Abstract
Alterations in mitochondrial function have long been considered a hallmark of cancer. We compared the lipidome and electron transport chain activities of non-synaptic brain mitochondria in two inbred mouse strains, the C57BL/6J (B6) and the VM/Dk (VM). The VM strain is unique in expressing a high incidence of spontaneous brain tumors (1.5%) that are mostly gliomas. The incidence of gliomas is about 210-fold greater in VM mice than in B6 mice. Using shotgun lipidomics, we found that the mitochondrial content of ethanolamine glycerophospholipid, phosphatidylserine, and ceramide was higher, whereas the content of total choline glycerophospholipid was lower in the VM mice than in B6 mice. Total cardiolipin content was similar in the VM and the B6 mice, but the distribution of cardiolipin molecular species differed markedly between the strains. B6 non-synaptic mitochondria contained 95 molecular species of cardiolipin that were symmetrically distributed over seven major groups. VM non-synaptic mitochondria contained only 42 species that were distributed asymmetrically. The activities of Complex I, I/III, and II/III enzymes were lower, whereas the activity of complex IV was higher in the mitochondria of VM mice than in B6 mice. The high glioma incidence and alterations in electron transport chain activities in VM mice compared to B6 mice could be related to the unusual composition of mitochondrial lipids in the VM mouse brain.
Introduction
Mitochondria are necessary for the maintenance of brain metabolism supporting neurological function. Alterations in brain mitochondria can impair bioenergetic efficiency resulting in disease pathology or neurological impairment. Although such alterations can exist at the DNA or protein level, little attention has focused on the role of mitochondrial lipids in neurological function. Mitochondrial membrane lipids can regulate numerous functions to include electron transport chain (ETC) activities, membrane fluidity, mitochondrial protein import, and ATP synthesis {Daum, 1985 #19; Hoch, 1992 #35; Petrushka, 1959 #97; Shinzawa-Itoh, 2007 #100; Jiang, 2000 #336}. Non-synaptic (NS) brain mitochondria are derived from the cell bodies of neurons and glia and represent the predominant mitochondrial population in brain {Davey, 1996 #721}. Alterations in the NS mitochondrial lipid composition could influence ETC activities resulting in a change in neural cell metabolism.
Shotgun lipidomics, using electrospray ionization mass spectrometry (ESI/MS), can now determine the total content and composition mitochondrial lipids (the lipidome). Using this approach, we recently characterized the lipid composition of mouse brain mitochondria (Kiebish et al 2008). The mitochondrial specific lipid cardiolipin (1,3-diphosphatidyl-sn-glycerol, Ptd2Gro), contains nearly 100 different molecular species in the C57BL/6J (B6) mouse brain (Kiebish et al Cheng et al 2008). Moreover, these molecular species are symmetrically distributed in 7 major groups when arranged according to mass to charge ratios (Kiebish et al 2008). This distribution in B6 mice is similar to that in several other mammalian species and reflects a conserved cardiolipin remodeling process for brain (Cheng et al 2008).
Our objective was to compare and contrast the lipidome of NS mitochondria and ETC activities in mice of the B6 and the VM/Dk (VM) strains. B6 mice are often used as a control strain for many existing neurological mutations that alter CNS function {Bedell, 1997 #237; Bedell, 1997 #236}. VM mice are unique in expressing a relatively high incidence (1.5%) of spontaneous brain tumors. Most of theses brain tumors were characterized as gliomas (astrocytomas and microgliomas) {Huysentruyt, 2008 #2062; Fraser, 1971 #1258; Fraser, 1986 #1263}. Indeed, the incidence in VM mice is about 210-fold greater than the incidence in B6 mice. Although altered mitochondrial energy metabolism has long been connected to tumorigenesis (Warburg hypothesis) {Warburg, 1931 #175; Warburg, 1956 #106; Cavalli, 1998 #126; Augenlicht, 2001 #433}, the role of mitochondrial lipids in this connection remains unclear. Ours is the first comparative analysis of the NS brain mitochondria lipidome in two mouse strains that differ in susceptibility to spontaneous gliomas.
Experimental Procedures
Animals
The VM mice were obtained from Professor H. Fraser, University of Edinburgh. The B6 mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice of both strains were propagated at the Boston College Animal Facility and were maintained by brother-sister inbreeding. All mice were housed in plastic cages with filter tops containing Sani-Chip bedding (P.J. Murphy Forest Products Corp., Montville, NJ). The room was maintained at 22 °C on a 12 hr light/dark cycle. Food (Prolab RMH 3000; PMI LabDiet, Richmond, IN) and water were provided ad libitum. This study was conducted with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and was approved by the Institutional Animal Care Committee.
