Abstract
The Ebola virus (EBOV) RNA-dependent RNA polymerase (RdRp) complex consists of the catalytic subunit of the polymerase, L, and its cofactor VP35. Using immunofluorescence analysis and coimmunoprecipitation assays, we mapped the VP35 binding site on L. A core binding domain spanning amino acids 280– 370 of L was sufficient to mediate weak interaction with VP35, while the entire N-terminus up to amino acid 380 was required for strong VP35–L binding. Interestingly, the VP35 binding site overlaps with an N-terminal L homo-oligomerization domain in a non-competitive manner. N-terminal L deletion mutants containing the VP35 binding site were able to efficiently block EBOV replication and transcription in a minigenome system suggesting the VP35 binding site on L as a potential target for the development of antivirals.
Keywords: Ebola virus, Ebolavirus, RNA-dependent RNA polymerase, L protein, VP35, Nucleocapsid, Filovirus, Negative sense RNA virus, Polymerase complex, Ribonucleoprotein complex, Replication
Introduction
Filoviruses cause a severe hemorrhagic fever in humans with case fatality rates up to 90%. The filovirus family is divided into two genera: Ebolavirus with five distinct species including Zaire ebolavirus (EBOV) and Marburgvirus (MARV) with a single species, Marburg marburgvirus. Along with the rhabdoviruses, paramyxo-viruses, and bornaviruses, the filovirus family belongs to the nonsegmented negative-sense (NNS) RNA viruses of the order Mononegavirales. The RNA genome of EBOV is about 19 kb in length and encodes seven structural proteins. Replication and transcription of the viral genome requires formation of a ribonu-cleoprotein complex, comprising the viral genome encapsidated by the nucleoprotein NP in association with the RNA-dependent RNA polymerase (RdRp) complex and a viral transcription factor, VP30. The RdRp complex consists of the catalytic subunit of the polymerase, L, and the polymerase cofactor VP35 (Mühlberger, 2007; Mühlberger et al., 1999). Minigenome assays revealed that NP, VP35, and L are essential and sufficient to support viral replication. For efficient transcription, the transcription factor VP30 is also required (Enterlein et al., 2006; Groseth et al., 2005; Martinez et al., 2008, 2011; Mühlberger et al., 1999; Watanabe et al., 2004).
The polymerase cofactor VP35 is the functional equivalent of the phosphoprotein P of other NNS RNA viruses (Mühlberger et al., 1999). It interacts with L and NP leading to the formation of trimeric complexes in which VP35 serves as a bridge between NP and L (Becker et al., 1998; Boehmann et al., 2005; Groseth et al., 2009). In addition, VP35 forms homo-oligomers mediated by an amino-terminally located coiled-coil motif (Reid et al., 2005; Zinzula et al., 2009). It was shown for the closely related MARV that homo-oligomerization of VP35 is a prerequisite for interaction with L, but is not required for NP binding (Möller et al., 2005).
In addition to its function as polymerase cofactor, VP35 plays an important role in antagonizing cellular antiviral responses. It acts as type I interferon antagonist by interfering with retinoic acid inducible gene I (RIG-I)-dependent activation of the interferon regulatory factor 3 (IRF-3) (reviewed in (Ramanan et al., 2011)), blocks activation of the double-stranded RNA-dependent protein kinase PKR (Feng et al., 2007; Schümann et al., 2009) and suppresses RNA silencing (Fabozzi et al., 2011; Haasnoot et al., 2007; Zhu et al., 2012).
Much less is known about the major component of the polymerase complex, the large protein L. The EBOV L protein is 2212 amino acids in length with an estimated molecular mass of 253 kDa (Volchkov et al., 1999). Comparative sequence analysis of the L proteins of representative members of the Mononegavirales revealed six conserved regions (Poch et al., 1990), which were also identified in EBOV L (Volchkov et al., 1999). The L proteins of NNS RNA viruses are thought to contain all catalytic functions required for transcription and replication, including RNA-dependent RNA polymerization, capping, and methyltransferase activities (Poch et al., 1990). To form the functional polymerase complex, the L proteins need to interact with P/VP35. The interaction domain for VP35 on MARV L resides within the first 530 amino acids (Becker et al., 1998). Similarly, the first 505 amino acids of EBOV L were shown to be sufficient to mediate binding to VP35 (Prins et al., 2010). However, the exact binding domain for VP35 on L has not been determined yet.
In this study, we mapped the VP35 binding domain on the EBOV L protein. We show that an amino-terminal fragment spanning amino acids 280–370 is sufficient to mediate weak L–VP35 binding, whereas strong binding activity was observed with L fragments spanning the first 380 amino acids. In addition to the VP35 binding domain, we identified an L homo-oligomerization domain located in the N-terminal 450 amino acids of L, which does not compete with VP35 binding. Finally, we used L fragments containing the VP35 binding domain to inhibit minigenome replication, potentially offering a new antiviral strategy against filovirus infection.
Results
VP35 interacts with L and mediates its relocation into NP-derived inclusions
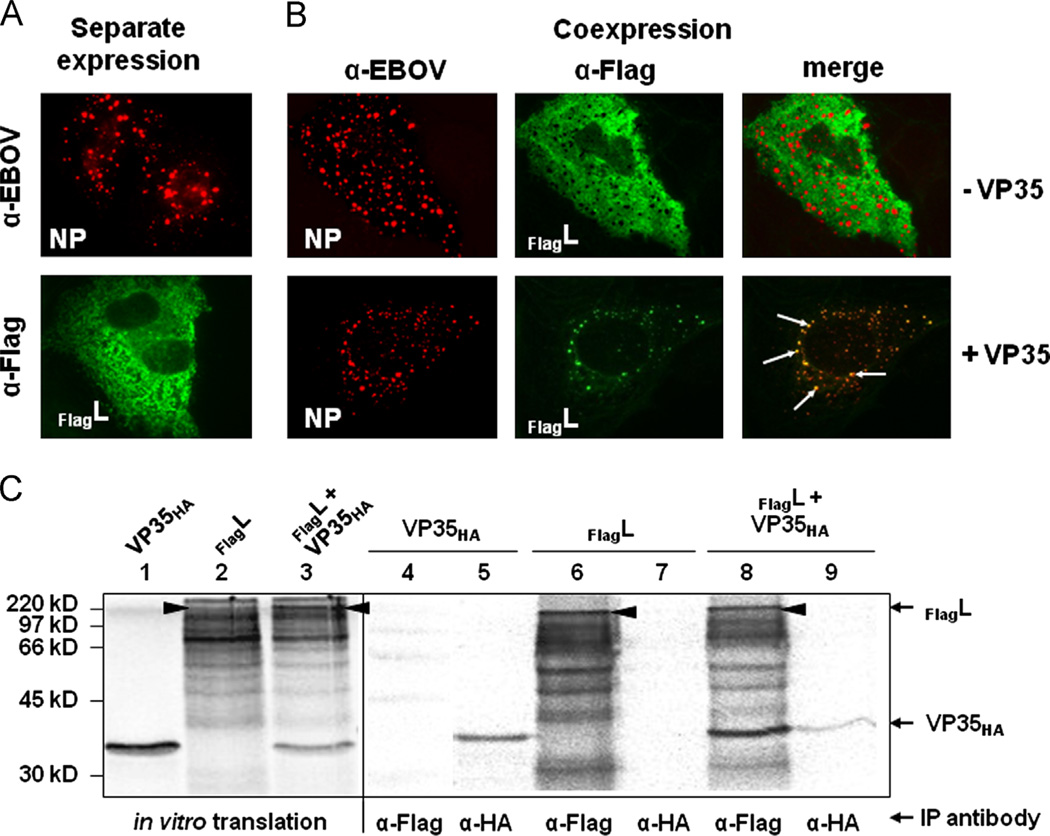
Interaction between L and VP35 was characterized in cell culture by immunofluorescence analysis (IFA) and in a cell-free transcription/translation system followed by coimmunoprecipitation (CoIP). Since no efficient VP35- or L-specific antibodies were available at the time of the experiments, L was expressed as a FLAG-tagged protein and VP35 was used with an HA-tag in the CoIP studies. For IFA, cells were stained with an anti-EBOV antiserum to detect NP or an anti-FLAG antibody for detection of FlagL proteins. The used anti-EBOV antibody recognized neither VP35 nor L in IFA (data not shown). Interaction between L and VP35 in IFA was determined indirectly by taking advantage of the colocalization of L with NP-derived inclusions via VP35 (Becker et al., 1998; Boehmann et al., 2005; Noda et al., 2011; Schmidt et al., 2011).
