Abstract
Positron emission tomography (PET) has become a vital imaging modality in the diagnosis and treatment of disease, most notably cancer. A wide array of small molecule PET radiotracers have been developed that employ the short half-life radionuclides 11C, 13N, 15O, and 18F. However, PET radiopharmaceuticals based on biomolecular targeting vectors have been the subject of dramatically increased research in both the laboratory and the clinic. Typically based on antibodies, oligopeptides, or oligonucleotides, these tracers have longer biological half-lives than their small molecule counterparts and thus require labeling with radionuclides with longer, complementary radioactive half-lives, such as the metallic isotopes 64Cu, 68Ga, 86Y, and 89Zr. Each bioconjugate radiopharmaceutical has four component parts: biomolecular vector, radiometal, chelator, and covalent link between chelator and biomolecule. With the exception of the radiometal, a tremendous variety of choices exists for each of these pieces, and a plethora of different chelation, conjugation, and radiometallation strategies have been utilized to create agents ranging from 68Ga-labeled pentapeptides to 89Zr-labeled monoclonal antibodies. Herein, the authors present a practical guide to the construction of radiometal-based PET bioconjugates, in which the design choices and synthetic details of a wide range of biomolecular tracers from the literature are collected in a single reference. In assembling this information, the authors hope both to illuminate the diverse methods employed in the synthesis of these agents and also to create a useful reference for molecular imaging researchers both experienced and new to the field.
Introduction
Over the course of the past fifty years, advances in medical imaging have revolutionized clinical practice, with a wide variety of imaging modalities playing critical roles in the diagnosis and treatment of disease. Today, clinicians have at their disposal a remarkable range of medical imaging techniques, from more conventional modalities like ultrasound, conventional radiography (X-rays), X-ray computed tomography (CT scans), and magnetic resonance imaging (MRI) to more specialized methodologies such as single-photon emission computed tomography (SPECT) and positron emission tomography (PET).
In recent years, medical imaging research has experienced a paradigm shift from its foundations in anatomical imaging towards techniques aimed at probing tissue phenotype and function.1 Indeed, both the cellular expression of disease biomarkers and fluctuations in tissue metabolism and microenvironment have emerged as extremely promising targets for imaging.2 Without question, the unique properties of radiopharmaceuticals have given nuclear imaging a leading role in this movement. The remarkable sensitivity of PET and SPECT combines with their ability to provide information complementary to the anatomical images produced by other modalities to make these techniques ideal for imaging biomarker- and microenvironment-targeted tracers.3,4 Both relatively young modalities, SPECT and PET have had an impact on medicine (and oncology in particular), which belies their novelty, and both have been the topic of numerous thorough and well-reasoned reviews.5–9 Both modalities have become extremely important in the clinic, and while PET is generally more expensive on both the clinical and pre-clinical levels, it also undoubtedly possesses a number of significant advantages over its single-photon cousin, most notably the ability to quantify images, higher sensitivity (PET requires tracer concentrations of ∼10−8 to 10−10 M, while SPECT requires concentrations approaching 10−6 M), and higher resolution (typically 6–8 mm for SPECT, compared to 2–3 mm or lower for PET). Therefore, in the interest of scope, the article at hand will limit itself to the younger and higher resolution of the techniques: positron emission tomography.
Regardless of the broader perspective, any discussion of PET benefits from a brief description of the underlying physical phenomena. Starting from the beginning, a positron released by a decaying radionuclide will travel in a tissue until it has exhausted its kinetic energy. At this point, it will encounter its antiparticle, an electron, and the two will mutually annihilate, completely converting their mass into two 511 keV γ-rays that must, due to conservation of momentum, have equal energies and travel 180° relative to one another. These γ-rays will then leave the tissue and strike waiting coincidence detectors; importantly, only when signals from two coincidence detectors simultaneously trigger the circuit is an output generated. The two principal advantages of PET thus lie in the physics: the short initial range of the positrons results in high resolution, and the coincidence detection methodology allows for tremendous sensitivity.
In the early 1950s, Brownell10 and Sweet11 developed the first devices for creating images using the coincident detection of γ-rays emitted from positron-electron annihilation events. At the same time, these researchers and others were pioneering the oncologic applications of positron imaging, specifically the imaging of brain tumors.10–14 Not until the 1970s, however, did the field take the next important practical step forward: tomographic systems and computer analysis were first applied to positron imaging, innovations which paved the way for the widespread clinical use of the modality.
Since the advent of PET in both the clinic and medical research laboratories, a number of positron-emitting isotopes have been developed for use in radiopharmaceuticals. For years, the field was dominated by small molecule tracers, radiopharmaceuticals whose short biological half-lives favor the use of non-metallic radionuclides with correspondingly short radioactive half-lives, such as 18F, 15O, 13N, and 11C (Table 1). In many ways, this is still true: [18F]-fluoride and the ubiquitous [18F]-fluorodeoxyglucose ([18F]-FDG) are the only FDA-approved PET radiopharmaceuticals commonly employed in oncology ([13N]-NH3 and [82Rb]-RbCl are FDA-approved but are used principally for myocardial perfusion scans). Further still, an examination of the list of PET radiotracers currently in NIH-sponsored clinical trials reveals an overwhelming majority of agents with non-metallic radionuclides, including among others the promising agents [18F]-FLT, [18F]-FES, [18F]-FDHT, [18F]-FMISO, [18F]-FACBC, [18F]-fluoroethylcholine, [18F]-deshydroxycholine, [18F]-FMAU, [11C]-acetate, [11C]-choline, [11C]-MeAIB, [11C]-MET, [124I]-IAZGP, and [124I]-FIAU.15
Table 1.
| Radionuclide | Half-life | Decay mode (% branching ratio) | Production route | E(β+)/keV | β+ end-point energy/keV | Abundance, Iβ+/% | Eγ/keV (intensity, Iγ/%) |
|---|---|---|---|---|---|---|---|
| 11C | 1223.1 (12) s | β+ (100) | 14N(p,a)11C | 385.6 (4) | 960.2 (9) | 99.759 (15) | 511.0 (199.5) |
| 13N | 9.965 (4) m | β+ (100) | 16O(p,a)13N | 491.82 (12) | 1198.5 (3) | 99.8036 (20) | 511.0 (199.6) |
| 15O | 122.24 (16) s | β+ (100) |
15N(p,n)15O 14N(d,n)15O |
735.28 (23) | 1732.0 (5) | 99.9003 (10) | 511.0 (199.8) |
| 18F | 109.77 (5) m | β+ (100) |
18O(p,n)18F 20Ne(d,a)18F |
249.8 (3) | 633.5 (6) | 96.73 (4) | 511.0 (193.5) |
| 124I | 4.1760 (3) d | ε + β+ (100) β+ (22.7 [13]) |
124Te(p,n)124I | 687.04 (85) 974.74 (85) |
1,534.9 (19) 2,137.6 (19) |
11.7 (10) 10.7 (9) |
511.0 (45) 602.7 (62.9) 722.8 (10.4) 1,691.0 (11.2) |
ε = electron capture; m = minutes; d = days; s = seconds. Where positrons or γ-rays of different energies are emitted, only those with abundances of greater than 10% are listed. Unless otherwise stated, standard deviations are given in parentheses.
Yet despite the significant successes of small molecule probes labeled with non-metallic isotopes, these radionuclides possess a few critical limitations. First, the short half-lives of the most common non-metallic radionuclides - approximately 20 min for 11C, 10 min for 13N, 2 min for 15O, and 110 min for 18F - allow only for investigations of biological processes on the order of minutes or a few hours using tracers with rapid pharmacokinetic profiles. Second, both the short half-lives of the radionuclides and the frequent necessity of incorporating the radioisotopes into the core structure of the tracer (rather than in an appended chelator or prosthetic group) often necessitate demanding and complex syntheses. Third, the clinical and pre-clinical use of short half-life, non-metallic radionuclides often requires a local cyclotron facility; in its absence, the radionuclide in question will undergo many half-lives of decay while in transit. Given the resources required for the construction and operation of medical cyclotrons, this is simply not an option in many locations.
These limitations have been brought into focus by the increasing study and development of biomolecular targeting agents for cancer, including short peptides, antibodies, antibody fragments, and natural and non-natural oligonucleotides. Given that Nature herself has designed or inspired these agents, they often show sensitivities and specificities for cancer cell biomarkers that far exceed those of their small molecule counterparts. However, these biomolecular tracers typically have biological half-lives that are much longer than the radioactive half-lives of the most common non-metallic positron-emitting radionuclides; further, though less pressing, many of these biomolecules are incompatible with the chemistry required for direct labeling with non-metallic radionuclides.16–19
Given the enormous potential of biomolecular imaging agents, significant effort has been dedicated to the production, purification, and radiochemistry of positron-emitting radioisotopes of the metals Zr, Y, Ga, and Cu. These isotopes, specifically 64Cu, 68Ga, 86Y, and 89Zr, have radioactive half-lives (roughly 12.7, 1.1, 14.7, and 78.4 h, respectively) that favorably complement the biological half-lives of many biomolecular targeting vectors (Table 2). Although all four radiometals emit positrons, each has a characteristic positron range, which is the principal factor in determining imaging resolution. 64Cu and 89Zr emit very low energy positrons, producing image resolution comparable to that of 18F. 86Y and 68Ga, in contrast, emit higher energy positrons, which can result in slightly lower imaging resolutions, though this can be corrected through the use of mathematical algorithms.20 Further still, and equally critical, all four metals form stable chelate complexes that may be employed for the radiolabeling of biomacromolecules. To be sure, not all biomolecular PET tracers are labeled with radiometals, nor are all radiometallated PET tracers biomolecules. An 18F-labeled variant of the integrin-targeting RGD peptide21 and an 124I-labeled carbonic anhydrase-targeting antibody22 have produced very exciting results and are currently being employed in human studies. Moreover, a few radiometal-based small molecule tracers have also proved extremely promising, most notably [64Cu]-Cu(PTSM)23 and [64Cu]-Cu(ATSM),24 with the latter currently in a multi-center clinical trial as an imaging agent for hypoxia.25–28 Yet, despite these exceptions, the single most important application of positron-emitting radiometals is the development of tracers based on peptides, antibodies, and oligonucleotides.
Table 2.
Physical decay characteristics of common PET radiometals40
| Radionuclide | Half-life | Decay mode (% branching ratio) | Production route | E(β+)/keV | β+ end-point energy/keV | Abundance, Iβ+/% | Eγ/keV (intensity, Iγ/%) | Ref. |
|---|---|---|---|---|---|---|---|---|
| 64Cu | 12.701(2) h | ε + β+ (61.5 [3]) β+ (17.6 [22]) β− (38.5 [3]) |
64Ni(p,n)64Cu | 278.21 (9) | 653.03 (20) | 17.60 (22) | 511.0 (35.2) | 84, 85 |
| 68Ga | 67.71 (9) m | ε + β+ (100) β+ (89.14 [12]) |
68Ge/68Ga | 836.02 (56) | 1889.1 (12) | 87.94 (12) | 511.0 (178.3) | 160, 70, 161 |
| 86Y | 14.74 (2) h | ε + β+ (100) β+ (31.9 [21]) |
86Sr(p,n)86Y | 535 (7) | 1221 (14) | 11.9 (5) | 443.1 (16.9) 511.0 (64) 627.7 (36.2) 703.3 (15) 777.4 (22.4) 1076.6 (82.5) 1153.0 (30.5) 1854.4 (17.2) 1920.7 (20.8) |
77 |
| 89Zr | 78.4 (12) h | ε + β+ (100) β+ (22.74 [24]) |
89Y(p,n)89Zr | 395.5 (11) | 902 (3) | 22.74 (24) | 511.0 (45.5) 909.2 (99.0 |
79, 83, 162–164 |
ε = electron capture; m = minutes; h = hours; s = seconds. Where positrons or γ-rays of different energies are emitted, only those with abundances of greater than 10% are listed. Unless otherwise stated, standard deviations are given in parentheses.
Importantly, the basic strategy for the incorporation of a radiometal into a biomolecule differs somewhat from the synthesis of a small molecule radiotracer containing a non-metallic PET radionuclide. In small molecule tracers, the radionuclide most often replaces an isotopologue (e.g. [11C]-acetate or [15O]-H2O) or is incorporated into the basic structure of a molecule with either the intent of strategically altering the behavior of the parent molecule (e.g. [18F]-FDG) or, more likely, disturbing the activity of the parent molecule as little as possible (e.g. [18F]-FDHT or [18F]-FES). In contrast, in biomolecular tracers, the radiometal is almost never directly attached to the biomolecule itself. Rather, the radionuclide is bound to a chelating moiety (e.g. DOTA29 or EDTA30), which is first covalently appended to the biomolecule with the intent of altering the vector's biochemical properties as little as possible.31,32
As new targets are described and radiometals become more available to the wider molecular imaging community, the amount of research into radiometal-based PET tracers has exploded in recent years. For example, over 60% of all publications describing 89Zr-PET have been published in the last four years (with well over 20% in 2010 alone).33 Indeed, the dramatic growth in this area and the expansion in the availability of radiometals have had the dual effects of broadening the appeal of biomolecular PET imaging and opening the field to investigators who previously may have left the development of PET probes to dedicated radiochemistry and molecular imaging laboratories. However, the frenetic pace of the field and the array of choices in chelation, conjugation, and metallation strategies may serve as an obstacle to those who are interested in the development of radiometallated PET tracers but lack significant bioconjugation or radiochemical experience.
This perspective aims at lowering this barrier. Here, we strive to create a practical guide to the synthesis of radiometal-based PET tracers. To this end, we have compiled the experimental details of chelator choice, conjugation strategy, and radiometallation conditions from the syntheses of a wide array of 64Cu-, 68Ga-, 86Y-, and 89Zr-labeled PET agents. Typically, reviews discuss the structure, behavior, biology, and imaging applications of these agents, with the experimental details touched upon only briefly or simply referenced.7,16,34–37 All too often, however, the search for a specific conjugation or metallation protocol results in an elongated, and in some cases circuitous, trek through the literature to find a simple incubation time or buffer concentration. Importantly, we do not strive for an exhaustive review of the radiochemistry or imaging applications of radiometal-based PET tracers. Others - most notably Carolyn Anderson and her coworkers at the Washington University School of Medicine and Martin Brechbiel and his coworkers at the National Cancer Institute - have produced well-written and remarkably thorough reviews on these topics.3,30,34,38–45
The core of this perspective lies not in the text but rather in the series of tables containing the practical details of chelator conjugation and radiometallation from a diverse collection of 64Cu-, 68Ga-, 86Y-, and 89Zr-labeled bioconjugates. We have elected not to include two types of macromolecular radiopharmaceuticals, bispecific antibodies and biomolecule-based nanoparticles, in the interest of space and scope, though these have been addressed well elsewhere.46–49 Further, it is important to note that some of the conjugation strategies described herein are now, for the most part, obsolete with respect to their original vector; for example, a number of syntheses for DOTATOC will be outlined, though this DOTA-modified somatostatin analogue is now widely commercially available. Yet we believe it is important to detail these conjugation methods nonetheless, for the synthetic routes themselves may prove useful in the future for the creation of conjugates with different biomolecular vectors. In collecting these techniques in one place, we hope not only to shed light upon the diverse methods employed in the synthesis of these agents but also, and perhaps more importantly, to create a useful reference for both experienced molecular imaging scientists and researchers new to the field.
The anatomy of a PET bioconjugate
A radiometallated PET bioconjugate has four component parts, each of which must be carefully considered during the design and synthesis of the tracer: (1) the biomolecular targeting vector, (2) the radiometal, (3) the chelator, and (4) the linker connecting the chelator and the biomolecule (Fig. 1). A detailed discussion of the possible targeting vectors lies outside the scope of this work, though biomolecules ranging from cyclic pentapeptides and short oligonucleotides to 40-amino acid peptides, antibody fragments, and full antibodies have been employed.30 Of course, the most important facet of the biomolecule moiety is its specificity for its biomarker target. Indeed, a wide array of biomarkers have been exploited. Most often, the chosen target is a cell surface marker protein or receptor, such as the somatostatin receptor family (SSTr),50 integrin family (e.g. αvβ3),51 gastrin-releasing peptide receptor (GRPR),52 and epidermal growth factor receptor (EGFR).53 In more specialized cases, disialogangliosides (e.g. GD2), mRNA gene products, and even the low pH environment of tumors have been targeted by antibodies,54 oligonucleotides,55 and short peptides,56 respectively. Targeting cytosolic proteins and enzymes with antibodies and oligopeptides is rare due to the considerable difficulty of getting large biomolecules into the cytoplasm. However, significant progress is being made in the development of cell- and nucleus-penetration strategies, and this technology may prove productive for intracellular or intranuclear PET imaging agents in the near future.
Fig. 1.

The anatomy of a PET bioconjugate.
Radiometals: properties and production
The principal radiometals employed for the labeling of biomolecular tracers are 64Cu, 68Ga, 86Y, and 89Zr. Of course, these are not the only positron-emitting radiometals. Some metallic radioisotopes, such as 60Cu, 61Cu, 62Cu, 82Rb, 52mMn, and 94mTc, have been used in PET studies to varying degrees, but their half-lives make them far better suited for small molecule tracers (e.g. [60Cu]-Cu(ATSM)).57–60 Other positron-emitting radiometals, including 45Ti ([45Ti]-transferrin61), 52Fe ([52Fe]-citrate/transferrin62), 55Co ([55Co]-antiCEA F(ab′)263,64), 66Ga ([66Ga]-octreotate65), 110mIn ([110mIn]-octreotate66), and 74As ([74As]-bavituximab67,68), have been employed in the synthesis of biomolecular radiopharmaceuticals.41 However, these will not receive more than a brief discussion here, due to either the lack of more than one or two radiotracers per isotope, the limited availability of the radionuclide in question, or decay characteristics that make the isotope sub-optimal for use in a clinical PET radiopharmaceutical.40
The selection of a radiometal from the four main candidates, 64Cu, 68Ga, 86Y, and 89Zr, is a critical factor in determining the ultimate properties of a PET bioconjugate. In this regard, one of the most important considerations is matching the radioactive half-life of the isotope to the biological half-life of the biomolecule. For example, 68Ga is an inappropriate choice for labeling fully intact IgG molecules, for the radionuclide will decay through a number of half-lives before the antibody reaches its fully optimal biodistribution within the body. Therefore, the longer lived radiometals 64Cu, 86Y, and especially 89Zr are most often employed for immunoPET with fully intact mAbs. That said, 68Ga has been used successfully in the construction of PET bioconjugates based on antibody fragments with shorter biological half-lives. Conversely, 89Zr would be an inappropriate choice for a short peptide radiotracer; in this case, the multi-day radioactive half-life of 89Zr would far exceed what is typically a multi-hour biological half-life of the peptide, resulting in poor PET counting statistics and unnecessarily increased radiation dose to the patient. Thus, 64Cu, 86Y, and 68Ga are most often employed for oligopeptide PET tracers. It is important to note that 64Cu and 86Y occupy a favorablef middle ground with respect to radioactive half-life, allowing these radionuclides to be utilized advantageously in both antibody- and peptide-based based tracers.
The production of radiometals in high radionuclidic purity and specific activity is essential to the development of effective bioconjugates for PET imaging, and while an in-depth understanding of the nuclear reactions and purification chemistry behind their production may not be necessary for the biomedical use of these isotopes, a brief overview of the processes surely has merit. The production methods for radionuclides fall into three general categories: generator, cyclotron, and nuclear reactor (Fig. 2). Of the positron-emitting radiometals addressed in this perspective, 68Ga is generator-produced, while 64Cu, 86Y, and 89Zr are produced using a medical cyclotron.
Fig. 2.
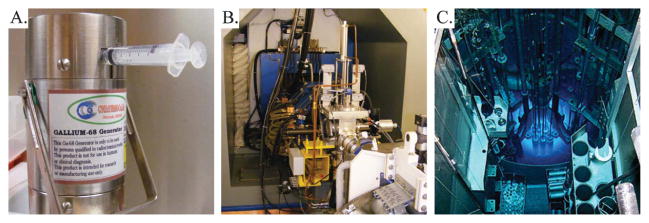
Three methods for the production of radionuclides: (A) 68Ga generator, (B) cyclotron, and (C) nuclear reactor. The authors acknowledge David Nickolaus of the Missouri University Research Reactor for the photo of the nuclear reactor.