Non-synaptic Brain Mitochondrial Isolation
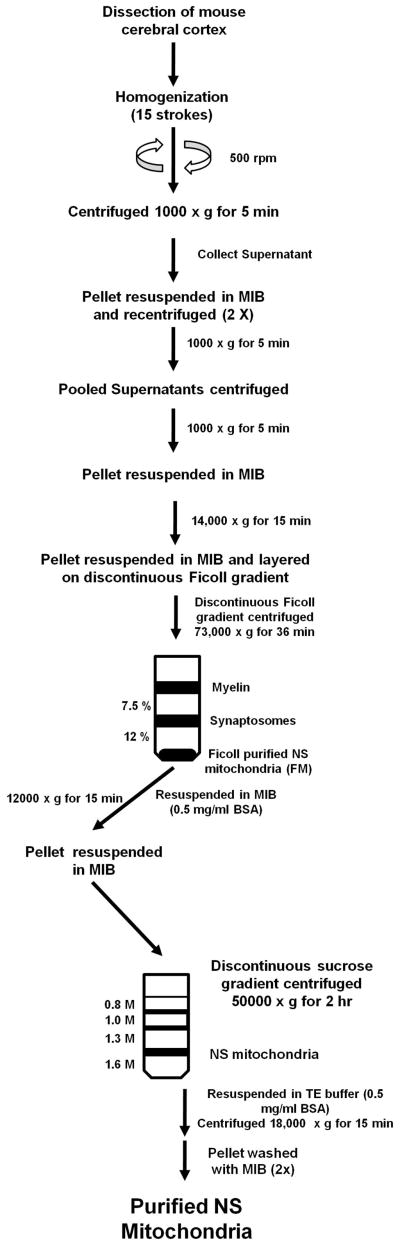
We recently described an improved procedure for isolation and purification of NS mitochondria from mouse brain for the purpose of lipid analysis and biochemical analysis of ETC activities (Kiebish, 2008). Briefly, aged matched B6 and VM mice (4 months) were sacrificed by cervical dislocation and the cerebral cortex was dissected. Mitochondria were isolated in a cold room (4 °C) and all reagents were kept on ice. The isolation procedure employed a combination of gradients and strategies as previously described {Lai, 1976 #45; Lai, 1977 #181; Mena, 1980 #93; Dagani, 1983 #792; Rendon, 1985 #60; Battino, 1991 #4; Brown, 2006 #14} (Fig. 1). The cerebral cortexes (a pool of 6/sample) were diced on an ice cold metal plate and then placed in 12 mL of mitochondria isolation buffer (MIB; 0.32 M sucrose, 10 mM Tris-HCl, and 1 mM EDTA-K (pH 7.4)). The pooled cerebral cortexes were homogenized using a Potter Elvehjem homogenizer with a Teflon coated pestle attached to a hand-held drill. Samples were homogenized using 15 up and down strokes at 500 rpm. The homogenate was centrifuged at 1,000 × g for 5 min. The supernatant was collected and the pellet was washed twice by centrifugation, collecting the supernatants each time. The supernatants were pooled and centrifuged at 1,000 × g for 5 min. The collected supernatant was then spun at 14,000 × g for 15 min. The supernatant was discarded and the pellet, which contained primarily NS mitochondria, synaptosomes and myelin, was resuspended in 12 mL MIB and was layered on a 7.5%/12% discontinuous Ficoll gradient. Each Ficoll gradient layer contained 12 mL for a total volume of 36 mL. The Ficoll gradients were made from a 20% Ficoll stock with MIB. The gradient was centrifuged at 73,000 × g for 36 min (4 °C) in a Sorvall SW 28 rotor with slow acceleration and deceleration (Optima L-90K Ultracentrifuge). The centrifugation time used permitted sufficient acceleration and deceleration to achieve maximum g force and to prevent synaptosomal contamination of the mitochondrial fraction below the 12% Ficoll layer {Battino, 1991 #5}. Crude myelin collected at the MIB/7.5% Ficoll interface was discarded. The Ficoll gradient purified NS mitochondria (FM) were collected as a pellet below the 12% Ficoll.
Fig 1.
Procedure used for the isolation and purification of NS mitochondria from mouse cerebral cortex
The FM pellet, containing NS mitochondria, was resuspended in MIB containing 0.5 mg/mL bovine serum albumin (BSA) and was centrifuged at 12,000 × g for 15 min. The resulting pellet was collected and resuspended in 6 mL of MIB. The resuspended FM pellet was layered on a discontinuous sucrose gradient containing 0.8 M/1.0 M/1.3 M/1.6 M sucrose. The volumes for the sucrose gradient were 6 mL/6 mL/10 mL/8 mL, respectively. The gradients were made from a 1.6 M sucrose stock containing 1 mM EDTA-K and 10 mM Tris-HCl (pH 7.4). The discontinuous sucrose gradient was centrifuged at 50,000 × g for 2 hr (4 °C) in a Sorvall SW 28 rotor using slow acceleration and deceleration to prevent disruption of the gradient. Purified NS mitochondria were collected at the interface of 1.3 M and 1.6 M sucrose. NS Mitochondria were collected and resuspended in (1:3, v/v) TE buffer (1 mM EDTA-K and 10 mM Tris-HCl, pH 7.4) containing 0.5 mg/mL BSA and centrifuged at 18,000 × g for 15 min. The pellet was then resuspended in MIB and centrifuged at 12,000 × g for 10 min. The pellet was again resuspended in MIB and centrifuged at 8,200 × g for 10 min.
