Abstract
Th17 cells are central to the pathogenesis of autoimmune disease, and recently specific noncoding microRNAs (miRNAs) have been shown to regulate their development. However, it remains unclear if miRNAs are also involved in modulating Th17 cell effector functions. Consequently, we examined the role of miR-155 in differentiated Th17 cells during their induction of Experimental Autoimmune Encephalomyelitis (EAE). Using adoptive transfer experiments, we found that highly purified, MOG antigen-specific Th17 cells lacking miR-155 were defective in their capacity to cause EAE. Gene expression profiling of purified miR-155−/− IL-17F+ Th17 cells identified a subset of effector genes that are dependent upon miR-155 for their proper expression through a mechanism involving repression of the transcription factor Ets1. Among the genes reduced in the absence of miR-155 was IL-23R, resulting in miR-155−/− Th17 cells being hypo-responsive to IL-23. Taken together, our study demonstrates a critical role for miR-155 in Th17 cells as they unleash autoimmune inflammation, and finds that this occurs through a signaling network involving miR-155, Ets1 and the clinically relevant IL-23-IL-23R pathway.
Introduction
Autoimmunity occurs when dysregulated, auto-reactive immune cells inappropriately respond to self-antigens and cause unwarranted inflammation that is destructive to sophisticated tissue systems (1). Recently, T helper 17 (Th17) cells, a subset of CD4+ T cells defined by their expression of interleukin 17 (IL-17) cytokines, have emerged as key drivers of tissue inflammation. Th17 cells promote both the onset and persistence of inflammatory responses during autoimmune disorders including Multiple Sclerosis (MS), Arthritis, Psoriasis, Lupus and Inflammatory Bowel Disease (IBD) (2, 3).
Because of their central roles in driving disease, significant effort has gone into understanding the genes and pathways that regulate Th17 cell development. Skewing of naïve CD4+ T cells towards the Th17 lineage is driven by the cytokines IL-6 and TGFβ, which induce Th17 cell signature genes through such factors as Stat3, RORγt, Ahr, Batf, and Irf4 (4–12). Furthermore, differentiated Th17 cells must receive additional signals from cytokines such as IL-23 to expand and achieve full inflammatory potential in vivo (13–17). Recently, noncoding microRNAs (miRNAs) have also been found to regulate Th17 cell development (18, 19). However, how miRNAs fit into the known regulatory circuits underlying Th17 cell biology remains an important area of investigation.
miRNAs are small, single-stranded RNA molecules that negatively regulate target gene expression post-transcriptionally. Specific miRNAs have been shown to support proper development of immune cells in mammals, and have just recently been implicated in autoimmunity (20, 21). Among the miRNAs expressed in immune cells is miR-155, which modulates the development of various inflammatory T cell subsets, including Th1, Th17 and Treg cells (18, 22–26). Demonstrating its importance during inflammation in vivo, we and others have recently shown that miR-155−/− mice are highly resistant to distinct mouse models of autoimmunity, including experimental autoimmune encephalomyelitis (EAE) – a model of human Multiple Sclerosis, T cell-dependent colitis, and collagen-induced arthritis (18, 24, 27). Furthermore, dysregulated expression of miR-155 is also observed in mice and people with various types of autoimmune disorders (24, 28–30). Although these reports reveal a prominent, clinically relevant role for miR-155 during autoimmunity, they have not determined if this is a consequence of reduced inflammatory T cell numbers or compromised effector cell function. Furthermore, it remains unclear how miR-155 functions at the molecular level to instruct Th17 cell biology.
In the present study, we have investigated the role of miR-155 in differentiated Th17 cells. This was accomplished by generating a novel mouse strain that allows for the isolation and analysis of viable, miR-155−/− IL-17F-expressing CD4+ T cells that specifically recognize the Myelin Oligodendrocyte Glycoprotein (MOG) 35–55 antigen. Our studies find that purified miR-155−/− Th17 cells are extremely defective in causing EAE following adoptive transfer when compared to Wt controls. Furthermore, we demonstrate that miR-155 directly targets the transcription factor Ets1 to regulate a subset of Th17 cell-effector genes, which includes the IL-23R. Consequently, miR-155−/− Th17 cells are hypo-responsive to IL-23, revealing a new link between miR-155 and the highly relevant IL-23-IL-23R pathway (31–36).
Materials and Methods
Mice
All experiments were approved by the University of Utah Institutional Animal Care and Use Committee (IACUC). miR-155+/+, miR-155−/−, Rag1−/−, IL-17F RFP+/−, 2D2 TCR Tg, and combinations of these mice are all on a C57BL/6 genetic background. miR-155−/− mice were crossed with IL-17F RFP+/+ to create miR-155+/− IL-17F RFP+/− mice, which were crossed further to create both miR-155+/+ and miR-155−/− IL-17F RFP+/− mice. miR-155+/+ IL-17F RFP+/+ and miR-155−/− IL-17F RFP+/+ mice were crossed with Wt 2D2+ to create miR-155+/+ IL-17F RFP+/− 2D2+ and miR-155−/− IL-17F RFP+/− 2D2+ mice, respectively. Genotyping was performed as described previously (22, 37, 38).
Mouse Models of EAE
EAE was induced in mice as described previously (18). Briefly, MOG35–55 peptide (BD) was emulsified in complete Freund's adjuvant (CFA) (100 µg/ml) and injected s.c. into the base of the mouse’s tail. Pertussis toxin was injected intraperitoneally into mice on days 0 and 2. Clinical symptoms of EAE were scored according to the following criteria: 0, no symptoms; 0.5, partially limp tail; 1, completely limp tail; 1.5, impaired righting reflex; 2, hind limb paresis; 2.5, hindlimb paralysis; 3, forelimb weakness; 4, complete paralysis; 5, death.
Cell Culture and Retrovirus Infection
For Th17 cell-skewing, CD4+ splenocytes were isolated by using a CD4+ T cell isolation kit (Miltenyi Biotec) per manufacturer’s instruction. The CD4+ T cells were cultured in RPMI supplemented with plate-bound αCD3e (5 µg/ml), soluble αCD28 (2 µg/ml), IL-6 (50 ng/ml) and TGF-β (3 ng/ml) (Biolegend) for 72 hrs. For some experiments αIL-4 (5 µg/ml) and αIFNγ antibodies (5 µg/ml) were supplemented to increase the differentiation of Th17 cells, or IMDM medium was used instead of RPMI. For delivery of the control or Ets1 shRNA-expressing retrovirus or MIG-IL-23R (Addgene 24066) (39) retrovirus, CD4+ T cells were cultured in RPMI with αCD3e and αCD28 for 24 hours. The next day cells were spin-infected with retrovector-containing medium (2500 rpm, 30°C) for 90 min and cultured in Th17-skewing conditions. The retrovirus was produced by transfecting 293T cells as described previously (40). Infected cells were GFP+ while IL-17F+ cells were RFP+ during FACS analyses.