When expressed in the absence of other viral proteins, NP forms cytoplasmic inclusions, while L is distributed homogenously in the cytoplasm (Fig. 1A). As mentioned above, L relocalizes into NP-derived inclusions when coexpressed with NP and VP35. In the absence of VP35, however, L does not colocalize with NP, indicating that VP35 serves as a linker between L and VP35. To show relocalization of full-length L into NP-derived inclusions mediated by VP35, a FLAG-tagged version of L (FlagL) was expressed in BSR-T7/5 or Huh-T7 cells along with NP in the absence or presence of VP35. NP formed characteristic cytoplasmic inclusions (Fig. 1B, top panel, red), while FlagL was homogeneously distributed throughout the cytoplasm (Fig. 1B, top panel, green) in the absence of VP35, indicating that L does not interact with NP. When NP, VP35, and FlagL were coexpressed, most of FlagL was recruited into the NP inclusions (Fig. 1B, bottom panel, white arrows), confirming that VP35 is required for relocation of L into the viral inclusions. Based on these data, we used the altered distribution pattern of L in cells co-expressing L, VP35, and NP as a readout to determine L–VP35 interaction.
Fig. 1.
Interaction of the polymerase subunit L, its cofactor VP35 and the nucleoprotein NP. (A) IFA of NP and the N-terminally FLAG-tagged L (FlagL). Huh-T7 cells were transfected with pTM1/FlagL or pTM1/NPEBO and analyzed 2 days after transfection by IFA using antibodies directed against NP (red) and FlagL (green). (B) Cellular localization of NP and FlagL in the absence (top panels) or presence (lower panels) of VP35. (C) Coimmunoprecipitation analysis of VP35 and L. VP35HA and FlagL were coexpressed in the presence of [35S]-Met using the TnT® T7 Coupled Reticulocyte Lysate System and subjected to CoIP. Radioactively labeled proteins were separated by SDS-PAGE and visualized on a Bio Imager plate.
The interaction of L with VP35 was also confirmed by CoIP. We first tried to perform CoIP analyses using lysates of cells transiently expressing VP35 and L. However, the combination of immunoprecipitation of cell lysates followed by Western blotting led to a high background due to unspecific precipitation and/or staining of cellular proteins. To keep the Co-IP assays as clean as possible, we finally used in vitro translated radioactively labeled proteins. In vitro translation followed by Co-IP was also used by Chandrika et al. (1995) to map the P binding site on Sendai virus L. FlagL and VP35HA were translated either individually or simultaneously in vitro in the presence of [35S]-methionine. Expression of both proteins was confirmed by SDS-PAGE and subsequent autoradiography (Fig. 1C, lanes 1–3). Strong expression of VP35HA was observed, which was diminished in the presence of L (Fig. 1C, lanes 1 and 3). Full-length FlagL was expressed in very low levels with several bands of lower molecular mass (Fig. 1C, lane 2; arrow head indicates full-length L). The lower molecular mass bands were also precipitated with the anti-FLAG antibody, suggesting that they represent N-terminal FlagL fragments (Fig. 1C, lane 6). Shorter in vitro translation products are frequently observed with proteins greater than 100 kDa and are likely due to incomplete translation of transcripts (Djavadi-Ohaniance and Friguet, 1996). Concurrent expression of VP35 and L resulted in a stronger band for full-length FlagL (Fig. 1C, lane 3), suggesting that either L is stabilized by VP35 or incomplete translation of the L gene occurs less frequently in the presence of other translation products. The latter hypothesis is supported by data showing that full-length L was also more abundant when coexpressed with EBOV NP, EBOV VP30, or EGFP (data not shown). Coexpression of FlagL and VP35HA followed by immunoprecipitation with anti-FLAG resulted in the detection of full-length FlagL, the FlagL fragments, and VP35HA, indicating that L and VP35 interact (Fig. 1C, lane 8). Precipitation with the anti-HA antibody resulted in a faint band for VP35HA but no detectable FlagL, likely caused by the low levels of precipitated VP35HA