68Ga is produced via the electron capture decay of its parent radionuclide, 68Ge. In the laboratory and clinic, 68Ga can be produced using a compact, cost-effective, and convenient 68Ge/68Ga generator system, which is capable of providing 68Ga for PET tracers for 1–2 years before being replaced.69 The 68Ga is eluted from the generator in 0.1 M HCl, providing a 68GaCl3 starting material for radiolabeling.70 Despite its convenience, the system does have some limitations, most notably high eluent volumes that often must be pH-adjusted prior to radiolabeling reactions, 68Ge break-through from the generator, and metal-based impurities. However, a number of purification techniques have been developed to circumvent the problems presented by the trace impurities in the 68Ga eluent.
86Y is the first of the three cyclotron-produced radiometals to be addressed here. 86Y is most often produced through the 86Sr(p,n)86Y reaction via bombardment of an isotopically enriched 86SrCO3 or 86SrO target with 8–15 MeV protons.71–74 A range of purification methods have been employed, including combinations of precipitation, ion exchange chromatography, chromatography with a Sr-selective resin, and electrolysis.75–77
89Zr has been produced via both the 89Y(p,n)89Zr and 89Y(d,2n)89Zr reactions. In the past, these methods have been used to successfully produce the radiometal using 13 MeV protons and 16 MeV deuterons, respectively, though both pathways have been complicated and limited by problematic purification protocols.78–80 A significant improvement upon these methods was provided by another production strategy that yielded 89Zr via the bombardment of 89Y on a copper target with 14 MeV protons, oxidation of Zr0 to Zr4+ with H2O2, and purification via anion exchange chromatography and subsequent sublimation steps.81,82 In the last few years, these methods have been improved upon further through the use of an 89Y thin-foil target (99% purity, 0.1 mm width), the optimization of bombardment conditions (15 MeV, 15 μA, 10° angle of incidence), and an improved solid phase hydroxamate resin purification to produce 89Zr reliably and reproducibly in very high specific activity (470–1195 μCi/mmol) and radionuclidic purity (>99.99%).83
Finally, 64Cu can be produced with either a nuclear reactor or a cyclotron via a variety of reaction pathways.3 In a nuclear reactor, 64Cu can be produced through the 63Cu(n,γ)64Cu and 64Zn(n,p)64Cu pathways. On a biomedical cyclotron, carrier-free 64Cu can be produced using the 64Ni(p,n)64Cu and 64Ni(d,2n)64Cu reactions.84–88 The former pathway has proven more successful and is currently used to provide 64Cu to research laboratories throughout the United States. In this method, the 64Cu is processed and purified via anion exchange chromatography to yield no carrier-added 64Cu2+. The expense of the enriched 64Ni target is a limitation of this production pathway, though a technique for the recycling of 64Ni has ameliorated this issue somewhat. In the last few years, a number of groups have worked to develop methods for the production of 64Cu using Zn targets through the 64Zn(d,2p)64Cu,66Zn(d,α)64Cu, and 68Zn(p,αn)64Cu reactions.89–92 These efforts have yielded some promising results but have failed to supplant the cyclotron-based 64Ni(p,n)64Cu pathway as the main route for 64Cu production.
Radiometal chelation chemistry
With both the targeting vectors and radiometals in hand, the spotlight next falls on how to combine these two essential parts of the PET bioconjugate. Indeed, both the formation of a kinetically inert metal chelate and the stable covalent attachment of the chelator moiety to the biomolecule are essential to the creation of an effective radiopharmaceutical. To this end, a wide variety of metal-chelating molecules have been synthesized, studied, and, in many cases, made bifunctional to facilitate their conjugation to a biomolecular vector (Fig. 3 and 4, vide infra). Transition metal chelators fall into two broad classes: macrocylic chelators and acyclic chelators. Each has its own unique set of advantages: while macrocyclic chelators typically offer greater kinetic stability, acyclic chelators usually have faster rates of metal binding. Generally, transition metal chelators offer at least four (and usually six or more) coordinating atoms, arrayed in a configuration that suits the preferred geometry of the oxidation state and d-orbital electron configuration of the metal in question. Yet simply having a generic chelator with well-organized and plentiful donor atoms is not enough; in every case, an appropriate chelator must be chosen to suit the selected radiometal (Table 3). Of course, however, some (e.g. DOTA) are more universally applicable than others (e.g. DiamSar). The most relevant oxidation states for the metals discussed here are Zr(IV), Ga(III), Y(III), and Cu(II); in vivo, only Cu(II) is at significant risk for reduction reactions. In terms of the commonly-employed ‘hard-soft’ system of classification, Zr(IV) is considered a very hard cation, with Y(III) and Ga(III) close behind on the spectrum. Cu(II), which is a borderline acid, straddles the hard/soft border and is thus easily the softest of the four.
Fig. 3.
Selected chelators and bifunctional chelators for 64Cu, 68Ga, 86Y, and 89Zr.
Fig. 4.
Selected chelators and bifunctional chelators for 64Cu, 68Ga, 86Y, and 89Zr.
Table 3.
Coordination number, donor set, and geometrya for selected complexes of Cu(II), Ga(III), Y(III), and Zr(IV)30
| Metal | Chelator | Ligand Donor Set | Total CN | Coordination Geometry | Ref. |
|---|---|---|---|---|---|
| Cu(II) | DTPA | N3O3 | 6 | — | 165 |
| NOTA | N3O3 | 6 | distorted trigonal prism | 166, 167 | |
| DOTA | N4O2 | 6 | distorted octahedron | 168–170 | |
| TETA | N4O2 | 6 | distorted octahedron | 168–170 | |
| CB-TE2A | N4O2 | 6 | distorted octahedron | 101, 171 | |
| EC | N2S2 | 4 | distorted square planar | 172 | |
| DIAMSAR | N6 | 6 | distorted octahedron or trigonal prism | 173 | |
| Ga(III) | EDTA | N2O4 | 6 | distorted octahedron | 174, 175 |
| HBED | N2O4 | 6 | — | 176, 177 | |
| DTPA | N3O3 | 6? | — | 175 | |
| NOTA | N3O3 | 6 | distorted octahedron | 168, 178 | |
| DOTA | N4O2 | 6 | distorted octahedron | 168 | |
| TETA | N4O2 | 6 | distorted octahedron | 168 | |
| CB-TE2A | N4O2 | 6 | distorted octahedron | 30, 179 | |
| DFO | O6 | 6 | — | 180 | |
| Y(III) | DTPA | N3O5 | 8 | monocapped square antiprism | 116, 181, 182 |
| DOTA | N4O4 | 8 | square antiprism | 114, 116, 182 | |
| TETA | N4O2 | 8? | distorted dodecahedron(?) | 183 | |
| Zr(IV) | DFO | O6 | 7 or 8 | — | 125 |
| DOTA | N4O4 | 8? | square antiprism? | 115 | |
| EDTA | N2O4 | 8 | distorted dodecahedron | 181, 184 | |
| DTPA | N3O5 | 8 | distorted dodecahedron | 181, 184 |
Question marks denote uncertainty in coordination number or geometry, and “—” denotes that the coordination geometry is not known.
Cu(II) has a rich chelation chemistry, capable of the formation of four-, five-, and six-coordinate complexes, with geometries ranging from square planar to trigonal bipyrimidal and octahedral.3,30,32,36,42,93 Due to its position on the border between hard and soft metals, Cu(II) exhibits a great affinity for nitrogen donors, though it is also known to bind either harder oxygen or softer sulfur donors as well. Generally, a copper chelator will feature a mixture of uncharged nitrogen and anionic oxygen or sulfur donors in order to neutralize the charge of the dicationic metal. Alone in solution, the metal forms a five-coordinate aquo-complex with rapid water-exchange rates that translate into facile substitution reactions with other ligands.94 Due to its 3d9 electronic structure, Cu(II) prefers a square planar coordination geometry. In consequence, both macrocyclic and acyclic tetradentate chelators have been developed for bioconjugation, including those with N4 (e.g. cyclam), N2O2, and N2S2 (e.g. bis(aminothiolate)-based ligands) donor sets.95,96 Due to the critical importance of kinetic stability, however, the complexation of Cu(II) with its maximum of six donor atoms has become more popular than the use of tetradentate chelators. To this end, both six-coordinate macro-cyclic and acyclic chelators have been employed with donor sets including N2O4 (e.g. EDTA), N3O3 (e.g. DTPA or NOTA), N4O2 (e.g. DOTA or CB-TE2A), and N6 (e.g. SarAr, DiamSar, and AmBaSar).54,97–101 Of these options, CB-TE2A and the SarAr family seem to be particularly promising, given their high kinetic and thermodynamic stability. The possibility of the reduction of Cu(II) to Cu(I) under physiological conditions with certain ligand sets must also be noted. In some cases (e.g. 64Cu-ATSM), this reduction may be essential to the pharmacodynamics of the radiotracer; however, in most situations, it is an extremely undesirable behavior that compromises the integrity of the radiopharmaceutical.24
Smaller and harder than Cu2+, the Ga3+ cation typically binds ligands containing multiple anionic oxygen donors and adopts a coordination number of six, though complexes with four or five donor atoms are also known.36,38,44,102,103 Aqueous pH is particularly important in Ga3+ chelation chemistry: the low pKa of the Ga(H2O)63+ complex results in low solubility at physiological pH, while under basic conditions the affinity of the metal for hydroxide anions can result in its dissociation from chelators to form gallium hydroxide species. Tetradentate chelators with NO3, NS3, and N2S2 donor sets have been used.104–106 These polydentate ligands often combine with one or two water molecules or halides to place the metal in a distorted octahedral or distorted square pyramidal geometry; however, in some cases, the Ga3+ can adopt a simple four-coordinate distorted tetrahedral geometry. Acyclic and macrocylic hexadentate chelators are more common for Ga3+, including those with N2O2S2 (e.g. bis(aminothiolate)-based ligands), N2O4 (HBED), N3O3 (NOTA), N3S3 (TACN-TM), N4O2 (DOTA), and O6 (DFO) donor sets.102,107–110 Complexes bearing these ligands almost always adopt a distorted octahedral geometry. Amongst these, DOTA is easily the most commonly employed in bioconjugates. However, the ligand has two drawbacks that limit its suitability for 68Ga3+.111 A central cavity that is too large for the cation limits the stability of the complex, and sluggish complexation kinetics require reaction times and temperatures that are less than ideally compatible with the short half-life of 68Ga and the stability of some biomolecular constructs, respectively. In contrast, TACN-TM and HBED- and NOTA-based ligands are particularly promising chelation systems for 68Ga due to their high thermodynamic and kinetic stability.103,112,113
The chemistry of Y(III) provides a significant change of pace from the previous two metals. Much larger than the three other common PET radiometals, the closed-shell, hard Y3+ cation often reaches coordination numbers of eight or nine. Donor sets of N2O4 (EDTA), N3O3 (NOTAM), N3O5 (DTPA), N4O4 (DOTA), and N4O2 (TETA) have all been used to chelate the metal, with water molecules or other exogenous ligands filling the remaining coordination sites.114–117 The DOTA and DTPA ligands, however, form much more stable complexes with the metal than TETA and EDTA, indicating better chelator-metal matches in the former cases. The higher coordination numbers also result in more exotic geometries: the DTPA complex adopts a monocapped square antiprism structure, the EDTA complex assumes a distorted dodecahedron geometry, and the DOTA complex results in a square antiprism structure. To date, DOTA- and DTPA-based chelators have been used in the vast majority of 86Y bioconjugate strategies, though future studies will no doubt expand the range of chelating moieties employed in these tracers.118–120
89Zr is easily the most recent addition to the family of common PET radiometals, and the relative scarcity of aqueous chelation chemistry studies reflects this fact. The highly cationic Zr4+ center exhibits a strong preference for ligands bearing multiple anionic oxygens and can accommodate up to nine coordinating atoms. The metal makes eight-coordinate, dodecahedral complexes with DTPA (N3O5), EDTA (N2O4 with two additional water ligands), and DOTA (N4O4, though the evidence here is less clear).121,122 However, the overwhelming majority of 89Zr-bioconjugates employ DFO as the chelating ligand.123,124 No solid state or NMR structural studies are available, though DFT calculations suggest that seven- or eight-coordinate species involving one or two water molecules in addition to the ligand's six oxygen donors are most likely.125 Given the considerable potential of 89Zr as a PET radiometal, the continued development of novel high-stability chelating systems is needed.
Conjugation strategies
The final piece of the anatomy of a radiometal PET bioconjugate is the covalent attachment of the chelator to the biomolecule. This link must be stable under physiological conditions and must not significantly compromise the binding strength and specificity of the biomolecule. Three bond-types comprise the overwhelming majority of chelator-biomolecule attachments: peptide, thiourea, and thioether bonds (Fig. 5). The first of these three attachments is formed through the reaction of an activated carboxylic acid and a primary amine, the second via an isothiocyanate and an amine, and the third via a thiol and a maleimide. These are not, however, the only options for the conjugation reaction; the reactions of vinylsulfones with thiols, bromoacetamides with amines, and bromoacetamides with thiols have also been employed in more unique cases. Further still, and more recently, the set of bioorthogonal cycloaddition reactions, broadly termed “click chemistry” reactions, have also been applied to chelator conjugations (vide infra).
Fig. 5.
The three principal types of bioconjugation reactions: (A) peptide bond formation via reaction of a primary amine with a carboxylic acid activated with a succinimidyl ester (NHS), a sulfosuccinimidyl ester (SNHS), tetrafluorophenol (TFP), or a peptide coupling reagent (e.g. HATU, HOBT, etc.); (B) thioether bond formation via reaction of a thiol and a maleimide; and (C) thiourea bond formation via reaction of an isothiocyanate and a primary amine.
To facilitate the formation of these covalent links, bifunctional chelators are often employed. Bifunctional chelators are molecules bearing both metal-binding moieties and either reactive bond-making functionalities or pendant linker arms (Fig. 3 and 4). Given the preponderance of available primary amines and free thiols on many biomolecules, the corresponding activated ester, isothiocyanate, and maleimide groups are usually incorporated into the bifunctional chelator. These molecules can be synthesized and isolated from known chelators (e.g. DOTA-NHS from DOTA), designed and synthesized de novo as bifunctional chelators (e.g. p-SCN-Bn-DOTA), or generated in situ prior to or during the conjugation reaction (e.g. DOTA(tBu)3-NHS from DOTA(tBu)3). In some cases, the modification to a chelator that confers bifunctionality is made at a point that otherwise may have been a metal donor site, for example the addition of an activated ester to a carboxylate arm in DOTA-NHS; in other situations, for example p-SCN-Bn-DOTA, a bifunctional linker is built into the backbone of the chelator so as to minimize any interference with the molecule's ability to bind to metal ions.
Both the number of chelates per biomolecule and the control over their placement can vary widely. The smaller size, well-established protecting group chemistry, and highly controlled and automated synthesis of peptide and nucleic acid vectors often allow for only a single chelator moiety, positioned at one terminus of the oligomer. In contrast, the method by which bifunctional chelators are typically conjugated to antibodies, i.e. the simple incubation of a given number of equivalents of bifunctional chelator with a solution of antibody, results in both a variable number of chelating moieties per antibody and their indeterminate placement on the macromolecule. The number of chelators per antibody can be determined fairly easily using isotopic dilution methods and can be controlled simply by altering the molar ratio of the bifunctional chelator in the conjugation reaction. Generally, more chelators per antibody is preferable, because higher specific activities can be attained. The control and knowledge of chelator placement, however, is harder to come by; the apprehension here, of course, is that the presence of a chelator in the binding region of the antibody can negatively effect its ability to bind to the antigen. Therefore, the goal in antibody conjugation is simple: attach as many chelators per antibody as possible, without compromising the immunoreactivty of the biomolecule.
Significantly, the conjugation of the chelator is almost always performed prior to radiometallation. Thus, the final step in the construction of a PET radiometal bioconjugate is the radiolabeling of the biomolecule-linker-chelator construct. The goal of this final step is the incorporation of as much activity as possible, as quickly as possible, without damaging the biomolecule. Therefore, temperature and pH conditions that favor rapid metallation reactions must be balanced against the concern for the integrity of the biomolecule. For example, while metallating a DOTA-conjugated antibody with 64Cu may proceed most quickly and efficiently at 90 °C, such high temperatures risk denaturing the antibody, and lower temperatures should be employed as a result.
In the preceding pages, it has become clear that the imaging scientist has many choices to make and factors to consider in the development and construction of a radiometal-based PET bioconjugate. In the final section of this perspective, we will provide a practical overview of the design and synthesis strategies used for PET bioconjugates currently described in the literature.
The construction of 68Ga bioconjugates
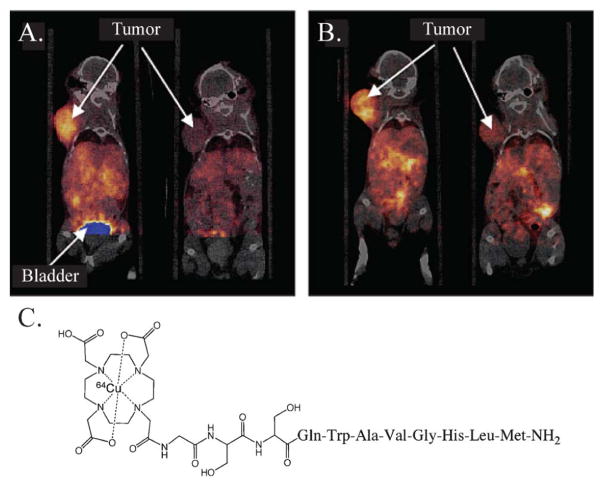
The short half-life and facile production of 68Ga have made it one of the radionuclides of choice for peptide-based PET bioconjugates.126 Tracers have been developed to target a wide array of cancer biomarkers, including epidermal growth factor receptor (EGFR), gastrin releasing peptide receptor (GRPR), integrin αvβ3, and melanocortin-1 receptor (MC1-R).127–130 However, the 68Ga peptide bioconjugates that have had the greatest impact in the clinic are without question the family of 68Ga-somatostatin analogues (SST).131–134 SST-receptors (SSTR) are over-expressed in neuroendorcrine tumors, prostate carcinomas, breast carcinomas, lymphomas, and small-cell lung cancers, among others, and 68Ga-somatostatin analogues, particularly 68Ga-DOTATOC, have been used to great effect in the imaging of these malignancies (Fig. 6).135
Fig. 6.
A 78-year-old woman with neuroendocrine tumor of unknown primary origin: (A) 68Ga-DOTATOC PET depicts diffuse bone metastases, (B) CT shows only part of widespread bone involvement, and (C) the structure of 68Ga-DOTATOC. Reprinted by permission of the Society of Nuclear Medicine from: D. Putzer, M. Gabriel, B. Henninger, D. Kendler, C. Uprimny, G. Dobrozemsky, C. Decristoforo, R. J. Bale, W. Jaschke and I. J. Virgolini, Journal of Nuclear Medicine, 2009, 50, 1214–1221. Fig. 2.155
A wide variety of chelators, conjugation strategies, and metallation procedures have been employed in the synthesis of 68Ga-labeled peptides (see Table 4 for experimental details and references). DOTA and NOTA-conjugated peptides are most common by a wide margin, though HBED and DFO have also been used. Given the solid-phase synthesis of many peptides, the conjugation of the chelator to the peptide is often performed while the peptide is still attached to a solid resin support. This can be achieved via the manual manipulation of the peptide-coated resin and subsequent incubation with a bifunctional chelator, or using an automated peptide synthesizer. In the latter scenario, a pre-prepared bifunctional chelator is not needed; rather, a monoreactive precursor is added to the automated synthesizer and is coupled to the growing peptide chain via an activated, bifunctional intermediate. Despite the preponderance of solid-phase methods, a number of in situ conjugations have also been reported using bifunctional chelators such as DOTA-NHS, HBED-CC-NHS, p-SCN-Bn-NOTA, and NH2-Bn-NOTA. Generally, peptide and isothiocyanate-based conjugations are performed at a slightly basic pH (8–9.5), due to the participation of a deprotonated primary amine in the bond-forming reactions. Further still, the peptide-chelator conjugates are almost always purified via RPHPLC or C18 cartridge prior to radiolabeling.
Table 4.