Intracellular Staining and Flow Cytometry
For analyses of IL-17F-expressing RFP+ Th17 cells, cells were stained with fluorophoreconjugated αCD4 antibodies followed by FACS analysis using a BD LSRFortessa machine and FlowJo software. To detect intracellular expression of IL-17A or IFNγ, splenocytes, LNs or brain cells (purified with Percoll) were isolated from mice and seeded in 96 well plates. Cells were treated with 750 ng/ml ionomycin, 50 ng/ml phorbol myristate acetate (PMA) (Calbiochem) and 1 µl/well of GolgiPlug (BD Biosciences) for 4–5 hr at 37°C. Cells were subsequently surface stained with fluorphore-conjugated CD4 and CD3e antibodies followed by permeabilization and fixation. After washing, cells were stained with fluorophore-conjugated IL-17A antibody (eBioscience) for 20 min at 4°C and further analyzed by FACS. In some experiments, control or Ets1 shRNA-infected GFP+RFP+ cells were FACS sorted and stained to detect the expression of IL-17A. For p-STAT3 staining, RFP+ Th17 cells were sorted after in vitro skewing and re-plated in 96 well plates. Cells were treated with IL-23 and subjected to p-STAT3 staining [BD biosciences phosflow mouse anti-Stat3 (pY705)] per manufacturer's instruction.
qPCR and Microarray
To analyze the relative levels of mRNAs encoding BIC, IL-17F, IL-17A, IL-23R, IL-22, Ets1, IL-2 or L32, sybrgreen-based quantitative real-time PCR (qPCR) was conducted with the LightCycler 480 PCR system (Roche) and gene-specific primers. Primer sequences are available upon request. For all experiments, mRNA was normalized to L32.
For the microarray analysis, CD4+ T cells were purified from both miR-155+/+ and miR-155−/− IL-17F RFP+/− mouse spleens and cultured in vitro with Th17 skewing conditions. CD4+RFP+ and CD4+RFP- cells were sorted for both genotypes. Total RNA was isolated using the RNeasy kit (Qiagen). Global mRNA levels were assayed using the SurePrint G3 Mouse GE 8×60K Microarray Kit (Agilent), which was carried out by the University of Utah core facility (https://bioserver.hci.utah.edu/microarrayweb/ordering.html). The data were analyzed further using Genesifter and Ingenuity software. All data have been deposited into the NCBI Geo database under accession number GSE45122 (http://www.ncbi.nlm.nih.gov/geo/).
ELISAs
ELISAs to detect expression of IL-17A and IFN-γ were performed with cytokine-specific kits (eBioscience) according to the manufacturer's instructions.
Immunoblotting
Cell pellets were lysed in 8M urea buffer. Protein extracts were subjected to gel electrophoresis and transferred onto a nitrocellulose membrane followed by antibody staining (Ets1 and β-actin, Santa Cruz Biotechnology) and detection as described (40). Expression levels were quantified using NIH ImageJ software.
Luciferase Reporter Assays
A region of the 3’ UTR of mouse Ets1 containing the conserved miR-155 binding site was cloned downstream from luciferase in the pmiReport plasmid. Site directed mutagenesis was used to disrupt the seed sequence. The forward and reverse primer sequences for cloning Ets1 are gtaactagtTACCCGAAACATGGAAGACTC and gttaagcttTGCACAGCATGGCTAGGA, respectively. The restriction enzyme sites are in lower case. Luciferase assays were carried out as described previously (40). In brief, 293T cells were transfected with combinations of a miR-155 expression plasmid, a luciferase expression construct with a 3’ UTR and a b-gal expressing plasmid for normalization.
Statistical Analysis
Statistical significance was determined by performing a Student’s two-tailed t test. p values < 0.05 were considered significant. For the Ingenuity Transcription Factor Analysis, the z-score is calculated by the Ingenuity Pathway Analysis (IPA) software and represents the confidence that a transcription factor is activated or repressed based on the consistency between the direction of change of known targets of the transcription regulator from the microarray data set and what is expected from literature.
Results
Generation and characterization of miR-155−/− IL-17F RFP reporter mice
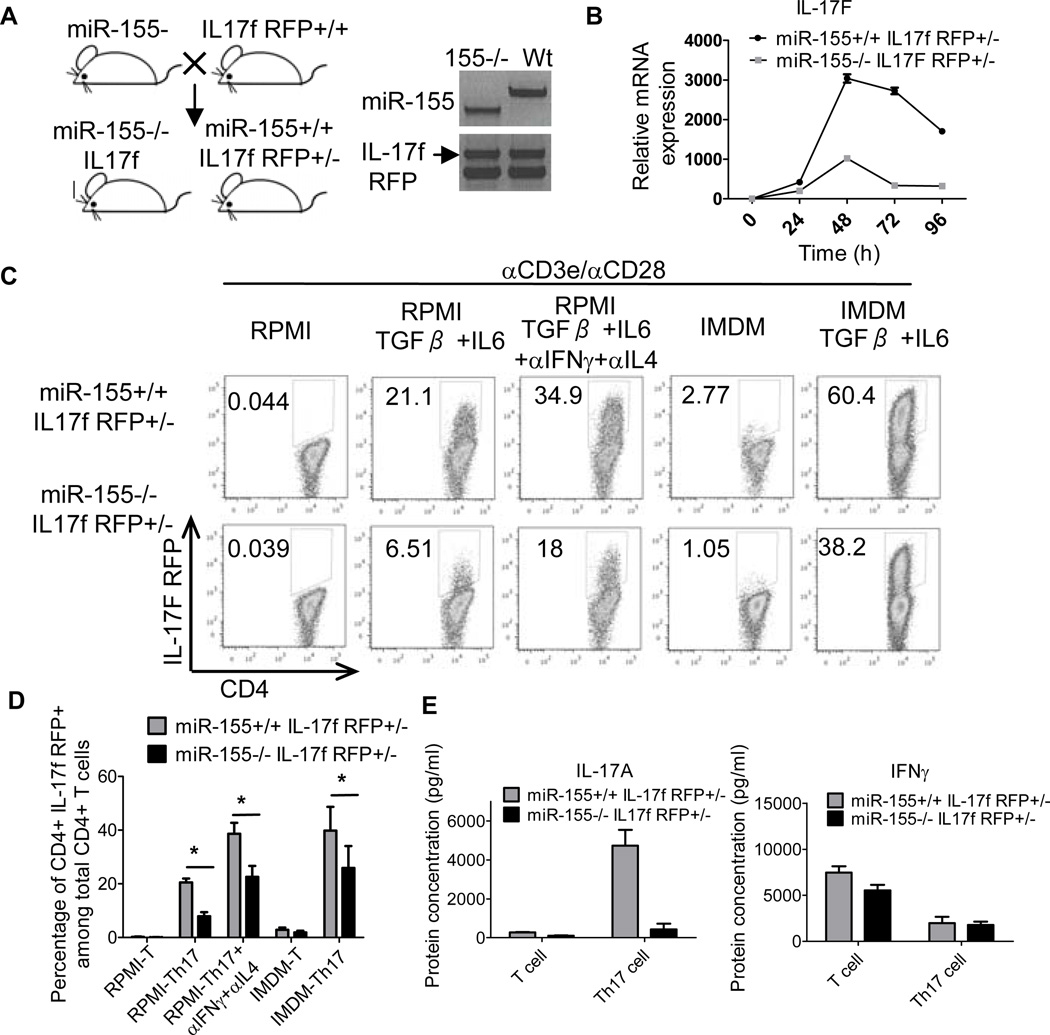
To investigate the role of miR-155 in Th17 cell function, and to circumvent the limitations of using intracellular staining to identify Th17 cells, we developed a mouse strain where miR-155−/− Th17 cells are marked with the red fluorescent protein (RFP) under the control of the IL-17F locus (Fig. 1A). IL-17F RFP reporter mice have been described previously and shown to faithfully mark Th17 cells both in vitro and in vivo (38). To retain IL-17F function, we created both miR-155+/+ and miR-155−/− IL-17F reporter mice that are heterozygous for the IL-17F RFP reporter gene. This approach disrupts one allele of the endogenous IL-17F gene and leaves the other intact to be expressed during Th17 cell development (Fig. 1B). We observed defective expression of IL-17F mRNA in miR-155−/− IL-17F+/− RFP reporter CD4+ T cells compared to miR-155+/+ control cells under Th17 cell skewing conditions (Fig. 1B).
