Summary
Structural and functional studies of many mammalian systems are critically dependent on abundant supplies of recombinant multi-protein complexes. Mammalian cells are often the most ideal, if not the only suitable host for such experiments. This is due to their intrinsic capability to generate functional mammalian proteins. This advantage is frequently countered by problems with yields in expression, time required to generate over-expressing lines, and elevated costs. Co-expression of multiple proteins adds another level of complexity to these experiments, as cells need to be screened and selected for expression of suitable levels of each component. Here we present an efficient fluorescence marking procedure for establishing stable cell lines that over-express two proteins in co-ordination, and we validate the method in the production of recombinant monoclonal antibody Fab fragments. This procedure may readily be expanded to systems of greater complexity, comprising more then two components.
1. Introduction
The importance of macromolecular assemblages over individual proteins in determining how eukaryotic cell function is becoming ever more clear, as evidenced by recent proteomic studies performed in yeast (1, 2). Justifiably as a result, macromolecular complexes are receiving an increasing amount of attention. Biophysical characterization and structure determination of ensembles of two or more protein components requires the capability to successfully generate stable complexes (3). These studies, more often than not, rest their chances of success on the ability to co-express every element of the complex in the same cell of an appropriate host. While re-constitution of the individually expressed components remains a viable option, frequently observed instability of the single, isolated proteins can only be overcome with a successful co-expression strategy.
Recombinant protein expression in bacteria, typically Escherichia coli, has been by far the most successful strategy for generating material to support biophysical and structural studies, and there are established methods for co-expression of genes in this organism (1, 3, 4). However, bacteria are often not suitable hosts for expression of functional eukaryotic proteins (5). Difficulties may arise due to toxicity of the foreign protein to the host, to differences in the protein folding machineries, or to the need for post-translational modifications, absent in bacteria.
Expression systems based on yeasts or on baculovirus-infected insect cells are viable, often successful choices for producing recombinant proteins of mammalian origin (6, 7). Undoubtedly though, mammalian cells provide the closest match to a native environment for ectopically expressed mammalian genes. Expression of recombinant proteins in mammalian cells can be achieved through transient transfection, viral infection or stable integration of expression constructs into the host’s genome. Transient co-transfection of multiple, separate plasmids carrying individual genes represents the most frequently and successfully employed technique for functional studies. This approach is unfortunately unsuitable for most structural studies, and other large-scale efforts, when abundant amounts of material have to be generated on a routine basis. These applications require a system based on viral infection (8) or on the generation of cell lines in which stable integration of the transfected DNA in the host’s genome has occurred. Stable integrants can be identified readily by antibiotic selection. However, the integration of the transfected constructs into the genome of the host cell inevitably leads to a wide spectrum in protein synthesis levels within the same batch of cells, depending primarily on the number of integrants and their sites of integration.
The most critical and often time-consuming step in generating a high-expressing cell line comes down to selecting those cells capable of producing the most protein. The task is made exponentially more complex when one needs to identify cells that can generate high levels of each of two or more protein components Coupling the expression of each polypeptide chain with that of different antibiotic-resistant markers, targeting a specific genomic locus for integration, or incorporating all the components in a single plasmid are valid strategies utilized to increase the frequency of integrants able to synthesize all the desired components. These methods however, offer little relief in the time consuming step of identifying cells with the required expression characteristics (9).