Guide to the construction of 68Ga-peptide bioconjugates
| Chelator | Target | Conjugation | Radiometallationa | Purification |
|---|---|---|---|---|
| DOTA | VAP-1185 | Peptide was coupled on the bead to DOTA(tBu)3 using an automated peptide synthesizer. Removal of the peptide from the support, deprotection, and RP-HPLC followed. | 68Ga eluent was adjusted to pH 5.5 with NaOAc, followed by addition of the peptide and incubation for 10–20 min at 90–100 °C. | None reported |
| EGFR127 | Peptide in borate buffer (0.08 M, pH 9.4) was added to dry DOTA-NHS. The pH of the resultant solution was adjusted to 9.0 with additional borate, and the mixture was allowed to stir overnight at RT, followed by RP-HPLC. | Peptide was incubated with 68Ga for 1 min at 90 °C via microwave in either NaOAc (pH 5.0, for use with non-concentrated eluent) or HEPES (pH 4.7, for use with concentrated eluent). | C18 cartridge | |
| GRPR128,186 | Peptide on resin was mixed with a pre-incubated solution of DOTA(tBu)3 and HATU in N-methylpyrrolidone (adjusted to pH 7–8 using DIPEA), followed by cleavage from resin, deprotection, and purification. | 68Ga eluent was dried and redissolved in NaOAc (0.1 M, pH 4.8), followed by addition of the peptide and incubation for 10 min at 90 °C. | C18 cartridge | |
| NTR187 | Peptide was coupled on the bead to DOTA(tBu)3 using an automated peptide synthesizer. Removal of the peptide from the support, deprotection, and purification followed. | Peptide was incubated with 68Ga in NaOAc (concentration not noted, pH 4.5) for 10 min at 95 °C. | None reported | |
| SSTR126,188,189 GRPR126 | Peptide was coupled on the bead to DOTA(tBu)3 using an automated peptide synthesizer. Removal of the peptide from the support, deprotection, and purification followed. | 68Ga eluent was adjusted to pH 3.5–3.8 with 1 M HEPES and incubated with the peptide for 4 min at 90 °C. | C18 cartridge | |
| GRPR190,191 | Peptide was coupled on the bead to DOTA(tBu)3 with HATU using an automated peptide synthesizer. Removal of the peptide from the support, deprotection, and purification followed. | 68Ga eluent was dried and redissolved in NaOAc (0.1 M, pH 4.8), followed by addition of peptide and incubation for 10 min at 90 °C. | C18 cartridge | |
| MC1-R192,193 | Deprotected peptide was dissolved in DMF with DIEA (1.5%) and added to a solution of DOTA(tBu)3 and HATU, which had beenincubated in DMF for 10 min at RT. The resultant solution was stirred at RT for 1 h, followed by precipitation, deprotection, and RP-HPLC. | Peptide was incubated with 68Ga in NaOAc (0.1 M, pH 4.8) for 15 min at 90 °C. | C18 cartridge | |
| SSTR132,133 | DOTA(tBu)3, HATU, and DIEA (1 : 1 : 1) were incubated with DMF for 10 min at RT, followed by addition of the peptide (in DMF with 1.5% DIEA) and stirring for 4 h at RT. Extraction, deprotection, and RP-HPLC followed. | Peptide was incubated with 68Ga in NaOAc (0.4 M, pH 4.8–5.5) for 15–25 min at 95 °C. | C18 cartridge | |
| SSTR134 | Conjugate was purchased from a commercial supplier. | Peptide was incubated with 68Ga in NaOAc (0.1 M, pH 4.5) for 5 min at 95 °C. | C18 cartridge | |
| SSTR135 | Conjugate was purchased from a commercial supplier. | 68Ga eluent in 0.05 M HCl/acetone (2 : 98) was added directly to the peptide in water and incubated for 10 min at 100 °C. | C18 cartridge | |
| SSTR148 | Hydroxylamine-modified peptide was incubated with deprotected acetyl-Bn-DOTA in 1 : 1 CH3CN : H2O (adjusted to pH 4 with TFA) for 18 h at RT, followed by RP-HPLC. | Experimental details were not given, though radiometallation was reported in publication. | None noted | |
| HBED | VEGFR194 |
|
Peptide solution (0.1 M phosphate buffer, pH 7.0) was combined with 10 μL 68Ga solution and 10 μL 2.1 M HEPES, for a final pH of 4.2. Time and temperature were not noted. | Size exclusion chromatography |
| NOTA | VEGFR194,195 | Peptide modified with NHS-PEG group was incubated with p-NH2-Bn-NOTA in carbonate buffer (pH 8.0) for 1 h, followed by C4 RP-HPLC. | Same as above | Size exclusion chromatography |
| αvβ3129,196 GRPR196 | Peptide was incubated with p-SCN-Bn-NOTA in NaHCO3 (0.1 M, pH 9.0) for 5 h at RT, followed by RP-HPLC. | Peptide was incubated with 68Ga in NaOAc (0.1 M, pH 5) for 10–15 min at 40–45 °C. | RP-HPLC | |
| αvβ3197 | Peptide was incubated with p-SCN-Bn-NOTA in NaHCO3 (0.1 M, pH 9.5) for 20 h at RT, followed by RP-HPLC. | 68Ga eluent was adjusted to pH 6.0 with 7% NaHCO3 solution, followed by addition of peptide for 10 min at RT. | RP-HPLC | |
| SSTR131 | Peptide on resin was mixed with a pre-incubated solution of NODAGA(tBu)3 and HATU in N-methylpyrrolidone (adjusted to pH 7–8 using DIPEA), followed by cleavage from resin, deprotection, and RP-HPLC. | Peptide was incubated with 68Ga in NaOAc (0.4 M, pH 5) or HEPES (0.1 M, pH 5.8) for 25 min at 95 °C | C18 cartridge | |
| DFO | SSTR198 | Peptide bearing an activated succinimidyl ester was incubated with DFO mesylate in DMF with DCC/HOBT. Time, temperature, and intermediate purification were not noted. | Peptide (in 0.1% AcOH) was incubated with 68Ga (in 0.1 M NH4OAc, pH 4.5) for 5 min at RT. | None reported |
| DTPA | LDL199,200 | Peptide was reacted with cyclic DTPA anhydride in HEPES buffer (0.1 M, pH 7) for 30 min at RT, followed by size exclusion chromatography. Other buffer types were reported to work for this reaction as well. | 68Ga eluent was dried and re-dissolved in NaOAc (0.4 M, pH 7.0), followed by addition of peptide and incubation for 1–30 min at RT. | Size exclusion chromatography |
| None (HSA)201 | Peptide was incubated with mixed acid anhydride DTPA in aqueous buffer (type and concentration not noted) for 12 h at 4 °C, followed by size exclusion chromatography. | DTPA-HSA solution pH was lowered to 3.1 with 1 M HCl, followed by addition of 68Ga solution (in 1 : 3 EtOH:0.9% NaCl), incubation at RT for 30 min, and final adjustment of pH to 5.5 with 0.1 M NaOH. | Size exclusion chromatography |
Some protocols call for the use of gentisic acid (typically 1–5 mg mL−1) to protect the biomolecule from radiolysis.
The metallation procedures for the peptide-chelator constructs follow the same general course, though the experimental details can vary considerably. The most common buffers for the metallation reaction are NaOAc, HEPES, NaH2PO4/Na2HPO4, and Na2CO3/NaHCO3, and these are used in concentrations ranging from 0.1 M to 0.5 M. The pH for the reaction depends on the chelator: 5–6 for NOTA-based chelators, 3.8–5.5 for DOTA-based chelators, 4–5 for HBED, and 4–5 for DFO. Reaction times and temperatures likewise vary depending on the chelator employed, ranging from 5 min at room temperature for DFO to 25 min at 95 °C and 20 min at 100 °C for DOTA. Often, the radiolabeling reaction is quenched by the addition of free chelator to scavenge excess unreacted radiometal. Finally, the purification of the resultant radiolabeled peptides is most often achieved using C18 cartridges (e.g. Waters Sep-PakTM), RP-HPLC, or size exclusion chromatography.
68Ga has also been used for the labeling of antibody fragments and affibody molecules, though far fewer examples exist than for 68Ga-peptides (see Table 5 for experimental details and references). In these cases, HBED, DOTA, and DTPA have been employed as the chelators of choice. The conjugation reactions are usually performed via the incubation of a solution of antibody fragment with a bifunctional chelator, such as DOTA-NHS or HBEDCC-TFP; however, in the case of one affibody construct, solid phase peptide synthesis and a monoreactive chelator precursor are used as described above for the peptide-based conjugates. Again, all of the macromolecules are purified subsequent to conjugation in order to remove excess chelator. Despite the relatively few examples, the metallation reactions are performed using an array of buffer types (HEPES, phosphate, and NH4OAc) and concentrations (0.1 M to 1.25 M). The time, temperature, and pH of the metallation reactions are all dependent on the identity of the chelator, though in a few cases, these conditions are not noted in the literature. After a suitable incubation, the radiolabeling reaction is often quenched with the addition of free chelator, and in all cases, the resultant radiometallated conjugate is purified with size exclusion chromatography. It thus becomes clear that the labeling of antibody fragments does not yet have standardized methodologies, which is a limitation that will be resolved as more examples of these extremely promising radiotracers come to light.
Table 5.
Guide to the construction of 68Ga-antibody bioconjugates
| Chelator | Target | Conjugation | Radiometallationa | Purification |
|---|---|---|---|---|
| HBED | EGFR112 EpCAM112,202 |
|
Antibody solution (0.1 M phosphate buffer, pH 7.0) was combined with 68Ga eluent and 2.1 M HEPES (for a final pH of 4.1–4.5) and incubated for 5–10 min at RT (40 °C). | Size exclusion chromatography |
| DOTA | HER2203–205 | Active DOTA ester was first formed via reaction of DOTA with NHS and subsequently EDC at RT, followed by cooling to 4 °C for 1 h and incubation with antibody fragment solution (0.1 M sodium phosphate, pH 7.0) overnight at 4 °C. Excess DOTA was removed with centrifugal filtration. | Antibody solution (in 1 M NH4OAc stock) was combined with 68Ga eluent (in 0.1 M HCl) and incubated for 15 min at 37 °C. Final reaction pH was not noted. | Size exclusion chromatography |
| HER2206–208 | Affibody was synthesized using standard solid phase synthesis and Fmoc chemistry, followed by conjugation on the bead with DOTA(tBu)3-NHS and subsequent deprotection, removal from the bead, and purification. | 68Ga eluent (in 0.1 M HCl) was combined (∼1 : 1) with affibody in NH4OAc (1.25 M, pH 4.2) and incubated for 10 min at 90 °C. | None noted | |
| DTPA | hPSP209,210 | Antibody was incubated with DTPA cyclic anhydride in aqueous solution (buffer conditions not described), followed by size exclusion chromatography to remove excess DTPA. | 68Ga eluent was evaporated to dryness, reconstituted in NH4OAc buffer (0.1 M), and incubated with antibody. Time, pH, and temperature were not noted. | Size exclusion chromatography |
| DTPA | CD45211 | Antibody was incubated with p-SCN-DTPA, mx-DTPA, or CHX-A″-DTPA in HEPES buffer (0.05 M, pH 9.5) for 20 h at RT, followed by size exclusion chromatography. | 68Ga eluent pH was adjusted to ∼5 with 1 M NaOAc, followed by addition of antibody for 10 min at RT. Reaction was quenched with DTPA. | Size exclusion chromatography |
Some protocols call for the use of gentisic acid (typically 1–5 mg mL−1) to protect the biomolecule from radiolysis.
Finally, a small number of oligonucleotide-based 68Ga-labeled bioconjugates have also been developed (see Table 6 for experimental details and references).136–138 2′-Deoxyphosphodiester (PO), 2′-deoxyphosphorthioate (PS), 2′-O-methyl phosphodiester (OMe), and locked nucleic acid (LNA) oligonucleotides have been synthesized and radiolabeled for gene expression imaging. In all cases, DOTA has been used as the chelator for 68Ga and is incorporated into the oligonucleotide using a DOTA-SNHS bifunctional chelate. Metallations have been performed in either NaOAc or HEPES buffer at pH 4.5–5.5, using short incubations at 90–100 °C (in some cases, microwave-assisted). Finally, the completed, radiolabeled oligonucleotides are typically purified with reverse-phase C4 or C18 cartridges (e.g. Waters Sep-PakTM).
Table 6.
Guide to the construction of radiometallated oligonucleotide bioconjugates
| Chelator | Nuclide | Type | Conjugation | Radiometallation | Purification |
|---|---|---|---|---|---|
| DOTA | 86Y | RNA oligomer120 | Amine-modified oligomer was incubated with p-SCN-Bn-DOTA in NaHCO3 buffer (0.7 M, pH 9.0) for 2 h at 40 °C, followed by purification via EtOH precipitation and size exclusion chromatography. | Oligomer was incubated with 86Y in NH4OAc buffer (0.5 M, pH 7.0) for 30–60 min at 90 °C. Reaction quenched with DTPA. | RP-HPLC |
| 86Y | RNA oligomer138 | Amine-modified oligomer was incubated with DOTA-NHS in NaHCO3 buffer (0.7 M, pH 8.1) for 2 h at 40 °C, followed by purification via size exclusion chromatography. | Oligomer was incubated with 86Y in NH4OAc buffer (0.5 M, pH 6.0) for 45 min at 90 °C. Reaction quenched with DTPA. | C18 cartridge | |
| 64Cu | PNA-peptide conjugate55,212 | Amine-modified PNA/peptide conjugate on a solid support was reacted with DOTA(tBu)3 and HATU on a peptide synthesizer for 60 min, followed by cleavage, deprotection, and RP-HPLC. | Conjugate was incubated with 64Cu in NH4OAc buffer (0.1 M, pH 5.5) for 15 min at 90 °C. | RP-HPLC | |
| 64Cu | PNA213 | DOTA(tBu)3 (in N-methylpyrrolidone) was combined with HATU (in DMF) and base (DIEA and 2,6-lutidine in DMF) and subsequently manually reacted with resin-bound PNA for 1 h at RT, followed by cleavage, deprotection, and RP-HPLC. | 64CuCl2 was converted to 64Cu-citrate with NH4-citrate (0.1 M, pH 7.0) and incubated with PNA in buffer for 1–2 h at 60 °C. Reaction quenched with DTPA. | Centrifugal column filtration | |
| 68Ga | RNA oligomer138 | Amine-modified oligomer was incubated with DOTA-NHS in NaHCO3 buffer (0.7 M, pH 8.1) for 2 h at 40 °C, followed by purification via size exclusion chromatography. | Oligomer was incubated with 68Ga in NH4OAc buffer (0.5 M, pH 4.1–4.7) for 45 min at 90 °C. Reaction quenched with DTPA. | C18 cartridge | |
| 68Ga | DNA, PS, and OMe oligomers137,214 | Activated ester was formed via the incubation of DOTA with SNHS and EDC in H2O from 4 °C to RT over 30 min. This mixture was added to amine-modified oligonucleotide solution in carbonate buffer (1 M, pH 9) overnight at 4 °C, followed by purification via size exclusion chromatography and C18 cartridge. | 68Ga eluent was adjusted to pH 5.5 with NaOAc, followed by addition of oligonucleotide for 10 min at 100 °C. | C18 cartridge | |
| 68Ga | DNA-LNA oligomer136,214 | Amine modified oligomer was incubated with DOTA-SNHS in Na2B4O7 buffer (adjusted to pH 8.5–10 with 5 M NaOH) overnight at 4 °C, followed by purification by centrifugal column filtration. | Oligomer was added to purified 68Ga eluent (HEPES, pH 4.6–5.0) and heated to 90 °C for 1 min in a microwave. | C4 cartridge | |
| SBTG2DAP | 64Cu | PNA-peptide conjugate215 | Using an automated peptide synthesizer, diaminopropanoate-modified PNA/peptide conjugate on a solid support was reacted with two equivalents of S-benzoyl thioglycolic acid using HATU and N-methylmorpholine in DMF. | Conjugate was incubated with 64Cu in NH4OAc buffer (0.1 M, pH 5.5) for 30 min at 90 °C. | RP-HPLC |
The construction of 64Cu bioconjugates
Given its intermediate half-life, favorable decay properties, relative accessibility, and well-established chelation chemistry, 64Cu has become a versatile and widely utilized radiometal for bioconjugate tracers. A variety of 64Cu-peptides have been developed, targeting biomarkers including SSTR, integrin αvβ3, GRPR, MC1-R, integrin α4β1, formyl peptide receptor (FPR), natriuretic peptide receptor (NPR), and vascular endothelial growth factor receptor (VEGFR), among many others (Fig. 7, see Table 7 for experimental details and references). The chelators employed in these conjugates are almost as diverse as the peptides themselves, with DOTA and CB-TE2A leading the way, but with TETA, NOTA, BPM-TACN, and DiamSar also used in some agents. As in the 68Ga peptides, both solid- and solution-phase chelator conjugation strategies have been used. For those involving bifunctional chelators, peptide and isothiocyanate-based conjugations are typically performed at slightly basic pH (8–9.5), because these reactions require a deprotonated primary amine to proceed. Also like the 68Ga cases, the post-conjugation purification of the peptide-chelator construct by RP-HPLC or C18 cartridge is a common practice. The buffers most often chosen for radiometallations are NH4OAc and NaOAc, typically utilized at concentrations ranging from 0.1 M to 0.5 M. However, incubation time, temperature, and pH vary according to the chelator. For example, the radiolabeling of DOTA-based conjugates is typically performed at pH 5–6.5 with 30–60 min incubations at temperatures ranging from room temperature to 95 °C. In contrast, the metallation of DiamSar-based conjugates can be performed at pH 8.0 with a 60 min incubation at room temperature. In many cases, unreacted 64Cu is scavenged after radiolabeling with free chelator (e.g. EDTA or DTPA), and after the successful radiometallation reaction, the overwhelming majority of the 64Cu-peptide conjugates are purified using RP-HPLC.
Fig. 7.
Coronal microPET images with co-registered CT of mice bearing PC-3 xenografts in the axillary thorax at (A) 1 h and (B) 24 h. The mice were injected i.v. with a GRPR-targeting 64Cu-bombesin analogue, 64Cu-DOTA-GSS-BN(7–14). The mice on the left (A) were not injected with blocking agent, while the mice on the right (B) received 100 μg of Tyr4-BN as an inhibitor. Adapted with permission from J. J. Parry, T. S. Kelly, R. Andrews and B. E. Rogers, Bioconjugate Chemistry, 2007, 18, 1110–1117.156 Copyright 2007 American Chemical Society.
Table 7.
