological prerequisites for transition of autotrophic plants from water to terrestrial life. Bioscience. 1968;18:694–701. [Google Scholar]
- Wilson GA, Bertrand N, Patel Y, Hughes JB, Feil EJ, Field D. Orphans as taxonomically restricted and ecologically important genes. Microbiology. 2005;151:2499–2501. doi: 10.1099/mic.0.28146-0. [DOI] [PubMed] [Google Scholar]
- Wodniok S, Brinkmann H, Glockner G, Heidel AJ, Philippe H, Melkonian M, Becker B. Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RH, Li SB, He S, Wassmann F, Yu CH, Qin GJ, Schreiber L, Qu LJ, Gu HY. CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell. 2011;23:3392–3411. doi: 10.1105/tpc.111.088625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, et al. (71 co-authors) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yang LA, Takuno S, Waters ER, Gaut BS. Lowly expressed genes in Arabidopsis thaliana bear the signature of possible pseudogenization by promoter degradation. Mol Biol Evol. 2011;28:1193–1203. doi: 10.1093/molbev/msq298. [DOI] [PubMed] [Google Scholar]
- Yang XH, Jawdy S, Tschaplinski TJ, Tuskan GA. Genome-wide identification of lineage-specific genes in Arabidopsis, Oryza and Populus. Genomics. 2009;93:473–480. doi: 10.1016/j.ygeno.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Yang ZH, Rannala B. Molecular phylogenetics: principles and practice. Nat Rev Genet. 2012;13:303–314. doi: 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhou YQ, Jiang HY, Li XY, Gan DF, Peng XJ, Zhu SW, Cheng BJ. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-zip gene family in maize. PLoS One. 2011;6: e28488. doi: 10.1371/journal.pone.0028488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler H. The evolution of stomata. In: Zeiger E, Farquar GD, Cowan IR, editors. Stomatal function. Stanford (CA): Stanford University Press; 1987. [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.