Taken together, interaction of L and VP35 was shown by IFA via the relocation of FlagL into NP inclusions only in the presence of VP35. In CoIP studies, an anti-FLAG antibody was able to pull down FlagL and VP35HA. These two assays were used to characterize the binding domain for VP35 on L.
The binding domain for VP35 is located within the first 370 amino acids of L
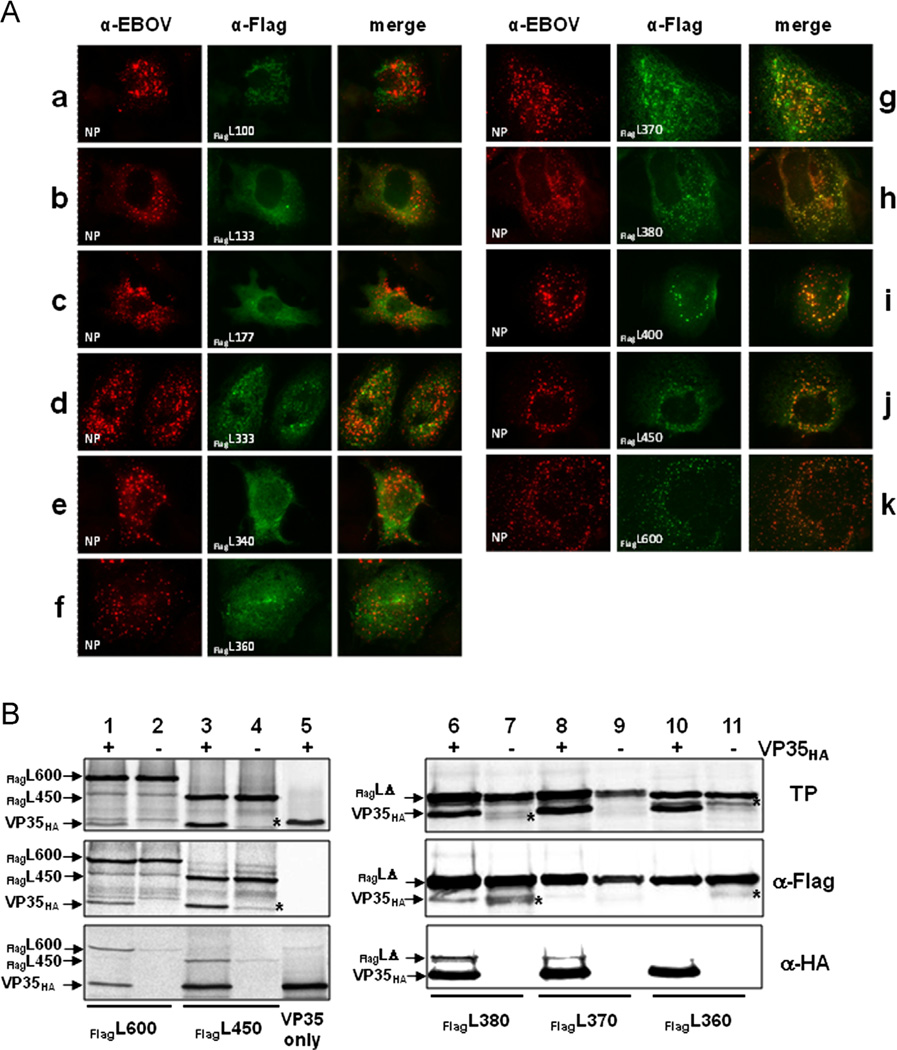
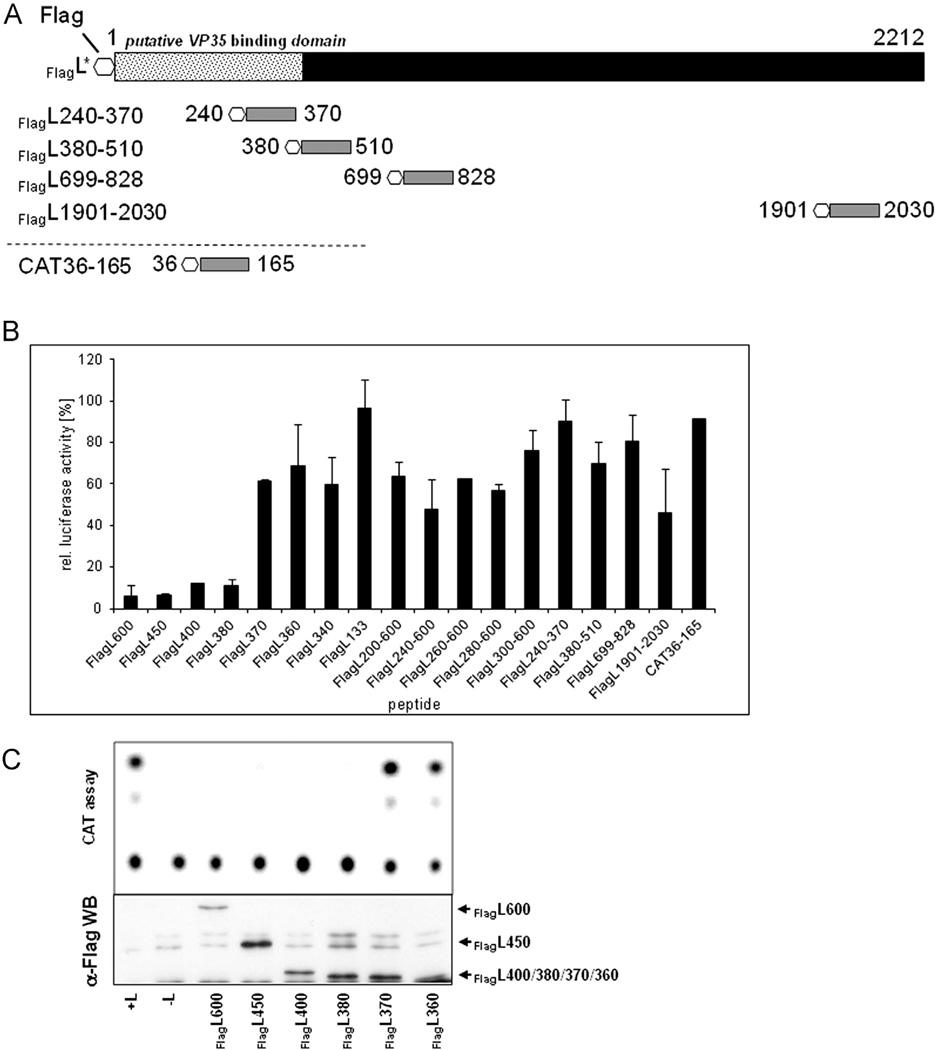
To determine the location of the binding domain for VP35 on L, we first created 11 C-terminal deletion constructs. These deletion mutants contained the first 100–600 amino acids of the N-terminus (Fig. 2A). A FLAG-epitope was added to the N-terminus of each construct allowing for specific detection by IFA or CoIP studies.
Fig. 2.
Deletion mutants of N-terminally tagged EBOV L. The EBOV L protein consists of 2212 amino acids with a putative VP35 binding domain located in the first 505 amino acids (dotted box; Prins et al., 2010). Due to the lack of an L-specific antibody, a FLAG-epitope was added to the N-terminus (octagon). Two classes of constructs were designed (gray boxes): Deletion mutants in which only the C-terminus was truncated (A) and dual deletion mutants that were lacking the C-terminal 1612 amino acids and had additional deletions at the N-terminus (B). For homo-oligomerization studies the FLAG-tag of constructs marked with an asterisk (*) was exchanged with an HA-tag. All constructs were cloned into the pTM1 vector under the control of the T7 RNA polymerase promoter.
For IFA, the FlagL deletion mutants were expressed in BSR-T7/5 or Huh-T7 cells along with NP in the presence or absence of VP35HA. At 48 h post transfection, the cells were fixed and stained with an anti-EBOV antiserum to detect NP, or an anti-FLAG antibody for detection of FlagL proteins. As mentioned above, the used anti-EBOV antibody recognized neither VP35 nor L. Colocalization of NP-derived inclusions and FlagL fragments, indicating an interaction of L with VP35, was observed when the first 370 amino acids of L were present (Fig. 3A, panels g–k). L fragments comprising the N-terminal 360 amino acids or less were distributed throughout the cytoplasm (Fig. 3A, panels a–f). Some aggregation of fragment FlagL333 was observed but the aggregates did not colocalize with NP-derived inclusions (Fig. 3A, panel d). None of the L fragments colocalized with NP when expressed in the absence of VP35 (Fig. S1), demonstrating that VP35-L interaction is essential for relocalization of L into NP inclusions.
Fig. 3.
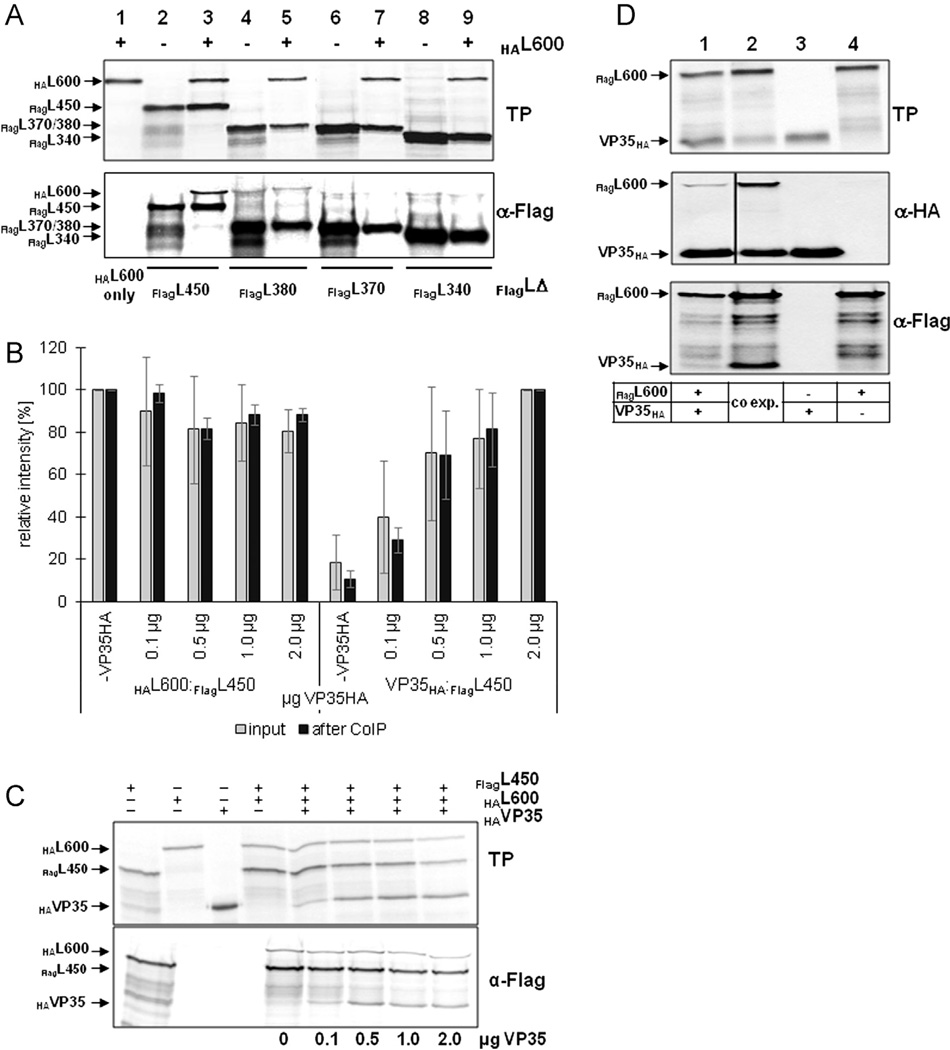
Interaction of C-terminal L deletion mutants with VP35. (A) Interaction of FlagL deletion mutants with VP35 was analyzed by IFA as described in Fig. 1. NP is stained red, FlagL mutants green. (B) The interaction of VP35HA and FlagL was confirmed by CoIP after in vitro translation of the proteins as described in Fig. 1. The positions of FlagL fragments 600 and 450 are indicated on the left; smaller FlagL fragments that were very similar in size (380, 370, and 360) are indicated by FlagLΔ. Lane 5 shows expression and precipitation of VP35HA only. The asterisk (*) in lanes 4, 7 and 11 indicates a FlagL fragment that runs close to the size of VP35HA. TP, in vitro translation products. Experiments were performed at least three times with similar outcome and representative images are shown.