Guide to the construction of 64Cu-peptide bioconjugates
| Chelator | Target | Conjugationa | Radiometallationb,c |
|---|---|---|---|
| DOTA | GRPR156,216,217 SSTR218 |
Peptide was coupled to DOTA(tBu)3 or DOTA(tBu)3-NHS using an automated peptide synthesizer and standard Fmoc chemistry, followed by cleavage from the resin, deprotection, and purification. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.5) for 30 min at RT. |
| MC1-R219,220 | Peptide was coupled to DOTA(tBu)3 using an automated peptide synthesizer and standard Fmoc chemistry, followed by cleavage from the resin, deprotection, and purification. | Peptide was incubated with 64Cu in NaOAc (0.1 M, pH 5.5) for 60 min at 65 °C. | |
| GRPR221 | Peptide was coupled to DOTA(tBu)3 while still on the peptide synthesizer, followed by cleavage from resin, deprotection, and purification. | Peptide was incubated with 64Cu in NH4OAc (0.4 M, pH 7) for 40 min at 70 °C. | |
| αvβ6222 | Peptide was coupled to DOTA(tBu)3 using HATU/DIEA while peptide was still on the resin, followed by cleavage from the resin, deprotection, and purification. | Peptide was incubated with 64Cu in NH4OAc (1 M, pH 7–8) for 60 min at RT. Reaction quenched with EDTA. | |
| GRPR223 | The method of DOTA incorporation method is unclear; however, a tributyl anhydride of DTPA was used for the acylation of prolines in similar DTPA-modified conjugates, so a similar strategy may have been used here. | Peptide was incubated with 64Cu in NH4OAc (0.5 M, pH 6.5) for 30 min at 80 °C. | |
| αvβ3224–228 GRPR229 VEGFR230,231 UPar232 IL-18R233,c |
DOTA was activated for coupling via reaction with EDC and N- hydroxysulfosuccinimide (SNHS) (10 : 5 : 4) in water (pH 5.5) for 30 min at 4 °C. Peptide (in water, buffer, or saline) was then added to the solution, the pH was adjusted to 8.5 with 0.1 M NaOH, and the resultant solution was stirred for 16 h at 4 °C. | Peptide was incubated with 64Cu in NaOAc (0.1 M, pH 5–6.5) for 30–60 min at 40–50 °C. In some cases the reaction was quenched with EDTA. | |
| MC1-R234,235 αvβ3235,c |
DOTA was activated for coupling via reaction with EDC and SNHS (1 : 1 : 0.8) in water (pH 5.5) for 30 min at 4 °C. Peptide (in phosphate buffer or water) was then added to the solution, the pH was adjusted to 8.5–9.0 with 0.1 M NaOH, and the resultant solution was stirred for 16 h at 4 °C. | Peptide was incubated with 64Cu in NaOAc (0.1 M, pH 5.5) for 60 min at 37–50 °C. | |
| αvβ3236 | DOTA was initially activated for coupling via 1 : 1 : 1 reaction with SNHS and EDC in water (pH 5.5) for 40 min at RT. Peptide in phosphate buffer (30 mM, pH 8.5) was then added to the solution, the pH was adjusted to 8.5 with 0.1 M NaOH, and the resultant solution was stirred for 1 h at RT and then 16 h at 4 °C. | Peptide was incubated with 64Cu in NaOAc (0.1 M, pH 6.3) for 60 min at 45 °C.c | |
| αvβ3224 | Peptide was incubated with (tBu)3 DOTA, HBTU, and Hunig's Base in DMF overnight at RT, followed by deprotection, and purification. | Peptide was incubated with 64Cu in NaOAc (0.5 M, pH 5.5) for 45 min at 50 °C. | |
| NPR237,238 | Peptide was incubated with DOTA-NHS overnight in Na2HPO4 buffer (0.1 M, pH 7.5) at RT. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.5) for 60 min at 43 °C. | |
| GC-C239 | Peptide was incubated with DOTA-NHS in HEPES buffer (0.3 M, pH 8.5) overnight at 4 °C. | Peptide was incubated with 64Cu in NH4OAc (0.4 M, pH 6.0) for 1 h at 80 °C. Reaction quenched with EDTA. | |
| FPR240 | Peptide was incubated with DOTA-NHS in water (pH 8.5) overnight at 4 °C. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.5) for 30 min at 40 °C. | |
| VEGFR195,241 | p-NH2-Bn-DOTA was activated via reaction with NHS-PEG-maleimide in buffer (15 mM NaOAc, 50 mM Na2CO3, 115 mM NaCl, pH 8.0) at RT for 1 h, followed by quenching of excess maleimide with Tris HCl (1 M, pH 8.0), the addition of peptide, incubation for 1 h at RT, and RP-HPLC with a C4 column.d | Peptide was incubated with 64Cu in NaOAc (0.1 M, pH 5.3–5.5) for 60 min at 55 °C. Reaction quenched with EDTA.d | |
| Low pH242 | Peptide was incubated with DOTA-maleimide in PBS (pH 7, with 2 mM EDTA) overnight at 4 °C.d | Peptide was incubated with 64Cu in NH4OAc (0.5 M, pH 5.5) for 30 min at RT. Reaction quenched with EDTA. | |
| CB-TE2A | MC1-R243 | Peptide was coupled to CB-TE2A using standard Fmoc/HBTU chemistry on a peptide synthesizer, followed by cleavage from the resin and deprotection. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 8) for 60 min at 95 °C. |
| SSTR244 | Peptide was reacted while still on the resin with CB-TE2A that had been pre-activated with DIEA and DCC, followed by cleavage from the resin, deprotection, and purification. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 8) for 90 min at 95 °C. | |
| SSTR50,245 | CB-TE2A was dissolved in DMF with DIEA and DIC and stirred for 25 min at RT before adding the solution to the peptide-containing resin. The resultant mixture was agitated for 3 h before filtration, washing, cleavage, and purification. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 8) for 60–90 min at 95 °C. | |
| α4β1246 | CB-TE2A was dissolved in DMF with DIEA and DIC and stirred for 25 min at RT before adding the solution to the peptide-containing resin. The resultant mixture was agitated overnight before filtration, washing, cleavage, and purification. | Peptide was incubated with 64Cu in NH4OAc (0.14 M, pH 7) for 60 min at 95 °C. | |
| GRPR221 | Peptide was coupled to CB-TE2A precursor on peptide synthesizer, followed by cleavage from resin, deprotection, and purification. | Peptide was incubated with 64Cu in NH4OAc (0.4 M, pH 7) for 40 min at 70 °C. | |
| αvβ6222 | CB-TE2A was pre-activated with DIC in DIEA and subsequently reacted with resin-bound peptide for 30 min at RT, followed by cleavage from the resin, deprotection, and purification. | Peptide was incubated with 64Cu in NH4OAc (1 M, pH 7.5–8.5) for 60 min at 95 °C. Reaction quenched with EDTA. | |
| αvβ399,247,248 | Peptide was reacted with CB-TE2A in the presence of DIC and HOBT in anhydrous DMF, followed by deprotection and purification. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 8) for 45–120 min at 95 °C. | |
| TETA | SSTR249,250 | Lys-protected peptide was conjugated in situ to TETA via reaction with DIEA, DIC/HOBT, and HBTU in DMF (time and temperature not noted), followed by deprotection and purification. | Peptide incubated with 64Cu in NH4OAc (0.1 M, pH 5.5) for 30 min at RT. |
| SSTR50 | Peptide was coupled to TETA(tBu)3 using standard Fmoc chemistry on an automated peptide synthesizer. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.5) for 1 h at RT. | |
| SSTR245,251–253 | Peptide was coupled to TETA(tBu)3 using standard Fmoc chemistry on an automated peptide synthesizer. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.5–6.5) for 1 h at RT-37 °C. | |
| GC-C239 | The activated ester of TETA was formed with EDC and SNHS in sodium phosphate buffer (0.2 M, pH 8.0), followed by the addition of peptide and incubation overnight at 4 °C. | Peptide was incubated with 64Cu in NH4OAc (0.4 M, pH 6.0) for 1 h at 80 °C. Reaction quenched with EDTA. | |
| α3β199,254 | Resin-bound peptide was combined with HBTU, DIEA, and TETA (from DMSO stock) and stirred for 4 h at RT, followed by cleavage from resin and deprotection. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.5, 1% BSA) for 15–30 min at RT. Reaction quenched with EDTA. | |
| NOTA | GRPR255,256 | NOTA was activated for coupling via reaction with SNHS and EDC in MES buffer (0.1 M, pH 4.7) for 10 min at RT. Peptide (in phosphate buffer, pH 7.4) was then added to the solution, the pH was adjusted to 7.4 with 10% NaOH, and the resultant solution was stirred for 16 h at RT. | Peptide was incubated with 64Cu in NH4OAc (0.4 M, pH 7.0) for 1 h at 70 °C. Reaction quenched with DTPA. |
| GC-C239 | The activated ester of NOTA was formed with EDC and SNHS in sodium phosphate buffer (0.2 M, pH 8.0), followed by the addition of peptide and incubation overnight at 4 °C. | Peptide was incubated with 64Cu in NH4OAc (0.4 M, pH 6.0) for 1 h at 80 °C. Reaction quenched with EDTA. | |
| GRPR196 αvβ3196 | Peptide was incubated with p-SCN-Bn-NOTA in NaHCO3 buffer (0.1 M, pH 9.0) for 5 h at RT. | Peptide was incubated with 64Cu in NaOAc (0.1 M, pH 6.5) for 15 min at 40 °C. | |
| CPTA | SSTR249 | Lys-protected peptide was conjugated in situ to CPTA via reaction with DIC and HOBT in DMF (time and temperature were not noted), followed by deprotection and purification. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.5). Time and temperature were not noted. |
| Bis(2-mercaptoacetamide) | VPAC257,258 | Protected chelator precursors were incorporated into the peptide via automated, solid-phase peptide synthesis using standard Fmoc/DIC/HOBT chemistry. | Peptide was incubated with 64Cu in glycine buffer (0.2 M, pH 9), SnCl2·2H2O (0.1 M), and HCl (0.05 M) for 20–45 min at 90 °C. |
| BPM-TACN | GRPR259 | Peptide was dissolved in DMF with chelator and HBTU. Subsequently, DIPEA was added, and the resultant solution was stirred for 20 h at RT. | 64Cu in NH4OAc (0.1 M) was added to peptide in 1 : 1 MeCN : H2O and incubated for 30 min at 40 °C. |
| DiamSar | αvβ3247 | Peptide was reacted with DiamSar in the presence of DIC and HOBT in anhydrous DMF, followed by deprotection. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 8.0) for 1 h at RT. |
| AmBaSar | αvβ3260 | AmBaSar was activated for coupling via reaction with EDC and SNHS (1 : 1 : 0.8) in water (pH 5.5) for 30 min at 4 °C. Peptide (in water) was then added to the solution, the pH was adjusted to 8.6 with 0.1 M NaOH, and the resultant solution was stirred for 16 h at RT. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.0) for 60 min at RT. |
| αvβ3261 | AmBaSar, HATU, HOAt, and DMSO were stirred at RT for 10 min, followed by the addition of DIPEA and peptide to the solution at 0 °C and incubation for 3 h at RT. | Peptide was incubated with 64Cu in NH4OAc (0.1 M, pH 5.0) for 30 min at RT. |
Unless otherwise noted, peptide-chelator constructs were purified with RP-HPLC or C18 cartridge prior to radiolabeling.
Unless otherwise noted, final radiometallated peptides were purified by RP-HPLC or C18 cartridge.
Some protocols call for the use of gentisic acid (typically 1–5 mg mL−1) to protect the biomolecule from radiolysis.
Purified via size exclusion chromatography
The 12.7 h half-life of 64Cu has allowed it to be utilized in antibody-based conjugates as well as those derived from peptides (see Table 8 for experimental details and references). Indeed, 64Cu-labeled antibody radiotracers have been developed against an array of biomarker antigens, for example human epidermal growth factor receptor 2 (HER2), prostate specific membrane antigen (PSMA), epidermal growth factor receptor (EGFR), and carcinoembryonic antigen (CEA). As with the 64Cu-peptides, a number of chelators have been used, including DOTA, CPTA, DO3A, TETA, and SarAr. The conjugation strategies for antibodies rely almost exclusively on incubation with bifunctional chelators, either generated in situ or synthesized and isolated (or purchased) beforehand. As is now clearly becoming a trend, the antibody-chelator constructs are almost always purified after conjugation by size exclusion chromatography or centrifugation with a high molecular weight filter membrane. The metallation procedures closely resemble those used for 64Cu-peptides: the most common buffers are NaOAc, NH4OAc, and NH4-citrate at concentrations of 0.1–0.25 M. The incubation time, temperature, and pH vary according to chelator; however, the incubation temperatures seldom rise above 43 °C due to concerns over antibody stability. Again, in many cases, unreacted 64Cu is scavenged after radiolabeling with free chelator (e.g. EDTA or DTPA). Finally, the radiometallated antibody bioconjugates are typically purified via size exclusion chromatography (e.g. HPLC, FPLC, or GE Life Sciences PD-10 columns) or centrifugal column filtration (e.g. Amicon Ultra-4 30,000 MWCO centrifugal filtration units).
Table 8.
Guide to the construction of 64Cu-antibody bioconjugates
| Chelator | Target | Conjugation | Radiometallationa | Purification |
|---|---|---|---|---|
| DOTA | HER2262 | Reduced affibody (in PBS, pH 7.4) was incubated with maleimide-modified DOTA (from a stock in DMSO) for 2 h at RT, followed by purification via overnight dialysis. | Affibody was incubated with 64Cu in NaOAc (0.1 M, pH 5.5) for 1 h at 40 °C. | Size exclusion chromatography |
| HER2263,264 CEA264 |
Activated DOTA was prepared via the combination of DOTA, SNHS and EDC (1 : 1 : 0.1) in water (pH 5.5) for 30 min at 4 °C. This solution was then adjusted to pH 7.3 with Na2HPO4 (0.2 M, pH 9.2), added to antibody (in NaH2PO4, 0.1 M, pH 7.5), and incubated overnight at 4 °C. Centrifugal column filtration followed. | Antibody was incubated with 64Cu in NH4-citrate (0.1 M, pH 5.5) for 50–60 min at 43 °C. Reaction quenched with EDTA. | Size exclusion chromatography | |
| αvβ351 EGFR53,265,266 |
Activated DOTA was prepared via the combination of DOTA, EDC, and SNHS (10 : 5 : 4) in water (pH 5.5) for 30 min at RT. This solution was then added to antibody, the pH of the reaction mixture was adjusted to 8.5 with 0.1 M NaOH, and the solution was incubated overnight at 4 °C. Size exclusion chromatography followed for purification. | Antibody was incubated with 64Cu in NaOAc (0.1 M, pH 6.5) for 1 h at 40 °C. | Size exclusion chromatography | |
| HER2267 | Antibody was incubated with DOTA-NHS (from DMSO stock) in borate-buffered saline (0.1 M pH 8.5) for 16 h at RT, followed by size exclusion chromatography. | Antibody was incubated with 64Cu in NaOAc (0.25 M, pH 6.0) for 1.5 h at 40 °C. Reaction quenched with EDTA. | Size exclusion chromatography | |
| CEA268 TAG-72269 |
Antibody was first dialyzed against PBS (pH 7.2) and NaHCO3 (0.1 M, pH 8.5). DOTA-NHS was then added to the antibody solution and incubated for 2 h at RT, followed by dialysis for purification. | Antibody was incubated with 64Cu in NH4-citrate (0.1 M, pH 5.5) for 45 min at 43 °C. | Size exclusion chromatography | |
| CD22270 | Antibody was incubated with DOTA-NHS in tetramethyl ammonium phosphate (0.1 M, pH 8) for 2.5 h at 37 °C, followed by centrifugal column filtration. | Antibody was incubated with 64Cu in NH4OAc (0.25 M, pH 7) for 1 h at 40 °C. Reaction quenched with EDTA. | Centrifugal column filtration | |
| PSMA271 | Antibody was incubated with DOTA-NHS in Na2HPO4 (0.1 M, pH 7.5) for 24 h at 4 °C, followed by dialysis for purification. | Antibody was incubated with 64Cu in NH4OAc (0.25 M, final pH 5.5) for 40 min at 40 °C. Reaction quenched with DTPA. | None listed | |
| EGFR272 | Antibody was incubated with (tBu)3 DOTA-NHS in Na2HPO4 buffer (0.1 M, pH 7.4) for 16 h at 4 °C, followed by centrifugal column filtration. | Antibody was incubated with 64Cu in NH4-citrate (0.1 M, pH 5.5) for 1 h at 40 °C. | Size exclusion chromatography | |
| L1-CAM273 | Antibody in phosphate buffer (0.1 M, pH 8) was added to DOTA-NCS variants. The pH was adjusted to 9–10 with Na3PO4, and the solution was incubated for 16 h at 4 °C, followed by centrifugal column filtration. | Antibody was incubated with 64Cu in NaOAc (0.1 M, pH 5.5) for 1 h at RT. Reaction quenched with EDTA. | Size exclusion chromatography | |
| CPTA | L1-CAM273,274 | Antibody was incubated with CPTA-NHS in sodium phosphate buffer (0.1 M, pH 7) for 2 h at RT, followed by centrifugal column filtration. | Antibody was incubated with 64Cu in NaOAc (0.1 M, pH 5.5) for 1 h at RT. Reaction quenched with EDTA. | Size exclusion chromatography |
| DO3A | L1-CAM273 | Antibody in phosphate buffer (0.1 M, pH 8) was added to DO3A-Bn-NCS. The pH was adjusted to 9–10 with Na3PO4, and the solution was incubated for 16 h at 4 °C, followed by centrifugal column filtration. | Antibody was incubated with 64Cu in NaOAc (0.1 M, pH 5.5) for 1 h at RT. Reaction quenched with EDTA. | Size exclusion chromatography |
| CEA268 |
|
Antibody was incubated with 64Cu in NH4-citrate (0.1 M, pH 5.5) for 45 min at 43 °C. | Size exclusion chromatography | |
| CEA268 | Antibody was incubated DO3A-VS in PBS (adjusted to pH 9.0 with 0.1 M NaOH) for 18 h at RT, followed by dialysis. | Antibody was incubated with 64Cu in NH4-citrate (0.1 M, pH 5.5) for 45 min at 43 °C. | Size exclusion chromatography | |
| TETA | CC17,275 | Antibody in ammonium phosphate (0.1 M, pH 8.0) was incubated with excess Br-benzyl-TETA and fresh 2-iminothiolane in triethanolamine (50 mM) for 30 min at 37 °C, followed by centrifugal column filtration. | Antibody was incubated with 64Cu in NH4-citrate (0.1 M, pH 5.5) for 15–30 min at RT. | Size exclusion chromatography |
| SarAr | GD254 | Antibody was incubated with SarAr and EDC in NaOAc (0.1 M, pH 5.0) for 30 min at 37 °C, followed by size exclusion HPLC. | Antibody was incubated with 64Cu in NaOAc (0.1 M, pH 5.0) for 30 min at 37 °C. | None reported |
Some protocols call for the use of gentisic acid (typically 1–5 mg mL−1) to protect the biomolecule from radiolysis.
SATA = S-acetylthioacetate
A small number of peptide nucleic acid (PNA) and hybrid PNA-oligopeptide 64Cu-labeled conjugates have also been created for mRNA-targeted imaging (see Table 6 for experimental details and references). These conjugates have employed either DOTA or SBTG2DAP as the chelating moieties, with solid-phase Fmoc synthesis techniques analogous to those for peptides used to incorporate the chelators into the oligomers. Radiolabeling reactions have been performed in either NH4OAc or NH4-citrate buffer (pH 5.5–6.0), with incubations of 15–120 min at temperatures ranging from 60 to 90 °C.
The construction of 86Y bioconjugates
The intermediate half-life, well-studied chelation chemistry, and presence of a radiotherapeutic isotopologue in 90Yall make 86Ya promising PET radiometal. However, decay properties that result in a lower image quality than 89Zr, 64Cu, and 68Ga and difficulties in its production and purification have limited the development of 86Y bioconjugates. Nevertheless, a number of 86Y-based antibody, peptide, and oligonucleotide PET radiopharmaceuticals have been successfully synthesized and evaluated (see Table 9 for experimental details and references). For example, radiolabeled antibodies against EGFR, human epidermal growth factor 1 (HER1), Lewis Y antigen, and mindin/RG1 have been developed (Fig. 8). All of these conjugates have been synthesized via incubation of antibody with the CHX-A″-DTPA bifunctional chelator under basic buffer conditions, followed by purification steps to separate the bioconjugate from unreacted chelator. Radiolabeling reactions are typically performed in NH4OAc buffer (0.1–3.0 M, pH 5–6) with incubations of 30–60 min at room temperature, followed by quenching with free chelator (e.g. DTPA or EDTA). The resultant completed bioconjugates are purified via size exclusion chromatography (e.g. HPLC, FPLC, or PD-10 columns) to remove any unbound 86Y.
Table 9.