Materials for Mass Spectrometry
Synthetic phospholipids including 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (14:1-14:1 PtdCho), 1,2-dipalmitoleoyl-sn-glycero-3phosphoethanolamine (16:1-16:1 PtdEtn), 1,2-dipentadecanoyl-sn-glycero-3-phosphoglycerol (15:0-15:0 PtdGro), 1,2-dimyristoyl-sn-glycero-3-phosphoserine (14:0-14:0 PtdSer), N-lauroryl sphingomyelin (N12:0 CerPCho), 1,1′,2,2′-tetramyristoyl cardiolipin (T14:0 Ptd2Gro), heptadecanoyl ceramide (N17:0 Cer), 1-heptadecanoyl-2-hydroxy-sn-Glycero-3-phosphocholine (17:0 LysoPtdCho) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). It should be noted that the prefix “N” denotes the amide-linked acyl chain. All the solvents were obtained from Burdick and Jackson (Honeywell International Inc., Burdick and Jackson, Muskegon, MI, USA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Sample Preparation for Mass Spectrometric Analysis
An aliquot of the mitochondrial preparation was transferred to a disposable culture borosilicate glass tube (16 × 100 mm). Internal standards were added based on protein concentration and included 16:1-16:1 PtdEtn (100 nmol/mg protein), 14:1-14:1 PtdCho (45 nmol/mg protein), T14:0 Ptd2Gro (3 nmol/mg protein), 15:0-15:0 PtdGro (7.5 nmol/mg protein), 14:0-14:0 PtdSer (20 nmol/mg protein), 17:0 LysoPtdCho (1.5 nmol/mg protein), N12:0 CerPCho (20 nmol/mg protein), N17:0 Cer (5 nmol/mg protein). This allowed the final quantified lipid content to be normalized to the protein content and eliminated potential loss from the incomplete recovery. The molecular species of internal standards were selected because they represent < 0.1% of the endogenous cellular lipid mass as demonstrated by ESI/MS lipid analysis.
A modified Bligh and Dyer procedure was used to extract lipids from each mitochondrial preparation as previously described {Cheng, 2006 #1279}. Each lipid extract was reconstituted with a volume of 500 mL/mg protein (which was based on the original protein content of the samples as determined from protein measurement) in CHCl3/MeOH (1:1, v/v). The lipid extracts were flushed with nitrogen, capped, and stored at −20 °C for ESI/MS analysis. Each lipid solution was diluted approximately 50-fold immediately prior to infusion and lipid analysis.
Instrumentation and Mass Spectrometry
A triple-quadrupole mass spectrometer (Thermo Scientific TSQ Quantum Ultra, Plus, San Jose, CA, USA), equipped with an electrospray ion source and Xcalibur system software, was utilized as previously described {Han, 2004 #32}. The first and third quadrupoles serve as independent mass analyzers using a mass resolution setting of peak width 0.7 Th while the second quadrupole serves as a collision cell for tandem MS. The diluted lipid extract was directly infused into the ESI source at a flow rate of 4 mL/min with a syringe pump. Typically, a 2-min period of signal averaging in the profile mode was employed for each mass spectrum. For tandem MS, a collision gas pressure was set at 1.0 mTorr, but the collision energy varied with the classes of lipids as described previously {Han, 2004 #32; Han, 2005 #30}. Typically, a 2- to 5-min period of signal averaging in the profile mode was employed for each tandem MS spectrum. All the mass spectra and tandem MS spectra were automatically acquired by a customized sequence subroutine operated under Xcalibur software. Data processing of 2D MS analyses including ion peak selection, data transferring, peak intensity comparison, and quantitation was conducted using self-programmed MicroSoft Excel macros {Han, 2004 #32}.
Electron Transport Chain Enzyme Activities
Purified mitochondrial samples were freeze-thawed three times before use in enzyme analysis to give substrate access to the inner mitochondrial membrane. All assays were performed on a temperature controlled SpectraMax M5 plate reader (Molecular Devices) and were done in triplicate. Specific enzyme activities were calculated using ETC complex inhibitors in order to subtract background activities.
Complex I (NADH-ubiquinone oxidoreductase) activity was determined by measuring the decrease in the concentration of NADH at 340 nm as previously described {Birch-Machin, 2001 #9; Ellis, 2005 #283}. The assay was performed in buffer containing 50 mM potassium phosphate (pH 7.4), 2 mM KCN, 5 mM MgCl2, 2.5 mg/mL BSA, 2 μM antimycin, 100 μM decylubiquinone, and 0.3 mM K2NADH. The reaction was initiated by adding purified mitochondria (20 μg). The enzyme activity was measured for 5 min and values were recorded 30 sec after the initiation of the reaction. Specific activities were determined by calculating the slope of the reaction in the linear range in the presence or absence of 1 μM rotenone (Complex I inhibitor).
Complex II (succinate decylubiquinone DCIP oxidoreductase) activity was determined by measuring the reduction of 2,6-dichloroindophenol (DCIP) at 600 nm as previously described {King, 1967 #41; Ellis, 2005 #283}. The Complex II assay was performed in buffer containing 25 mM potassium phosphate (pH 7.4), 20 mM succinate, 2 mM KCN, 50 μM DCIP, 2 μg/mL rotenone, and 2 μg/mL antimycin. Purified mitochondria (10 μg) were added prior to initiation of the reaction. The reaction was initiated by adding 56 μM decylubiquinone. Specific activities were determined by calculating the slope of the reaction in the linear range in the presence or absence of 0.5 mM Thenoyltrifluoroacetone (Complex II inhibitor).
Complex III (ubiquinol cytochrome c reductase) activity was determined by measuring the reduction of oxidized cytochrome c at 550 nm. The Complex III assay was performed in buffer containing (25 mM potassium phosphate (pH 7.4), 1 mM EDTA, 1 mM KCN, 0.6 mM dodecyl maltoside, 32 μM oxidized cytochome c) using purified mitochondria (2.5 μg). The reaction was initiated by adding 35 μM decylubiquinol. The reaction was measured following the linear slope for 1 min in the presence or absence of 2 μM antimycin (Complex III inhibitor). Decylubiquinol was made by dissolving decylubiquinone (10 mg) in 2 mL acidified ethanol (pH 2) and using sodium dithionite as a reducing agent. Decylubiquinol was further purified by cyclohexane {Degli Esposti, 2001 #21; Birch-Machin, 2001 #9; Ellis, 2005 #283}.