CoIP studies were performed with select FlagL deletion mutants (FlagL600, 450, 380, 370, and 360). The mutants were transcribed and translated in a cell-free system in the presence or absence of VP35HA. The resulting proteins were precipitated with an anti-HA or anti-FLAG antibody conjugated to protein-A agarose. VP35HA was coprecipitated with FlagL600, 450, and 380 (Fig. 3B, lanes 1, 3, and 6, middle panel) but not with shorter fragments (FlagL370 and FlagL360; Fig. 3B, lanes 8 and 10, middle panel). In some precipitation reactions, a protein with a size similar to that of VP35HA was detected (indicated by an asterisk *). Due to the appearance of the band in the absence of VP35HA, it is assumed to be a FlagL degradation fragment or a prematurely terminated translation product (e.g. Fig. 3B, lane 7, middle panel). Precipitation of the translation products with an anti-HA antibody targeting VP35HA showed similar results (Fig. 3B, bottom panels). Interaction of VP35HA with FlagL fragments 600, 450, and 380 was confirmed, but additionally FlagL370 was detectable (Fig. 3B, lane 8, lower panel). The discrepancy between the HA and FLAG precipitation results is likely due to only weak binding of the FlagL370 fragment to VP35HA, resulting in less stable complexes which were not recovered in the CoIP assay. However, the protein interaction was strong enough to recruit FlagL370 into NP inclusions (Fig. 3A, panel g).
In summary, IFA and CoIP data indicate strong VP35–L interaction with FlagL fragments containing the N-terminal 380 amino acids and weak interaction when only 370 amino acids are present. Binding to VP35 was not observed for FlagL fragments containing 360 or less N-terminal amino acids.
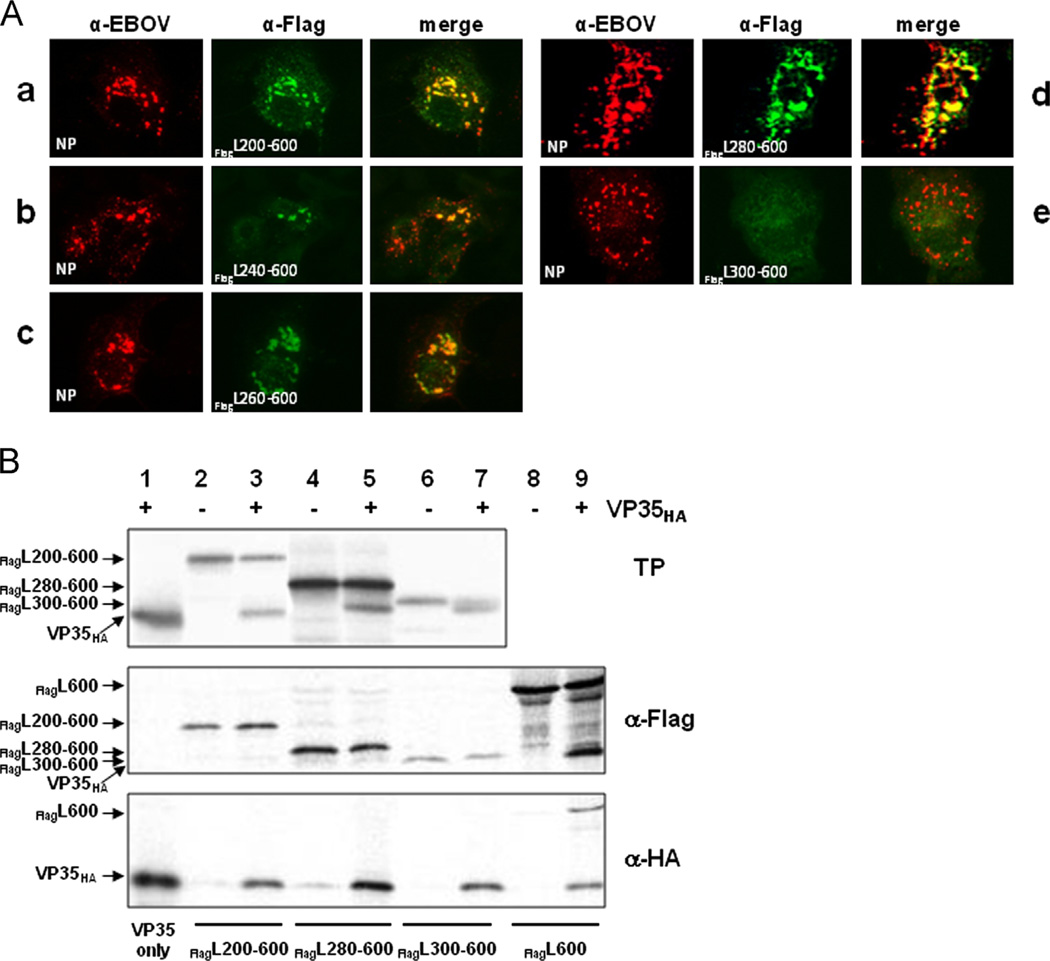
The N-terminal 280 amino acids of L are not required for VP35 binding
Next we deleted N-terminal amino acids from FlagL constructs that already had truncations in the C-terminus (dual deletions) to further narrow down the VP35 binding domain on L. The first set of constructs was based on FlagL600 that contained the 600 N-terminal amino acids (Fig. 2A). The first 200–300 amino acids (in 20-aa iterations) were deleted to generate the mutants depicted in Fig. 2B. To confirm protein expression, the constructs were in vitro translated using the TnT® translation system and detected by autoradiography. All constructs except FlagL220–600 were correctly expressed, leading to protein products of the estimated molecular mass (data not shown). The constructs were analyzed for interaction with VP35HA in IFA and CoIP studies. In IFA studies, FlagL constructs FlagL200–600 to FlagL280–600 colocalized with NP (Fig. 4A, panels a–d). Construct FlagL300–600, comprising amino acids 300–600, was not relocalized into NP-derived inclusions (Fig. 4A, panel e). In the CoIP analysis, however, none of the in vitro translated C- and N-terminally truncated FlagL constructs were able to precipitate VP35HA (Fig. 4B, middle panel, lanes 3, 5, and 7), while the FlagL600 control interacted strongly with VP35HA (Fig. 4B, middle panel, lane 9). Vice versa, after precipitation of VP35HA none of the C- and N-terminal FlagL deletion constructs were detected (Fig. 4B, bottom panel, lanes 3, 5, and 7). Again, the control construct FlagL600 coprecipitated with VP35HA (Fig. 4B, lower panel, lane 9). This indicated only weak interactions of the dual deletion mutants with VP35HA that were disrupted during the CoIP procedures.
Fig. 4.
Interaction of N- and C-terminally truncated FlagL mutants with VP35. (A) Interactions of FLAG-tagged L mutants containing deletions at the C- and N-terminus with VP35 were analyzed by IFA. Red: NP; green: FlagL. (B) CoIP analysis of radioactively labeled FlagL fragments and VP35HA. Proteins were expressed using the TnT® T7 system, precipitated with either an anti-HA or anti-FLAG antibody and resolved on an SDS-polyacrylamide gel. The resulting autoradiograph is shown. TP, in vitro translation products. Experiments were performed at least three times with similar outcome and representative images are shown.
The combined results from N-terminal and dual, i.e. C- and N-terminal, deletion mutants of FlagL suggested that the core binding domain of L with VP35 resides within aa 280 and 370. However, for strong binding of VP35 to L amino acids 1–380 were required.
An L homo-oligomerization domain located in the N-terminus does not compete with VP35 binding
To our knowledge homo-oligomerization of filovirus L proteins has not been described yet. It has been shown for Sendai virus (SeV), also a member of the order Mononegavirales, that the large polymerase subunit possesses a homo-oligomerization domain in the N-terminus (Smallwood et al., 2002). The formation of L homo-oligomers was essential for proper polymerase function (Cevik et al., 2003). Based on our data, the interaction domain of L with VP35 is located within the N-terminus between aa 280 and 370. We therefore analyzed the N-terminus for the presence of an L homo-oligomerization domain and potential competition between L–L and L–VP35 complex formation by CoIP studies.