Figure 1. Creation of a miR-155−/− IL-17F RFP reporter mouse strain.
(A) Generation of miR-155+/+ and miR-155−/− IL-17F RFP reporter mouse strains. Left: Schematic diagram showing the mouse cross. Right: Genotyping results demonstrating completion of the desired mouse strains. (B) Expression of IL-17F mRNA by CD4+ T cells from the indicated genotypes cultured under Th17 cell-skewing conditions was assayed by qPCR over a timecourse. (C-E) CD4+ T cells were isolated from miR-155+/+ and miR-155−/− CD4+ IL17F RFP+/− mice and cultured in vitro under different Th17 cell-skewing conditions. (C) After 72 hours, expression of RFP was assayed by FACS. (D) Average percentage of IL-17F+ cells in different in vitro culture conditions (n=4). (E) ELISA of IL-17A from in vitro cultured CD4+ T cells. Error bars represent +/− SEM. * denotes a p value of <0.05.
Next, we analyzed the capacity of CD4+ T cells from both miR-155+/+ and miR-155−/− IL-17F RFP+/− reporter transgenic mice to express the RFP reporter gene under a variety of different Th17 cell-skewing conditions. Impaired Th17 cell differentiation by miR-155−/− IL-17F+ RFP+/− CD4+ T cells compared to miR-155+/+ controls was observed in all cases, as determined by monitoring the formation of RFP-expressing T cells by FACS (Fig. 1C, 1D). Defective IL-17F RFP expression in the absence of miR-155 also occurred when cells were cultured in Iscove's Modified Dulbecco's Media (IMDM), which is a medium rich in aromatic amino acids that promotes higher levels of Th17 cell differentiation by Wt CD4+ T cells compared to more conventional RPMI medium (Fig. 1C, 1D) (41). We observed very few RFP+ cells in both groups under neutral, non-Th17 cell skewing conditions, highlighting the specificity of this reporter system. Impaired Th17 cell development by the miR-155−/− IL-17F RFP+/− CD4+ T cells, compared to miR-155+/+ controls, was further confirmed by ELISA. Results demonstrated defective concentrations of IL-17A, but not IFNγ, in the culture supernatants from miR-155−/− vs. miR-155+/+ IL-17F RFP+/− CD4+ T cells growing under Th17 cell-skewing conditions (Fig. 1E).
We also observed the formation of some IL-17F RFP+ Th17 cells in the absence of miR-155. These data demonstrate an important role for miR-155 in Th17 cell development, and validate a novel reagent to be used for experiments with purified and viable miR-155+/+ and miR-155−/− IL-17F+ Th17 cells.
miR-155 regulates Th17-effector genes in purified IL17F+ Th17 cells
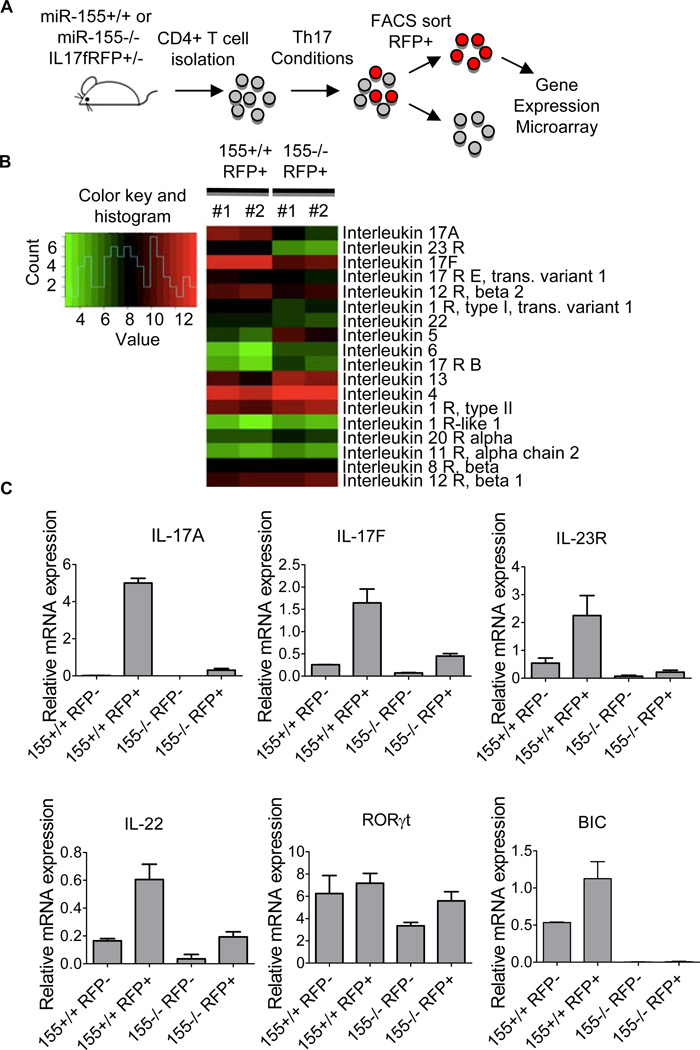
Although naïve miR-155−/− CD4+ T cells are defective in their output of IL-17-expressing CD4+ T cells, a portion of IL-17F+ cells are still produced. However, it is not known if these cells are functional on a per cell basis. To begin answering this question, we performed a gene expression analysis on purified miR-155+/+ and miR-155−/− IL-17F+ Th17 cells (Fig. 2A). Global gene expression profiling was performed using a microarray analysis. Genes differentially regulated by miR-155 in purified IL-17F+ Th17 cells were identified, and this subset contained several genes involved in T cell lineage skewing and Th17 cell effector functions (Supplemental Table S1).
Figure 2. miR-155 regulates Th17-related genes in purified IL17F+ Th17 cells.