Guide to the construction of 86Y bioconjugates
| Chelator | Target | Vector | Conjugation | Radiometallationa | Purification |
|---|---|---|---|---|---|
| CHX-A″-DTPA | EGFR157,276 HER1277,278 |
Antibody | Antibody (in 0.05 M bicarbonate buffer and 0.001 M EDTA, pH 8.0) was incubated with CHX-A″-DTPA (from stock in same buffer) for 4 h at 37 °C, followed by removal of the unbound chelate by dialysis against 0.15 M NH4OAc. | 86Y solution (in 0.1 M nitric acid) was adjusted to pH 5–6 with NH4OAc buffer (5 M, pH 7.0), followed by addition of antibody (in 0.15 M NH4OAc) and incubation for 30 min at RT. Reaction quenched with EDTA. | Size exclusion chromatography |
| Lewis Y antigen279–281 | Antibody and antibody fragments | Antibody was dialyzed against sodium bicarbonate buffer (0.05 M, pH 8.6) containing 0.15 M NaCl for 6 h. CHX-A″-DTPA was added to the mAb solution and incubated at RT overnight in the dark. Unbound chelate was removed by dialysis for 8 h against 20 mM NH4OAc with 0.15 M NaCl (pH 6.3). | 86Y-acetate was prepared by mixing stock 86Y solution with NH4OAc (3 M, final pH 5.0), followed by addition of antibody and incubation for 30 min at RT. Reaction quenched with EDTA. | Size exclusion chromatography | |
| Mindin/RG1282 | Antibody | Antibody (in 0.05 M sodium bicarbonate, 0.15 M NaCl, pH 8.5) was incubated with CHX-A″-DTPA (from DMSO stock) overnight at RT, followed by size exclusion chromatography. | 86Y solution (0.1 M HCl) was diluted three-fold with NH4OAc (0.1 M, pH 5.6) and subsequently added to antibody for incubation at RT for 1 h. Reaction quenched with EDTA. | Size exclusion chromatography | |
| MC1-R283 | Peptide | Peptide was incubated with NHS ester of CHX-A″-DTPA glutarate ligand in DMF in the presence DIEA for 2 h at RT, followed by diethyl ether precipitation, deprotection, and RP-HPLC purification. | 86Y (in 0.1 M HCl) was incubated with peptide (in 0.5 M NH4OAc, pH 5.5) at 75 °C for 30 min. | RP-HPLC | |
| SSTR118 | Peptide | Peptide was incubated with NHS ester of CHX-A″-DTPA(tBu)5 glutarate in DMF for 6–8 h at RT, followed by diethyl ether precipitation, deprotection, and RP-HPLC purification. | 86Y (in 0.1 M HCl) was incubated with peptide (in 0.5 M NH4OAc, pH 5.5) at RT for 1 h. | None reported | |
| DOTA | GRPR223,284,285 | Peptide | Peptide was synthesized via standard Fmoc solid phase chemistry. DOTA incorporation method is unclear; however, a tributyl anhydride of DTPA was used for the acylation of prolines in similar DTPA-modified conjugates, so a similar strategy may have been used here. | 86Y was incubated with peptide in NH4OAc (0.5 M, pH 6.5) for 30 min at 80 °C. | RP-HPLC |
| MC1-R219,220,286 | Peptide | Peptide was coupled to DOTA(tBu)3 using an automated peptide synthesizer and standard solid-phase Fmoc chemistry, followed by cleavage from the resin, deprotection, and RP-HPLC purification. | 86Y was incubated with peptide in NaOAc (0.1 M, pH 5.5) for 30 min at 85 °C. | RP-HPLC | |
| SSTR287,288 | Peptide | A solution of peptide, NHS, and DCC in DMF was added to a solution of DOTA in H2O, followed by incubation at RT for 24–72 h, deprotection, and RP-HPLC purification. | Peptide in NH4OAc (0.15 M, pH 4.5, 0.3% BSA) was incubated with 86Y (from α-hydroxyisobutyric acid stock) for 15 min at 100 °C. | RP-HPLC | |
| none (HSA)119 | Peptide Microspheres | Human serum albumin microspheres were suspended in Na2B4O7 (0.1 M, pH not noted) and incubated with p-SCN-Bn-DOTA for 24 h at 50 °C, followed by repeated centrifugation and washing steps for purification. | 86Y (in 0.04 M HCl) was added to DOTA-HSAM in NH4OAc (0.5 M, pH 7) and incubated for 15 min at 90 °C at a final pH of 6.5. | Centrifugation |
Some protocols call for the use of gentisic acid (typically 1–5 mg mL−1) to protect the biomolecule from radiolysis.
Fig. 8.
Representative reconstructed and processed maximum intensity projections of female athymic (NCr) nu/nu mice bearing (A) SHAW, (B) HT29, (C) DU145, and (D) SKOV3 tumor xenografts injected i.v. with 3.8–4.0 MBq of 86Y-CHX-A″-DTPA-cetuximab. Arrows indicate tumors. The scaling is based on % maximum and minimum threshold intensity without normalization to absolute value. With kind permission from Springer Science + Business Media: T. K. Nayak, C. A. S. Regino, K. J. Wong, D. E. Milenic, K. Garmestani, K. E. Baidoo, L. P. Szajek and M. W. Brechbiel, European Journal of Nuclear Medicine and Molecular Imaging, 37, 1368–1376. Fig. 3.157
In addition to the antibody-based tracers, a variety peptide-based agents have been synthesized, including agents targeting MC1-R, SSTR, and GRPR. In these conjugates, CHX-A″-DTPA and DOTA have been the predominant chelators employed, with both solid-phase synthesis and bifunctional chelator conjugation routes utilized. Not surprisingly, the radiometallation conditions are dependent upon the chelator, though NaOAc and NH4OAc buffers and pH values of 5–7 are most common. To complete the synthesis, the radiometallated peptide conjugates are almost all purified using RP-HPLC with C18 or C4 columns.
An isolated few 86Y-labeled RNA-based conjugates have also been synthesized for mRNA targeted imaging (see Table 6 for experimental details and references). These conjugates all utilize DOTA for 86Y chelation, and the chelator is incorporated into the oligomer via reaction with p-SCN-Bn-DOTA. Radiolabeling is accomplished via incubation of the chelator-RNA construct with 86Y in NH4OAc (0.5 M, pH 7.0) for 30–60 min at 90 °C. Unfortunately, however, only two publications on 86Y-labeled oligonucleotide radiotracers currently exist in the literature, so more general procedures and guidelines cannot be presented here.
The construction of 89Zr bioconjugates
Due to its long half-life, 89Zr has been used almost exclusively in the formation of antibody bioconjugates. Yet despite this narrow range of application, a wide array of antibody-based radiopharmaceuticals have been developed, including those targeting EGFR, VEGFR, carbonic anhydrase IX (CAIX), HER2, PSMA, and B-lymphocyte antigen CD20 (CD20) (Fig. 9, see Table 10 for experimental details and references). Interestingly, one chelator, DFO, has been employed in the overwhelming majority of these bioconjugates. Two routes have dominated the reported chelator conjugations: (1) the peptide coupling of a TFP ester of an FeIII(DFO) complex to the antibody of choice, followed by the removal of the Fe3+ cation and (2) the incubation of the antibody with a bifunctional DFO-SCN chelator. More recently, however, promising site-specific conjugation routes using maleimide- and halide-modified DFO have been reported. Regardless of the route, the resultant conjugate is typically purified via size exclusion chromatography to remove unbound chelate and subsequently metallated with 89Zr at pH 6.7–8.5 in buffer (HEPES and/or carbonate), with incubations of 30–120 min at room temperature. In all cases, the resultant radiolabeled bioconjugate is purified via size exclusion chromatography (most often GE Life Sciences PD-10 columns).
Fig. 9.
Temporal immunoPET images of 89Zr-DFO-J591 recorded in (A) LNCaP tumor–bearing (PSMA-positive) and (B) PC-3 tumor–bearing (PSMA-negative) mice between 3 and 144 h after injection. Transverse and coronal planar images intersect the center of the tumors and the mean tumor-to-muscle ratios derived from volume-of-interest analysis of immunoPET images are given. Upper thresholds of immunoPET have been adjusted for visual clarity, as indicated by scale bars. Reprinted by permission of the Society of Nuclear Medicine from: J. P. Holland, V. Divilov, N. H. Bander, P. M. Smith-Jones, S. M. Larson and J. S. Lewis, Journal of Nuclear Medicine, 2010, 51, 1293–1300. Fig. 4.125
Table 10.
Guide to the construction of 89Zr bioconjugates
| Chelator | Target | Vector | Conjugation | Radiometallationa | Purification |
|---|---|---|---|---|---|
| DFO | EpCAM289 E48 antigen289 | Antibody |
|
After removal of oxalic acid in vacuo, 89Zr solution was added to a solution of mAb (0.1 M NH4OAc, pH not noted) and incubated for 1 h at RT. | Size exclusion chromatography |
| CD4481,290,291 Met292 EGFR293,294 VEGFR295,296 CD20297 HER2298,299 |
Antibody |
|
Antibody solution (in water, buffer, or sterile saline) was incubated with 89Zr in 0.5 M HEPES buffer for 30–60 min at RT (89Zr originally in 1 M oxalic acid, adjusted to final pH 6.7–7.4 with 2 M Na2CO3 and/or 0.5 M HEPES). | Size exclusion chromatography | |
| CAIX300,301 | Antibody | Same as immediately above. | Same as above, but with incubation at 37 °C for 60 min. | Size exclusion chromatography | |
| HER2302 PSMA125,303 |
Antibody | Same as immediately above. | Antibody solution (in sterile saline) was incubated with 89Zr for 1–2 h at RT (89Zr stock in 1 M oxalic acid adjusted to pH 7.7–8.5 with 1 M Na2CO3). | Size exclusion chromatography | |
| CD44124,304 EGFR124 CD20124 |
Antibody | DFO-SCN (in DMSO) was incubated with antibody in NaHCO3 buffer (0.1 M, pH 8.9–9.1) for 30 min at 37 °C, followed by size exclusion chromatography. | Antibody solution was incubated with 89Zr in 0.5 M HEPES buffer for 60 min at RT (89Zr originally in 1 M oxalic acid, adjusted to final pH 6.8–7.2 with 2 M Na2CO3 and 0.5 M HEPES) | Size exclusion chromatography | |
| HER2305 | Antibody | Thiol-modified antibody was incubated with DFO-Chx-Mal in buffer (50 mM Tris, 150 mM NaCl, pH 7.5) for 1 h at RT, followed by size exclusion chromatography. | Antibody solution was incubated for 1 h at RT with 89Zr solution (originally in 1 M oxalic acid, adjusted to final pH 7.0–8.0, with 2 M Na2CO3 and 0.5 M HEPES). | Size exclusion chromatography | |
| HER2305 | Antibody | Thiol-modified antibody was incubated with DFO-BAC in buffer (0.05 M sodium borate, pH 9.0) for 5 h at RT, followed by size exclusion chromatography. | Same as immediately above | Size exclusion chromatography | |
| HER2304,305 | Antibody | Thiol-modified antibody was incubated with DFO-IAC in buffer (50 mM Tris, 150 mM NaCl, 0.0125 M sodium carbonate buffer pH 9) for 2 h at RT, followed by size exclusion chromatography. | Same as immediately above | Size exclusion chromatography |
Gentisic acid (typically 1–5 mg mL−1) is often included in metallation solutions to protect against radiolysis.
SMCC = succinimidyl 4-(N-maleimdomethyl) cyclohexane-1-carboxylate
Frontiers in bioconjugate development
Both in the laboratory and in the clinic, the field of radiometallated PET bioconjugates is progressing at an exciting rate. Indeed, researchers are currently pushing back the frontiers for all of the components of the bioconjugate anatomy. To be sure, the arena with the most limited prospects is the choice of radiometal, however, efforts do exist to expand the family of PET radiometals used for bioconjugates to include new possibilities, such as 45Ti and 74As.39,40,61,68 The development of new biomolecular vectors is a particularly fertile area, with the discovery of new cancer biomarkers and advances in protein engineering fueling this growth. Interestingly, an increasing number of vectors are being studied, which target not specific cell-surface proteins or gene products but rather characteristics of the tumor microenvironment.56
Yet most relevant to the discussion at hand is the forefront of research on chelation, metallation, and conjugation strategies. New chelating architectures and bifunctional chelators are being designed and synthesized at a tremendous rate, often out-stripping the pace of the development of new bioconju-gates themselves.30–32,139 The advent of rapid, bioorthogonal, and chemoselective ‘click chemistry’ reactions represents a particularly exciting new approach to chelator conjugation.140–144 While the exact definition of click chemistry can vary, the most common example is the copper-catalyzed 1,3-dipolar Huisgen cycloaddition between an azide and an alkyne. The reaction is rapid, high-yielding, clean, and chemoselective and has already been extensively employed in 18F-based PET radiotracers.142,145,146 However, the application of click chemistry to radiometal probes has lagged behind somewhat, perhaps due to concern over Cu(I) contamination from the cycloaddition catalyst. Nonetheless, a small number of ‘clickable’ bifunctional chelators and their resultant bioconjugates have begun to appear in the literature, including DOTA and CB-TE2A examples.147,148 Further, the development of new click reactions that do not require a copper catalyst, including [3+2] cycloadditions between azides and strained alkynes,149,150 inverse electron demand Diels–Alder cycloadditions between tetrazines and strained dienophiles,151,152 and azaelectrocyclizations,153 have begun to capture the interest of radiochemists and will surely soon occupy an important place in the synthesis of novel bioconjugates. Finally, the most exciting developments in radiolabelling lie in the full automation of bioconjugate radiometallation and, perhaps ultimately, chelator conjugation as well.154 Such developments will increase standardization and reproducibility while concomitantly decreasing radiation dose rates to researchers.
Conclusions
In the preceding pages, we have detailed the synthesis of radiotracers using four different radiometals, tens of biomolecular vectors, over thirty chelating scaffolds, and a myriad of different conjugation and metallation strategies. This diversity is the hallmark of an important and rapidly growing field, one that is increasingly attracting the attention of scientists from a wide variety of other specialities. Indeed, during the writing of this perspective, three new bioconjugates were published that required inclusion; between submission and publication, even more will likely appear in the literature. This influx of new interest is a boon to the molecular imaging community, bringing with it new expertise and perspectives. The rapid pace of development in this area, however, may inadvertently act as an obstacle to new researchers, simply due to the sheer number of conjugation and metallation protocols, which can vary by as much as the identity of a radiometal or by as a little as a tenth of a pH unit. Therefore, we believe it is extremely important not only to encourage the development of diverse strategies for the synthesis of PET bioconjugates but also to make these experimental methods widely accessible and straightforward to the field as a whole. Our hope is that this perspective will aid in this effort.
Acknowledgments
First and foremost, the authors would like to thank all of those researchers whose work has contributed to the imaging agents discussed in these pages. Compiling this perspective has made us ever more aware of the intellectual vibrancy of this field and the scientists within it. More specifically, we would like to thank Dr Jason Holland and Dr NagaVaraKishore Pillarsetty for helpful discussions. We would also like to thank the NIH for their generous funding [NIH R01 CA138468 (JSL) and NIH F32 CA144138 (BMZ)].
Biographies

Brian M. Zeglis, Ph.D.
Dr. Brian Zeglis received his B.S. in chemistry summa cum laude from Yale University (2004), where he worked under the guidance of Professor Robert H. Crabtree. For his graduate studies, he attended the California Institute of Technology as an NSF pre-doctoral fellow. At Caltech, Brian worked under the mentorship of Professor Jacqueline K. Barton, studying the synthesis and development of DNA-binding octahedral metal complexes. After receiving his Ph.D. (2009), Brian moved to Memorial Sloan-Kettering Cancer Center, where he works as an NIH post-doctoral fellow in the laboratory of Professor Jason S. Lewis. Currently, his research is focused on the design, synthesis, and evaluation of 64Cu- and 89Zr-based PET radiopharmaceuticals.

Jason S. Lewis, Ph.D.
Professor Lewis earned a B.Sc. Hons (1992) and an M.Sc. (1993) in Chemistry from the University of Essex. He received a Ph.D. in Biochemistry (1996) from the University of Kent at Canterbury. Following his postdoctoral study at the Washington University School of Medicine, he joined the Radiology faculty (2003). In 2008, he moved to Memorial Sloan-Kettering Cancer Center (New York) where he is currently a Member (with tenure) and Vice Chairman for Basic Research, Chief Attending Radiochemist, and Director of the Cyclotron Core. He holds joint appointments at the Sloan-Kettering Institute, Weill Cornell Medical College and Gerstner Sloan-Kettering Graduate School. Professor Lewis has co-authored over 110 peer-reviewed journal articles, reviews and book chapters.