To analyze the formation of L homo-oligomers, two L proteins based on FLAG-tagged constructs were generated, in which the FLAG-tag was replaced by an HA-tag: HAL (full length) and HAL600 (Fig. 2A, marked with asterisk). All proteins were detected after expression in the TnT® T7 system and in Western blot analysis of transfected cells. L homo-oligomerization is examined by CoIP of HA- and FLAG-tagged proteins. Despite many attempts, the precipitation of full-length HAL was unsuccessful. Since it is known for the L proteins of various paramyxoviruses that the L–L oligomerization domain resides in the N-terminus (see discussion), further studies were performed using HAL600. Construct HAL600 was coexpressed in the TnT® T7 system with FLAG-tagged L deletion mutants described earlier (FlagL450, FlagL380, FlagL370, and FlagL340) (Fig. 5A, top panel). Precipitation of the reaction products with an anti-FLAG antibody revealed that only the longest construct, FlagL450, was able to efficiently precipitate HAL600 (Fig. 5A, bottom panel, lane 3). A faint protein band migrating at the size of HAL600 was consistently observed with FlagL380 (Fig. 5A, bottom panel, lane 5). However, since there was a background band at approximately the same size in the sample without HAL600 (Fig. 5A, bottom panel, lane 4), it remained elusive whether or not fragment FlagL380 weakly interacted with HAL600. Shorter deletion mutants containing the N-terminal 370 or 340 amino acids failed to precipitate HAL600. These results indicate that EBOV L harbors an L homo-oligomerization domain in the N-terminal 450 amino acids.
Fig. 5.
The homo-oligomerization domain in the N-terminus of EBOV L does not compete with VP35 binding. (A) In vitro translation products (TP) and CoIP analysis of HAL600 with various FLAG-tagged L fragments using an anti-FLAG antibody. (B) Impact of VP35HA on L homo-oligomerization. HAL600 and FlagL450 were in vitro translated in the presence of increasing amounts of VP35HA. The ratio of HAL600 and VP35HA relative to FlagL450 was determined before (gray bars) and after (black bars) CoIP with an anti-FLAG antibody. The average of three experiments is shown with standard deviations. (C) Representative gels used for the quantification shown in Fig. 5B. Upper panel, in vitro translation products (TP); bottom panel, CoIP using an anti-FLAG antibody. (D) Comparison of expression and coprecipitation levels of FlagL600 and VP35HA when expressed separately (lane 1) or concurrently (lane 2). Single expression is shown in lanes 3 and 4. The experiment was performed three times and a representative gel is shown.
It was now of interest to investigate whether the L homo-oligomerization and the VP35 binding domains in the N-terminus overlap in a way that L and VP35 binding is competitive. To address the question of competition for L binding, HAL600 and FlagL450 were coexpressed in the TnT® T7 system with increasing amounts of VP35HA. Translation products were precipitated with an anti-FLAG antibody. The intensities of the protein bands after SDS-PAGE were determined in TINA 2.09 to calculate the ratio of HAL600 to FlagL450 and VP35HA to FlagL450 before and after precipitation. Competition for the binding site in the N-terminus of L would result in less coprecipitation of HAL600 in the presence of increasing amounts of VP35HA (or an increase of the VP35HA to FlagL450 ratio). As shown in Fig. 5B and C no difference in the ratios of precipitated HAL600 and VP35HA relative to FlagL450 was observed. These data indicate that binding domains for VP35 and L, although located in overlapping regions in the N-terminus of L, do not compete.
Formation of the functional polymerase complex could occur either by binding of VP35 to preformed L oligomers or by cotranslational L–VP35 interaction. To distinguish these possibilities, VP35 and L were either expressed separately and mixed prior to CoIP or concomitantly synthesized in the in vitro translation system before they were subjected to CoIP. Although VP35 still bound to L when the proteins were mixed after synthesis, binding was strongly reduced, indicating that cotranslation of the two proteins is required for efficient binding (Fig. 5D, lanes 1 and 2).
In summary, EBOV L forms homo-oligomers where the interaction site resides within the N-terminal 450 amino acids. CoIP studies showed no competition between L and VP35 binding to L. Additionally, VP35–L binding is enhanced when the two proteins are concurrently expressed.
EBOV transcription and replication can be inhibited by L peptides
Since our results revealed that the N-terminal domain of EBOV L plays a crucial role in both L–L oligomerization and L–VP35 interaction, the question arose of whether it would be possible to inhibit EBOV transcription and replication by disrupting the polymerase complex using N-terminal L peptides. First, we tested the tagged L proteins for their ability to mediate transcription and replication in a modified minigenome assay (Mühlberger et al., 1999). Constructs HAL, FlagL and FlagL600, respectively, were transfected into BSR-T7/5 cells along with the CAT-expressing mini-genome 3E–5E and supporter plasmids for expression of VP35, VP30, and NP. Untagged full-length L was transfected as positive control. The HA-tag did not negatively impact polymerase activity whereas the FLAG-tag reduced transcription and replication to 16% (data not shown). FlagL600 was not able to support transcription and replication, confirming the importance of the C-terminus for polymerase function (data not shown).
For inhibition studies, we tested most of the previously described C-terminal L deletion mutants and the dual deletion mutants (Fig. 2A and B). Additionally, we designed L peptides which were 130 amino acids in length from various positions inside the L protein; a 130 amino acid long peptide based on the cat gene was designed as negative control (Fig. 6A). Inhibition analysis of L fragments was performed in a quantitative dual luciferase minigenome assay. BSR-T7/5 cells were transfected with one of the L peptide plasmids or the CAT peptide plasmid along with plasmids encoding EBOV NP, VP35, VP30, L, and the mini-genome. Inhibition was calculated relative to samples without additional peptides. L fragments containing at least the N-terminal 380 amino acids were able to suppress transcription and replication to less than 15% (Fig. 6B). These fragments were also able to interact with VP35 as shown in IFA (Fig. 3A) and CoIP (Fig. 3B). Interestingly, peptides that interacted with VP35 in IFA but not CoIP (e.g. FlagL370 or FlagL200–600) did not impair polymerase function. This strongly supports the notion that the interactions of these fragments with VP35 are weaker than interactions between full-length L and VP35, resulting in displacement of the peptides with higher-affinity full-length L. The results shown in Fig. 6B were confirmed by cat gene-based minigenome assays using a selection of L fragments. Only fragments spanning at least the first 380 aa of L were able to block replication and transcription of the minigenome, while shorter fragments were not (Fig. 6C).
Fig. 6.
Inhibition of polymerase activity by expression of L fragments. (A) Schematic of peptides used for polymerase inhibition studies in addition to L deletion mutants described in Fig. 2. (B) Dual luciferase assay results. BSR-T7/5 cells were transfected with plasmids encoding L, NP, VP35, and VP30 to enable transcription and replication of the firefly luciferase-expressing minigenome 3E–5E_F-luc. Additionally, 0.5 µg of the L deletion mutant was transfected to evaluate the impact on polymerase function as indicated by firefly luciferase activity. As transfection efficiency control 0.3 µg of a renilla luciferase-expressing plasmid was co-transfected. Cells were lysed after 24 h and renilla and firefly luciferase activity were measured. Results were normalized to the levels of renilla luciferase and graphed as relative expression setting the sample without additional L fragment as 100%. The average values of 3 independent experiments are shown with standard deviation as error bars. (C) Representative result using a CAT-expressing minigenome (3E–5E) instead of 3E–5E_F-luc to evaluate inhibition of replication (top). Expression control of L fragments (bottom). Cell lysates used for CAT assay were subjected to Western blot analysis using an anti-Flag antibody.
In conclusion, we show that the EBOV L protein harbors both the VP35 binding and L homo-oligomerization domain in the N-terminal 450 amino acids. More specifically, weak VP35 binding was observed with fragments spanning amino acids 280–370. However, only larger fragments containing at least the first 380 amino acids were able to coprecipitate VP35HA indicating stronger interactions. Finally we were able to demonstrate that polymerase function could be inhibited by these L peptides. This finding might be helpful to design specific inhibitors of the EBOV polymerase for use as a therapeutic.