Complex IV (cytochrome c oxidase) activity was determined by measuring the oxidation of reduced ferrocytochrome c at 550 nm. The Complex IV assay was performed in buffer containing (10 mM Tris-HCl and 120 mM KCl (pH 7.0)) using purified mitochondria (5 μg). The reaction was initiated by adding 11 μM reduced ferrocytochrome c and monitoring the slope for 30 sec in the presence or absence of 2.2 mM KCN (Complex IV inhibitor) {Yonetan, 1967 #80; Ellis, 2005 #283}.
Complex I/III (NADH cytochrome c reductase) activity was determined by measuring the reduction of oxidized cytochrome c at 550 nm. The Complex I/III assay was performed in buffer (50 mM potassium phosphate (pH 7.4), 1 mM EDTA, 2 mM KCN, 32 μM oxidized cytochrome c, and 105 μM K2NADH) and was initiated by adding purified mitochondria (10 μg). The reaction was measured for 30 sec with a linear slope in the presence or absence of 1 μM rotenone and 2 μM antimycin (Complex I and III inhibitors) {Birch-Machin, 2001 #9; Degli Esposti, 2001 #21; Ellis, 2005 #283}.
Complex II/III (succinate cytochrome c reductase) activity was measured following the reduction of oxidized cytochrome c at 550 nm. The Complex II/III assay was performed in buffer (25 mM potassium phosphate (pH 7.4), 20 mM succinate, 2 mM KCN, 2 μg/mL rotenone) using purified mitochondria (10 μg). The reaction was initiated by adding 40 μM oxidized cytochrome c in the presence or absence of 2 μM antimycin (Complex III inhibitor) {Birch-Machin, 2001 #9; Ellis, 2005 #283}.
Results
Lipid Composition of B6 and VM NS Brain Mitochondria
The differences between the B6 and VM mice for the content NS mitochondrial lipids are shown in Table 1. The lipid classes were listed according to their relative abundance. The mass content of phosphatidylinositol (PtdIns), PtdSer, and Cer was higher, whereas the content of total choline glycerophospholipids (ChoGpl), in particular PtdCho, was lower in VM mice than in B6 mice. Plasmenylethanolamine (plasmalogen, PlsEtn) was significantly higher in VM mice than in B6 mice. Plasmanylethanolamine (PakEtn) was detected only in the NS mitochondria of VM mice. No differences were found between the strains for the mass content of PtdEtn, Ptd2Gro, CerPCho, PlsCho, or PakCho. Previously, it was reported that the ratios of PC/PE and SM/PE were accurate indicators of a change in mitochondrial membrane fluidity. Bangur et al. 1995 showed that differences in these ratios correlated to changes in membrane fluidity as determined by fluorescence polarization {Bangur, 1995 #3}. The ratio of ChoGpl to EtnGpl in the B6 and VM mice was 0.693 and 0.333, respectively, whereas the ratio of CerPCho to EtnGpl in these strains was 0.028 and 0.010, respectively. Myelin-enriched lipids, sulfatides and cerebrosides, were not detected in B6 or VM NS mitochondria (data not shown). Our previous findings also showed that gangliosides are not present in NS mitochondria from B6 mice (Kiebish et al 2008). Corresponding lipid molecular species mass content can be found in supplementary Tables 4–12.
Table 1.
Lipid composition of non-synaptic brain mitochondria in B6 and VM mice
| Lipid | B6 | VM |
|---|---|---|
| Ethanolamine glycerophospholipids | 187.4 ± 12.1 | 264.0 ± 4.8* |
| Phosphatidylethanolamine | 164.9 ± 10.0 | 186.1 ± 5.9 |
| Plasmenylethanolamine | 22.5 ± 2.2 | 72.2 ± 3.7* |
| Plasmanylethanolamine | N.D. | 5.7 ± 0.9* |
| Choline glycerophospholipids | 129.9 ± 7.7 | 87.9 ± 12.9* |
| Phosphatidylcholine | 119.6 ± 5.3 | 74.9 ± 13.2* |
| Plasmenylcholine | 1.2 ± 0.1 | 4.2 ± 4.2 |
| Plasmanylcholine | 9.1 ± 3.2 | 8.8 ± 4.1 |
| Cardiolipin | 52.7 ± 4.5 | 50.0 ± 2.4 |
| Phosphatidylinositol | 9.3 ± 0.8 | 12.5 ± 0.8* |
| Phosphatidylglycerol | 7.1 ± 0.5 | 8.2 ± 0.5 |
| Sphingomyelin | 5.3 ± 1.2 | 2.7 ± 0.2 |
| Phosphatidylserine | 4.6 ± 1.5 | 18.6 ± 2.0** |
