Abstract
In the last 10 years it has become increasingly apparent that the gut microbiome has profound effects on the immune system to which it is juxtaposed, the mucosal immune system. Here I explore recent studies in which the effects of the microbiota expand or facilitate anti-inflammatory or regulatory immunologic machinery or which favor development of pro-inflammatory immunologic machinery in this system. I then focus on how these opposing processes play out in inflammatory bowel disease, a disease in which normal immune homeostasis is disturbed and inflammation takes hold.
The Gut Microbiota Drives Inflammmatory Bowel Disease
In recent years, studies probing the composition and function of the endogenous microbiota in the normal GI tract have greatly expanded our appreciation for and understanding of how the microbiota shape mucosal immune responses, as well as more global GI tract activities. To some extent, these studies have been driven by the desire to better understand the inflammatory bowel diseases (IBD) Crohn’s disease (CD) and ulcerative colitis (UC), CD and UC are thought to result from a breakdown in mucosal unresponsiveness to gut commensal organisms (1–3) (see Figure 1) This concept is based first on the fact that in the many existent mouse models of colonic inflammation, either those induced by various external agents or those occurring spontaneously in genetically altered mice, one does not see inflammation in the absence of colonic microbiota (1). In addition, there is now solid evidence that the most prominent genetic polymorphisms associated with IBD cause disease (or prevent disease) by affecting responsiveness of the mucosal immune system. For example, NOD2 deletion in mice or Crohn’s disease-associated NOD2 polymorphisms in humans lead to increased TLR responses because such responses are regulated by prolonged or repeated stimulation of NOD2 (4). Similarly, deletion of the ATG16L1 gene in mice, another gene with Crohn’s disease-associated polymorphisms, results in hyperactivity of the NLRP3 inflammasome and a polymorphism in the IL-23R gene in humans that is associated with decreased risk for developing IBD leads to decreased T cell IL-17 responses (5,6). Finally, as discussed below, while colitogenic microbiota can be demonstrated in certain mouse models of colonic inflammation, there is as yet little evidence that such microbiota can cause persistent disease in the normal host.
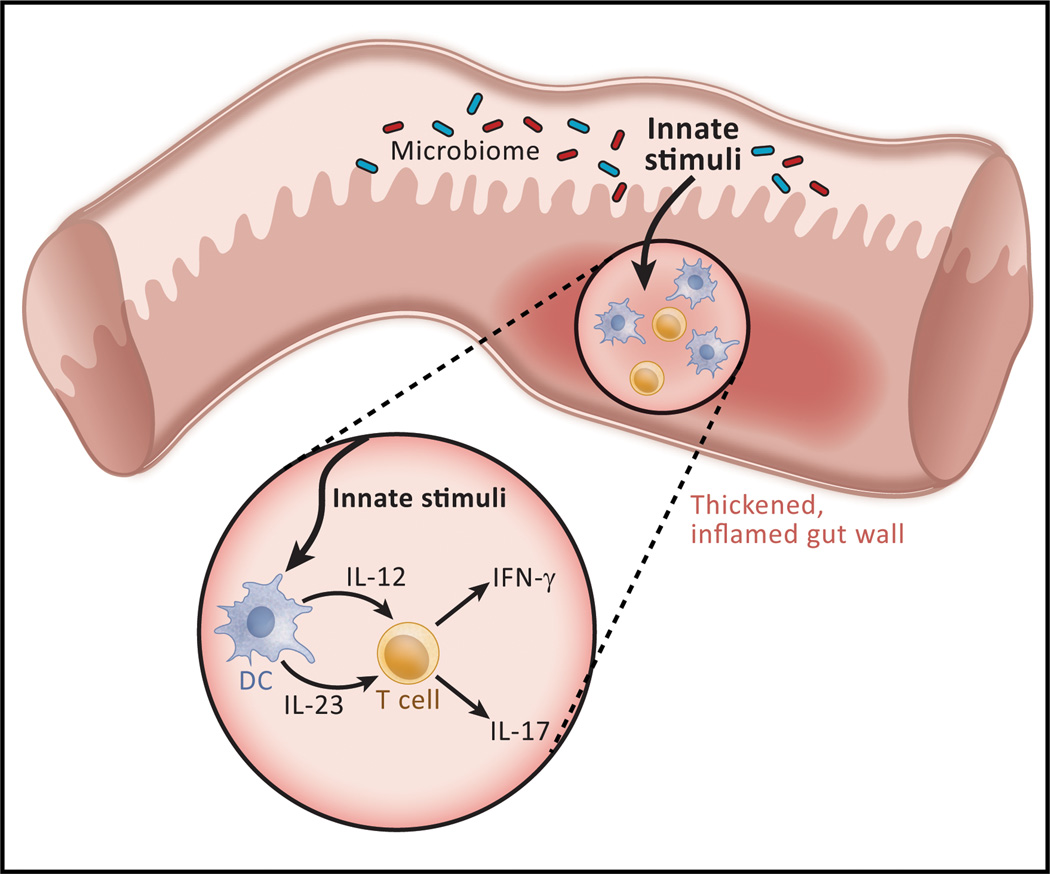
Figure 1.
The basis of Crohn’s disease, a major form of inflammatory bowel disease. The consensus view of Crohn’s disease pathogenesis is that the latter consists of, at least in part, an excessive immunologic response to some component of the microbiota existing in the bowel lumen, most likely a MAMP (microbial-associated molecular pattern) interacting with either a TLR or an NLR. Thus, a MAMP stimulation gives rise to production of cytokines such as IL-12 or IL-23 that induce T cell differentiation into Th1 or Th17 cells, the ultimate effectors of the inflammation. The excessive response could be due to a direct disturbance in the induction of effector cells or to an indirect disturbance in the regulatory cells that control such induction. The mechanism underlying ulcerative colitis is thought to be similar although the cellular processes resulting from the excessive response is different.
In the review below I will first summarize current information on how the microbiota of the GI tract either controls or prevents gut inflammation by the induction of regulatory T cells and then discuss data suggesting that changes in the microbiota can also result in the opposite, namely the induction or aggravation colitis. I will then review the now extensive information on microbiota changes in IBD patients and its possible relation to the causation of this disease.