References
- 1.Rudin M, Weissleder R. Nat Rev Drug Discovery. 2003;2:123–131. doi: 10.1038/nrd1007. [DOI] [PubMed] [Google Scholar]
- 2.Gillies RJ. J Cell Biochem. 2002;87(S39):231–238. doi: 10.1002/jcb.10450. [DOI] [PubMed] [Google Scholar]
- 3.Smith SV. J Inorg Biochem. 2004;98:1874–1901. doi: 10.1016/j.jinorgbio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Blower P. Dalton Trans. 2006:1705–1711. doi: 10.1039/b516860k. [DOI] [PubMed] [Google Scholar]
- 5.Mettler JFA, Guiberteau MJ. Essentials of Nuclear Medicine Image. 5. 2007. [Google Scholar]
- 6.Dilworth JR, Parrott SJ. Chem Soc Rev. 1998;27:43–55. [Google Scholar]
- 7.Verel I, Visser GWM, van Dongen GA. J Nucl Med. 2005;46S1:164S–171S. [PubMed] [Google Scholar]
- 8.Mahmood A, Jones AG. Handbook of Radiopharmaceuticals. 2003:323–362. [Google Scholar]
- 9.Liuand S, Edwards DS. Chem Rev. 1999;99:2235–2268. doi: 10.1021/cr980436l. [DOI] [PubMed] [Google Scholar]
- 10.Brownell GL, Sweet WH. Nucleonics. 1953;11:40–45. [Google Scholar]
- 11.Sweet WH. N Engl J Med. 1951;245:875–878. doi: 10.1056/NEJM195112062452301. [DOI] [PubMed] [Google Scholar]
- 12.Sweet WH, Brownell GL. Journal of the American Medical Association. 1955;157:1183–1188. doi: 10.1001/jama.1955.02950310009002. [DOI] [PubMed] [Google Scholar]
- 13.Wrenn FRJ, Good ML, Handler P. Science. 1951;113:525–527. doi: 10.1126/science.113.2940.525. [DOI] [PubMed] [Google Scholar]
- 14.Bagnall HJ, Benda P, Brownell GL, Sweet WH. J Neurosurg. 1958;15:411–426. doi: 10.3171/jns.1958.15.4.0411. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Scollard D, Chen P, Wang J, Holloway C, Reilly RM. Nucl Med Commun. 2005;26:427–432. doi: 10.1097/00006231-200505000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Knight LC. Handbook of Radiopharmaceuticals. 2003:643–684. [Google Scholar]
- 17.Anderson CJ, Connett JM, Schwarz SW, Rocque PA, Guo LW, Philpott GW, Zinn KR, Meares CF, Welch MJ. J Nucl Med. 1992;33:1685–1691. [PubMed] [Google Scholar]
- 18.Schrama D, Reisfeld RA, Becker JC. Nat Rev Drug Discovery. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 19.Zalutsky MR, Lewis JS. Handbook of Radiopharmaceuticals. 2003:685–714. [Google Scholar]
- 20.Ruangma A, Bai B, Lewis JS, Sun X, Welch MJ, Leahy R, Laforest R. Nucl Med Biol. 2006;33:217–226. doi: 10.1016/j.nucmedbio.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Beer AJ, Grosu AL, Carlsen J, Kolk A, Sarbia M, Stangier I, Watzlowik P, Wester HJ, Haubner R, Schwaiger M. Clin Cancer Res. 2007;13:6610–6616. doi: 10.1158/1078-0432.CCR-07-0528. [DOI] [PubMed] [Google Scholar]
- 22.Divgi CR, Pandit-Taskar N, Jungbluth AA, Reuter VE, Gönen M, Ruan S, Pierre C, Nagel A, Pryma DA, Humm J, Larson SM, Old LJ, Russo P. Lancet Oncol. 2007;8:304–310. doi: 10.1016/S1470-2045(07)70044-X. [DOI] [PubMed] [Google Scholar]
- 23.Wong TZ, Lacy JL, Petry NA, Hawk TC, Sporn TA, Dewhirst MW, Vlahovic G. AJR, Am J Roentgenol. 2008;190:427–432. doi: 10.2214/AJR.07.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis JS, McCarthy DW, McCarthy TJ, Fujibayashi Y, Welch MJ. J Nucl Med. 1999;40:177–183. [PubMed] [Google Scholar]
- 25.Vavere AL, Lewis JS. Dalton Trans. 2007:4893–4902. doi: 10.1039/b705989b. [DOI] [PubMed] [Google Scholar]
- 26.Dehdashti F, Grigsby PW, Lewis JS, Laforest R, Siegel BA, Welch M. J Nucl Med. 2008;49:201–205. doi: 10.2967/jnumed.107.048520. [DOI] [PubMed] [Google Scholar]
- 27.Lewis JS, Laforest R, Dehdashti F, Grigsby PW, Welch MJ, Siegel BA. J Nucl Med. 2008;49:1177–1182. doi: 10.2967/jnumed.108.051326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland JP, Lewis JS, Dehdashti F. Q J Nucl Med Mol Imag. 2009;53:193–200. [PMC free article] [PubMed] [Google Scholar]
- 29.Viola-Villegas N, Doyle RP. Coord Chem Rev. 2009;253:1906–1925. [Google Scholar]
- 30.Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Chem Rev. 2010;110:2858–2902. doi: 10.1021/cr900325h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mewis RE, Archibald SJ. Coord Chem Rev. 2010;254:1686–1712. [Google Scholar]
- 32.Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Curr Pharm Des. 2007;13:3–16. doi: 10.2174/138161207779313768. [DOI] [PubMed] [Google Scholar]
- 33.ISI Web of Knowledge v.4.10. [Accessed October 26, 2010]. [Google Scholar]
- 34.Boswell CA, Brechbiel MW. Nucl Med Biol. 2007;34:757–778. doi: 10.1016/j.nucmedbio.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunphy MPS, Lewis JS. J Nucl Med. 2009;50:106S–121S. doi: 10.2967/jnumed.108.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S. Chem Soc Rev. 2004;33:445–461. doi: 10.1039/b309961j. [DOI] [PubMed] [Google Scholar]
- 37.van Dongen GAMS, Visser GWM, Lub-de Hooge MN, de Vries EG, Perk LR. Oncologist. 2007;12:1379–1389. doi: 10.1634/theoncologist.12-12-1379. [DOI] [PubMed] [Google Scholar]
- 38.Anderson CJ, Welch MJ. Chem Rev. 1999;99:2219–2234. doi: 10.1021/cr980451q. [DOI] [PubMed] [Google Scholar]
- 39.Lewis JS, Singh RK, Welch Michael J. Long Lived and Unconventional PET Radionuclides. 2009 [Google Scholar]
- 40.Holland JP, Williamson MJ, Lewis JS. Mol Imaging. 2010;9:1–20. [PMC free article] [PubMed] [Google Scholar]
- 41.Nayak TK, Brechbiel MW. Bioconjugate Chem. 2009;20:825–841. doi: 10.1021/bc800299f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shokeen M, Anderson CJ. Acc Chem Res. 2009;42:832–841. doi: 10.1021/ar800255q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson CJ, Ferdani R. Cancer Biother Radiopharm. 2009;24:379–393. doi: 10.1089/cbr.2009.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Nahhas A, Win Z, Szyszko T, Singh A, Nanni C, Fanti S, Rubello D. Anticancer Res. 2007;27:4087–4094. [PubMed] [Google Scholar]
- 45.Brechbiel MW. Q J Nucl Med Mol Imag. 2008;52:166–173. [PMC free article] [PubMed] [Google Scholar]
- 46.Goldenberg DM, Rossi EA, Sharkey RM, McBride WJ, Chang CH. J Nucl Med. 2008;49:158–163. doi: 10.2967/jnumed.107.046185. [DOI] [PubMed] [Google Scholar]
- 47.Schoffelen R, Sharkey RM, Goldenberg DM, Franssen G, McBride WJ, Rossi EA, Chang CH, Laverman P, Disselhorst JA, Eek A, Van Der Graaf WTA, Oyen WJG, Boerman OC. Mol Cancer Ther. 2010;9:1019–1027. doi: 10.1158/1535-7163.MCT-09-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai WB, Chen XY. J Nucl Med. 2008;49:113S–128S. doi: 10.2967/jnumed.107.045922. [DOI] [PubMed] [Google Scholar]
- 49.Cheon J, Lee JH. Acc Chem Res. 2008;41:1630–1640. doi: 10.1021/ar800045c. [DOI] [PubMed] [Google Scholar]
- 50.Eiblmaier M, Andrews R, Laforest R, Rogers BE, Anderson CJ. J Nucl Med. 2007;48:1390–1396. doi: 10.2967/jnumed.107.039990. [DOI] [PubMed] [Google Scholar]
- 51.Cai WB, Wu Y, Chen K, Cao QZ, Tice DA, Chen XY. Cancer Res. 2006;66:9673–9681. doi: 10.1158/0008-5472.CAN-06-1480. [DOI] [PubMed] [Google Scholar]
- 52.Biddlecombe GB, Rogers BE, de Visser M, Parry JJ, de Jong M, Erion JL, Lewis JS. Bioconjugate Chem. 2007;18:724–730. doi: 10.1021/bc060281l. [DOI] [PubMed] [Google Scholar]
- 53.Cai WB, Chen K, He LN, Cao QH, Koong A, Chen XY. Eur J Nucl Med Mol Imaging. 2007;34:850–858. doi: 10.1007/s00259-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 54.Voss SD, Smith SV, DiBartolo N, McLntos LJ, Cyr EM, Bonab AA, Dearling JLJ, Carter EA, Fischman AJ, Treves ST, Gillies SD, Sargeson AM, Huston JS, Packard AB. Proc Natl Acad Sci USA. 2007;104:17489–17493. doi: 10.1073/pnas.0708436104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian XB, Winter R, Aruva MR, Zhang KJ, Cardi CA, Thakur ML, Wickstrom E. Clin Cancer Res. 2005;11:9130S–9130S. [Google Scholar]
- 56.Vavere AL, Biddlecombe GB, Spees WM, Garbow JR, Wijesinghe D, Andreev OA, Engelman DM, Reshetnyak YK, Lewis JS. Cancer Res. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dehdashti F, Mintun MA, Lewis JS, Bradley J, Govindan R, Laforest R, Welch MJ, Siegel BA. Eur J Nucl Med Mol Imaging. 2003;30:844–850. doi: 10.1007/s00259-003-1130-4. [DOI] [PubMed] [Google Scholar]
- 58.Bigott HM, Parent E, Luyt LG, Katzenellenbogen JA, Welch MJ. Bioconjugate Chem. 2005;16:255–264. doi: 10.1021/bc049770g. [DOI] [PubMed] [Google Scholar]
- 59.Buck A, Nguyen N, Burger C, Ziegler S, Frey L, Weigand G, Erhardt W, Senekowitsch-Schmidtke R, Pellikka R, Blauenstein P, Locher JT, Schwaiger M. Eur J Nucl Med Mol Imaging. 1996;23:1619–1627. doi: 10.1007/BF01249625. [DOI] [PubMed] [Google Scholar]
- 60.Haynes NG, Lacy JL, Nayak N, Martin CS, Dai D, Mathias CJ, Green MA. J Nucl Med. 2000;41:309–314. [PubMed] [Google Scholar]
- 61.Vavere AL, Welch MJ. J Nucl Med. 2005;46:683–690. [PubMed] [Google Scholar]
- 62.Bruehlmeier M, Leenders KL, Vontobel P, Calonder C, Antonini A, Weindl A. J Nucl Med. 2000;41:781–787. [PubMed] [Google Scholar]
- 63.Srivastava SC, Kolsky KL, Mease RC, Joshi V, Meinken GE, Pyatt B, Wolf AP, Schlyder DJ, Levy AV, Fowler JS. J Labelled Comp Rad. 1994;35:389–391. [Google Scholar]
- 64.Wallberg H, Ahlgren S, Widstrom C, Orlova A. Mol Imaging Biol. 2010;12:54–62. doi: 10.1007/s11307-009-0238-8. [DOI] [PubMed] [Google Scholar]
- 65.Ugur O, Kothari PJ, Finn RD, Zanzonico P, Ruan S, Guenther I, Maecke HR, Larson SM. Nucl Med Biol. 2002;29:147–157. doi: 10.1016/s0969-8051(01)00290-6. [DOI] [PubMed] [Google Scholar]
- 66.Lubberink M, Tolmachev V, Widstrom C, Bruskin A, Lundqvist H, Westlin JE. J Nucl Med. 2002;43:1391–1397. [PubMed] [Google Scholar]
- 67.Jennewein M, Hermanne A, Mason RP, Thorpe PE, Rosch F. Nucl Instrum Methods Phys Res, Sect A. 2006;569:512–517. [Google Scholar]
- 68.Jennewein M, Lewis MA, Zhao D, Tsyganov E, Slavine N, He J, Watkins L, Kodibagkar VD, O'Kelly S, Kulkarni P, Antich PP, Hermanne A, Rosch F, Mason RP, Thorpe PE. Clin Cancer Res. 2008;14:1377–1385. doi: 10.1158/1078-0432.CCR-07-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ehrhardt GJ, Welch MJ. J Nucl Med. 1978;19:925–929. [PubMed] [Google Scholar]
- 70.Zhernosekov KP, Filosofov DV, Baum RP, Aschoff P, Bihl H, Razbash AA, Jahn M, Jennewein M, Rosch F. J Nucl Med. 2007;48:1741–1748. doi: 10.2967/jnumed.107.040378. [DOI] [PubMed] [Google Scholar]
- 71.Sadeghi M, Aboudzadeh M, Zali A, Mirzaii M, Bolourinovin F. Appl Radiat Isot. 2009;67:7–10. doi: 10.1016/j.apradiso.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 72.Sadeghi M, Aboudzadeh M, Zali A, Zeinali B. Appl Radiat Isot. 2009;67:1392–1396. doi: 10.1016/j.apradiso.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 73.Sadeghi M, Zali A, Aboudzadeh M, Sarabadani P, Aslani G, Majdabadi A. Appl Radiat Isot. 2009;67:2029–2032. doi: 10.1016/j.apradiso.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 74.Park LS, Szajek LP, Wong KJ, Plascjak PS, Garmestani K, Googins S, Eckelman WC, Carrasquillo JA, Paik CH. Nucl Med Biol. 2004;31:297–301. doi: 10.1016/j.nucmedbio.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 75.Avila-Rodriguez MA, Nye JA, Nickles RJ. Appl Radiat Isot. 2008;66:9–13. doi: 10.1016/j.apradiso.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 76.Lukic D, Tamburella C, Buchegger F, Beyer GJ, Comor JJ, Seimbille Y. Appl Radiat Isot. 2009;67:523–529. doi: 10.1016/j.apradiso.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 77.Yoo J, Tang L, Perkins TA, Rowland DJ, Laforest R, Lewis JS, Welch MJ. Nucl Med Biol. 2005;32:891–897. doi: 10.1016/j.nucmedbio.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 78.DeJesus OT, Nickles RJ. Int J Radiat Appl Instrum, Part A. 1990;41:789–790. doi: 10.1016/0883-2889(90)90001-w. [DOI] [PubMed] [Google Scholar]
- 79.Zweit J, Downey S, Sharma HL. Int J Radiat Appl Instrum, Part A. 1991;42:199–201. [Google Scholar]
- 80.Link JM, Krohn KA, Eary JF, Kishore R, Lewellen TK, Johnson MW, Badger CC, Richter KY, Nelp WB. J Labelled Comp Rad. 1986;23:1297–1298. [Google Scholar]
- 81.Verel I, Visser GWM, Boellaard R, Stigter-van Walsum M, Snow GB, van Dongen G. J Nucl Med. 2003;44:1271–1281. [PubMed] [Google Scholar]
- 82.Meijs WE, Herscheid JDM, Haisma HJ, Wijbrandts R, van Langevelde F, van Leuffen PJ, Mooy R, Pinedo HM. Appl Radiat Isot. 1994;45:1143–1147. [Google Scholar]
- 83.Holland JP, Sheh YC, Lewis JS. Nucl Med Biol. 2009;36:729–739. doi: 10.1016/j.nucmedbio.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Obata A, Kasamatsu S, McCarthy DW, Welch MJ, Saji H, Yonekura Y, Fujibayashi Y. Nucl Med Biol. 2003;30:535–539. doi: 10.1016/s0969-8051(03)00024-6. [DOI] [PubMed] [Google Scholar]
- 85.McCarthy DW, Shefer RE, Klinkowstein RE, Bass LA, Margeneau WH, Cutler CS, Anderson CJ, Welch MJ. Nucl Med Biol. 1997;24:35–43. doi: 10.1016/s0969-8051(96)00157-6. [DOI] [PubMed] [Google Scholar]
- 86.Avila-Rodriguez MA, Nye JA, Nickles RJ. Appl Radiat Isot. 2007;65:1115–1120. doi: 10.1016/j.apradiso.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 87.Kim JY, Park H, Lee JC, Kim KM, Lee KC, Ha HJ, Choi TH, An GI, Cheon GJ. Appl Radiat Isot. 2009;67:1190–1194. doi: 10.1016/j.apradiso.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 88.Le VS, Howse J, Zaw M, Pellegrini P, Katsifis A, Greguric I, Weiner R. Appl Radiat Isot. 2009;67:1324–1331. doi: 10.1016/j.apradiso.2009.02.069. [DOI] [PubMed] [Google Scholar]
- 89.Kozempel J, Abbas K, Simonelli F, Zampese M, Holzwarth U, Gibson N, Leseticky L. Radiochim Acta. 2007;95:75–80. [Google Scholar]
- 90.Abbas K, Kozempel J, Bonardi M, Groppi F, Alfarano A, Holzwarth U, Simonelli F, Hofman H, Horstmann W, Menapace E, Leseticky L, Gibson N. Appl Radiat Isot. 2006;64:1001–1005. doi: 10.1016/j.apradiso.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 91.Hilgers K, Stoll T, Skakun Y, Coenen HH, Qaim SM. Appl Radiat Isot. 2003;59:343–351. doi: 10.1016/s0969-8043(03)00199-4. [DOI] [PubMed] [Google Scholar]
- 92.Van So L, Pellegrini P, Katsifis A, Howse J, Greguric I. J Radioanal Nucl Chem. 2008;277:451–466. [Google Scholar]
- 93.Bakaj M, Zimmer M. J Mol Struct. 1999;508:59–72. [Google Scholar]
- 94.Pasquarello A, Petri I, Salmon PS, Parisel O, Car R, Toth E, Powell DH, Fischer HE, Helm L, Merbach AE. Science. 2001;291:856–859. doi: 10.1126/science.291.5505.856. [DOI] [PubMed] [Google Scholar]
- 95.Delgado R, Felix V, Lima LMP, Price DW. Dalton Trans. 2007:2734–2745. doi: 10.1039/b704360k. [DOI] [PubMed] [Google Scholar]
- 96.Sun X, Kim J, Martell AE, Welch MJ, Anderson CJ. Nucl Med Biol. 2004;31:1051–1059. doi: 10.1016/j.nucmedbio.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 97.Smith SV. Q J Nucl Med Mol Imag. 2008;52:193–202. [PubMed] [Google Scholar]
- 98.Sun X, Wuest M, Weisman GR, Wong EH, Reed DP, Boswell CA, Motekaitis R, Martell AE, Welch MJ, Anderson CJ. J Med Chem. 2002;45:469–477. doi: 10.1021/jm0103817. [DOI] [PubMed] [Google Scholar]
- 99.Wadas TJ, Anderson CJ. Nat Protoc. 2006;1:3062–3068. doi: 10.1038/nprot.2006.431. [DOI] [PubMed] [Google Scholar]
- 100.Li WP, Meyer LA, Capretto DA, Sherman CD, Anderson CJ. Cancer Biother Radiopharm. 2008;23:158–171. doi: 10.1089/cbr.2007.0444. [DOI] [PubMed] [Google Scholar]
- 101.Wong EH, Weisman GR, Hill DC, Reed DP, Rogers ME, Condon JS, Fagan MA, Calabrese JC, Lam KC, Guzei IA, Rheingold AL. J Am Chem Soc. 2000;122:10561–10572. [Google Scholar]
- 102.Bandoli G, Dolmella A, Tisato F, Porchia M, Refosco F. Coord Chem Rev. 2009;253:56–77. [Google Scholar]
- 103.Weiner RE, Thakur ML. Handbook of Radiopharmaceuticals. 2003:363–399. [Google Scholar]
- 104.Francesconi LC, Liu BL, Billings JJ, Carroll PJ, Graczyk G, Kung HF. J Chem Soc, Chem Commun. 1991:94–95. [Google Scholar]
- 105.Jarjayes O, Mortini F, d'Hardemare AD, Philouze C, Serratrice G. Eur J Inorg Chem. 2005:4417–4424. [Google Scholar]
- 106.Motekaitis RJ, Martell AE, Koch SA, Hwang JW, Quarless DA, Welch MJ. Inorg Chem. 1998;37:5902–5911. [Google Scholar]
- 107.Blend MJ, Stastny JJ, Swanson SM, Brechbiel MW. Cancer Biother Radiopharm. 2003;18:355–363. doi: 10.1089/108497803322285107. [DOI] [PubMed] [Google Scholar]
- 108.Li YJ, Martell AE, Hancock RD, Reibenspies JH, Anderson CJ, Welch MJ. Inorg Chem. 1996;35:404–414. doi: 10.1021/ic941330l. [DOI] [PubMed] [Google Scholar]
- 109.Ma R, Motekaitis RJ, Martell AE. Inorg Chim Acta. 1994;224:151–155. [Google Scholar]
- 110.Martell AE, Motekaitis R, Clarke ET, Delgado R, Sun YZ, Ma R. Supramol Chem. 1997;8:253–253. [Google Scholar]
- 111.Fani M, Andre JP, Maecke HR. Contrast Media Mol Imaging. 2008;3:53–63. doi: 10.1002/cmmi.232. [DOI] [PubMed] [Google Scholar]