| Lysophosphatidylcholine | 2.7 ± 0.6 | 4.7 ± 0.9 |
| Ceramide | 0.7 ± 0.2 | 2.0 ± 0.1* |
Values are expressed as nmol/mg protein ± S.D.M. of three independent samples
Astericks indicate that the value in VM mice differ from B6 mice at
: P < 0.02;
: P < 0.001 as determined from the two-tailed t-test
N.D. - Not detected
Cardiolipin Molecular Species Distribution in B6 and VM NS Brain Mitochondria
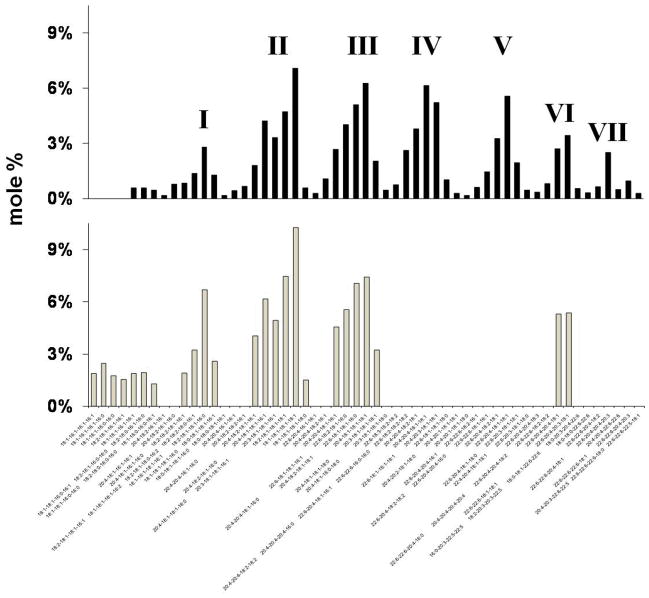
B6 mice contained 95 molecular species of Ptd2Gro, whereas VM mice contained only 45 molecular species (suppl Table 6). In addition, some Ptd2Gro molecular species found in the VM mice (18:1-16:1-16:1-16:1, 18:1-16:1-16:1-16:0, 18:1-16:1-16:0-16:0, 18:2-18:1-16:1-16:1) were not found in the B6 mice. These species may represent newly synthesized forms of Ptd2Gro based on their shorter chain lengths and degree of unsaturation. These fatty acids are also similar to those found in PtdGro, the precursor of Ptd2Gro (suppl Table 8). When presented as mole percentage relative to the distribution of molecular species based on mass to charge ratios, B6 Ptd2Gro forms a unique pattern consisting of seven major molecular species groups (Fig 2). Group I has the shortest fatty acid chain lengths and the least degree unsaturation, whereas group VII has the longest chains and the greatest unsaturation (Kiebish 2008). The Ptd2Gro pattern found for VM mice differed markedly from that of B6 mice (Fig 2 and suppl Table 6). The Ptd2Gro in VM mice had elevated amounts of short chain fatty acids and lacked the molecular species found in groups IV, V, and VII. These groups are generally enriched in 20:4 and 22:6 fatty acids, which are reduced in VM Ptd2Gro (Table 2). Cardiolipin molecular species in VM NS mitochondria also contained a higher 16:0, 16:1, 16:2, and 18:1 content compared to B6 NS mitochondria (Table 2). EtnGpl fatty acids varied in VM NS compared to B6 NS mitochondria and only 22:6 fatty acids were lower in VM NS ChoGpl compared to B6 NS ChoGpl.
Fig 2. Distribution of brain cardiolipin molecular species in B6 NS (black bar) and VM NS (grey bar) mitochondria.
Cardiolipin molecular species were arranged according to mass charge. Corresponding molecular species mass content can be found in supplementary Table 6. Molecular species can be arranged into seven groups (I – VII) based on chain length and degree of unsaturation.
Table 2.
Fatty acid percent distribution in phospholipids involved in cardiolipin remodeling
| Fatty Acid | B6
|
VM
|
||||
|---|---|---|---|---|---|---|
| PE
|
PC
|
CL
|
PE
|
PC
|
CL
|
|
| 14:0 | N.D. | 0.2 ± 0.1 | N.D. | 0.7 ± 0.3 | 2.6 ± 3.8 | N.D. |
| 14:1 | N.D. | 0.6 ± 0.1 | N.D. | 0.1 ± 0.0 | 0.7 ± 0.5 | N.D. |
| 16:0 | 12.3 ± 0.3 | 34.3 ± 1.7 | 3.9 ± 0.0 | 13.7 ± 0.1** | 32.8 ± 2.4 | 7.1 ± 0.5** |
| 16:1 | N.D. | 0.7 ± 0.1 | 5.4 ± 0.0 | 1.7 ± 0.1** | 2.1 ± 2.5 | 11.5 ± 1.1* |
| 16:2 | N.D. | N.D. | 0.2 ± 0.0 | 0.1 ± 0.0 | N.D. | 0.4 ± 0.0*** |
| 18:0 | 20.8 ± 0.2 | 7.6 ± 0.2 | 2.0 ± 0.0 | 17.3 ± 0.2*** | 10.7 ± 2.2 | 1.9 ± 0.3 |
| 18:1 | 17.0 ± 0.1 | 18.0 ± 0.7 | 44.7 ± 1.0 | 19.2 ± 0.3** | 15.9 ± 1.9 | 51.3 ± 0.9*** |
| 18:2 | 1.3 ± 0.1 | 16.1 ± 0.4 | 8.2 ± 0.2 | 4.3 ± 0.2*** | 13.3 ± 2.2 | 7.9 ± 0.1 |
| 18:3 | N.D. | 0.1 ± 0.0 | 0.3 ± 0.0 | N.D. | 0.1 ± 0.0 | N.D. |
| 20:0 | 2.6 ± 0.1 | 3.2 ± 1.1 | N.D. | 3.0 ± 0.4 | 3.5 ± 1.4 | N.D. |
| 20:1 | N.D. | 0.1 ± 0.0 | 0.1 ± 0.0 | 3.0 ± 0.3** | 0.2 ± 0.1 | N.D. |
| 20:2 | 1.5 ± 0.2 | 2.7 ± 0.5 | N.D. | 1.1 ± 0.2 | 1.4 ± 1.0 | N.D. |
| 20:3 | 1.9 ± 0.3 | 0.4 ± 0.1 | 2.7 ± 0.1 | 3.7 ± 0.1** | 0.5 ± 0.6 | 1.9 ± 0.0*** |
| 20:4 | 7.8 ± 0.2 | 11.1 ± 0.3 | 21.0 ± 0.3 | 7.7 ± 0.4 | 6.9 ± 2.5 | 12.9 ± 1.5* |
| 22:2 | N.D. | N.D. | N.D. | 0.6 ± 0.3 | N.D. | N.D. |
| 22:3 | 2.7 ± 0.3 | N.D. | N.D. | 0.9 ± 0.2** | N.D. | N.D. |
| 22:4 | 7.2 ± 0.1 | N.D. | 0.1 ± 0.0 | 3.9 ± 0.1*** | N.D. | N.D. |