Mucosal Homeostasis and Regulatory T Cells Induced by the Gut Microbiota
In recent years, considerable evidence has accumulated supporting the notion that the gut microbiota induces mucosal regulatory T cells that then play a vital role in maintaining gut homeostasis under normal conditions or in controlling inflammatory responses that would lead to disease. Evidence of this type was first obtained in studies in which the gut epithelial barrier is transiently perturbed by the intra-rectal administration of ethanol or Vibrio cholerae zonula occludens toxic hexapeptide, agents which cause increased epithelial permeability and increased exposure of lamina propria cells to luminal commensal microbiota (7). Such treatment was shown to result in barely perceptible and transient inflammation accompanied by IL-10-dependent induction of Foxp3-negative, cellsurface TGF-p-positive CD4+ regulatory T cells that could be shown to protect mice from induction of TNBS-colitis. Importantly, the development of these regulatory T cells requires the presence of the gut microbiota and the presence of TLR2; it was thus established that innate TLR2 responses initiated by the microbiota are necessary for the Treg development.
Further work confirming and expanding on these results utilized germ-free mice re-colonized with an “altered Schaedler’s flora,” a non-pathogenic mixture of commensal organisms (8). Here one observes induction of CD4+ regulatory T cells (Tregs) that in this case are Foxp3-positive and IL-10-independent. The development of these Tregs was dependent on both innate and adaptive immune responses as re-colonized MyD88/Ticam-1(TRIF) double-deficient mice whose T cells bear a TCR-transgene specific for lymphocytic choriomeningitis virus (SMARTA mice) that do not respond to commensal organisms, exhibit greatly impaired Treg responses. In addition, in the absence of Treg development or IL-10, the mice manifest robust IL-17 and IFN-γ responses in the colonic lamina propria, albeit in the absence of tangible inflammatory changes. Finally, in studies parallel to those involving mice treated with ethanol, re-colonized mice pre-exposed to a low dose of dextran sulfate to cause injury to the intestinal barrier, develop heightened Treg responses and minimal cytokine responses, whereas re-colonized SMARTA or MyD88/Ticam1 deficient mice failed to develop heightened Treg cell responses and exhibited vigorous Clostridial cytokine responses, the latter associated with high mortality.
These studies, taken together, lead to the view that intestinal homeostasis, i.e., the non-inflamed state of the normal intestinal, is dependent on regulatory T cells induced by commensal microbiota that gain entry into the lamina propria. Thus, while unfettered entry of commensals into the lamina propria due to gross epithelial damage (9) may cause severe inflammation, low level and transient entry has the opposite effect of girding the lamina propria from inflammatory influences. These concepts condition our understanding of inflammatory bowel diseases as they make it likely that the inflammation of the GI tract that define these diseases must initially overcome two anti-inflammatory barriers: the barrier imposed by regulatory T cells induced by commensal microbiota and the barrier created by regulatory T cells that are generated by the inflammation itself (see Figure 2).
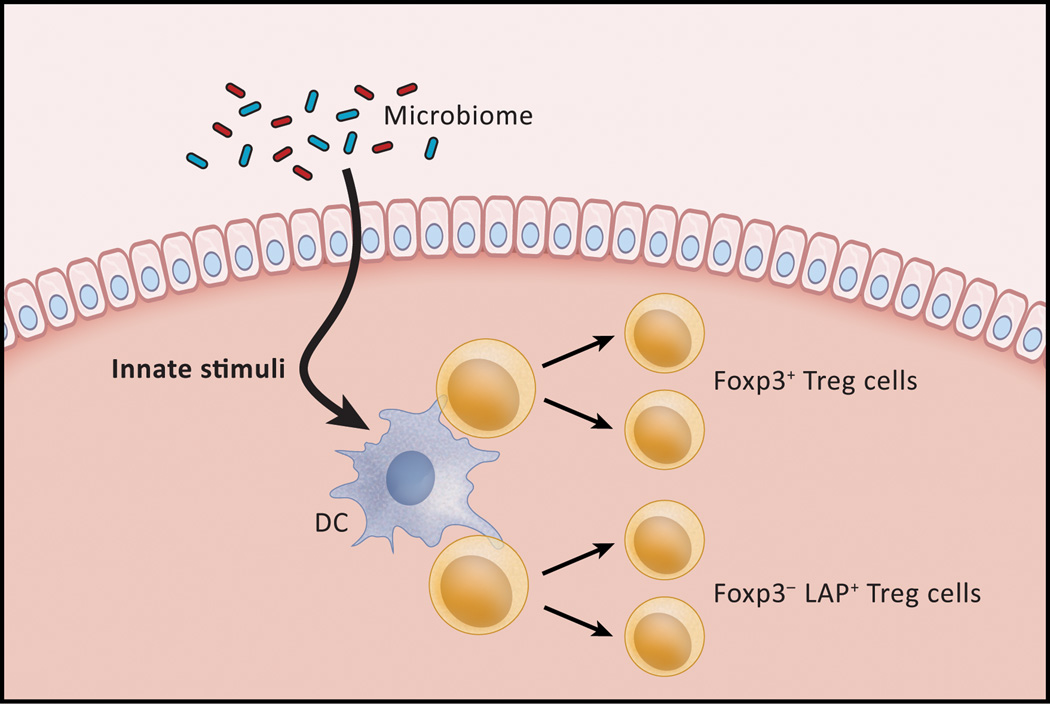
Figure 2.
Homeostatic regulation of mucosal responses. Although the epithelial cell layer of the intestine is a barrier to the entry of luminal materials, it is porous enough to allow entry of microbiome-derived components that mediate the development of regulatory T cells. As shown in this Figure, innate stimuli (TLR or NLR ligands) can interact with dendritic cells in the lamina propria and these, in turn, activate regulatory cells that maintain a quiescent state in the unperturbed intestine despite the presence of adjacent microbiota. In addition, innate stimuli may interact directly with the regulatory cell as well. Two types of regulatory cells have been described: a Tr1-like (IL-10-dependent) cell that bears surface TGF-β and an (IL-10-independent) Foxp3+ cell. Under conditions in which the epithelial barrier is made more porous these cells increase in number, but in this case their function is overwhelmed by a strong pro-inflammatory stimulus.