- 112.Eder M, Wangler B, Knackmuss S, LeGall F, Little M, Haberkorn U, Mier W, Eisenhut M. Eur J Nucl Med Mol Imaging. 2008;35:1878–1886. doi: 10.1007/s00259-008-0816-z. [DOI] [PubMed] [Google Scholar]
- 113.Clarke ET, Martell AE. Inorg Chim Acta. 1991;181:273–280. [Google Scholar]
- 114.Broan CJ, Cox JPL, Craig AS, Kataky R, Parker D, Harrison A, Randall AM, Ferguson G. J Chem Soc, Perkin Trans 2. 1991:87–99. [Google Scholar]
- 115.Parker D, Pulukkody K, Smith FC, Batsanov A, Howard JAK. J Chem Soc, Dalton Trans. 1994:689–693. [Google Scholar]
- 116.Kumar K, Chang CA, Francesconi LC, Dischino DD, Malley MF, Gougoutas JZ, Tweedle MF. Inorg Chem. 1994;33:3567–3575. [Google Scholar]
- 117.Amin S, Marks C, Toomey LM, Churchill MR, Morrow JR. Inorg Chim Acta. 1996;246:99–107. [Google Scholar]
- 118.Clifford T, Boswell CA, Biddlecombe GB, Lewis JS, Brechbiel MW. J Med Chem. 2006;49:4297–4304. doi: 10.1021/jm060317v. [DOI] [PubMed] [Google Scholar]
- 119.Schiller E, Bergmann R, Pietzsch J, Noll B, Sterger A, Johannsen B, Wunderlich G, Pietzsch HJ. Nucl Med Biol. 2008;35:227–232. doi: 10.1016/j.nucmedbio.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 120.Schlesinger J, Koezle I, Bergmann R, Tamburini S, Bolzati C, Tisato F, Noll B, Klussmann S, Vonhoff S, Wuest F, Pietzsch HJ, Steinbach J. Bioconjugate Chem. 2008;19:928–939. doi: 10.1021/bc700453h. [DOI] [PubMed] [Google Scholar]
- 121.Davidovich RL, Logvinova VB, Teplukhina LV. Koordinatsionnaya Khimiya. 1992;18:580–584. [Google Scholar]
- 122.Pozhidaev AI, Poraikoshits MA, Polynova TN. J Struct Chem. 1974;15:548–553. [Google Scholar]
- 123.Meijs WE, Haisma HJ, Klok RP, van Gog FB, Kievit E, Pinedo HM, Herscheid JDM. J Nucl Med. 1997;38:112–118. [PubMed] [Google Scholar]
- 124.Perk LR, Vosjan M, Visser GWM, Budde M, Jurek P, Kiefer GE, van Dongen G. Eur J Nucl Med Mol Imaging. 2010;37:250–259. doi: 10.1007/s00259-009-1263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Holland JP, Divilov V, Bander NH, Smith-Jones PM, Larson SM, Lewis JS. J Nucl Med. 2010;51:1293–1300. doi: 10.2967/jnumed.110.076174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meyer GJ, Macke HR, Schuhmacher J, Knapp WH, Hoffman TJ. Eur J Nucl Med Mol Imaging. 2003;31:1097–1104. doi: 10.1007/s00259-004-1486-0. [DOI] [PubMed] [Google Scholar]
- 127.Velikyan I, Sundberg AL, Lindhe O, Hoglund AU, Eriksson O, Werner E, Carlsson J, Bergstrom M, Langstrom B, Tolmachev V. J Nucl Med. 2005;46:1881–1888. [PubMed] [Google Scholar]
- 128.Dimitrakopoulou-Strauss A, Hohenberger P, Haberkorn U, Macke HR, Eisenhut M, Strauss LG. J Nucl Med. 2007;48:1245–1250. doi: 10.2967/jnumed.106.038091. [DOI] [PubMed] [Google Scholar]
- 129.Liu Z, Niu G, Shi J, Liu S, Wang F, Liu S, Chen X. Eur J Nucl Med Mol Imaging. 2009;36:947–957. doi: 10.1007/s00259-008-1045-1. [DOI] [PubMed] [Google Scholar]
- 130.Wei L, Zhang X, Gallazzi F, Miao Y, Jin X, Brechbiel MW, Xu H, Clifford T, Welch MJ, Lewis JS, Quinn TP. Nucl Med Biol. 2009;36:345–354. doi: 10.1016/j.nucmedbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Eisenwiener KP, Prata MIM, Buschmann I, Zhang HW, Santos AC, Wenger S, Reubi JC, Macke HR. Bioconjugate Chem. 2002;13:530–541. doi: 10.1021/bc010074f. [DOI] [PubMed] [Google Scholar]
- 132.Henze M, Schuhmacher J, Hipp P, Kowalski J, Becker DW, Doll J, Macke HR, Hofmann M, Debus J, Haberkorn U. J Nucl Med. 2001;42:1053–1056. [PubMed] [Google Scholar]
- 133.Heppeler A, Froidevaux S, Macke HR, Jermann E, Behe M, Powell P, Hennig M. Chem–Eur J. 1999;5:1974–1981. [Google Scholar]
- 134.Hofmann M, Maecke H, Borner AR, Weckesser E, Schoffski P, Oei ML, Schumacher J, Henze M, Heppeler A, Meyer GJ, Knapp WH. Eur J Nucl Med Mol Imaging. 2001;28:1751–1757. doi: 10.1007/s002590100639. [DOI] [PubMed] [Google Scholar]
- 135.Asti M, De Pietri G, Fraternali A, Grassi E, Sghedoni R, Floroni F, Roesch F, Versari A, Salvo D. Nucl Med Biol. 2008;35:721–724. doi: 10.1016/j.nucmedbio.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 136.Lendvai G, Monazzam A, Velikyan I, Eriksson B, Josephsson R, Langstrom B, Bergstrom M, Estrada S. Oligonucleotides. 2009;19:223–231. doi: 10.1089/oli.2009.0192. [DOI] [PubMed] [Google Scholar]
- 137.Roivainen A, Tolvanen T, Salomaki S, Lendvai G, Velikyan I, Numminen P, Valila M, Sipila H, Bergstrom M, Harkonen P, Lonnberg H, Langstrom B. J Nucl Med. 2004;45:347–355. [PubMed] [Google Scholar]
- 138.Schlesinger J, Bergmann R, Klussmann S, Wuest F. Lett Drug Des Discovery. 2006;3:330–335. [Google Scholar]
- 139.Anderson CJ, Green MA, Fujibayashi Y. Handbook of Radiopharmaceuticals. 2003:401–422. [Google Scholar]
- 140.Mindt TL, Muller C, Stuker F, Salazar JF, Hohn A, Mueggler T, Rudin M, Schibli R. Bioconjugate Chem. 2009;20:1940–1949. doi: 10.1021/bc900276b. [DOI] [PubMed] [Google Scholar]
- 141.Nwe K, Brechbiel MW. Cancer Biother Radiopharm. 2009;24:289–301. doi: 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Glaser M, Robins EG. J Labelled Compd Radiopharm. 2009;52:407–414. [Google Scholar]
- 143.Waengler C, Schirrmacher R, Bartenstein P, Waengler B. Curr Med Chem. 2010;17:1092–1116. doi: 10.2174/092986710790820615. [DOI] [PubMed] [Google Scholar]
- 144.Wang C, Wang N, Zhou W, Shen YM, Zhang L. Progress in Chemistry. 2010;22:1591–1602. [Google Scholar]
- 145.Glaser M, Solbakken M, Turton DR, Pettitt R, Barnett J, Arukwe J, Karlsen H, Cuthbertson A, Luthra SK, Arstad E. Amino Acids. 2008;37:717–724. doi: 10.1007/s00726-008-0200-0. [DOI] [PubMed] [Google Scholar]
- 146.Marik J, Sutcliffe JL. Tetrahedron Lett. 2006;47:6681–6684. [Google Scholar]
- 147.Lebedev AY, Holland JP, Lewis JS. Chem Commun. 2009;46:1706–1708. doi: 10.1039/b924784j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Knor S, Modlinger A, Poethko T, Schottelius M, Wester HJ, Kessler H. Chem–Eur J. 2007;13:6082–6090. doi: 10.1002/chem.200700231. [DOI] [PubMed] [Google Scholar]
- 149.Martin ME, Parameswarappa SG, O'Dorisio MS, Pigge FC, Schultz MK. Bioorg Med Chem Lett. 2010;20:4805–4807. doi: 10.1016/j.bmcl.2010.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schultz MK, Parameswarappa SG, Pigge FC. Org Lett. 2010;12:2398–2401. doi: 10.1021/ol100774p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Devaraj NK, Upadhyay R, Hatin JB, Hilderbrand SA, Weissleder R. Angew Chem, Int Ed. 2009;48:7013–7016. doi: 10.1002/anie.200903233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tanaka K, Masuyama T, Hasegawa K, Tahara T, Mizuma H, Wada Y, Watanabe Y, Fukase K. Angew Chem, Int Ed. 2008;47:102–105. doi: 10.1002/anie.200702989. [DOI] [PubMed] [Google Scholar]
- 154.Velikyan I, Beyer GJ, Langstrom B. Bioconjugate Chem. 2004;15:554–560. doi: 10.1021/bc030078f. [DOI] [PubMed] [Google Scholar]
- 155.Putzer D, Gabriel M, Henninger B, Kendler D, Uprimny C, Dobrozemsky G, Decristoforo C, Bale RJ, Jaschke W, Virgolini IJ. J Nucl Med. 2009;50:1214–1221. doi: 10.2967/jnumed.108.060236. [DOI] [PubMed] [Google Scholar]
- 156.Parry JJ, Kelly TS, Andrews R, Rogers BE. Bioconjugate Chem. 2007;18:1110–1117. doi: 10.1021/bc0603788. [DOI] [PubMed] [Google Scholar]
- 157.Nayak TK, Regino CAS, Wong KJ, Milenic DE, Garmestani K, Baidoo KE, Szajek LP, Brechbiel MW. Eur J Nucl Med Mol Imaging. 2010;37:1368–1376. doi: 10.1007/s00259-009-1370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. [Accessed October 11, 2010];Brookhaven National Laboratory: National Nuclear Decay Center. 2010 http://www.nndcbnl.gov.
- 159.Welch MJ, Redvanly CS, editors. Handbook of Radiopharmaceuticals: Radiochemistry and Applications. Wiley; New York: 2003. [Google Scholar]
- 160.Maecke HR, Andre JP. Ernst Schering Res Found Workshop. 2007;62:215–242. doi: 10.1007/978-3-540-49527-7_8. [DOI] [PubMed] [Google Scholar]
- 161.Ehrhardt GJ, Welch MJ. J Nucl Med. 1978;19:925–929. [PubMed] [Google Scholar]
- 162.Dejesus OT, Nickles RJ. Int J Radiat Appl Instrum, Part A. 1990;41:789–790. doi: 10.1016/0883-2889(90)90001-w. [DOI] [PubMed] [Google Scholar]
- 163.Eary JF, Link JM, Kishore R, Johnson MW, Badger CC, Richter KY, Krohn KA, Nelp WB. J Nucl Med. 1986;27:983–983. [Google Scholar]
- 164.Meijs WE, Herscheid JDM, Haisma HJ, Wijbrandts R, Vanlangevelde F, Vanleuffen PJ, Mooy R, Pinedo HM. Appl Radiat Isot. 1994;45:1143–1147. [Google Scholar]
- 165.Martell AE, Smith RM. Critical Stability Constants. Plenum Press; New York: 1982. [Google Scholar]
- 166.Martell AE, Hancock RD. Metal Complexes in Aqueous Solutions. Plenum Press; New York: 1996. [Google Scholar]
- 167.Bevilacqua A, Gelb RI, Hebard WB, Zompa LJ. Inorg Chem. 1987;26:2699–2706. [Google Scholar]
- 168.Martell AE, Motekaitis RJ, Clarke ET, Delgado R, Sun YZ, Ma R. Supramol Chem. 1996;6:353–363. [Google Scholar]
- 169.Delgado R, Dasilva JJR. Talanta. 1982;29:815–822. doi: 10.1016/0039-9140(82)80251-8. [DOI] [PubMed] [Google Scholar]
- 170.Chaves S, Delgado R, Dasilva J. Talanta. 1992;39:249–254. doi: 10.1016/0039-9140(92)80028-c. [DOI] [PubMed] [Google Scholar]
- 171.Woodin KS, Heroux KJ, Boswell CA, Wong EH, Weisman GR, Niu WJ, Tomellini SA, Anderson CJ, Zakharov LN, Rheingold AL. Eur J Inorg Chem. 2005:4829–4833. [Google Scholar]
- 172.Bharadwaj PK, Potenza JA, Schugar HJ. J Am Chem Soc. 1986;108:1351–1352. [Google Scholar]
- 173.Bernhardt PV, Bramley R, Engelhardt LM, Harrowfield JM, Hockless DCR, Korybutdaszkiewicz BR, Krausz ER, Morgan T, Sargeson AM, Skelton BW, White AH. Inorg Chem. 1995;34:3589–3599. [Google Scholar]
- 174.Delgado R, Figueira MD, Quintino S. Talanta. 1997;45:451–462. doi: 10.1016/s0039-9140(97)00157-4. [DOI] [PubMed] [Google Scholar]
- 175.Smith RM, Martell AE. Critical Stability Constants. Plenum Press; New York: 1989. [Google Scholar]
- 176.Ma R, Motekaitis RJ, Martell AE. Inorg Chim Acta. 1994;224:151–155. [Google Scholar]
- 177.Motekaitis RJ, Sun YZ, Martell AE. Inorg Chem. 1991;30:1554–1556. [Google Scholar]
- 178.Delgado R, Sun YZ, Motekaitis RJ, Martell AE. Inorg Chem. 1993;32:3320–3326. [Google Scholar]
- 179.Terova O. University of New Hampshire; 2008. [Google Scholar]
- 180.Evers A, Hancock RD, Martell AE, Motekaitis RJ. Inorg Chem. 1989;28:2189–2195. [Google Scholar]
- 181.Martell AE, Smith RM. Critical Stability Constants, Vol 1: Amino Acids. Plenum Press; New York: 1974. [Google Scholar]
- 182.Koudelkova M, Vinsova H, Jedinakova-Krizova V. Czech J Phys. 2003;53:A769–A775. [Google Scholar]
- 183.Kodama M, Koike T, Mahatma AB, Kimura E. Inorg Chem. 1991;30:1270–1273. [Google Scholar]
- 184.Bottari E, Anderegg G. Helv Chim Acta. 1967;50:2349–2355. [Google Scholar]
- 185.Ujula T, Salomaki S, Virsu P, Lankinen P, Makinen TJ, Autio A, Yegutkin GG, Knuuti J, Jalkanen S, Roivainen A. Nucl Med Biol. 2009;36:631–641. doi: 10.1016/j.nucmedbio.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 186.Schuhmacher J, Zhang HW, Doll J, Macke HR, Matys R, Hauser H, Henze M, Haberkorn U, Eisenhut M. J Nucl Med. 2005;46:691–699. [PubMed] [Google Scholar]
- 187.Maschauer S, Einsiedel J, Hocke C, Hubner H, Kuwert T, Gmeiner P, Prante O. ACS Med Chem Lett. 2010;1:224–228. doi: 10.1021/ml1000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wild D, Schmitt JS, Ginj M, Macke HR, Bernard BF, Krenning E, de Jong M, Wenger S, Reubi JC. Eur J Nucl Med Mol Imaging. 2003;30:1338–1347. doi: 10.1007/s00259-003-1255-5. [DOI] [PubMed] [Google Scholar]
- 189.de Jong J, Bakker W, Krenning E, Breeman WAP, Van Der Pluijm ME, Bernard BF, Visser TJ, Jermann E, Behe M, Powell P, Macke HR. Eur J Nucl Med Mol Imaging. 1997;24:368–371. doi: 10.1007/BF00881807. [DOI] [PubMed] [Google Scholar]
- 190.Zhang HW, Chen JH, Waldherr C, Hinni K, Waser B, Reubi JC, Maecke HR. Cancer Res. 2004;64:6707–6715. doi: 10.1158/0008-5472.CAN-03-3845. [DOI] [PubMed] [Google Scholar]
- 191.Zhang HW, Schuhmacher J, Waser B, Wild D, Eisenhut M, Reubi JC, Maecke HR. Eur J Nucl Med Mol Imaging. 2007;34:1198–1208. doi: 10.1007/s00259-006-0347-4. [DOI] [PubMed] [Google Scholar]
- 192.Froidevaux S, Calame-Christe M, Schuhmacher J, Tanner H, Saffrich R, Henze M, Eberle AN. J Nucl Med. 2004;45:116–123. [PubMed] [Google Scholar]
- 193.Froidevaux S, Calame-Christe M, Tanner H, Sumanovski L, Eberle AN. J Nucl Med. 2002;43:1699–1706. [PubMed] [Google Scholar]
- 194.Eder M, Krivoshein AV, Backer M, Backer JM, Haberkorn U, Eisenhut M. Nucl Med Biol. 2010;37:405–412. doi: 10.1016/j.nucmedbio.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 195.Backer MV, Levashova Z, Patel V, Jehning BT, Claffey K, Blankenberg FG, Backer JM. Nat Med. 2007;13:504–509. doi: 10.1038/nm1522. [DOI] [PubMed] [Google Scholar]
- 196.Liu ZF, Yan YJ, Liu SL, Wang F, Chen XY. Bioconjugate Chem. 2009;20:1016–1025. doi: 10.1021/bc9000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jeong JM, Hong MK, Chang YS, Lee YS, Kim YJ, Cheon GJ, Lee DS, Chung JK, Lee MC. J Nucl Med. 2008;49:830–836. doi: 10.2967/jnumed.107.047423. [DOI] [PubMed] [Google Scholar]
- 198.Smithjones PM, Stolz B, Bruns C, Albert R, Reist HW, Fridrich R, Macke HR. J Nucl Med. 1994;35:317–325. [PubMed] [Google Scholar]
- 199.Hnatowich DJ, Layne WW, Childs RL. Int J Appl Radiat Isot. 1982;33:327–332. doi: 10.1016/0020-708x(82)90144-2. [DOI] [PubMed] [Google Scholar]
- 200.Moerlein SM, Daugherty A, Sobel BE, Welch MJ. J Nucl Med. 1991;32:300–307. [PubMed] [Google Scholar]
- 201.Wagner SJ, Welch MJ. J Nucl Med. 1979;20:428–433. [PubMed] [Google Scholar]
- 202.Eder M, Knackmuss S, Le Gall F, Reusch U, Rybin V, Little M, Haberkorn U, Mier W, Eisenhut M. Eur J Nucl Med Mol Imaging. 2010;37:1397–1407. doi: 10.1007/s00259-010-1392-6. [DOI] [PubMed] [Google Scholar]
- 203.Smith-Jones PM, Vallabahajosula S, Goldsmith SJ, Navarro V, Hunter CJ, Bastidas D, Bander NH. Cancer Res. 2000;60:5237–5243. [PubMed] [Google Scholar]
- 204.Smith-Jones PM, Solit DB, Akhurst T, Alfroze F, Rosen N, Larson SM. Nat Biotechnol. 2004;22:701–706. doi: 10.1038/nbt968. [DOI] [PubMed] [Google Scholar]
- 205.Smith-Jones PM, Solit DB, Afroze F, Rosen N, Larson SM. J Nucl Med. 2006;47:793–796. [PMC free article] [PubMed] [Google Scholar]
- 206.Baum RP, Prasad V, Muller D, Schuchardt C, Orlova A, Wennborg A, Tolmachev V, Feldwisch J. J Nucl Med. 2010;51:892–897. doi: 10.2967/jnumed.109.073239. [DOI] [PubMed] [Google Scholar]
- 207.Orlova A, Tolmachev V, Pehrson R, Lindborg M, Tran T, Sandstrom M, Nilsson FY, Wennborg A, Abrahmsen L, Feldwisch J. Cancer Res. 2007;67:2178–2186. doi: 10.1158/0008-5472.CAN-06-2887. [DOI] [PubMed] [Google Scholar]
- 208.Tolmachev V, Velikyan I, Sandstrom M, Orlova A. Eur J Nucl Med Mol Imaging. 2010;37:1356–1367. doi: 10.1007/s00259-009-1367-7. [DOI] [PubMed] [Google Scholar]
- 209.Otsuka FL, Welch MJ, Kilbourn MR, Dence CS, Dilley WG, Wells SA. Int J Radiat Appl Instrum, Part A. 1991;18:813–816. doi: 10.1016/0883-2897(91)90023-e. [DOI] [PubMed] [Google Scholar]
- 210.Otsuka FL, Cance WG, Dilley WG, Scott RW, Davie JM, Wells SA, Welch MJ. Int J Radiat Appl Instrum, Part A. 1988;15:305–311. doi: 10.1016/0883-2897(88)90111-0. [DOI] [PubMed] [Google Scholar]
- 211.Koop B, Reske SN, Neumaier B. Radiochim Acta. 2007;95:39–42. [Google Scholar]
- 212.Tian XB, Aruva MR, Zhang KJ, Shanthly N, Cardi CA, Thakur ML, Wickstrom E. J Nucl Med. 2007;48:1699–1707. doi: 10.2967/jnumed.107.042499. [DOI] [PubMed] [Google Scholar]
- 213.Sun XK, Fang HF, Li XX, Rossin R, Welch MJ, Taylor JS. Bioconjugate Chem. 2005;16:294–305. doi: 10.1021/bc049783u. [DOI] [PubMed] [Google Scholar]
- 214.Lendvai G, Velikyan I, Bergstrom M, Estrada S, Laryea D, Valila M, Salomaki S, Langstrom B. Eur J Pharm Sci. 2005;26:26–38. doi: 10.1016/j.ejps.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 215.Tian XB, Chakrabarti A, Amirkhanov NV, Aruva MR, Zhang KJ, Mathew B, Cardi C, Qin WY, Sauter ER, Thakur ML, Wickstrom E. Tumor Progression and Therapeutic Resistance. 2005;1059:106–144. doi: 10.1196/annals.1339.038. [DOI] [PubMed] [Google Scholar]
- 216.Rogers BE, Bigott HM, McCarthy DW, Della Manna D, Kim J, Sharp TL, Welch MJ. Bioconjugate Chem. 2003;14:756–763. doi: 10.1021/bc034018l. [DOI] [PubMed] [Google Scholar]
- 217.Parry JJ, Andrews R, Rogers BE. Breast Cancer Res Treat. 2007;101:175–183. doi: 10.1007/s10549-006-9287-8. [DOI] [PubMed] [Google Scholar]