Discussion
Replication and transcription of NNS RNA viruses are complex events that require several viral (and presumably host) proteins. The L protein of all NNS RNA viruses studied so far must interact with the phosphoprotein (P/VP35) to form a functional replicase and transcriptase complex. Despite this general requirement, there are virus-specific differences regarding the mechanisms of L–P interaction and the location of the binding sites on both proteins. The VP35 binding domain on L of both MARV and EBOV is located in the N-terminal part of the protein. In our study, the VP35 binding domain on EBOV L was mapped to the first 380 N-terminal amino acids. Since VP35–L interaction was observed with a MARV L deletion mutant spanning the first 530 amino acids, but not with a mutant comprising the first 309 amino acids of L (Becker et al., 1998), it is assumed that the VP35 binding domain on MARV L is located in the same region as shown for EBOV L. Similar to filoviruses, the P binding site of several members of the Paramyxoviridae family was found to be located in the N-terminal part of the L proteins, including measles virus (MeV), rinderpest virus (RPV), SeV, simian virus 5 (SV5), and human parainfluenza viruses (hPIV) 2 and 3 (Cevik et al., 2003, 2004; Chandrika et al., 1995; Chattopadhyay and Shaila, 2004; Holmes and Moyer, 2002; Horikami et al., 1994; Nishio et al., 2011; Parks, 1994; Smallwood and Moyer, 2004). The identified binding regions ranged from 1305 amino acids in the case of hPIV3 to 380 amino acids for RPV, which is exactly the size of the VP35 binding region on EBOV L (Chattopadhyay and Shaila, 2004; Smallwood and Moyer, 2004).
Sequence comparison analysis of multiple NNS RNA L proteins revealed four highly conserved aa stretches in the N-terminal part of the proteins (Poch et al., 1990). Mutations in any of these regions abrogated P binding of SeV L (Holmes and Moyer, 2002). The identified aa stretches are also conserved in EBOV L. Notably, region 4 spans amino acids 368–383 in EBOV L which have been shown to be crucial to stabilize VP35–L binding. Three of the four highly conserved aa stretches are located in domain I, a conserved region present in the L proteins of NNS RNA viruses (Poch et al., 1990). Domain I spans aa 226–426 of EBOV L and overlaps with the VP35 and L interaction domains ((Volchkov et al., 1999), Fig. 7). Mutational analysis of SeV L domain I resulted in a spectrum of diverse phenotypes, including deficiency in P binding and uncoupling of transcription and replication, indicating that domain I is involved in multiple functions of the L protein (Chandrika et al., 1995).
Fig. 7.
Proposed binding model of L and VP35. (A) Linear representation of the L protein. The core and complete binding domain for VP35 on L are shown as boxes with diagonal stripes and dots, respectively. The homo-oligomerization domain is represented by a box with horizontal stripes. Six regions that are conserved within the order Mononegavirales are indicated by I–VI (Poch et al., 1990).
In contrast to filo- and paramyxoviruses, amino acids important for P binding of rhabdoviral L proteins were mapped within the C-terminal part (Canter and Perrault, 1996; Chenik et al., 1998). CoIP analysis of vesicular stomatitis virus (VSV) L and P showed that both an N-terminal L fragment comprising the first 1593 amino acids of L and a C-terminal fragment spanning amino acids 1594– 2109 bound P, although less efficiently than full-length L, suggesting that an intact L tertiary structure is a prerequisite for P binding (Rahmeh et al., 2010). This is clearly different to EBOV, corroborating L gene-based phylogenetic analyses showing that filoviruses are more closely related to paramyxoviruses than to rhabdoviruses (Mühlberger et al., 1992; Volchkov et al., 1999).
While P–L interactions were examined extensively, few paramyxoviral L proteins have been analyzed for L–L interaction. Of the ones analyzed, the identified L–L interaction domains are located in the N-terminal part of L, overlapping with the P binding domains similar to EBOV L (Cevik et al., 2004; Nishio et al., 2011; Smallwood et al., 2002; Smallwood and Moyer, 2004). Despite the overlap of the EBOV L homo-oligomerization domain with the VP35 binding site, no competition between L–L and L–VP35 binding was observed (Fig. 5). Similarly, non-competing overlapping L–L and L–P interaction domains were mapped on MeV and SeV L proteins. L homo-oligomerization and binding to P are mediated by different amino acids located in the N-terminus of L (Cevik et al., 2003, 2004). Also, oligomerization of MeV and SeV L proteins does not require P, which has also been observed for EBOV L.
To date, the order of L–L, L–P, and P–P binding events for most viruses is not completely understood. Coexpression studies revealed that SV5 L and P did not coimmunoprecipitate when expressed separately and mixed prior to CoIP, indicating that simultaneous expression of the two proteins in the same cell is required for L–P complex formation (Parks, 1994). This is different for rabies virus L and P which are able to interact when combined after separate expression (Chenik et al., 1998). Similarly, recombinant purified VSV or SeV L and P proteins formed transcriptionally functional complexes when mixed after purification (Ogino et al., 2005; Rahmeh et al., 2010). Our data indicate that FlagL600–VP35 interaction is strongly enhanced upon coexpression, although weak co-precipitation was still observed when the two proteins were expressed separately and mixed prior to CoIP (Fig. 5D).
A regulatory impact of P binding to L to prevent unspecific L–L homo-oligomerization has been proposed for hPIV3 (Chattopadhyay and Banerjee, 2009). Although oligomerization of SeV and MeV L proteins takes place in the absence of P, P has been implicated in the stabilization of L for SeV, SV5, MeV, and VSV when coexpressed in cells, most likely by preventing misfolding and degradation (Canter and Perrault, 1996; Horikami et al., 1992, 1994; Smallwood et al., 1994). Recombinant purified VSV, SeV, and Chandipura virus L proteins are stable in the absence of P, whereas purified respiratory syncytial virus L seems to require P for stabilization (Morin et al., 2012; Noton et al., 2012; Ogino and Banerjee, 2010; Ogino et al., 2005; Rahmeh et al., 2009). In our hands in vitro transcribed full-length EBOV FlagL was more abundant in the presence of VP35 (Fig. 1C, lanes 2 and 3). However, the same effect was observed when L was co-expressed with EBOV NP, EBOV VP30, or EGFP (data not shown), suggesting that co-translational activity leads to enhanced synthesis or stabilization of full-length L in an in vitro translation system.
A common feature of NNS RNA virus P proteins is that they form homo-oligomers, typically tetramers. Oligomerization of filoviral VP35 proteins and the P proteins of many members of the order Mononegavirales is mediated by coiled-coil motifs (Bousse et al., 2001; Curran et al., 1995; Möller et al., 2005; Reid et al., 2005). For MARV it was shown that an intact coiled-coil motif on VP35 is a prerequisite for VP35–L interaction (Möller et al., 2005). A possible interpretation of these data is that L binds to preformed VP35 oligomers. It is also conceivable that the coiled-coil domain is not only involved in VP35 oligomerization but also in L binding.
The requirement of EBOV L to oligomerize and interact with VP35 in order to form functional polymerase complexes could be exploited to develop antiviral drugs. L fragments binding to VP35 and/or L inhibited replication in an EBOV minigenome system (Fig. 6). A similar observation was reported for SeV. An L fragment comprising the first 895 amino acids of SeV L blocked viral transcription (Cevik et al., 2003). In conclusion, our data show that blocking the formation of the EBOV replication complex is a promising approach for antiviral interventions.
Materials and methods
Cells
The baby hamster kidney cell line BSR-T7/5 constitutively expressing the T7 RNA polymerase (kindly provided by K. K. Conzelmann, Max von Pettenkofer Institute and Gene Center, Munich, Germany) was cultured as described in Buchholz et al. (1999). The human hepatoma cell line Huh-T7 constitutively expressing the T7 RNA polymerase (kindly provided by V. Gaussmüller, Department of Medical Molecular Biology, University of Lübeck, Germany) was grown in DMEM supplemented with 10% FCS and 1 mg/ml geneticin (Schultz et al. 1996).
Construction of L mutants
Generation of plasmids containing nucleocapsid genes of Zaire ebolavirus, Mayinga isolate, has been described elsewhere (Mühlberger et al., 1999). Plasmid pTM1/LEBO (Mühlberger et al., 1999) was used as template for the L mutants. All L fragments used for immunofluorescence and immunoprecipitation studies contained either an N-terminal FLAG- or HA- epitope. We chose to N-terminally tag the proteins because this is a well established and widely used approach in the field. Full-length pTM1/FlagL was obtained by ligation of an annealed oligonucleotide pair coding for the FLAG-epitope into the NotI restriction site of pTM1/LEBO. Mutants lacking the C-terminus were obtained by PCR amplification using pTM1/FlagL as template. The forward primer contained an NcoI restriction site and the start codon of the FLAG-epitope while the reverse primers were complementary to the last 24 nucleotides of the L-coding sequence followed by a stop codon and a SacI restriction site. The PCR fragments were cloned into the pTM1 plasmid using the NcoI and SacI restriction sites. For homo-oligomerization studies, the FLAG-tag of constructs marked with an asterisk (*) was exchanged with an HA-tag (Fig. 2A). The FLAG-tag was excised with NotI and an annealed oligonucleotide encoding the HA-tag was ligated into the vector. Mutants with deletions in both the N- and C-terminus were obtained by PCR amplification of the respective L fragment flanked by the coding sequence of the FLAG-epitope and a stop codon and cloned into pTM1. All positive clones were verified by DNA sequencing.