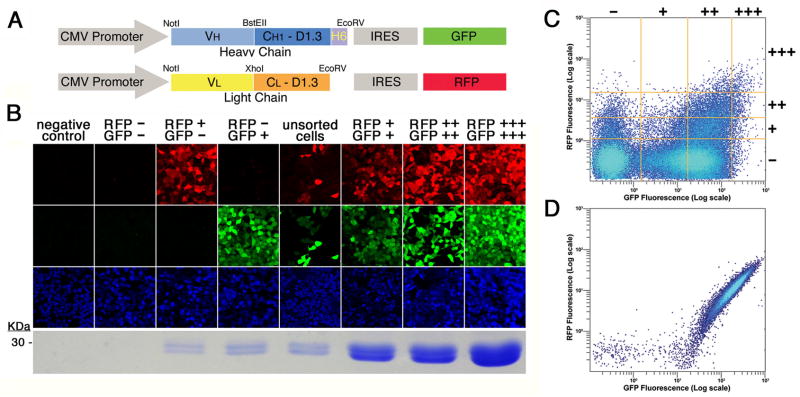
Discussed here in detail is a fluorescence-based method for the rapid selection of mammalian cells that co-express two associated proteins (10). The expression of each protein chain is coupled to a separate fluorescent marker via an internal ribosome entry site element (11) present in the vector. One chain is linked to green fluorescent protein (GFP; (12)), the other to red fluorescent protein (RFP; (13)). Gene expression is driven by a constitutively active strong promoter, derived from cytomegalovirus (CMV; (14)). Figure 1A shows a schematic of the expression constructs, adapted for the production of recombinant antibody Fab fragments, the example discussed here. Expression levels of the two chains are directly proportional to GFP and RFP fluorescence levels (Figure 1B). The task of identifying cells expressing the highest levels of both components is thus reduced to simple detection and isolation of the most double fluorescent cells. These can be picked manually when colonies grow out of individual cells after antibiotic selection. Alternatively, the process can be automated with a fluorescence activated cell sorter (FACS), where individual cells, bright for both GFP and RFP can be identified and deposited in single wells of 96-well plates (Figure 1C). The selected, double fluorescent colony or cell can then be expanded into a production cell line. The fluorescence profile obtained through the FACS, of a production cell line secreting high-levels (> 20 mg/L) of an Fab fragment, is shown in Figure 1D. For the example discussed here, a hexa-histidine tag genetically fused to the C-terminus of the H chain (Figure 1A), allows for rapid purification of the Fab complex. The entire procedure is schematized in Figure 2. By working with additional fluorophores, we expect this method to be readily expandable to cells simultaneously expressing more then two polypetide chains.
Figure 1.
Co-expression of two polypeptide chains with fluorescent markers. (a) Schematic of the expression constructs. Expression is driven by the cytomegalovirus (CMV) promoter. The gene for the heavy chain of the Fab is in blue colors (VH and CH1 D1.3), followed by a hexa-histidine tag (H6) fused to its C-terminus. The light chain, in a separate vector is shown in yellow (VL) and orange (CL D1.3). Expression of the heavy and light chains is coupled to that of green (GFP) and red (RFP) protein, respectively, by an internal ribosome entry site (IRES). (b) Correlation of Fab 2A11 expression levels with fluorescence. Cells were sorted according to their fluorescence profile, and the pools photographed by confocal microscopy in the emission channels corresponding (from top to bottom) to RFP, GFP and the nuclear stain TOTO-3. The resulting Fab is shown under the respective columns, run out, after purification, on a Coomassie blue-stained SDS–PAGE gel. (c) Fluorescence profile of sorted cells. Fluorescence data from 100,000 viable HEK293-T cells expressing 2A11 after antibiotic selection are shown. Fluorescence is plotted in logarithmic scale for RFP (vertical axis) and GFP (horizontal axis). The orange lines are representative of the 4 × 4 grid used to select negative, +, ++ and +++ cells, for RFP fluorescence (horizontal lines from bottom to top, in increasing order) and for GFP fluorescence (vertical lines left to right, in increasing order). (d) Fluorescence profile of a production cell line. A clonal cell line expressing 2A11 Fab was analyzed by FACS as described for panel (c). Reprinted from Protein Expression and Purification, volume 55, Zahra Assur, Ira Schieren, Wayne A. Hendrickson and Filippo Mancia, Two-color selection for amplified co-production of proteins in mammalian cells, pages 319–324, copyright (2007), with permission from Elsevier.
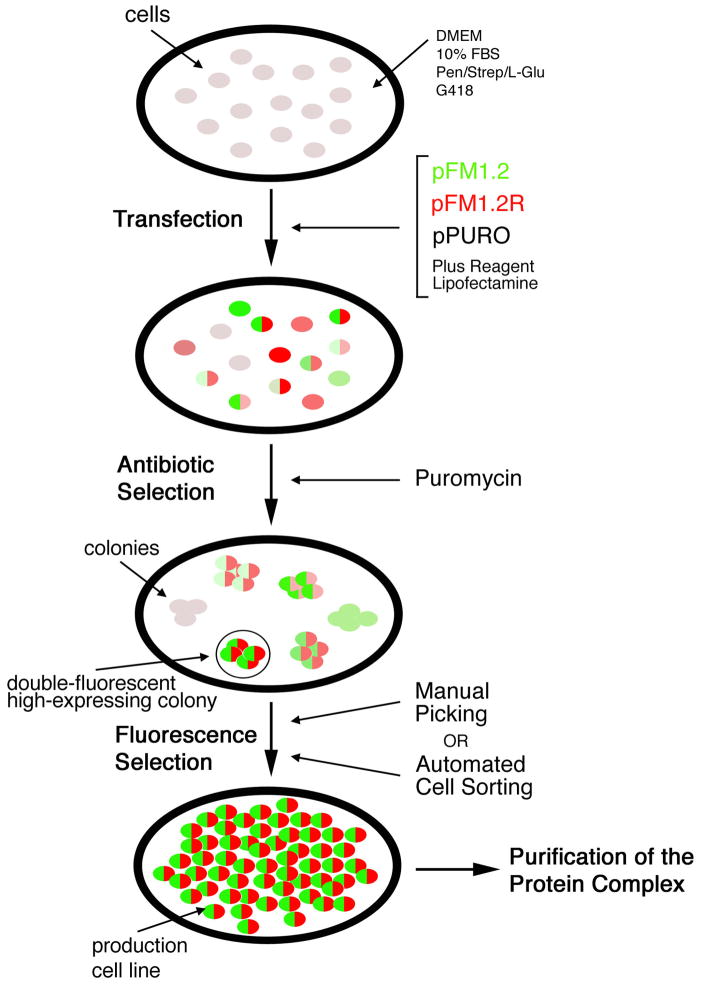
Figure 2.