Induction of Tregs and Prevention of Colitis by Specific Commensal Organisms
So far, I have focused on the ability of the intestinal commensal microbiota as a whole to induce regulatory effects rather than on the ability of individual members of the commensal microbial community with a special propensity to induce such effects. However, there are, in fact, studies that show that certain bacteria are more effective than others in inducing regulatory T cell activity in the GI tract. Perhaps the most complete of these studies relate to the regulatory function of non-enterotoxigenic Bacteroides fragilis, a commensal organism existing within the colonic microflora of both mice and humans. Early studies showed that mice mono-colonized with B. fragilis give rise to IL-10-producing regulatory T cells that can protect RAG2-deficient mice from H. hepaticus-induced colitis; furthermore, these cells were induced by a polysaccharide produced by B. fragilis known as polysaccharide A (PSA)(10). Later studies expanded on these findings by showing that colonization of germ-free mice with PSA-producing B. fragilis but not PSA non-producing B. fragilis elicits induction of Foxp3+ T cells that produce IL-10 and exhibited a PSA-specific profile as they produce TGF-β2 and not TGF-β1 and do not manifest increased expression of CTLA4 or GITR when stimulated by PSA (11). The regulatory function of the PSA-induced Foxp3+ T cells was then revealed in studies of mice with TNBS-colitis treated with PSA that showed that PSA elicits Foxp3 Tregs that suppress effector cell responses and ameliorates the colitis when administered before and after TNBS administration. On this basis, PSA has been proposed as a possible treatment of human inflammatory bowel disease, but this possibility will have to await studies showing that it can be given in a way that does not also induce Th1 effector cell responses as was shown in some studies (12). In addition, PSA may be antigenic and lose effectiveness when repeatedly administered.
An interesting aspect of B. fragilis PSA regulatory activity is that it does not increase the number of Foxp3+ regulatory cells in TLR2-deficient mice and is thus TLR2-dependent (11). This initially posed a problem in explaining its mechanism of action inasmuch as other bacteria also expressing TLR2 ligands did not have a similar capacity to induce regulatory cell activity. This problem was resolved in additional studies that showed that PSA, but not conventional TLR2 ligands, interacts directly with TLR2 on Foxp3+ T cells rather than on dendritic cells to induce production of IL-10 and to establish the profile of PSA-induced Foxp3+ cells mentioned above (12). The possible biological significance of PSA stimulation of regulatory T cells via TLR2 was revealed in studies that showed that colonization of the gut with B. fragilis reduces IL-17 production via PSA-induction of regulatory T cells and that B. fragilis expressing PSA (but not those not expressing PSA) are capable of colonizing colonic crypts in close proximity to the mucosal immune system presumably because of their capacity to reduce local IL-17 production (12). This leads to the concept that PSA is representative of a new class of TLR ligands that induce regulatory responses rather than inflammatory responses.
While the ability of PSA to induce regulatory T cell activity is now well-established by the studies discussed above, its overall function in mucosal homeostasis remains unclear. It may be that under normal homeostatic conditions it has a limited role in enabling B. fragilis to occupy a particular niche in the colonic crypts rather than a more global role in inducing suppressor T cells. This conclusion is suggested by compelling recent evidence that clostridium organisms rather than bacteroides organisms are the primary drivers of Foxp+ T cell development in the colon.
Turning now to these clostridium-related regulatory activity, it was shown initially that germ-free mice provided a specific-pathogen-free (SPF) flora exhibit an increase in the numbers of Foxp3+ T cells in the colon and that this change depends on the gram-positive, spore-forming fraction of the SPF (i.e., a fraction that excludes B. fragilis) (13). In subsequent studies designed to identify which bacterial species induce regulatory T cells, the approach taken was to reconstitute germ-free mice with various cocktails of specific organisms and determine the effect of each cocktail on Foxp3+ cell levels in the colon. While segmented filamentous bacteria (SFB), and large collections of Bacteroides or lactobacillus organisms, had no capacity to increase the number of Foxp3+ cells, clostridium organism groups, particularly those belonging to clusters IV and XIVa, had a striking capacity to increase the number of Foxp3+ T cells. In addition, colonization with clostridium organisms enhanced colonic TGF-p concentration as well as other Treg-inducing molecules. These effects could also be observed in MyD88-, RIP2- and CARD9-deficient mice and were thus independent of TLR, NOD or Dectin receptor signaling. Finally, it was shown that Clostridium organisms but not other bacteria (including Bacteroides organisms) induced IL-10-producing cells in the colon. One caveat to these various findings is that they apply to colonic rather than small intestinal Foxp3+ T cells as reconstitution with Clostridial organisms affected neither Foxp+ T cells nor IL-10 producing T cells in the latter location.
Finally, in studies of the clinical significance of the above findings concerning Clostridial organisms, it was shown that mice colonized with Clostridium at an early age (clostridium-abundant mice) developed less severe DSS-colitis than control mice and also exhibited a reduced tendency to mount Th2 cytokine and IgE antibody responses (13). These findings mesh with the fact that Clostridium clusters IV and XIVa are proportionally reduced in patients with IBD, as discussed in greater detail below. Thus, overall, Clostridial organisms emerge as a major inducer of regulatory T cells, albeit by mechanisms that are as yet undefined (see Figure 2).
Colitogenic Bacterial Microbiota
The mirror image (or rather, negative image) of bacterial microbiota that induce regulatory cells and thus protect organisms from inflammation are the so-called colitogenic organisms mentioned at the outset of this review that either cause de novo intestinal inflammation in normal mice. The first convincing evidence that such colitogenic organisms exist came from studies of RAG2-deficient, T-bet deficient mice that develop a spontaneous colitis and have been termed “TRUC” mice reflecting both the immunodeficiency (“TR”) and the resemblance of the colitis to human inflammatory bowel disease (UC)(14). The origin of the colitis in these mice can be traced to the fact that T-bet is a transcriptional repressor of TNF-α in dendritic cells and its absence leads to excessive TNF-α production by these cells which then synergizes with IL-23 to drive IL-17 production by innate intestinal cells; in addition, the mice lack regulatory T cells due to the RAG2 deficiency (14,15). The TRUC model of colitis would not have been especially remarkable were it not for the fact that studies of this model showed for the first time that mice with experimental colitis could develop an “colitogenic” flora that transmitted colitis vertically to wild-type pups nursed by TRUC mice and horizontally to co-housed wild-type mice which then exhibited some level of colitis for considerable lengths of time even when separated from TRUC mice (see Figure 3).