(A) Schematic diagram of the experimental design. (B) Heatmap of the differentially expressed interleukin genes in miR-155+/+ versus miR-155−/− IL-17F+ Th17 cells (n=2). The colors represent normalized fluorescence values on a log scale. The color key is next to the heatmap. (C) Relative expression levels of bic ncRNA and IL-17A, IL-17F, IL-23R, RORgT and IL-22 mRNA in FACS sorted T cells from (B) were assayed by qPCR (n=2). Error bars represent +/− SEM.
Next, we used Ingenuity software to mine our microarray results and uncover canonical pathways most impacted by miR-155. This approach identified T helper cell differentiation as the most significant biological pathway impacted by miR-155, and altered B and T cell signaling in Rheumatoid Arthritis as the second most effected process. Both of these are consistent with a critical role for miR-155 in Th17 cell biology during autoimmunity.
Many of these pathway differences are represented by the expression of specific interleukin genes, which are either elevated or decreased in miR-155−/− IL-17F+ Th17 cells (Fig. 2B). Within this gene subset, we detected several that have been linked to Th17 cell biology, including IL-17A, IL-17F, IL-23R and IL-22, that were decreased in miR-155−/− IL-17F+ Th17 cells compared to miR-155+/+ controls. Alternatively, cytokines representing other Th lineages, such as IL-4, IL-5 and IL-13, were increased in miR-155−/− IL-17F+ Th17 cells (Fig. 2B). qPCR was performed to validate these defects in Th17-related genes expression profiles observed in miR-155−/− IL-17F+ Th17 cells, and among those assayed were IL-17A, IL-17F, IL-23R and IL-22 (Fig. 2C and Supplemental Fig. S1). We also assessed RORγT mRNA levels, and found minimal differences between the groups (Fig 2C). These data demonstrate that miR-155 is not only important for the development of IL-17F-expressing cells, but also critical for the expression of Th17 cell effector genes in differentiated, IL-17F-expressing Th17 cells.
miR-155 directly targets Ets1 in IL-17F+ Th17 cells
We next sought to identify mRNA targets of miR-155 with relevance to Th17 cell biology. To systematically search for potential miR-155 targets, we further analyzed our microarray data from Fig. 2 using an Ingenuity transcription factor analysis. Transcription factors predicted to be activated or inhibited in miR-155−/− compared to miR-155+/+ IL-17F+ Th17 cells based upon differential gene expression signatures between these two cell groups were identified (Fig. 3A). Since miR-155 inhibits expression of its targets, those transcription factors with a predicted increase in their levels in miR-155−/− cells were of interest (Fig. 3A, red line). On this list were several factors containing a conserved miR-155 binding site in their 3’ UTRs, according to Targetscan software, and these include Ets1, AP1 (cFos), C/EBPβ and cMAF. Among these, our attention was drawn to Ets1 for several reasons: 1) Ets1 was the most likely transcription factor to be activated in miR-155−/− IL-17F+ Th17 cells according to our analysis (Fig. 3A), 2) it has a highly conserved bonding site for miR-155 in its 3’ UTR (Fig. 3B), and 3) it has an established function as an inhibitor of Th17 cells (42).
Figure 3. miR-155 directly targets Ets1 in Th17 cells.
(A) Predicted transcription factors targeted by miR-155 according to an Ingenuity transcription factor analysis of the microarray data from Fig. 2. (B) Conserved miR-155 binding sites in the Ets1 3’UTR, and predicted interaction with the conserved 8-mer seed found within miR-155. (C) Luciferase assays were performed and found that the Ets1 3’ UTR has a functional miR-155 target site. The Picalm 3’ UTR and the miR-155 2-mer were included as positive controls, and the TRAF6 3’ UTR was a negative control. (D) miR-155+/+ or miR-155−/− IL-17F RFP+/− CD4+ T cells were cultured under Th17 skewing conditions. 72 hours later, Ets1 protein levels were assayed by Western blotting (left), which was quantified for multiple samples (right) (n=5). Error bars represent +/− SEM. * denotes a p value of <0.05.
To determine if miR-155 directly targets mouse Ets1, we cloned a region of the Ets1 3’ UTR containing the conserved miR-155 binding site downstream from luciferase and performed luciferase assays using 293T cells. Results indicated that miR-155 overexpression repressed luciferase in an Ets1 3’ UTR-dependent manner (Figure 3C). Furthermore, mutation of the miR-155 binding site in the 3’ UTR abolished repression by miR-155, indicating direct targeting. We next tested if miR-155 represses endogenous Ets1 in Th17 cells by assaying Ets1 expression in miR-155+/+ and miR-155−/− IL-17F RFP+/− CD4+ T cells under Th17 cell-skewing conditions. Expression of Ets1 was elevated at the protein level in miR-155−/− compared to controls (Fig. 3D). Together, these results identify Ets1 as a direct target of miR-155 in Th17 cells.
Ets1 is a functionally relevant target of miR-155 in IL-17F+ Th17 cells
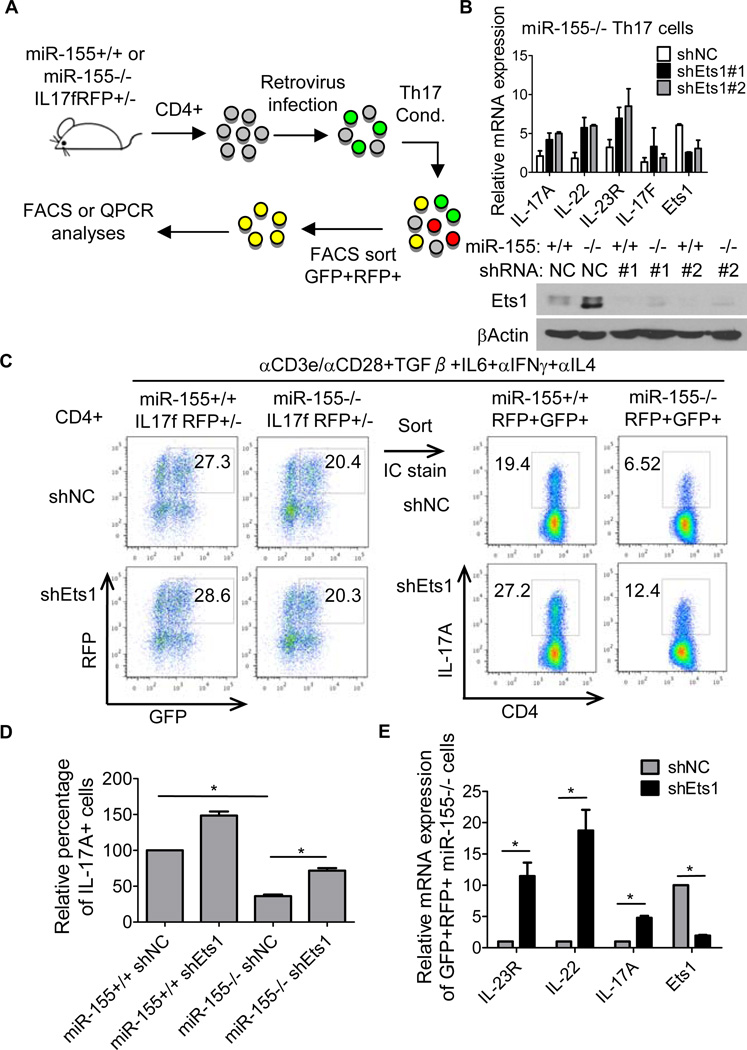
Following the identification of Ets1 as a direct target of miR-155 during Th17 development, we next determined if reducing its levels would impact expression of Th17-related genes in miR-155−/− CD4+ Th17 cells. To accomplish this, we built a retroviral vector that expresses an Ets1 shRNA based upon a format that we have described previously (43). Using 2 different Ets1 shRNAs or a control vector, we transduced miR-155−/− CD4+ T cells and assayed the expression of Ets1 and Th17-related genes under Th17 cell-skewing conditions (Fig. 4A). Ets1 protein levels were reduced in cells expressing the shRNA cassette when compared to the control vector (Fig. 4B). Ets1 knockdown in miR-155−/− Th17 cells led to up-regulation of several Th17 related genes, including IL-17A, IL-23R and IL-22 (Fig. 4B). Of note, Ets1 knockdown did not affect IL-17F expression.