- 218.Edwards WB, Xu B, Akers W, Cheney PP, Liang K, Rogers BE, Anderson CJ, Achilefu S. Bioconjugate Chem. 2008;19:192–200. doi: 10.1021/bc700291m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen JQ, Cheng Z, Owen NK, Hoffman TJ, Miao YB, Jurisson SS, Quinn TP. J Nucl Med. 2001;42:1847–1855. [PubMed] [Google Scholar]
- 220.McQuade P, Miao YB, Yoo J, Quinn TP, Welch MJ, Lewis JS. J Med Chem. 2005;48:2985–2992. doi: 10.1021/jm0490282. [DOI] [PubMed] [Google Scholar]
- 221.Garrison JC, Rold TL, Sieckman GL, Figueroa SD, Volkert WA, Jurisson SS, Hoffman TJ. J Nucl Med. 2007;48:1327–1337. doi: 10.2967/jnumed.107.039487. [DOI] [PubMed] [Google Scholar]
- 222.Hausner SH, Kukis DL, Gagnon MKJ, Stanecki CE, Ferdani R, Marshall JF, Anderson CJ, Sutcliffe JL. Mol Imaging. 2009;8:111–121. [PMC free article] [PubMed] [Google Scholar]
- 223.Biddlecombe GB, Rogers BE, de Visser M, Parry JJ, de Jong M, Erion JL, Lewis JS. Bioconjugate Chem. 2007;18:724–730. doi: 10.1021/bc060281l. [DOI] [PubMed] [Google Scholar]
- 224.Chen XY, Liu S, Hou YP, Tohme M, Park R, Bading JR, Conti PS. Mol Imaging Biol. 2004;6:350–359. doi: 10.1016/j.mibio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 225.Chen XY, Hou YP, Tohme M, Park R, Khankaldyyan V, Gonzales-Gomez I, Bading JR, Laug WE, Conti PS. J Nucl Med. 2004;45:1776–1783. [PubMed] [Google Scholar]
- 226.Lee HY, Li Z, Chen K, Hsu AR, Xu CJ, Xie J, Sun SH, Chen XY. J Nucl Med. 2008;49:1371–1379. doi: 10.2967/jnumed.108.051243. [DOI] [PubMed] [Google Scholar]
- 227.Wu Y, Zhang XZ, Xiong ZM, Cheng Z, Fisher DR, Liu S, Gambhir SS, Chen XY. J Nucl Med. 2005;46:1707–1718. [PubMed] [Google Scholar]
- 228.Li ZB, Cai WB, Cao QZ, Chen K, Wu ZH, He LN, Chen XY. J Nucl Med. 2007;48:1162–1171. doi: 10.2967/jnumed.107.039859. [DOI] [PubMed] [Google Scholar]
- 229.Chen XY, Park R, Hou YP, Tohme M, Shahinian AH, Bading JR, Conti PS. J Nucl Med. 2004;45:1390–1397. [PubMed] [Google Scholar]
- 230.Cai WB, Chen K, Mohamedali KA, Cao QZ, Gambhir SS, Rosenblum MG, Chen XY. J Nucl Med. 2006;47:2048–2056. [PubMed] [Google Scholar]
- 231.Wang H, Cai WB, Chen K, Li ZB, Kashefi A, He L, Chen XY. Eur J Nucl Med Mol Imaging. 2007;34:2001–2010. doi: 10.1007/s00259-007-0524-0. [DOI] [PubMed] [Google Scholar]
- 232.Li ZB, Niu G, Wang H, He L, Yang L, Ploug M, Chen XY. Clin Cancer Res. 2008;14:4758–4766. doi: 10.1158/1078-0432.CCR-07-4434. [DOI] [PubMed] [Google Scholar]
- 233.Cao Q, Cai W, Niu G, He L, Chen X. Clin Cancer Res. 2008;14:6137–6145. doi: 10.1158/1078-0432.CCR-08-0049. [DOI] [PubMed] [Google Scholar]
- 234.Cheng Z, Xiong ZM, Subbarayan M, Chen XY, Gambhir SS. Bioconjugate Chem. 2007;18:765–772. doi: 10.1021/bc060306g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jiang L, Kimura RH, Miao Z, Silverman AP, Ren G, Liu HG, Li PY, Gambhir SS, Cochran JR, Cheng Z. J Nucl Med. 2010;51:251–258. doi: 10.2967/jnumed.109.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Cancer Res. 2009;69:2435–2442. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liu YJ, Abendschein D, Woodard GE, Rossin R, McCommis K, Zheng J, Welch MJ, Woodard PK. J Nucl Med. 2010;51:85–91. doi: 10.2967/jnumed.109.066977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rossin R, Muro S, Welch MJ, Muzykantov VR, Schuster DP. J Nucl Med. 2008;49:103–111. doi: 10.2967/jnumed.107.045302. [DOI] [PubMed] [Google Scholar]
- 239.Liu DJ, Overbey D, Watkinson LD, Smith CJ, Daibes-Figueroa S, Hoffman TJ, Forte LR, Volkert WA, Giblin MF. Bioconjugate Chem. 2010;21:1171–1176. doi: 10.1021/bc900513u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Locke LW, Chordia MD, Zhang Y, Kundu B, Kennedy D, Landseadel J, Xiao L, Fairchild KD, Berr SS, Linden J, Pan DF. J Nucl Med. 2009;50:790–797. doi: 10.2967/jnumed.108.056127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Levashova Z, Backer MV, Horng G, Felsher D, Backer JM, Blankenberg FG. Bioconjugate Chem. 2009;20:742–749. doi: 10.1021/bc800443w. [DOI] [PubMed] [Google Scholar]
- 242.Vavere AL, Biddlecombe GB, Spees WM, Garbow JR, Wijesinghe D, Andreev OA, Englenman DM, Reshetnyak YK, Lewis JS. Cancer Res. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wei LH, Butcher C, Miao YB, Gallazzi F, Quinn TP, Welch MJ, Lewis JS. J Nucl Med. 2007;48:64–72. [PubMed] [Google Scholar]
- 244.Wadas TJ, Eiblmaier M, Zheleznyak A, Sherman CD, Ferdani R, Liang K, Achilefu S, Anderson CJ. J Nucl Med. 2008;49:1819–1827. doi: 10.2967/jnumed.108.054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sprague ES, Peng Y, Sun X, Weisman GR, Wong EH, Achilefu S, Anderson CJ. Clin Cancer Res. 2004;10:8674–8682. doi: 10.1158/1078-0432.CCR-04-1084. [DOI] [PubMed] [Google Scholar]
- 246.DeNardo SJ, Liu R, Albrecht H, Natarajan A, Sutcliffe JL, Anderson C, Peng L, Ferdani R, Cherry SR, Lam KS. J Nucl Med. 2009;50:625–634. doi: 10.2967/jnumed.108.056903. [DOI] [PubMed] [Google Scholar]
- 247.Wei L, Ye Y, Wadas TJ, Lewis JS, Welch MJ, Achilefu S, Anderson CJ. Nucl Med Biol. 2009;36:277–285. doi: 10.1016/j.nucmedbio.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sprague JE, Kitaura H, Zou W, Ye YP, Achilefu S, Weilbaecher KN, Teitelbaum SL, Anderson CJ. J Nucl Med. 2007;48:311–318. [PMC free article] [PubMed] [Google Scholar]
- 249.Anderson CJ, Pajeau TS, Edwards WB, Sherman ELC, Rogers BE, Welch MJ. J Nucl Med. 1995;36:2315–2325. [PubMed] [Google Scholar]
- 250.Anderson CJ, Jones LA, Bass LA, Sherman ELC, McCarthy DW, Cutler PD, Lanahan MV, Cristel ME, Lewis JS, Schwarz SW. J Nucl Med. 1998;39:1944–1951. [PubMed] [Google Scholar]
- 251.Lewis JS, Lewis MR, Cutler PD, Srinivasan A, Schmidt MA, Schwarz SW, Morris MM, Miller JP, Anderson CJ. Clin Cancer Res. 1999;5:3608–3616. [PubMed] [Google Scholar]
- 252.Lewis JS, Lewis MR, Srinivasan A, Schmidt MA, Wang J, Anderson CJ. J Med Chem. 1999;42:1341–1347. doi: 10.1021/jm980602h. [DOI] [PubMed] [Google Scholar]
- 253.Lewis JS, Srinivasan A, Schmidt MA, Anderson CJ. Nucl Med Biol. 1999;26:267–273. doi: 10.1016/s0969-8051(98)00105-x. [DOI] [PubMed] [Google Scholar]
- 254.Edwards WB, Anderson CJ, Fields GB, Welch MJ. Bioconjugate Chem. 2001;12:1057–1065. doi: 10.1021/bc015520d. [DOI] [PubMed] [Google Scholar]
- 255.Prasanphanich AF, Nanda PK, Rold TL, Ma LX, Lewis MR, Garrison JC, Hoffman TJ, Sieckman GL, Figueroa SD, Smith CJ. Proc Natl Acad Sci USA. 2007;104:12462–12467. doi: 10.1073/pnas.0705347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Prasanphanich AF, Retzloff L, Lane SR, Nanda PK, Sieckman GL, Rold TL, Ma LX, Figueroa SD, Sublett SV, Hoffman TJ, Smith CJ. Nucl Med Biol. 2009;36:171–181. doi: 10.1016/j.nucmedbio.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Thakur ML, Aruva MR, Gariepy J, Acton P, Rattan S, Prasad S, Wickstrom E, Alavi A. J Nucl Med. 2004;45:1381–1389. [PMC free article] [PubMed] [Google Scholar]
- 258.Zhang K, Aruva MR, Shanthly N, Cardi CA, Rattan S, Patel C, Kim C, McCue PA, Wickstrom E, Thakur ML. J Nucl Med. 2008;49:112–121. doi: 10.2967/jnumed.107.043703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Gasser G, Tjioe L, Graham B, Belousoff MJ, Juran S, Walther M, Kunstler JU, Bergmann R, Stephan H, Spiccia L. Bioconjugate Chem. 2008;19:719–730. doi: 10.1021/bc700396e. [DOI] [PubMed] [Google Scholar]
- 260.Cai HC, Fissekis J, Conti PS. Dalton Trans. 2009:5395–5400. doi: 10.1039/b902210d. [DOI] [PubMed] [Google Scholar]
- 261.Cai HC, Li ZB, Huang CW, Shahinian AH, Wang H, Park R, Conti PS. Bioconjugate Chem. 2010;21:1417–1424. doi: 10.1021/bc900537f. [DOI] [PubMed] [Google Scholar]
- 262.Cheng Z, De Jesus OP, Kramer DJ, De A, Webster JM, Gheysens O, Levi J, Namavari M, Wang S, Park JM, Zhang R, Liu H, Lee B, Syud FA, Gambhir SS. Mol Imaging Biol. 2010;12:316–324. doi: 10.1007/s11307-009-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Olafsen T, Kenanova VE, Sundaresan G, Anderson AL, Crow D, Yazaki PJ, Li L, Press MF, Gambhir SS, Williams LE, Wong JYC, Raubitschek AA, Shively JE, Wu AM. Cancer Res. 2005;65:5907–5916. doi: 10.1158/0008-5472.CAN-04-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lewis MR, Kao JY, Anderson ALJ, Shively JE, Raubitschek A. Bioconjugate Chem. 2001;12:320–324. doi: 10.1021/bc0000886. [DOI] [PubMed] [Google Scholar]
- 265.Niu G, Cai WB, Chen K, Chen XY. Mol Imaging Biol. 2008;10:99–106. doi: 10.1007/s11307-007-0123-2. [DOI] [PubMed] [Google Scholar]
- 266.Niu G, Li Z, Xie J, Le QT, Chen X. J Nucl Med. 2009;50:1116–1123. doi: 10.2967/jnumed.109.061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Paudyal P, Paudyal B, Hanaoka H, Oriuchi N, Iida Y, Yoshioka H, Tominaga H, Watanabe S, Ishioka NS, Endo K. Cancer Sci. 2010;101:1045–1050. doi: 10.1111/j.1349-7006.2010.01480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Li L, Bading J, Yazaki PJ, Ahuja AH, Crow D, Colcher D, Williams LE, Wong JYC, Raubitschek A, Shively JE. Bioconjugate Chem. 2008;19:89–96. doi: 10.1021/bc700161p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Li L, Turatti F, Crow D, Bading JR, Anderson AL, Poku E, Yazaki PJ, Williams LE, Tamvakis D, Sanders P, Leong D, Raubitschek A, Hudsony PJ, Colcher D, Shively JE. J Nucl Med. 2010;51:1139–1146. doi: 10.2967/jnumed.109.074153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Martin SM, O'Donnell RT, Kukis DL, Abbey CK, McKnight H, Sutcliffe JL, Tuscano JM. Mol Imaging Biol. 2008;11:79–87. doi: 10.1007/s11307-008-0148-1. [DOI] [PubMed] [Google Scholar]
- 271.Elsasser-Beile U, Reischl G, Wiehr S, Buhler P, Wolf P, Alt K, Shively J, Judenhofer MS, Machulla HJ, Pichler BJ. J Nucl Med. 2009;50:606–611. doi: 10.2967/jnumed.108.058487. [DOI] [PubMed] [Google Scholar]
- 272.Li WP, Meyer LA, Capretto DA, Sherman CD, Anderson CJ. Cancer Biother Radiopharm. 2008;23:158–171. doi: 10.1089/cbr.2007.0444. [DOI] [PubMed] [Google Scholar]
- 273.Zimmermann K, Grunberg J, Honer M, Ametamey S, Schubiger PA, Novak-Hofer I. Nucl Med Biol. 2003;30:417–427. doi: 10.1016/s0969-8051(03)00019-2. [DOI] [PubMed] [Google Scholar]
- 274.Smith-Jones P, Fridrich R, Kaden TA, Novak-Hofer I, Siebold K, Tschudin D, Maecke H. Bioconjugate Chem. 1991;2:415–421. doi: 10.1021/bc00012a006. [DOI] [PubMed] [Google Scholar]
- 275.Anderson CJ, Schwarz SW, Connett JM, Cutler PD, Guo LW, Germain CJ, Philpott GW, Zinn KR, Greiner DP, Meares CF, Welch MJ. J Nucl Med. 1995;36:850–858. [PubMed] [Google Scholar]
- 276.Milenic DE, Wong KJ, Baidoo KE, Ray GL, Garmestani K, Williams M, Brechbiel MW. Cancer Biother Radiopharm. 2008;23:619–631. doi: 10.1089/cbr.2008.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Nayak TK, Garmestani K, Baidoo KE, Milenic DE, Brechbiel MW. J Nucl Med. 2010;51:942–950. doi: 10.2967/jnumed.109.071290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ray GL, Baidoo KE, Wong KJ, Williams M, Garmestani K, Brechbiel MW, Milenic DE. Br J Pharmacol. 2009;157:1541–1548. doi: 10.1111/j.1476-5381.2009.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lovqvist A, Humm JL, Sheikh A, Finn RD, Koziorowski J, Ruan S, Pentlow KS, Jungbluth A, Welt S, Lee FT, Brechbiel MW, Larson SM. J Nucl Med. 2001;42:1281–1287. [PubMed] [Google Scholar]
- 280.Clarke K, Lee FT, Brechbiel MW, Smyth FE, Old LJ, Scott AM. Cancer Res. 2000;60:4804–4811. [PubMed] [Google Scholar]
- 281.Nikula TK, Curcio MJ, Brechbiel MW, Gansow OA, Finn RD, Scheinberg DA. Nucl Med Biol. 1995;22:387–390. doi: 10.1016/0969-8051(94)00126-5. [DOI] [PubMed] [Google Scholar]
- 282.Schneider DW, Heitner T, Alicke B, Light DR, McLean K, Satozawa N, Parry G, Yoo J, Lewis JS, Parry R. J Nucl Med. 2009;50:435–443. doi: 10.2967/jnumed.108.055608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Wei L, Zhang X, Gallazzi F, Miao Y, Jin X, Brechbiel MW, Xu H, Clifford T, Welch MJ, Lewis JS, Quinn TP. Nucl Med Biol. 2009;36:345–354. doi: 10.1016/j.nucmedbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Breeman WAP, De Jong M, Bernard BF, Kwekkeboom DJ, Srinivasan A, Van Der Pluijm ME, Hofland LJ, Visser TJ, Krenning EP. Int J Cancer. 1999;83:657–663. doi: 10.1002/(sici)1097-0215(19991126)83:5<657::aid-ijc15>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 285.Breeman WAP, de Jong M, Erion JL, Bugaj JE, Srinivasan A, Bernard BF, Kwekkeboom DJ, Visser TJ, Krenning EP. J Nucl Med. 2002;43:1650–1656. [PubMed] [Google Scholar]
- 286.Cheng Z, Chen JQ, Miao YB, Owen NK, Quinn TP, Jurisson SS. J Med Chem. 2002;45:3048–3056. doi: 10.1021/jm010408m. [DOI] [PubMed] [Google Scholar]
- 287.Albert R, Smith-Jones P, Stolz B, Simeon C, Knecht H, Bruns C, Pless J. Bioorg Med Chem Lett. 1998;8:1207–1210. doi: 10.1016/s0960-894x(98)00187-5. [DOI] [PubMed] [Google Scholar]
- 288.Rosch F, Herzog H, Stolz B, Brockmann J, Kohle M, Muhlensiepen H, Marbach P, Muller-Gartner HW. Eur J Nucl Med Mol Imaging. 1999;26:358–366. doi: 10.1007/s002590050398. [DOI] [PubMed] [Google Scholar]
- 289.Meijs WE, Haisma HJ, Klok RP, van Gog FB, Kievit E, Pinedo HM, Herscheid JDM. J Nucl Med. 1997;38:112–118. [PubMed] [Google Scholar]
- 290.Verel I, Visser GWM, Boellaard R, Boerman OC, van Eerd J, Snow GB, Lammertsma AA, van Dongen G. J Nucl Med. 2003;44:1663–1670. [PubMed] [Google Scholar]
- 291.Borjesson PKE, Jauw YWS, de Bree R, Roos JC, Castelijns JA, Leemans CR, van Dongen G, Boellaard R. J Nucl Med. 2009;50:1828–1836. doi: 10.2967/jnumed.109.065862. [DOI] [PubMed] [Google Scholar]
- 292.Perk LR, Walsum MSV, Visser GWM, Kloet RW, Vosjan M, Leemans CR, Giaccone G, Albano R, Comoglio PM, van Dongen G. Eur J Nucl Med Mol Imaging. 2008;35:1857–1867. doi: 10.1007/s00259-008-0774-5. [DOI] [PubMed] [Google Scholar]
- 293.Perk LR, Visser GWM, Vosjan M, Stigter-van Walsum M, Tijink BM, Leemans CR, van Dongen G. J Nucl Med. 2005;46:1898–1906. [PubMed] [Google Scholar]
- 294.Aerts H, Dubois L, Perk L, Vermaelen P, van Dongen G, Wouters BG, Lambin P. J Nucl Med. 2009;50:123–131. doi: 10.2967/jnumed.108.054312. [DOI] [PubMed] [Google Scholar]
- 295.Nagengast WB, de Vries EG, Hospers GA, Mulder NH, de Jong JR, Hollema H, Brouwers AH, van Dongen GA, Perk LR, Lub-de Hooge MN. J Nucl Med. 2007;48:1313–1319. doi: 10.2967/jnumed.107.041301. [DOI] [PubMed] [Google Scholar]
- 296.Nagengast WB, de Korte MA, Munnink THO, Timmer-Bosscha H, den Dunnen WF, Hollema H, de Jong JR, Jensen MR, Quadt C, Garcia-Echeverria C, van Dongen GAMS, Lub-de Hooge MN, Schroder CP, de Vries EGE. J Nucl Med. 2010;51:761–767. doi: 10.2967/jnumed.109.071043. [DOI] [PubMed] [Google Scholar]
- 297.Perk LR, Visser OJ, Walsum MSV, Vosjan M, Visser GWM, Zijlstra JM, Huijgens PC, van Dongen G. Eur J Nucl Med Mol Imaging. 2006;33:1337–1345. doi: 10.1007/s00259-006-0160-0. [DOI] [PubMed] [Google Scholar]
- 298.Munnink THO, de Korte MA, Nagengast WB, Timmer-Bosscha H, Schroder CP, de Jong JR, van Dongen G, Jensen MR, Quadt C, Lub-de Hooge MN, de Vries EGE. Eur J Cancer. 2010;46:678–684. doi: 10.1016/j.ejca.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 299.Munnink TO, Dijkers E, Hooge ML, Kosterink J, Brouwers A, de Jong J, van Dongen G, de Vries E. J Clin Oncol. 2009;27:1045. [Google Scholar]
- 300.Brouwers A, Verel I, Van Eerd J, Visser G, Steffens M, Oosterwijk E, Corstens F, Oyen W, Van Dongen G, Boerman O. Cancer Biother Radiopharm. 2004;19:155–163. doi: 10.1089/108497804323071922. [DOI] [PubMed] [Google Scholar]
- 301.Hoeben BAW, Kaanders J, Franssen GM, Troost EGC, Rijken P, Oosterwijk E, van Dongen G, Oyen WJG, Boerman OC, Bussink J. J Nucl Med. 2010;51:1076–1083. doi: 10.2967/jnumed.109.073189. [DOI] [PubMed] [Google Scholar]
- 302.Holland JP, Caldas-Lopes E, Divilov V, Longo VA, Taldone T, Zatorska D, Chiosis G, Lewis JS. Plos One. 2010;5 doi: 10.1371/journal.pone.0008859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ruggiero A, Holland JP, Lewis JS, Grimm J. J Nucl Med. 2010;51:1123–1130. doi: 10.2967/jnumed.110.076521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Vosjan M, Perk LR, Visser GWM, Budde M, Jurek P, Kiefer GE, van Dongen G. Nat Protoc. 2010;5:739–743. doi: 10.1038/nprot.2010.13. [DOI] [PubMed] [Google Scholar]
- 305.Tinianow JN, Gill HS, Ogasawara A, Flores JE, Vanderbilt AN, Luis E, Vandlen R, Darwish M, Junutula JR, Williams SP, Marik J. Nucl Med Biol. 2010;37:289–297. doi: 10.1016/j.nucmedbio.2009.11.010. [DOI] [PubMed] [Google Scholar]