| 22:5 | 11.3 ± 0.2 | 1.4 ± 0.3 | 0.2 ± 0.1 | 7.9 ± 0.8* | 2.1 ± 0.9 | N.D. |
| 22:6 | 13.6 ± 0.3 | 3.5 ± 0.5 | 11.4 ± 0.4 | 11.1 ± 0.8* | 7.2 ± 1.0* | 5.1 ± 0.5*** |
Values represented as the mean mole percentage distribution of fatty acids ± S.D.M. from 3 independent analyses as determined by electrospray MS Significantly different values from B6 NS mitochondria at
: P < 0.05;
: P< 0.01;
P < 0.001 as determined by the two-tailed t -test
N.D. - not detected
ETC activities in NS mitochondria of B6 and VM mice
The activities of the individual ETC complexes (Complex I, II, III, and IV) as well as those of the linked enzyme complexes (Complex I/III and II/III) are shown in (Table 4). The activities of Complex I, I/III, and II/III were 22%, 51%, and 25% lower and Complex IV was 22% higher in VM mitochondria than in B6 mitochondria (Table 3).
Table 3.
Electron transport chain activities of non-synaptic brain mitochondriaa
| B6 | VM | |
|---|---|---|
|
|
||
|
Complex I
NADH ubiquinone oxidoreductase |
975 ± 64 | 761 ± 48** |
|
Complex II
succinate DCIP oxidoreductase |
292 ± 18 | 316 ± 42 |
|
Complex III
ubiquinol cytochrome c oxidoreductase |
437 ± 71 | 308 ± 46 |
|
Complex IV
cytochrome c oxidase |
179 ± 25 | 230 ± 19* |
|
Complex I/III
NADH cytochrome c oxidoreductase |
269 ± 59 | 133 ± 26* |
| Complex II/III succinate cytochrome c reductase | 309 ± 30 | 230 ± 8* |
Enzyme activities are expressed as (nmol/min/mg protein) ± S.D.M. (N=3–4)
Signficantly different values from B6 NS mitochondria at
: P < 0.05;
: P < 0.01 as determined from the two-tailed t-test
Discussion
We found that the NS mitochondrial lipidome differed markedly between the B6 and VM mice. Mitochondrial membrane lipids are known to influence membrane fluidity/permeability, electron transport chain activities, membrane fusion, calcium homeostasis, and ATP synthesis {Daum, 1985 #19; Brand, 2003 #12; Petrushka, 1959 #97; Fleischer, 1962 #99}. EtnGpl and ChoGpl were the most abundant phospholipids in the NS mitochondria of both mouse strains. Previous studies showed that changes in the ChoGpl/EtnGpl ratio and the CerPCho/EtnGpl ratio could significantly influence mitochondrial membrane fluidity {Bangur, 1995 #3}. We found that these lipid ratios were markedly lower in the VM NS mitochondria than in the B6 mitochondria indicating that mitochondrial membrane fluidity is greater in VM mice than in B6 mice.
The most remarkable difference in NS mitochondrial lipid composition found between the strains was for the distribution of Ptd2Gro molecular species. While B6 mice expressed 95 molecular species that were symmetrically distributed over seven major groups, the VM mice expressed only 45 molecular species that were asymmetrically distributed and contained species not found in the B6 mice. The distribution of brain Ptd2Gro molecular species in B6 NS mitochondria is similar to that in the brains of other mammalian species indicating that the distribution in B6 mitochondria is a conserved pattern (Cheng et al 2008, Kiebish et al 2008). It is also important to mention that the distribution of Ptd2Gro molecular species in brain differs significantly from the distribution in non-neural tissues, which contain predominantly tetralinoleic Ptd2Gro (18:2-18:2-18:2-18:2). The unusual Ptd2Gro distribution in VM brain mitochondria could result from a strain difference in the activities or specificities of phospholipases and/or acyltransferases, which regulate Ptd2Gro remodeling. Also, the observed Ptd2Gro distribution for VM NS mitochondria could not be explained by the random incorporation of fatty acids during Ptd2Gro remodeling as previously shown for B6 NS Ptd2Gro (data not shown) (Kiebish 2008). Nevertheless, the NS mitochondrial lipid differences found between the strains would indicate differences in mitochondrial function.