Figure 4. Ets1 is a functionally relevant target of miR-155 in Th17 cells.
(A) Schematic of experimental design. (B) miR-155+/+ or miR-155−/− IL-17F RFP+/− CD4+ T cells were isolated and infected with a control (NC) or Ets1 shRNA expressing retrovirus. After culturing under Th17 cell-skewing conditions for 72 hrs, RNA or protein was extracted and expression of the indicated Th17-related genes were analyzed by qPCR (Upper panel) or Ets1 by Western blotting (Lower panel). (C) miR-155+/+ or miR-155−/−RFP+GFP+ T cells were sorted by FACS and subjected to intracellular staining for IL-17A. (D) Graph of the average relative percentage of IL-17A+ cells in sorted miR-155+/+ or miR-155−/− RFP+GFP+ cells transduced with the control or Ets1 shRNA expressing retroviral vector (n=4, 2 independent experiments). (E) Expression of Th17 cell effector genes in sorted miR-155−/− RFP+GFP+ cells transduced with the control or Ets1 shRNA expressing retroviral vector were analyzed by qPCR (n=4). Error bars represent +/− SEM. * denotes a p value of <0.05.
To further define the role of Ets1 in purified Th17 cells, we took advantage of our IL-17F reporter system and sorted out RFP+ (IL-17F+) GFP+ (vector+) cells from both miR-155+/+ and miR-155−/− CD4+T cells under Th17 cell-skewing conditions (Fig. 4A). Analysis of these RFP+GFP+ cells by intracellular staining and FACS revealed defective expression of IL-17A in the absence of miR-155 compared to Wt controls, and this was statistically significant (Fig. 4C, 4D). Notably, knockdown of Ets1 in miR-155−/− cells led to increased amounts of IL-17A+ cells, albeit this did not reach control levels (Fig. 4C, 4D). Moreover, Ets1 knockdown significantly increased the expression of other Th17-related genes in miR-155−/− RFP+GFP+ cells, and these included IL-17A, IL-23R and IL-22 (Fig. 4E). Taken together, these results indicate that Ets1 is a functionally relevant target of miR-155 in Th17 cells.
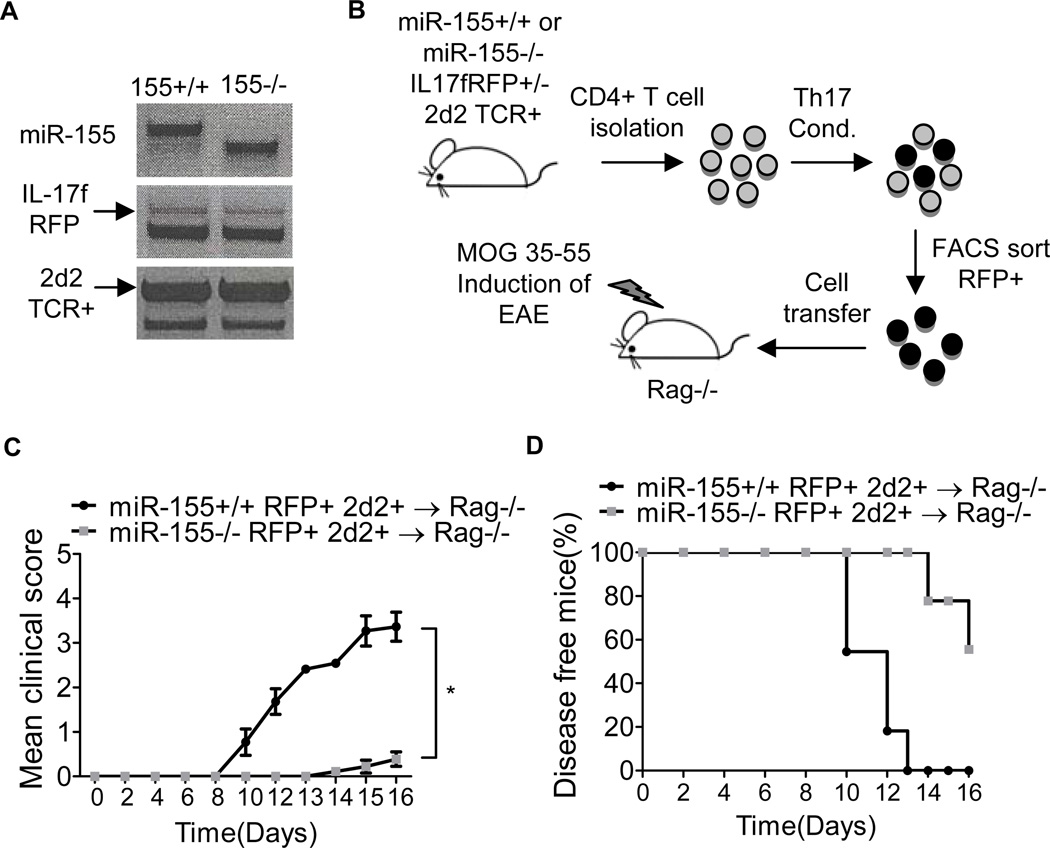
miR-155 promotes the function of IL-17F+ Th17 cells during EAE
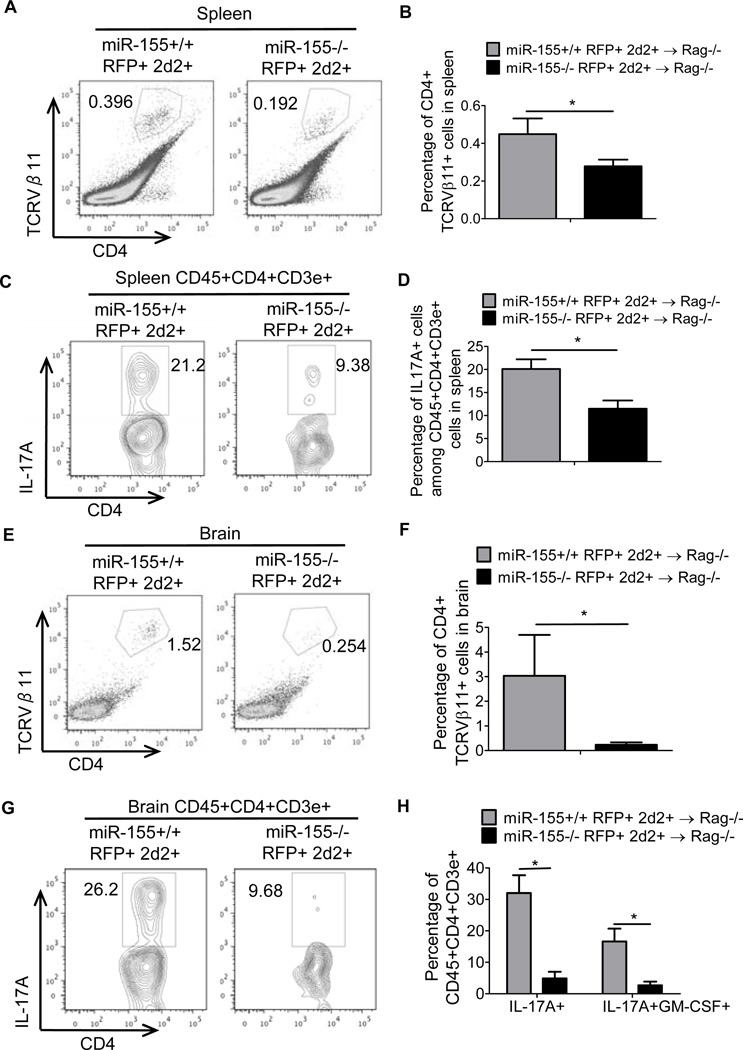
To determine whether miR-155 is required for the functionality of differentiated IL-17F+ Th17 cells, we generated miR-155+/+ and miR-155−/− IL-17F RFP+/− 2d2 TCR Tg+ mice (Fig. 5A). The 2D2 TCR specifically recognizes the MOG35–55 peptide, and T cells expressing this TCR can elicit EAE in mice on a C57BL/6 genetic background (37). By using this new mouse strain, we were able to isolate miR-155+/+ or miR-155−/− IL-17F+ Th17 cells that specifically recognize MOG35–55 and test the importance of miR-155 during Th17 cell encephalogenic function in vivo (Fig. 5B). We adoptively transferred 2 × 104 in vitro generated miR-155+/+ or miR-155−/− IL-17F RFP+ 2d2+ CD4+ T cells into Rag1−/− mice followed by the immunization with MOG35–55 and administration of pertussis toxin (Fig. 5B). Mice receiving miR-155+/+ IL-17F+ 2d2+ Th17 cells had a substantially more severe disease course, with higher clinical scores, compared to mice given miR-155−/− IL-17F+ 2D2+ Th17 cells (Fig. 5C). Rag1−/− mice receiving the miR-155+/+ IL-17F+ 2d2+ Th17 cells also had an earlier onset of disease symptoms, which began on day 9 compared to day 14, and a higher disease incidence compared to mice receiving miR-155−/− IL-17F+ 2D2+ Th17 cells (Fig. 5C, 5D). We also analyzed the brains and spleens from both groups for the presence of transferred IL-17F+ Th17 cells. Consistent with