Flow chart of the two-color selection system. Required procedures, from transfection, to antibiotic selection, to fluorescence selection, are schematized. The intensity of the colors is intended to represent the intensity of the emitted fluorescence for GFP (in green) and RFP (in red). Double fluorescent cells are drawn with two colored halves. Cells expressing only one fluorescent marker are represented with the matching color. Un-transfected cells, and cells expressing neither GFP nor RFP are drawn in light grey.
2. Materials
2.1 Construction of expression vectors
pIRES2-DsRed-Express (Clontech, cat. no. 632463) (see Note 2).
pASK84 (16), pBluescript (Stratagene, cat. nos. 212205-8), and Fv fragments cloned in pGEM-T easy (Promega, cat.no A1360) (see Note 3).
pPURO (see Note 4).
Standard, commercially available molecular biology reagents (Stratagene, Promega, New England Biolabs, Roche, Qiagen) (see Note 5).
DH5α Max Efficiency Competent Cells (Invitrogen, cat. no. 18258-012) (see Note 6).
Ampicillin Resistant LB-Agar plates.
LB liquid medium with 100ug/ml of Ampicillin.
2.2 Preparation of DNA for transfection
Maxi prep kit (Qiagen, cat. no. 12263).
S-200 DNA Microspin Columns (GE Healthcare cat. no. 27-5120-01).
2.3 Cell culture and generation of stable lines
T-antigen transformed Human Embryonic Kidney 293 cells (HEK293-T; ATCC, cat. no. CRL-11268) (see Note 7).
Dulbecco’s Modified Eagle’s Medium (DMEM; Chemicon, cat. no. SLM-020-B) (see Note 8).
Fetal bovine serum (FBS; Hyclone, cat. no. SH30071.03).
Penicillin, streptomycin and L-glutamine (Pen/Strep/L-Glu; SIGMA, cat. no. G1146).
Puromycin (Puro, prepared as a 0.5 mg/mL 100x stock with ultrapure H20 and filtered; Sigma, cat. no. P8833).
Geneticin (G418, prepared as a 50 mg/mL 100x stock with ultrapure H20 and filtered; Gibco, cat.no. 11811-031).
Plus® Reagent (Invitrogen, cat. no. 11514-015) (see Note 9).
Lipofectamine (Invitrogen, cat. no 18324-012).
Phosphate buffer saline, without Ca and Mg (PBS; Chemicon, cat. no. BSS-1006-B).
Ultrapure H20 (Chemicon, cat. no. TMS-006-B).
100 mm tissue culture treated circular dish (Corning, cat. no.).
150 cm2 tissue culture treated flasks (Corning, cat. no.).
2.4 Selecting high-expressing cells
2.4.1 Manual picking of double-fluorescent colonies
Trypsin-EDTA (Chemicon, cat. no. SM-2005-C).
0.1–10 μL sterile filter tips (USA Scientific, cat. no. 1121-4810).
0.1–10 μL pipette.
96 well round bottom tissue culture treated plates (Corning, cat. no.).
48 well flat bottom tissue culture treated plates (Corning, cat. no.).
Inverted fluorescence microscope (see Note 10).
2.4.2 FACS sorting
Beckman Coulter Altra flow cytometer equipped with Autoclone functionality (see Note 11).
Leibovitz’s L-15 medium (L15; Gibco, cat. no. 11415-064).
5 mL snap-cap plastic sterile tubes (BD Falcon cat. no.).