Consistent with this notion, we found significant differences in the ETC enzyme activities between the strains. Changes in Ptd2Gro molecular speciation can influence the activity of complex I and may affect the mitochondrial membrane environment influencing linked enzyme activities (I/III and II/III) or supercomplex formation {Hoch, 1992 #35; Chicco, 2007 #16; Fry, 1981 #92; Pfeiffer, 2003 #344; Zhang, 2002 #351}. Increased docosahexaenoic (DHA) containing Ptd2Gro results in decreased complex IV activity {Kraffe, 2002 #43; Yamaoka, 1988 #420; Fry, 1980 #330}. A loss of DHA containing Ptd2Gro might therefore produce increased Complex IV activity as we found in the VM strain. As mitochondrial membrane lipids can influence ETC activities {Fry, 1981 #92; Hoch, 1992 #35; Gohil, 2004 #335; Daum, 1985 #19}, it is likely that the differences in lipid composition underlie the differences in ETC activities.
Alterations in brain mitochondrial lipid composition should affect mitochondrial energy production and neurological function. Additionally, disturbances in the brain mitochondrial lipidome could produce a bioenergetic state conducive to brain tumorigenesis. Further studies will be needed to determine if the differences in brain mitochondrial lipid composition between the B6 and VM mouse strains are related to the differences between these strains for the incidence of spontaneous brain tumors.
Supplementary Material
Acknowledgments
We would like to thank Purna Mukherjee, Rena Baek, and John Mantis for helpful discussions. This work was supported by grants from NIH (HD39722), NCI (CA102135), and NIA (AG23168).
References
- 1.Bowling AC, Beal MF. Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sci. 1995;56:1151–1171. doi: 10.1016/0024-3205(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 2.Calabrese V, Scapagnini G, Giuffrida Stella AM, Bates TE, Clark JB. Mitochondrial involvement in brain function and dysfunction: relevance to aging, neurodegenerative disorders and longevity. Neurochem Res. 2001;26:739–764. doi: 10.1023/a:1010955807739. [DOI] [PubMed] [Google Scholar]
- 3.Pope S, Land JM, Heales SJ. Oxidative stress and mitochondrial dysfunction in neurodegeneration: cardiolipin a critical target? Biochim Biophys Acta. 2008 doi: 10.1016/j.bbabio.2008.030011. [DOI] [PubMed] [Google Scholar]
- 4.Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 5.Hoch FL. Cardiolipins and biomembrane function. Biochim Biophys Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 6.Petrushka E, Quastel JH, Scholefield PG. Role of phospholipids in oxidative phosphorylation and mitochondrial structure. Can J Biochem Physiol. 1959;37:989–998. [PubMed] [Google Scholar]
- 7.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- 9.Davey GP, Clark JB. Threshold effects and control of oxidative phosphorylation in nonsynaptic rat brain mitochondria. J Neurochem. 1996;66:1617–1624. doi: 10.1046/j.1471-4159.1996.66041617.x. [DOI] [PubMed] [Google Scholar]
- 10.Han X, Gross RW. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert Rev Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 11.Kiebish MA, Han X, Cheng H, Lunceford A, Clarke CF, Moon H, Chuang JH, Seyfried TN. Lipidomic analysis and electron transport chain activities in C57BL/6J mouse brain mitochondria. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng H, Mancuso DJ, Jiang X, Guan S, Yang J, Yang K, Sun G, Gross RW, Han X. Shotgun lipidomics reveals the temporally dependent, highly diversified cardiolipin profile in the mammalian brain: temporally coordinated postnatal diversification of cardiolipin molecular species with neuronal remodeling. Biochemistry. 2008;47:5869–5880. doi: 10.1021/bi7023282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedell MA, Jenkins NA, Copeland NG. Mouse models of human disease. Part I: techniques and resources for genetic analysis in mice. Genes Dev. 1997;11:1–10. doi: 10.1101/gad.11.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Bedell MA, Largaespada DA, Jenkins NA, Copeland NG. Mouse models of human disease. Part II: recent progress and future directions. Genes Dev. 1997;11:11–43. doi: 10.1101/gad.11.1.11. [DOI] [PubMed] [Google Scholar]
- 15.Fraser H. Astrocytomas in an inbred mouse strain. J Pathol. 1971;103:266–270. doi: 10.1002/path.1711030410. [DOI] [PubMed] [Google Scholar]
- 16.Huysentruyt LC, Mukherjee P, Banerjee D, Shelton LM, Seyfried TN. Metastatic cancer cells with macrophage properties: Evidence from a new murine tumor model. Int J Cancer. 2008;123:73–84. doi: 10.1002/ijc.23492. [DOI] [PubMed] [Google Scholar]
- 17.Fraser H. Brain tumours in mice, with particular reference to astrocytoma. Food Chem Toxicol. 1986;24:105–111. doi: 10.1016/0278-6915(86)90344-3. [DOI] [PubMed] [Google Scholar]
- 18.Warburg O. The metabolism of tumours. Richard R. Smith Inc; New York: 1931. [Google Scholar]
- 19.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 20.Cavalli LR, Liang BC. Mutagenesis, tumorigenicity, and apoptosis: are the mitochondria involved? Mutat Res. 1998;398:19–26. doi: 10.1016/s0027-5107(97)00223-6. [DOI] [PubMed] [Google Scholar]
- 21.Augenlicht LH, Heerdt BG. Mitochondria: integrators in tumorigenesis? Nat Genet. 2001;28:104–105. doi: 10.1038/88800. [DOI] [PubMed] [Google Scholar]