the reduced EAE symptoms, we observed significantly fewer miR-155−/− compared to miR-155+/+ Vb11+CD4+ T cells in the spleens and brains of mice immunized with MOG35–55 (Fig. 6A, 6B, 6E and 6F). Intracellular staining of the recovered T cells revealed that miR-155−/− cell were defective in their expression of IL-17A (Fig. 6C, 6D, 6G and 6H). In addition, we also observed decreased IL17A+GM-CSF+ cells in brain with miR-155−/− cell transfer (Fig. 6H). It is also of note that there were still reduced EAE symptoms in the absence of mIR-155 despite the TCR having a fixed specificity. These data demonstrate that miR-155 is required for both the development and proper function of IL-17F+ Th17 cells during EAE (Supplemental Fig. S2).
Figure 5. miR-155 promotes the function of IL-17F+ 2d2 TCR Tg+ CD4+ T cells during EAE.
(A) Genotyping results confirming the creation of miR-155+/+ or miR-155−/− IL-17F+/− 2D2 TCR Tg+ mice. (B) Schematic of the experimental design. (C-D) CD4+ T cells were isolated from miR-155+/+ and miR-155−/− IL-17F RFP+/− 2D2 TCR Tg+ mice and cultured under Th17 cell-skewing conditions. 72 hrs later, IL-17F RFP+ 2D2 TCR Tg+ CD4+ Th17 cells were sorted by FACS and equal numbers of miR-155+/+ or miR-155−/− cells were injected into Rag1−/− mice. EAE was induced in both groups by immunizing with 100 µg of the MOG35–55 peptide. (C) The disease severity was scored regularly based on clinical symptoms (n=10). (D) Disease incidence was assessed for each group. Error bars represent +/− SEM. * denotes a p value of <0.05.
Figure 6. miR-155 promotes brain infiltration and cytokine production by IL-17F+ 2d2 TCR Tg+ CD4+ T cells during EAE.
CD4+ T cells were isolated from the spleens and brains of mice following experiments as described in Figure 5, and (A–H) FACS analysis of the splenocytes (A–D) or brain cells (E–H) was performed. (A) and (E) show the percentage of transferred miR-155+/+ and miR-155−/− IL-17F+ TCR Vb11+ CD4+ T cells in each organ, while (B) and (F) show data for multiple mice in each group. (C) and (G) are representative FACS plots of IL-17A expression by the recovered T cells, and (D) and (H) show data for multiple mice. The percentage of brain IL-17A+GM-CSF+ cells are also shown in (H). n=5 for the miR-155+/+ groups and n=8 for the miR-155−/− groups. Error bars represent +/− SEM. * denotes a p value of <0.05.
miR-155 is necessary for proper IL-23 responsiveness by IL-17F+ Th17 cells
The IL-23/IL-23R signaling pathway is critical for the expansion of Th17 cells in vivo and the induction of EAE in mice, and polymorphisms in the IL-23R gene have been linked to human autoimmunity (17, 31–33). Since we observed defective expression of the IL-23R in miR-155−/− IL-17F+ Th17 cells (Fig. 2D), and reduced numbers of miR-155−/− Th17 cells during EAE, we speculated that miR-155 is important for the responsiveness of Th17 cells to IL-23. To test this, we sorted Wt and miR-155−/− IL-17F reporter cells following their development in vitro and examined their responsiveness to recombinant IL-23. Following 20 minutes of treatment, miR-155+/+ IL-17F+ CD4+ T cells had significantly higher levels of activated Stat3 compared to miR-155−/− IL-17F+ CD4+ T cells as determined by intracellular staining with an anti-phosphorylated Stat3 antibody (pY705) and FACS (Fig. 7A, 7B). These data indicate that miR-155−/− IL-17F+ Th17 cells are hyporesponsive to IL-23, likely due to reduced IL-23R expression.