In vitro translation
Proteins were translated in a cell-free system (TnT® T7 Coupled Reticulocyte Lysate System, Promega) in the presence of L-[35S]-methionine (Readyvue™ Pro-mix; GE Healthcare Europe or Easy Tag 35S-Methionine; Perkin Elmer) as described by the manufacturer. Briefly, 0.1–1. 5 µg of each expression plasmid and 1.0 µL of L-[35S]-met were added to 20–40 µL of TnT® T7 Quick Master Mix on ice. The volume was adjusted to 25–50 µL with nuclease-free H2O. The transcription/translation reaction was performed at 30 1C for 90 min.
Coimmunoprecipitation
Protein-A sepharose (Sigma-Aldrich) was equilibrated and washed three times with Tris/KCl wash buffer (10 mM Tris, 150 mM KCl, 0.5% NP-40). Five µL of the in vitro translation products were diluted with Tris/KCl buffer (wash buffer with 3% BSA) and centrifuged at 13,000 rpm for 20 min at 4 °C. The clarified supernatant was pre-adsorbed to 20 µL of protein-A sepharose for 1 h. After brief centrifugation the supernatants were incubated with the specific antibody for 1 h at 4 °C. FLAG-tagged proteins were incubated with a mouse monoclonal anti-FLAG M2 antibody (1:500; Sigma-Aldrich) and HA-tagged proteins with a mouse monoclonal anti-HA-7 antibody (1:5000; Sigma-Aldrich). Protein–antibody complexes were incubated overnight with 20 µL of protein-A sepharose and precipitated by low-speed centrifugation. Alternatively, cell lysates were incubated directly with anti-FLAG M2 or anti-HA clone7 agarose without pre-adsorption. Proteins were precipitated by incubation with the agarose for 2 h at 4 °C followed by low-speed centrifugation. The pellets were washed three times with Tris/KCl buffer and once with Tris/KCl wash buffer. Bound proteins were eluted with 4× Laemmli buffer at 95 °C for 5 min and separated using SDS-PAGE. Radioactively labeled proteins were detected using a BioImager Analyzer (Fuji BAS-1000) and the Raytest TINA software.
Transfection of cells
BSR-T7/5 or Huh-T7 cells were grown in 6-well plates to 60–70% confluence and transfected using FUGENE 6 (Roche Molecular Applied Science). Transfection was carried out as previously described (Modrof et al., 2002). For minigenome assays, 1.0 µg pTM1/LEBO, 0.5 µg pTM1/NPEBO, 0.5 µg pTM1/VP35EBO, 0.1 µg pTM1/VP30EBO, and 0.5 µg of pC-T7/Pol expressing the T7 RNA polymerase (kindly provided by T. Takimoto, St. Jude Children’s Research Hospital, Memphis, TN and Y. Kawaoka, University of Wisconsin, Madison, WI) were transfected (Mühlberger et al., 1998; Neumann et al., 2002). To analyze minigenome activity based on the chloramphenicol acetyltransferase (CAT) reporter gene, 1.0 µg minigenome DNA (3E–5E) was co-transfected. For analysis of minigenome activity based on firefly luciferase expression, 1.0 µg of 3E–5E_F-luc was transfected with 0.3 µg of pRL-SV40 (expressing Renilla luciferase as transfection control; Promega). For inhibition assays, 0.5 µg of L peptide plasmid DNA or CAT peptide plasmid DNA was transfected along with the EBOV minigenome plasmids. For immunofluorescence analysis, BSR-T7/5 or Huh-T7 cells were seeded on glass coverslips in 6-well plates, allowed to grow to 60–70% confluence overnight, and transfected the next day with the plasmids of interest (0.25 µg pTM1/NPEBO, pTM1/VP35EBO, and 1.0 µg for L-expressing plasmids), either individually or in combination. All transfections were adjusted to the same total amount of DNA using empty pTM1 plasmid.
Enzymatic CAT assay
BSR-T7/5 cells were transfected as described above. At 2 days post transfection, cell pellets were washed twice with PBS and lysed in 150 µl of reporter lysis buffer (Promega). CAT assays were performed using either a standard protocol (Modrof et al., 2002) or using a fluorescent substrate (FAST CAT) following the manufacturer’s protocol (Molecular Probes). Quantification of radioactive chloramphenicol was done with a BioImager Analyzer (Fuji BAS-1000) and the Raytest TINA software. Fluorescent samples were detected in the GelDoc system using Quantity One-4.0.0 software (Bio-Rad).
Dual luciferase assay
Dual luciferase activity of cell lysates was determined using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s protocol. Luminescence was measured using the LUMIstar luminometer (BMG).
Western blot analysis
Cell lysates in reporter lysis buffer (Promega) obtained for dual luciferase or CAT reporter assay were mixed with SDS loading buffer and separated by SDS PAGE. Proteins were then transferred to a PVDF membrane via semi-dry blotting. FlagL fragments were detected using anti-Flag M2 antibody (Sigma-Aldrich) followed by staining with an Alexa Fluor 680-conjugated secondary antibody (Molecular Probes).
Immunofluorescence analysis
BSR-T7/5 or Huh-T7 cells seeded on glass coverslips were transfected with plasmids as described previously. At 48 h post transfection, cells were fixed and permeabilized with a 1:1 mixture of acetone:methanol for 5 min at −20 1C. After washing three times with PBS the coverslips were first blocked with 0.1 M glycine for 10 min then with IFA blocking solution (PBS with 2% BSA, 0.2% Tween−20, 3% glycerol, 0.05% NaN3) for another 10 min. A goat anti-EBOV antiserum (1:00; kindly provided by S. Becker, University of Marburg, Marburg, Germany) was used to detect NP. This antiserum recognized neither VP35 nor L in IFA. A monoclonal mouse anti-FLAG M2 antibody (Sigma-Aldrich; 1:200) was used to detect FLAG-tagged proteins. A donkey anti-goat antibody coupled with Alexa Fluor® 594 (Invitrogen, 1:500) and a FITC-conjugated donkey anti-mouse antibody (Dianova; 1:100) were used as secondary antibodies. Antibodies were diluted in IFA blocking solution as indicated.
Supplementary Material
Acknowledgments
The authors thank T. Takimoto, Y. Kawaoka, K. K. Conzelmann, S. Becker, and V. Gaussmüller for providing material. This work was supported by National Institutes of Health (NIH) grants AI057159 (New England Regional Center of Excellence-Kasper, subaward 149047-0743) and U01-AI082954 (to E.M.), start-up funds from Boston University (to E.M.), and funds from the Deutsche Forschungsgemeinschaft (SFB 535; to E.M.), the Daimler and Benz Foundation (to D.C.), the FAZIT Foundation (to D.C.), the Jürgen Manchot Foundation (to J.O. and E.M.), and the Cusanuswerk (to K. B.).
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2013.03.013.