- 22.Lai JC, Clark JB. Preparation and properties of mitochondria derived from synaptosomes. Biochem J. 1976;154:423–432. doi: 10.1042/bj1540423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai JC, Walsh JM, Dennis SC, Clark JB. Synaptic and nonsynaptic mitochondria from rat brain: isolation and characterization. J Neurochem. 1977;28:625–631. doi: 10.1111/j.1471-4159.1977.tb10434.x. [DOI] [PubMed] [Google Scholar]
- 24.Mena EE, Hoeser CA, Moore BW. An improved method of preparing rat brain synaptic membranes. Elimination of a contaminating membrane containing 2′, 3′-cyclic nucleotide 3′-phosphohydrolase activity. Brain Res. 1980;188:207–231. doi: 10.1016/0006-8993(80)90569-7. [DOI] [PubMed] [Google Scholar]
- 25.Dagani F, Gorini A, Polgatti M, Villa RF, Benzi G. Synaptic and non-synaptic mitochondria from rat cerebral cortex. Characterization and effect of pharmacological treatment on some enzyme activities related to energy transduction. Farmaco (Sci) 1983;38:584–594. [PubMed] [Google Scholar]
- 26.Rendon A, Masmoudi A. Purification of non-synaptic and synaptic mitochondria and plasma membranes from rat brain by a rapid Percoll gradient procedure. J Neurosci Methods. 1985;14:41–51. doi: 10.1016/0165-0270(85)90113-x. [DOI] [PubMed] [Google Scholar]
- 27.Battino M, Bertoli E, Formiggini G, Sassi S, Gorini A, Villa RF, Lenaz G. Structural and functional aspects of the respiratory chain of synaptic and nonsynaptic mitochondria derived from selected brain regions. J Bioenerg Biomembr. 1991;23:345–363. doi: 10.1007/BF00762227. [DOI] [PubMed] [Google Scholar]
- 28.Brown MR, Sullivan PG, Geddes JW. Synaptic mitochondria are more susceptible to Ca2+ overload than nonsynaptic mitochondria. J Biol Chem. 2006;281:11658–11668. doi: 10.1074/jbc.M510303200. [DOI] [PubMed] [Google Scholar]
- 29.Cheng H, Guan S, Han X. Abundance of triacylglycerols in ganglia and their depletion in diabetic mice: implications for the role of altered triacylglycerols in diabetic neuropathy. J Neurochem. 2006;97:1288–1300. doi: 10.1111/j.1471-4159.2006.03794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han X, Yang J, Cheng H, Ye H, Gross RW. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Birch-Machin MA, Turnbull DM. Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell Biol. 2001;65:97–117. doi: 10.1016/s0091-679x(01)65006-4. [DOI] [PubMed] [Google Scholar]
- 32.Ellis CE, Murphy EJ, Mitchell DC, Golovko MY, Scaglia F, Barcelo-Coblijn GC, Nussbaum RL. Mitochondrial lipid abnormality and electron transport chain impairment in mice lacking alpha-synuclein. Mol Cell Biol. 2005;25:10190–10201. doi: 10.1128/MCB.25.22.10190-10201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King TE. Preparation of succinate dehydrogenase and reconstitution of succinate oxidase. In: Estabrook RW, Pullman ME, editors. Methods enzymol. Academic Press; New York: 1967. pp. 322–331. [Google Scholar]
- 34.Degli Esposti M. Assessing functional integrity of mitochondria in vitro and in vivo. Methods Cell Biol. 2001;65:75–96. doi: 10.1016/s0091-679x(01)65005-2. [DOI] [PubMed] [Google Scholar]
- 35.Yonetan T. Cytochrome oxidase: beef heart. In: Estabrook RW, Pullman ME, editors. Methods enzymol. Academic Press; New York: 1967. pp. 332–335. [Google Scholar]
- 36.Bangur CS, Howland JL, Katyare SS. Thyroid hormone treatment alters phospholipid composition and membrane fluidity of rat brain mitochondria. Biochem J. 1995;305(Pt 1):29–32. doi: 10.1042/bj3050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brand MD, Turner N, Ocloo A, Else PL, Hulbert AJ. Proton conductance and fatty acyl composition of liver mitochondria correlates with body mass in birds. Biochem J. 2003;376:741–748. doi: 10.1042/BJ20030984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleischer S, Brierley G, Klouwen H, Slautterback DB. Studies of the electron transfer system. 47. The role of phospholipids in electron transfer. J Biol Chem. 1962;237:3264–3272. [PubMed] [Google Scholar]
- 39.Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chem Phys lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2007 doi: 10.1194/JLR.R700018JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 42.Fry M, Green DE. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J Biol Chem. 1981;256:1874–1880. [PubMed] [Google Scholar]
- 43.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 45.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 46.Kraffe E, Soudant P, Marty Y, Kervarec N, Jehan P. Evidence of a tetradocosahexaenoic cardiolipin in some marine bivalves. Lipids. 2002;37:507–514. doi: 10.1007/s11745-002-0925-z. [DOI] [PubMed] [Google Scholar]
- 47.Yamaoka S, Urade R, Kito M. Mitochondrial function in rats is affected by modification of membrane phospholipids with dietary sardine oil. J Nutr. 1988;118:290–296. doi: 10.1093/jn/118.3.290. [DOI] [PubMed] [Google Scholar]
- 48.Fry M, Blondin GA, Green DE. The localization of tightly bound cardiolipin in cytochrome oxidase. J Biol Chem. 1980;255:9967–9970. [PubMed] [Google Scholar]
- 49.Gohil VM, Hayes P, Matsuyama S, Schagger H, Schlame M, Greenberg ML. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J Biol Chem. 2004;279:42612–42618. doi: 10.1074/jbc.M402545200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.