Figure 7. miR-155 promotes IL-23 signaling by IL-17F+ Th17 cells.
CD4+ T cells were isolated from miR-155+/+ and miR-155−/− IL-17F RFP+/− mice and cultured in Th17 condition for 72 hrs. RFP+ IL-17F+ cells were sorted by FACS and re-plated in 96 well plates followed by restimulation with IL-23 or iL-6 for 20 min. Cells were subjected to p-STAT3 staining. (A) Representing FACS plot of p-STAT3 staining after IL-23 restimulation. Black lines represent cells without cytokine treatment and red lines represent cells with IL-23. (B) Graphs of the mean fluorescence intensity (MFI) of p-STAT3 (n=4) before and after IL-23 treatment. (C) Schematic of the experimental design. (D) Representative FACS plots of IL-17A expression by MOG35–55 restimulated CD4+ T cells from the brains Wt and miR-155−/− EAE mice with and without IL-23 treatment. (E) Data from (D) are shown graphically for multiple mice. (F) Schematic of the experimental design. CD4+ T cells were isolated from miR-155+/+ and miR-155−/− IL-17F RFP+/− 2D2 TCR Tg+ mice and infected with a control (MIG) or MIG-IL23R expressing retrovirus. After culturing under Th17 cell-skewing conditions for 72 hrs, miR-155+/+ or miR-155−/− IL-17F+ 2D2 TCR Tg+ GFP+ CD4+ T cells were sorted by FACS and equal numbers injected into Rag1−/− mice. (G) EAE was induced following the adoptive cell transfer, and disease severity was scored regularly based on clinical symptoms (n=5 for the miR-155+/+ groups and n=8 for the miR-155−/− groups). (H) Disease incidence was also analyzed for each group (n=5 for miR-155+/+ groups and n=8 for miR-155−/− groups). Error bars represent +/− SEM. * denotes a p value of <0.05.
In another experiment, brain mononuclear cells were collected from Wt or miR-155−/− mice 13 days post immunization with MOG35–55 and re-stimulated with recombinant IL-23 for 3 days. The expansion of IL-17A-expressing cells was determined by FACS. Results demonstrated that Wt CD4+ T cells responded to IL-23 by expressing more IL-17A, while miR-155−/− CD4+ T cells did not have a response to IL-23 (Figure 7C–E). These results further support a role for miR-155 in promoting IL-23 responsiveness by Th17 cells.
Next, we attempted to complement the miR-155−/− Th17 cell phenotype by expressing the IL-23R in miR-155−/− IL-17F+ Th17 cells before their adoptive transfer into Rag1−/− mice. Overexpression of IL-23R was achieved by transducing miR-155+/+ and miR-155−/− 2D2+CD4+ T cells with MIG-IL-23R or MIG control retrovectors, which have been described previously (39). After retroviral transduction and in vitro skewing, IL-17F+ GFP+ (marking MIG-IL-23R or MIG positive cells) 2D2+ Th17 cells were sorted and injected into Rag1−/− mice followed by induction of EAE (Fig. 7F). Rag1−/− mice given MIG-IL23R+ miR-155−/− IL-17F+ 2d2+ Th17 cells had an earlier onset of disease symptoms, which began on day 12 compared to day 15, and significantly higher disease scores compared to mice receiving MIG+ miR-155−/− IL-17F+ 2D2+ Th17 cells (Fig. 7G, 7H). Mice receiving MIG+ or MIG-IL23R+ miR-155+/+ IL-17F+ 2d2+ Th17 cells had similar disease courses, with an earlier disease onset and increased severity of symptoms compared to mice having received MIG+ or MIG-IL-23R+ miR-155−/− IL-17F+ 2D2+ Th17 cells (Fig. 7G, 7H). These data provide evidence that one important function of miR-155 during tissue inflammation is the promotion of IL-23R expression by IL-17F+ Th17 cells.
Discussion
In this report, we evaluated the role of miR-155 in developed Th17 cells and made several important discoveries. First, miR-155 is required for expression of several effector genes in differentiated IL-17F-expressing Th17 cells. Second, miR-155 directly targets the transcription factor Ets1, a known negative regulator of Th17 cell formation. Thus, elevated expression of Ets1 in miR-155−/− Th17 cells inhibits effector gene expression. Third, IL-17F+ Th17 cells are functionally defective if they lack miR-155, and thus largely incapable of causing severe EAE symptoms like miR-155+/+ controls. Fourth, defective IL-23R expression in miR-155−/− Th17 cells results in hypo-responsiveness to IL-23, a pivotal step during Th17 cell pathogenesis. Taken together, these data significantly advance our understanding of how miR-155 impacts autoimmune inflammation during Th17-dependent disorders.
Previous studies have found reduced levels of Th17 cells in miR-155−/− mice correlating with decreased autoimmune symptoms. However, it has been unclear if the Th17 cells that are present in the absence of miR-155 retain normal function on a per cell basis. From a clinical perspective, it is important to determine if therapeutic targeting of miR-155 in differentiated, pathogenic Th17 cells that have accumulated in inflamed patient tissues can diminish their functionality and thus disease symptoms. Our current study demonstrates that miR-155−/− IL-17F+ Th17 cells are incapable of causing severe EAE symptoms like their miR-155+/+ counterparts, and this phenotype is consistent with the defective expression of several effector genes including IL-23R. These results predict that inhibiting miR-155 could reduce Th17 cell function during human autoimmune disease.
The identification of Ets1 as a target of miR-155 in Th17 cells provides insight into the ability of miR-155 to regulate Th17 cell effector function. Ets1 is a well-established negative regulator of Th17 cell development, and also targeted by another miRNA involved in the formation of Th17 cells, called miR-326 (19, 42). Thus, Ets1 appears to be a common target of miRNAs in the context of inflammatory Th17 cell biology. Ets1 is mutated in patients with certain types of autoimmune conditions, underscoring its connection to pathological tissue inflammation (44, 45). Taken together, Ets1 is emerging as an important node in the cellular networks that regulate human autoimmunity, and its therapeutic manipulation should be examined as a possible treatment.
A previous study found that Ets1 functions downstream of IL-2 to block expression of Th17-related genes (42). Co-culture experiments and exogenous IL-2 delivery demonstrated that Ets1 regulates the responsiveness of CD4+ T cells to the inhibitory effects of IL-2 on Th17 cell development (which appeared in this study to be more relevant than its role in promoting IL-2 expression). The study did not detect Ets1 binding to the IL-17A gene promoter, or Ets1-dependent effects on Stat5 activation. Thus, the mechanisms by which Ets1 regulates IL-17A and other Th17 genes (such as IL-23R) in response to IL-2 are unclear. Our experiments were done without IL-2 blocking antibodies or addition of exogenous IL-2. The only IL-2 in the system was produced by the activated T cells themselves, and we did not observe differences in the amount of IL-2 expressed by miR-155−/− versus Wt T cells (Supplemental Fig. S3). Thus, the approximately 2 fold increase in Ets1 in miR-155−/− cells is not sufficient to increase IL-2 expression in this context. Taken together with the study described above, it is possible that miR-155 functions to reduce the Ets1-dependent repressive effects of IL-2 signaling on Th17 cell effector gene expression. This will be an area of future investigation.