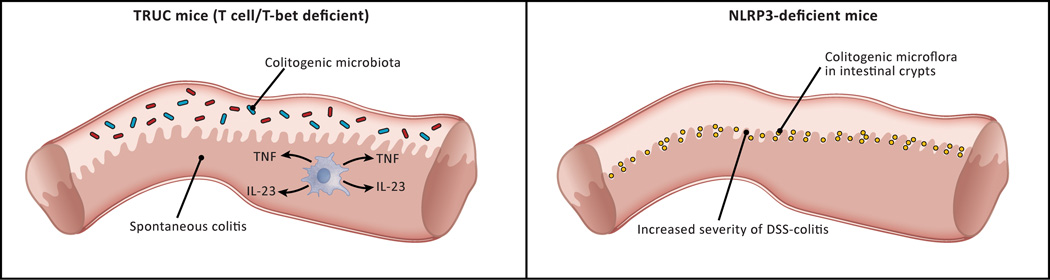
Figure 3.
Two types of “colitogenic” microbiota. Studies of murine models of GI inflammation have identified two types of “colitogenic” microbiota, i.e., microbiota that cause colitis in a normal host. Truly colitogenic microbiota, so far only identified in TRUC mice (see text) are organisms that cause spontaneous colitis in co-housed normal mice. Partially, colitogenic microbiota are represented by Prevotella organisms that proliferate in the intestinal crypts of mice lacking epithelial cells that produce NLRP3 and therefore are deficient in IL-18 production. These organisms are partially colitogenic because they cause intensification of a pre-existing colitis and not de novo colitis
Subsequently, molecular characterization of fecal flora (based on 16s rRNA analyses) was conducted to define the organism or organisms that were colitogenic in TRUC mice (16). The most important findings to emerge from this analysis were evidence that two bacterial species, Klebsiella pneumoniae and Proteus mirabilis were likely to be components of the colitogenic flora. This evidence consisted of the fact that these organisms were present in TRUC fecal flora as well as in the flora of wild type mice fostered by TRUC mice and antibiotic treatment of the TRUC mice that ameliorated the colitis reduced the numbers of these bacteria in the feces to levels below the limit of detection. However, these bacteria appeared to require interaction with other bacteria in the normal flora to cause colitis since they did not cause colitis in germ-free TRUC mice free of colitis but did cause colitis in wild-type mice or RAG2−/− mice with a normal (pathogen-free) flora. Thus, whether these organisms are true colitogenic bacteria that require “help” from other bacteria or are instead necessary “helpers” of true colitogenic bacteria awaits additional study. This question is relevant to findings in a more recent study of bacteria in TRUC mice in which it was found that a substrain of TRUC that does not develop colitis differs from mice that develop colitis by the fact that their microbiota lacked Helicobacter typhlonius and develop colitis if they are administered the latter organism (15). Thus. It is possible that H. typhlonius are the most “proximal” colitogenic organism in TRUC mice.
In a final set of studies TRUC mice were administered either anti-TNF-a neutralizing antibody or regulatory T cells to determine if restoration of immune function influenced colitis in the mice (15). Both therapies ameliorated colitis and, while administration of anti-TNF-α diminished K. pneumoniae or P. mirabilis levels, administration of regulatory T cells did not. The latter fact suggests that these organisms and/or other organisms in the colitogenic microbiota require the presence of an appropriately abnormal mucosal immunologic microenvironment to cause disease and are not “intrinsically” colitogenic despite their ability to cause disease in WT recipients for some period of time; it is possible, for instance, that the colitogenic potential of these organisms are enhanced by interaction with pro-inflammatory cytokines.
Another set of studies revealing the presence of organisms that are associated with colitis involves studies of mice with deficiencies of NLRP6. The latter is a member of an NLR family of intra-cellular microbial recognition molecules that activate inflammasomes, i.e., molecular complexes that result in the activation of caspase 1 and thus the proteolytic cleavage and then secretion of active IL-1 β and IL-18. NLRP6 is a unique member of the NLR inflammasome family because its expression is localized to epithelial cells and thus it is particularly likely to be activated by microbial flora inhabiting the microbial-epithelial interface.
Initial studies of NLRP6 showed that mice deficient in this molecule manifested more severe induced colitis (DSS-colitis) or even mild spontaneous colitis (17–18). Later studies provided evidence that NLRP3 deficiency led to a change in the bacterial microbiota that mediated these susceptibilities (19). This consisted of the fact that WT mice co-housed for prolonged periods developed DSS-colitis of equal severity to their NLRP6-deficient cohorts. Similarly, WT mice co-housed with mice deficient in ASC, a key inflammasome component necessary for caspase I cleavage or co-housed with mice deficient in IL-18 also developed colitis, establishing that the NLRP6 deficiency was in fact an inflammasome-related defect. It is important to point out, however, that the increased susceptibility to DSS-colitis in WT mice is a transient abnormality and WT mice separated from NLRP6-deficient mice eventually lose the increased susceptibility.
Subsequent studies of the microbiota in NLRP6- ASC- and IL-18-deficient mice revealed that these mice or indeed WT mice co-housed with these mice exhibited a microflora enriched for the anaerobic taxa, Prevotellaceae and TM7 and that treatment of these mice with antibiotics abolished the transferability of DSS-colitis susceptibility to co-housed WT mice (19). Of interest, the Prevotella organisms in deficient mice were located adjacent to epithelial cells in the intestinal crypts indicating that lack of epithelial inflammasome activity led to a defect in the ability of the deficient mouse to control the proliferation of certain potentially pathogenic intestinal organisms normally occupying this micro-niche. The mechanism of this defect is, however, not yet known.