References
- Becker S, Rinne C, Hofsäss U, Klenk H-D, Mühlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- Boehmann Y, Enterlein S, Randolf A, Mühlberger E. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology. 2005;332:406–417. doi: 10.1016/j.virol.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Bousse T, Takimoto T, Matrosovich T, Portner A. Two regions of the P protein are required to be active with the L protein for human parainfluenza virus type 1 RNA polymerase activity. Virology. 2001;283:306–314. doi: 10.1006/viro.2001.0881. [DOI] [PubMed] [Google Scholar]
- Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter DM, Perrault J. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology. 1996;219:376–386. doi: 10.1006/viro.1996.0263. [DOI] [PubMed] [Google Scholar]
- Cevik B, Smallwood S, Moyer SA. The L-L oligomerization domain resides at the very N-terminus of the sendai virus L RNA polymerase protein. Virology. 2003;313:525–536. doi: 10.1016/s0042-6822(03)00342-8. [DOI] [PubMed] [Google Scholar]
- Cevik B, Holmes DE, Vrotsos E, Feller JA, Smallwood S, Moyer SA. The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology. 2004;327:297–306. doi: 10.1016/j.virol.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Chandrika R, Horikami SM, Smallwood S, Moyer SA. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology. 1995;213:352–363. doi: 10.1006/viro.1995.0008. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A, Shaila MS. Rinderpest virus RNA polymerase subunits: mapping of mutual interacting domains on the large protein L and phosphoprotein p. Virus Genes. 2004;28:169–178. doi: 10.1023/B:VIRU.0000016855.25662.95. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Banerjee AK. Phosphoprotein, P of human parainfluenza virus type 3 prevents self-association of RNA-dependent RNA polymerase, L. Virology. 2009;383:226–236. doi: 10.1016/j.virol.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenik M, Schnell M, Conzelmann KK, Blondel D. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J. Virol. 1998;72:1925–1930. doi: 10.1128/jvi.72.3.1925-1930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 1995;69:849–855. doi: 10.1128/jvi.69.2.849-855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavadi-Ohaniance L, Friguet B. Incomplete polypeptides of in vitro translation for epitope localization. Methods Mol. Biol. 1996;66:355–361. doi: 10.1385/0-89603-375-9:355. [DOI] [PubMed] [Google Scholar]
- Enterlein S, Volchkov V, Weik M, Kolesnikova L, Volchkova V, Klenk HD, Mühlberger E. Rescue of recombinant Marburg virus from cDNA is dependent on nucleocapsid protein VP30. J. Virol. 2006;80:1038–1043. doi: 10.1128/JVI.80.2.1038-1043.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Cerveny M, Yan Z, He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J. Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Feldmann H, Theriault S, Mehmetoglu G, Flick R. RNA polymerase I-driven minigenome system for Ebola viruses. J. Virol. 2005;79:4425–4433. doi: 10.1128/JVI.79.7.4425-4433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Charton JE, Sauerborn M, Feldmann F, Jones SM, Hoenen T, Feldmann H. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res. 2009;140:8–14. doi: 10.1016/j.virusres.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, Moyer SA. The phosphoprotein (P) binding site resides in the N terminus of the L polymerase subunit of sendai virus. J. Virol. 2002;76:3078–3083. doi: 10.1128/JVI.76.6.3078-3083.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikami SM, Curran J, Kolakofsky D, Moyer SA. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro . J. Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikami SM, Smallwood S, Bankamp B, Moyer SA. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology. 1994;205:540–545. doi: 10.1006/viro.1994.1676. [DOI] [PubMed] [Google Scholar]
- Martinez MJ, Biedenkopf N, Volchkova V, Hartlieb B, Alazard-Dany N, Reynard O, Becker S, Volchkov V. Role of Ebola virus VP30 in transcription reinitiation. J. Virol. 2008;82:12569–12573. doi: 10.1128/JVI.01395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MJ, Volchkova VA, Raoul H, Alazard-Dany N, Reynard O, Volchkov VE. Role of VP30 phosphorylation in the Ebola virus replication cycle. J. Infect. Dis. 2011;204(Suppl 3):S934–S940. doi: 10.1093/infdis/jir320. [DOI] [PubMed] [Google Scholar]
- Modrof J, Mühlberger E, Klenk HD, Becker S. Phosphorylation of VP30 impairs Ebola virus transcription. J. Biol. Chem. 2002;277:33099–33104. doi: 10.1074/jbc.M203775200. [DOI] [PubMed] [Google Scholar]
- Möller P, Pariente N, Klenk HD, Becker S. Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J. Virol. 2005;79:14876–14886. doi: 10.1128/JVI.79.23.14876-14886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin B, Rahmeh AA, Whelan SP. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 2012;31:1320–1329. doi: 10.1038/emboj.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E. Filovirus replication and transcription. Future Virol. 2007;2:205–215. doi: 10.2217/17460794.2.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E, Sanchez A, Randolf A, Will C, Kiley MP, Klenk HD, Feldmann H. The nucleotide sequence of the L gene of Marburg virus, a filovirus: homologies with paramyxoviruses and rhabdoviruses. Virology. 1992;187:534–547. doi: 10.1016/0042-6822(92)90456-y. [DOI] [PubMed] [Google Scholar]
- Mühlberger E, Lötfering B, Klenk H-D, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E, Weik M, Volchkov VE, Klenk H-D, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J. Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Feldmann H, Watanabe S, Lukashevich I, Kawaoka Y. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 2002;76:406–410. doi: 10.1128/JVI.76.1.406-410.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Tsurudome M, Garcin D, Komada H, Ito M, Le Mercier P, Nosaka T, Kolakofsky D. Human parainfluenza virus type 2 L protein regions required for interaction with other viral proteins and mRNA capping. J. Virol. 2011;85:725–732. doi: 10.1128/JVI.01226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kolesnikova L, Becker S, Kawaoka Y. The importance of the NP: VP35 ratio in Ebola virus nucleocapsid formation. J. Infect. Dis. 2011;204(Suppl 3):S878–S883. doi: 10.1093/infdis/jir310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton SL, Deflube LR, Tremaglio CZ, Fearns R. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 2012;8:e1002980. doi: 10.1371/journal.ppat.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Banerjee AK. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J. Gen Virol. 2010;91:1311–1314. doi: 10.1099/vir.0.019307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Kobayashi M, Iwama M, Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- Parks GD. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein. P. J. Virol. 1994;68:4862–4872. doi: 10.1128/jvi.68.8.4862-4872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J. Virol. 2010;84:10581–10591. doi: 10.1128/JVI.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh AA, Schenk AD, Danek EI, Kranzusch PJ, Liang B, Walz T, Whelan SP. Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20075–20080. doi: 10.1073/pnas.1013559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Shabman RS, Brown CS, Amarasinghe GK, Basler CF, Leung DW. Filoviral immune evasion mechanisms. Viruses. 2011;3:1634–1649. doi: 10.3390/v3091634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Cardenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KM, Schümann M, Olejnik J, Krähling V, Mühlberger E. Recombinant Marburg virus expressing EGFP allows rapid screening of virus growth and real-time visualization of virus spread. J. Infect. Dis. 2011;204(Suppl 3):S861–S870. doi: 10.1093/infdis/jir308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DE, Honda M, Whetter LE, McKnight KL, Lemon SM. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J. Virol. 1996;70:1041–1049. doi: 10.1128/jvi.70.2.1041-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümann M, Gantke T, Mühlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J. Virol. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S, Moyer SA. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology. 2004;318:439–450. doi: 10.1016/j.virol.2003.09.045. [DOI] [PubMed] [Google Scholar]
- Smallwood S, Ryan KW, Moyer SA. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology. 1994;202:154–163. doi: 10.1006/viro.1994.1331. [DOI] [PubMed] [Google Scholar]
- Smallwood S, Cevik B, Moyer SA. Intragenic complementation and oligomerization of the L subunit of the sendai virus RNA polymerase. Virology. 2002;304:235–245. doi: 10.1006/viro.2002.1720. [DOI] [PubMed] [Google Scholar]
- Volchkov VE, Volchkova VA, Chepurnov AA, Blinov VM, Dolnik O, Netesov SV, Feldmann H. Characterization of the L gene and 5′ trailer region of Ebola virus. J. Gen Virol. 1999;80:355–362. doi: 10.1099/0022-1317-80-2-355. [DOI] [PubMed] [Google Scholar]
- atanabe S, Watanabe T, Noda T, Takada A, Feldmann H, Jasenosky LD, Kawaoka Y. Production of novel ebola virus-like particles from cDNAs: an alternative to ebola virus generation by reverse genetics. J. Virol. 2004;78:999–1005. doi: 10.1128/JVI.78.2.999-1005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Celebi Cherukuri N, Jackel JN, Wu Z, Crary M, Buckley KJ, Bisaro DM, Parris DS. Characterization of the RNA silencing suppression activity of the Ebola virus VP35 protein in plants and mammalian cells. J. Virol. 2012 doi: 10.1128/JVI.05741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L, Esposito F, Mühlberger E, Trunschke M, Conrad D, Piano D, Tramontano E. Purification and functional characterization of the full length recombinant Ebola virus VP35 protein expressed in E. coli. Protein Expression Purif. 2009;66:113–119. doi: 10.1016/j.pep.2009.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.