Our study also suggests that multiple targets of miR-155, in addition to Ets1, are relevant during Th17 cell development and function. Individual miRNAs have the potential to repress many mRNAs. This multi-target nature of miRNAs, in addition to the inability of Ets1 knockdown to fully restore expression of Th17-related genes in miR-155−/− Th17 cells, indicate that miR-155 likely targets multiple relevant mRNAs in this cellular compartment. Further experiments will be needed to test whether other targets predicted from our Ingenuity analysis, or presently unidentified, are being regulated by miR-155 during Th17 cell development and/or function.
In order for Th17 cells to expand during autoimmunity, they must be able to respond to IL-23 that is produced by a variety of immune cells within secondary lymphoid organs and lesions that are created during autoimmunity (31). Certain genetic variants of the IL-23R gene that promote IL-23R function have been associated with human autoimmune disorders, including Crohn’s disease and psoriasis, as determined through genome-wide association studies. This further highlights the relevance of this pathway during human autoimmunity (34–36). Importantly, we found IL-23R to be among the effector genes that is regulated by the miR-155-Ets1 axis in Th17 cells, and provide evidence that miR-155−/− Th17 cells are unresponsive to IL-23 ex vivo. Furthermore, recovery of IL-23R expression by miR-155−/− Th17 cells partially restored EAE disease symptoms. The regulation of this signaling pathway by miR-155 is a promising component of miR-155 biology that has not been recognized previously, yet may turn out to be a central mechanism by which miR-155 promotes autoimmune inflammation.
Supplementary Material
Acknowledgements
We would like to thank Allan Bradley for providing miR-155−/− mice, Dong Chen for providing the IL-17F RFP reporter mice, and the microarray and FACS core facilities at University of Utah. We also thank Angela Presson for her help with the Ingenuity software.
This study was supported by the National Institutes of Health grant 5R00HL102228-04.
Footnotes
Competing financial interests
The authors declare that they have no competing financial interests.
References
- 1.Mackay IR. Travels and travails of autoimmunity: a historical journey from discovery to rediscovery. Autoimmun Rev. 2010;9:A251–A258. doi: 10.1016/j.autrev.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Oukka M. Th17 cells in immunity and autoimmunity. Ann Rheum Dis. 2008;67(67 Suppl 3):ii26–ii29. doi: 10.1136/ard.2008.098004. [DOI] [PubMed] [Google Scholar]
- 3.Marwaha AK, Leung NJ, McMurchy AN, Levings MK. TH17 Cells in Autoimmunity and Immunodeficiency: Protective or Pathogenic? Front Immunol. 2012;3:129. doi: 10.3389/fimmu.2012.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 5.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 6.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 8.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 10.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 11.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 13.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, Zurawski S, Wiekowski M, Lira SA, Gorman D, Kastelein RA, Sedgwick JD. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 14.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 16.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O'Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 20.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 21.Ha TY. MicroRNAs in Human Diseases: From Cancer to Cardiovascular Disease. Immune Netw. 2011;11:135–154. doi: 10.4110/in.2011.11.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, Svan Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 24.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, McSharry C, Hueber AJ, Baxter D, Hunter J, Gay S, Liew FY, McInnes IB. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci U S A. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 27.Chen DF, Gong BD, Xie Q, Ben QW, Liu J, Yuan YZ. MicroRNA155 is induced in activated CD4(+) T cells of TNBS-induced colitis in mice. World J Gastroenterol. 2010;16:854–861. doi: 10.3748/wjg.v16.i7.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, Wekerle H, Hohlfeld R, Meinl E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain. 2009;132:3342–3352. doi: 10.1093/brain/awp300. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Tam LS, Kwan BC, Li EK, Chow KM, Luk CC, Li PK, Szeto CC. Expression of miR-146a and miR-155 in the urinary sediment of systemic lupus erythematosus. Clin Rheumatol. 2012;31:435–440. doi: 10.1007/s10067-011-1857-4. [DOI] [PubMed] [Google Scholar]
- 30.Paraboschi EM, Solda G, Gemmati D, Orioli E, Zeri G, Benedetti MD, Salviati A, Barizzone N, Leone M, Duga S, Asselta R. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int J Mol Sci. 2011;12:8695–8712. doi: 10.3390/ijms12128695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwakura Y, Ishigame H. The IL-23/IL-17 axis in inflammation. J Clin Invest. 2006;116:1218–1222. doi: 10.1172/JCI28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakker P, Leach MW, Kuang W, Benoit SE, Leonard JP, Marusic S. IL-23 is critical in the induction but not in the effector phase of experimental autoimmune encephalomyelitis. J Immunol. 2007;178:2589–2598. doi: 10.4049/jimmunol.178.4.2589. [DOI] [PubMed] [Google Scholar]
- 33.Awasthi A, Riol-Blanco L, Jager A, Korn T, Pot C, Galileos G, Bettelli E, Kuchroo VK, Oukka M. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amre DK, Mack D, Israel D, Morgan K, Lambrette P, Law L, Grimard G, Deslandres C, Krupoves A, Bucionis V, Costea I, Bissonauth V, Feguery H, D'Souza S, Levy E, Seidman EG. Association between genetic variants in the IL-23R gene and early-onset Crohn's disease: results from a case-control and family-based study among Canadian children. Am J Gastroenterol. 2008;103:615–620. doi: 10.1111/j.1572-0241.2007.01661.x. [DOI] [PubMed] [Google Scholar]
- 35.Newman WG, Zhang Q, Liu X, Amos CI, Siminovitch KA. Genetic variants in IL-23R and ATG16L1 independently predispose to increased susceptibility to Crohn's disease in a Canadian population. J Clin Gastroenterol. 2009;43:444–447. doi: 10.1097/MCG.0b013e318168bdf0. [DOI] [PubMed] [Google Scholar]
- 36.Sarin R, Wu X, Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc Natl Acad Sci U S A. 2011;108:9560–9565. doi: 10.1073/pnas.1017854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettelli E, Pagany M, Weiner HL, Linington C, Sobel RA, Kuchroo VK. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197:1073–1081. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, Feng XH, Jetten AM, Dong C. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007;204:2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 45.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, Hirankarn N, Ying D, Pan HF, Mok CC, Chan TM, Wong RW, Lee KW, Mok MY, Wong SN, Leung AM, Li XP, Avihingsanon Y, Wong CM, Lee TL, Ho MH, Lee PP, Chang YK, Li PH, Li RJ, Zhang L, Wong WH, Ng IO, Lau CS, Sham PC, Lau YL. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.