The “dysbiosis” occurring in NLRP6-deficient mice may apply to mice with other inflammasome defects affecting hematopoietic cells rather than epithelial cells, and indeed to mice with other defects in innate immune responses; this possibility is in fact suggested by the finding that NLRP3-deficient mice and NOD2-deficient mice also exhibit increased severity of induced colitis and also develop an altered microbiota (20, 21). In addition, there is evidence in the case of NOD2-deficiency that this altered microbiota causes increased susceptibility to induced colitis in WT mice. In any case, it is important to point out that the organisms occurring in mice with NLRP6 inflammasome defects (and possibly other defects) differ from those associated with TRUC mice discussed above in that they pre-dispose normal (or deficient) mice to an induced colitis rather than causing spontaneous colitis; thus these organisms are more properly called colitis pre-disposing organisms rather than colitogenic organisms. The significance of this lies in the fact that these organisms, even more than the TRUC organisms, probably do not cause intestinal inflammation in themselves and thus cannot initiate colitis in humans in the absence of an intrinsic immune defect (see Figure 3). A final point relating to the above findings in NLRP6-deficient mice concerns a recent comprehensive study of the intestinal microbiota of mice deficient in various TLRs or MyD88 that disclosed that whereas the microbiota in the various mice differed markedly from each other they generally did not differ greatly from WT littermates (22). This suggested that the differences originated from an “extended husbandry in isolation” rather from differences in innate immune responsivity. It can be argued that these findings call into question the idea that NLRP6 deficiency was actually causing changes in the microbiota associated with the epithelium as implied above; however, this is not likely since the appearance of colitis–predisposing organisms in the intestine occurred in mice with ASC and IL-18 deficiency as well as NLRP6 indicating that a particular inflammasome-mediated mechanism in a variety of mouse colonies was leading to the proliferation of a colitis-predisposing organism even in mice of differing origins.
The Gastrointestinal Microbiome in Inflammatory Bowel Disease
In the light of the above studies detailing either the anti- or pro-inflammatory effects of gut microbiota, it becomes of great interest to define the microbiota of patients with inflammatory bowel disease and thus to determine if the organisms contained within this microbiota contributes to the occurrence of such disease.
A considerable number of surveys of the microbiota in patients with IBD and control individuals using metagenomic analyses of 16S rRNA in extracted gut specimens or fecal material have now been conducted. While these molecular techniques are generally superior to culture-based techniques because of inherent difficulties in culturing many members of the microbial community they still have some limitations. Chief among these is that they rely on PCR-based techniques that may not detect bacterial species present in low abundance and they quantify copy number of 16S rRNA species rather than true bacterial number. Other difficulties in such surveys which adhere to culture-based surveys as well is that patient populations are both environmentally and genetically heterogeneous so that results obtained with studies (especially of small numbers of patients) may not reflect general abnormalities or may pertain to only a subclass of patients.
One landmark study conducted by Frank et al in 2007 in large groups of patients provides a general framework for microbiota abnormalities observed in IBD patients and serves as a basis for evaluation of changes found in studies of smaller patient groups (23). These investigators analyzed surgically-obtained gut-wall biopsy specimens and found that in colons of patients with IBD (both CD and UC) Bacteroidetes and Firmacutes (Lachnospiraceae family) were depleted whereas Proteobacteria (which contain the E. coli species) and the Bacillus group of Firmacutes were increased. In small intestines of patients, the Bacillus group of Firmacutes were decreased and Proteobacteria were increased relative to controls whereas Bacteroidetes were unchanged. Upon principle component analysis, however, these overall differences were due to microbiota changes in only one-third of CD patients and one quarter of UC patients, the remainder exhibiting a normal microbiota pattern. In these sub-groups, the decrease in Firmacutes (Lachnospiraceae family) and Bacteroidetes were particularly apparent in both colon and small intestine.
A notable feature of this analysis is that it did not disclose a particular bacterium present at levels expected of a pathologic agent; in particular, the analysis detected few if any copies of Mycobacterium avium ssp. paratuberculosis rRNA, an agent that has been linked to Crohn’s disease in some studies (24). In addition, the subset of IBD patients with abnormal intestinal microbiota were younger and were more likely to have disease associated with abscess formation. These associations suggested that the abnormal microbiota were a feature of more severe disease and could thus be a factor that aggravates disease. This view fits with the fact that an abnormal microbiota was not found in the majority of patients and is thus unlikely to be a primary etiologic factor. Finally, this analysis revealed that IBD small intestinal microbiota was characterized by reduced diversity in the Bacteroidetes and Firmacutes phyla, meaning that the number of distinctly different bacterial clones in these phyla was decreased.
Other groups of investigators also using molecular methodology to assess bacterial populations associated with gut specimens obtained results that were similar in some ways and different in others (Reviewed in 25). In summary, these studies of smaller and mostly CD patient populations provide data that add to that obtained in the study by Frank et al., in that they emphasize that decreases in the population of Firmacutes include Faecalibacterium prausnitizii decreases and increases in the population of Proteobacteria include E. coli increases; in addition, they suggest that these Firmacutes/Proteobacteria abnormalities may be limited to CD patients with small bowel disease (26–31). The picture with respect to Bacteroidetes is somewhat less clear in that several studies found decreased number of these bacteria whereas in other studies, most notably those ilia examining the iliae, increased concentrations of Bacteroidetes (B. fragilis) were found (29,32,33).
The molecular analysis of the gut microbiome in patients with IBD summarized above, as well as previous culture-based studies of the microbiota not discussed here, offers several insights into the role of commensal bacteria in the etiology of this disease. They provide strong evidence that a single pathogenic bacterial organism is not the root cause of IBD-related inflammation. This evidence begins with the fact that the molecular analyses did not reveal the presence of any known pathogen in sufficient numbers to cause inflammation (23), but also includes the fact that in several studies distortions in the microbiota were present in uninvolved tissue and/or were absent from involved tissue (such as colonic tissue), and that distortions tended to disappear when patients were administered agents that ameliorated immunologic abnormalities, suggesting that they were secondary effects of underlying immunologic defects (34).
Distortions in the gut microbiota were not found in all patients and tended to occur in only certain parts of the intestine and in patients with more severe disease (21,26). In addition, such changes in microbiota were much more evident in Crohn’s disease than in ulcerative colitis (31). These facts argue against the idea that the distortions are a universal driver of the IBD disease process and for the idea that are secondary and situational. However, these views need to be tempered by the realization that the methods so far used to analyze the gut microbiota may be grossly deficient in identifying the presence of more subtle bacterial abnormalities.
A consistent finding in Crohn’s disease is the selective loss of Firmacutes and Bacteriodetes organisms that conceivably could be members of the microbiota important in the induction of regulatory cells, as suggested by the murine studies of B. fragilis and Clostridial organisms, as discussed above. The most striking of these findings relates to F. prausnitzii, which, as discussed above, is a Clostridial organism that is consistently decreased in patients with Crohn’s disease, particularly those with small bowel inflammation (29). In direct studies of the immunoregulatory function of this organism Sokol et al showed that human PBMC’s cultured in the presence of F prausnitzii exhibited a higher ratio of IL-10 to IL-12 production than cells cultured in the presence of several other commensal organisms; moreover, supernatants from F. prausnitzii cultures suppressed IL-1β-induced IL-8 secretion and NF-κB reporter gene activity in CaCo-2 cells (35). Perhaps more importantly, intra-gastric administration of F prausnitzii or a supernatant obtained from its culture reduced the severity of TNBS-colitis and lowered colonic IL-12 production; in addition, such treatment tended to correct the dysbiosis observed in mice with TNBS-colitis. These studies support the idea that changes in F prausnitzii in the microbiota are a significant factor in the severity of Crohn’s disease, but this requires further investigation as the cytokine changes observed in vivo, while statistically significant, were nevertheless rather marginal. Finally, with respect to B. fragilis, one can hardly claim that lack of this bacterium is playing an immunoregulatory role in IBD, since, as noted above, these organisms are greatly increased in the biofilm of IBD patients.
A final insight of the analyses of bacterial populations in IBD concerns the finding that E.coli organisms are frequently increased in patients with Crohn’s disease (27). This finding coincides with a series of studies showing that a sub-type of E.coli with “semi-pathogenic” properties called adherent-invasive E.coli (AIEC) are frequently found in patients with Crohn’s disease and play some role in the pathogenesis of this inflammation (36,37). AIEC occurs in the inflamed ileum of about 22% of CD patients with chronic inflammation and a somewhat higher percentage in the terminal small bowel in post-surgical patients, but in only 6% of the ilia of control patients (36). However, it is usually not found in affected colons of patients and is found in 22% of the iliae of patients without ileal inflammation; thus, AIEC is by no means a universal accompaniment of Crohn’s inflammation. As indicated by how they have been named, AIEC adhere to and colonizes small intestinal epithelial cells (37). This important property of the organism is explained by the fact that AIEC express a unique type I pilus that has the capacity to bind to CEACAM6 on the surface of epithelial cells and thus facilitate AIEC attachment to such cells (38). Not unexpectedly, such binding is dependent on the level of CEACAM6 expression. This is well shown by the fact that mice bearing a CEACAM (CEABAC) transgene in epithelial cells and expressing large amount of epithelial CEACAM6 exhibit massive AIEC colonization (39). This relationship between CEACAM6 expression and colonization might explain the occurrence of AIEC colonization in Crohn’s disease inasmuch as the secretion of inflammatory cytokines such as TNF- α up-regulates CEACAM6 expression.
Two important questions relating to the role of AIEC in Crohn’s disease concerns first their capacity to penetrate the epithelial barrier and thus invade the mucosa and second their inflammatory properties once such penetration is achieved.
Regarding their invasive properties, AIEC organisms are found in epithelial cells and in the lamina propria of inflamed Crohn’s disease patients, but it is not clear if such penetration is occurring through an already damaged (and possibly ulcerated) epithelium or through an intact epithelium. The fact that there are no reports showing that AIEC occur within cells or tissues of un-inflamed mucosa would suggest that some damage to the epithelium is antecedent to such invasion. The latter conclusion is concordant with the fact that while the invasive (and pro-inflammatory) properties of AIEC are impressive, these organisms do not manifest the invasive properties of true pathogens such as Shigella and Salmonella organisms since, if this were the case, they would cause inflammation throughout the bowel not just in areas of Crohn’s inflammation. These caveats concerning the invasive capacity of AIEC are counter-balanced by the fact that mice expressing a transgene expressing human CEACAM6 mentioned above which do exhibit AIEC translocation and passage into the lamina propria (39). In addition, these mice express increased amounts of claudin in the plasma membrane (a pore forming molecule) and manifest increased epithelial permeability (40). Thus, when CEACAM6 is very highly expressed penetration of pre-existing normal epithelium may occur and this may are be the case in inflamed IBD tissue exhibiting up-regulated CEACAM6.
Related to the fact that AIEC can gain access to the mucosa via epithelial cells is recent evidence that AIEC, in common with several gut bacterial pathogens, express “long polar fimbriae” that allow them to bind to an M cell glycoprotein called GP2 and thus enter M cells (41). Inasmuch as M cells are specialized epithelial cells that take up and translocate bacteria and soluble molecules in to the Peyer’s patches, this property of AIEC is a means by which these bacteria can interact with the “organized” mucosal immune system.
AIEC taken up by both epithelial cells and macrophages, tend to replicate and survive within these cells and induce the macrophages to produce inflammatory cytokines such as TNF-α and IL-6 (42,43). Recently it has been shown that such survival in macrophages is controlled, at least in part, by autophagic machinery and cells with various autophagic defects due to impaired expression of genes associated with Crohn’s disease that adversely affect autophagy such as the ATG16L1 and NOD2 genes support increased intra-cellular AIEC survival and cytokine induction (44). This raises the possibility that CD patients with these genetic abnormalities are more prone to AIEC colitogenic effects.
The adherent, invasive and pro-inflammatory properties of AIEC discussed above have led to the suggestion that these organisms are primary causative factors in Crohn’s disease, at least in some patients. However, several observations mitigate against this possibility. First, as alluded to above, AIEC colonization is dependent of CEACAM6 expression and it is known that the latter can occur as a result of inflammatory cytokine secretion; thus, it is possible or even likely that AIEC colonization is secondary to an underlying inflammation rather than its cause. To counter this argument one would have to have evidence that AIEC can induce CEACAM6 expression in cells that cannot themselves produce cytokines that would have this effect. Second, as already mentioned, penetration of AIEC organisms in the Crohn’s mucosa lacks the uniformity that one might expect of a colitogenic organism and the inflammation of the lamina propria in CEACAM6 transgenic mice where penetration is more uniform is a neutrophil-dominant inflammation that does not resemble the granulomatous inflammation of Crohn’s disease (39). Third and finally, if AIEC were playing a major role in Crohn’s disease there should be evidence that those patients with AIEC colonization exhibit responsiveness to treatment with antibiotics to which AIEC are sensitive. To date, antibiotic treatment of Crohn’s disease patients in general with a variety of agents has had only marginal success and, perhaps more importantly, no patient sub-groups have emerged (presumably those with AIEC colonization) who are particularly responsive to such treatment (45).
On the basis of the above, AIEC is most likely a secondary and aggravating factor in Crohn’s disease whose colitogenic potential is dependent on the presence of pre-existent and genetically determined pro-inflammatory immune abnormalities (46) (see Figure 4). This view obtains additional support from a recent study of mice with TLR5 deficiency, i.e., mice that do not bind flagellated organisms on the basolateral surface of epithelial cells due to absence of the TLR5 flagellin receptor (47). It was found that such mice exhibit spontaneous colitis and that the latter is associated with the development of a microbiota that includes organisms that bind to the intestinal surface and contains increased numbers of proteobacteria and enterobacteria such as E.coli. Evidence that such organisms were initiating the colitis was suggested by the fact that germ-free TLR5-deficient mice, but not WT mice were subject to the development of chronic colitis when exposed to a reference strain of adherent-invasive E.coli (AIEC) and that such colitis persisted after this organisms was no longer present. One explanation of these findings is that bacteria that occasionally penetrate the mucosal barrier interact with TLR5 and thus stimulate epithelial cell production of factors (such as chemokines causing neutrophil infiltration) that ordinarily limit proliferation of such penetrant bacteria; thus, in the absence of such TLR5 function local proliferation of bacteria in the lamina propria is more likely to occur and this causes general changes in epithelial function that cause colitis. This may include loss of barrier function that leads on the one hand to the excessive TLR responses that define the colitic state and on the other to changes in the microbiota such as the appearance of AIEC-like organisms that aggravate the pro-inflammatory process.
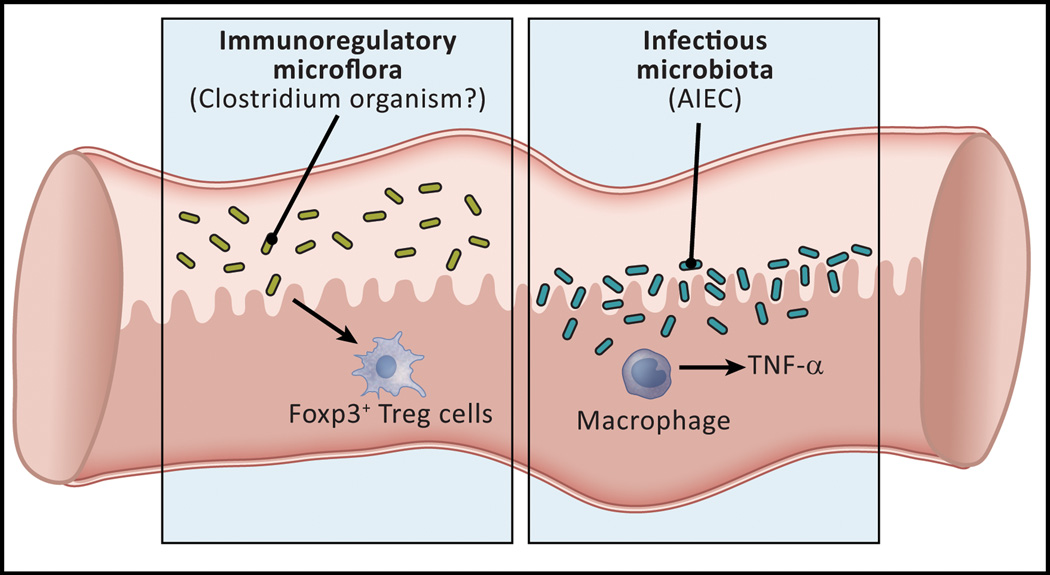
Figure 4.
The gut microflora may cause or aggravate Crohn’s disease by defective induction of regulatory T cells or by infection of the mucosa and the induction of inflammatory cytokines. Studies of mice and humans with GI inflammation have led to the identification of two kinds of microflora that may cause or aggravate the inflammation of Crohn’s disease. In the first category are organisms in the Firmacute phylum represented by Faecalibacterium prausnitzii, a clostridial organism that may be related to those shown in murine systems to induce regulatory T cells in the gut. In the second category is an E.coli organism known as “adherent-invasive” E. coli (AIEC) that lacks pathogenic properties in normals but that colonizes the small intestine of some patients with Crohn’s disease. This organism invades macrophages and induces the latter cells to produce pro-inflammatory cytokines.
Overall then, these studies of TLR5-deficient mice again suggest that organisms with the properties of AIEC are colitogenic but only in the sense that they trigger inflammation in the presence of an underlying genetic defect (in this case involving epithelial cell function). Thus, we come back to the conclusion that whereas colitogenic organisms capable of initiating de novo inflammation in a normal host have been identified in experimental (murine) models of GI inflammation, such as TRUC mice, they haven’t yet been identified in human inflammatory bowel disease.
Concluding remarks
The study of the relation of the gut microbiota to the development and maintenance of the mucosal immune system is a dynamic and rapidly expanding area of research. We know that the organisms comprising the gut microbiome act severally and singly to shape both the anti-inflammatory (i.e., regulatory) as well as the pro-inflammatory aspects of mucosal function. In addition, we know that the gut microbiome can be altered by disease and that such alterations may be a prime or secondary factors in disease pathogenesis. Finally, we know that genetic factors underlying the development of disease in many or all instances affect the latter because of defective responses to organisms in the gut microbiome. Thus, one of the major research challenges in the area of gut inflammation is to define the precise mechanisms that underlie these defective responses.
Organisms in the microbiome of the non-inflamed gut induce Tregs that maintain homeostasis.
Colitogenic organisms can arise in an inflamed gut that causes inflammation in a normal gut.
IBD can be associated with an abnormal microbiome that may affect the severity of the disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 2001;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 2.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu. Rev. Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J. Clin. Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strober W, Watanabe T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn’s disease. Mucosal Immunol. 2011;4:484–495. doi: 10.1038/mi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujmura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 6.Sarin R, Wu X, Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ amd CD8+ human T-cell functional responses. Proc Natl Acad Sci USA. 2011;108:9560–9565. doi: 10.1073/pnas.1017854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boirivant M, Amendola A, Butera A, Sanchez M, Xu L, Marinaro M, Kitani A, Di Giacinto C, Strober W, Fuss IJ. A transient breach in the epithelial barrier leads to regulatory T-cell generation and resistance to experimental colitis. Gastroenterology. 2008;135:1612–1623. doi: 10.1053/j.gastro.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 8.Geuking MB, Cahenzi J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 9.Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science. 1995;270:1203–1207. doi: 10.1126/science.270.5239.1203. [DOI] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 11.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov I, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridiumspecies. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmania SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell N, Walker AW, Stolarczyk E, Canavam JB, Refik Gokman M, Marks E, Jackson I, Hashim A, Curtis MA, Jenner RG, Howard JK, Parkhill J, MacDonald TT, Lord G. The transcription factor T-bet regulates intestinal inflammation mediated by Interleukin-7 Receptor+ Innate Lymphoid Cells. Immunity. 2012;37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen GY, Liu M, Wang F, Berlin J, Núñez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Normand S, Delanoye-Crupin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, Lemoine Y, Hot D, Chamaillard M. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial selfrenewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elinav E, Strowig T, Kau AL, Henao-Meija J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirota SA, Ng J, Lueng A, Khaja M, Parhar K, Li Y, Lam V, Potentier MS, Ng K, Bawa M, McCafferty DM, Rioiux KP, Ghosh S, Xavier RJ, Colgan SP, Tschopp J, Muruve D, MacDonald JA, Beck PL. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm. Bowel Disease. 2011;17:1359–1372. doi: 10.1002/ibd.21478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couturier-Mallard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, Gaillot O, Schreiber S, Lemoine Y, Ryffel B, Hot D, Núñez G, Chen G, Rosenstiel P, Chamaillard M. NOD2-mediated dysbiosis pre-disposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, Pace NR, Li EL. Disease phenotype and genotype are associated with shifts in intestinalassociated microbiota in inflammatory bowel diseases. Proc. Nat’l. Acad Sci. USA. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiodini RJ, Chamberlin WM, Sarosiek J, McCallum RW. Crohn’s disease and the mycobacterioses: a quarter century later. Causation or simple association? Crit. Rev. Microbiology. 2012;36:52–93. doi: 10.3109/1040841X.2011.638273. [DOI] [PubMed] [Google Scholar]
- 25.Vanderploeg R, Panaccione R, Ghosh S, Rioux K. Influence of intestinal bacteria in human inflammatory bowel disease. Infect Dis Clin N Am. 2010;24:977–993. doi: 10.1016/j.idc.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. The ISME Journal. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 28.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Järnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm. Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 29.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 30.Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 31.Schwiertz A, Jacobi M, Frick J, Richter M, Rusch K, Köhler H. Microbiota in pediatric inflammatory bowel disease. J. Pediatr. 2010;157:240–244. doi: 10.1016/j.jpeds.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 32.Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, McSweeney CS. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm. Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- 33.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 35.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A, Barnich N, Bringer M, Swidsinski A, Beaugerie L, Columbel J. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 37.Chambrun G, Columbel J, Poulain D, Darfeuille-Michaud A. Pathogenic agents in inflammatory bowel diseases Curr. Opin. Gastroenterol. 2008;24:440–447. doi: 10.1097/MOG.0b013e3283023be5. [DOI] [PubMed] [Google Scholar]
- 38.Barnich N, Carvalho FA, Glasser A, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Columbel J, Darfeuille-Michaud A. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, Darfeuille-Michaud A. Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 2009;206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denizot J, Sivignon A, Barreau F, Darcha C, Chan HF, Stanners CP, Hofman P, Darfeuille-Michaud A, Barnich N. Adherent-invasive Escherichia coli induce claudin-2 expression and barrier defect in CEABAC10 mice and Crohn’s disease patients. Inflamm. Bowel Dis. 2012;18:294–304. doi: 10.1002/ibd.21787. [DOI] [PubMed] [Google Scholar]
- 41.Chaissaing B, Rohlion N, de Vallée A, Salim SY, Prorok-Hamon M, Neut C, Campbell BJ, Söderholm JD, Hugot JP, Columbel JF, Darfeuille-Michaud A. Crohn disease-associated adherent-invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. J. Clin. Invest. 2011;121:966–975. doi: 10.1172/JCI44632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glasser AL, Boudeau J, Barnich N, Perruchot MH, Columbel JF, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn’s disease survive and replicate within macrophages without inducing host cell death. Infect. Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bringer MA, Billard E, Glasser AL, Columbel JF, Darfeuille-Michaud A. Replication of Crohn’s disease-associated AIEC within macrophages is dependent on TNF-α secretion. Lab Invest. 2012;92:411–419. doi: 10.1038/labinvest.2011.156. [DOI] [PubMed] [Google Scholar]
- 44.Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defectsin autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 45.Khan KJ, Ullman TA, Ford AC, Abreu MT, Abadir A, Marshall JK, Talley NJ, Moayyedi P. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 46.Strober W. Adherent-invasive E. coli in Crohn disease: bacterial “agent provocateur”. J. Clin. Invest. 2011;121:841–844. doi: 10.1172/JCI46333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho FA, Koren O, Goodrich JK, Johansson MEV, Nalbantoglu I, Aitken JD, Yueju S, Chaissaing B, Walters WA, Gonzalez A, Clemente JC, Cullender TC, Barnich N, Darfeille-Marchaud A, Vijay-Kumar M, Knight R, Ley RE, Gewirtz AT. Transient inability to manage Proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host and Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]