96-well tissue culture treated flat bottom plates (Corning cat. no.).
BD-Filters
2.5 Large-scale protein expression
Hyper-Flasks (Corning, cat. no.) (see Note 12).
2.6 His-Tag based Fab purification
Ni-NTA agarose resin (Qiagen, cat. no. 30210).
Disposable columns (Bio-Rad, cat. no) fitted with 22–1.5 gauge needles (Fisher Scientific, cat. no.).
Na/Hepes 1M pH7.5, NaCl 5M and Imidazole/HCl 2M pH 7.5 stock solutions (see Note 13).
Slide-a-lyzer dialysis cassettes 10,000 KDa cut-off, 3–12 mL capacity (Pierce, cat. no. 66810) (see Note 14).
10 mL syringe (Fisher Scientific, cat. no.).
Bio-Rad Protein Assay (Bradford Dye; Bio-Rad, cat. no. 500-0006).
3. Methods
3.1 Cloning and construction of expression vectors
To generate the RFP expressing pFM1.2R from the GFP expressing pFM1.2, the region coding for the IRES-RFP elements was excised from pIRES2-DsRed-Express to replace IRES-GFP in pFM1.2. This was achieved in 5 steps, and ultimately an XhoI-SpeI fragment (IRES-RFP, after the introduction of a 3′ SpeI site by site-directed mutagenesis, and a 5′ XhoI site via a short oligonucleotide linker) was cloned XhoI-XbaI into pFM1.2 (see Note 15).
FvH and FvL regions of anti-serotonin receptor subtype 2c (17) monoclonal antibodies (18) were cloned into pBluescript containing the CH1 and CL regions of anti-lysozyme D1.3 (19) monoclonal antibody respectively. The genes were fused using BstEII (H chain) and XhoI (L chain) as reported for the expression of Fab fragments in E. coli (16). The two chains were then cloned as NotI/EcoRV fragments into A1.2 (H chain) and A1.2R (L chain) prepared by digestion with the same restriction enzymes (Figure 1A).
3.2 Preparation of DNA for transfection
Retransform mini-prep quality DNA and plate on LB/Amp plates.
Inoculate a 250 mL LB/Amp culture with a single colony.
Purify plasmid DNA using a maxi prep kit (see Note 16).
Spin 50 μL of plasmid DNA through an S-200 column, prepared following the manufacturers instructions, for 1 minute at 3000 RPM in a bench-top centrifuge (see Note 17).
3.3 Cell culture and generation of stable lines
HEK293-T cells are grown in DMEM supplemented with 10% FBS, 1:100 Pen/Strep/L-Glu and 500 μg/mL G418 are maintained at all times in temperature controlled incubators at 37°C, in a humidified environment enriched with 5% CO2.
For transfection of these cells to generate stable lines:
Plate HEK293-T cells on a 100 mm tissue culture dish to 30–40% confluency the evening before the transfection (see Note 18).
The next morning, to a 15 mL sterile tube add 1 μg of pPURO and 5 μg of the two expression plasmids containing the genes to be co-expressed (see Note 19).
Add 750 μL of serum-free DMEM (without any supplements) and 20 μL of Plus® Reagent to the DNA. Mix or vortex gently, and incubate at room temperature for a minimum of 15–20 minutes.
Add 750 μL of serum-free DMEM (without any supplements) and 30 μL of Lipofectamine to the same tube. Mix or vortex gently, and incubate at RT for at least 15–20 minutes.
Replace media from the cells with 5 mL of serum-free DMEM (without any supplements). Perform this step immediately before or after step 4.
Add 5 mL of serum-free DMEM (without any supplements) to the transfection mixture, mix well and add to the dish containing the cells after having removed their previous media.
Allow cells to incubate with the transfection mixture for a minimum of 5 hours at 37 °C in the humidified incubator, enriched with 5% CO2.
Add 10 mL of fully supplemented media and leave overnight.
The next morning, replace with 10 mL of fresh media (see Note 20).
The following day, supplement the media by adding 5 μg/mL of puromycin to the growth medium (see Note 21).
Replace media every 3–4 days (see Note 22).
Monitor for the formation of puromycin-resistant, double fluorescent colonies by visual inspection of the cells under the fluorescence microscope (see Note 23).
3.4 Selecting high-expressing cells
After antibiotic selection, the only cells to survive are those in which stable integration of the transfected DNA in the host’s genome has occurred. Cells that failed to transfect, and cells in which the uptake of plasmid has only been transient will not survive the antibiotic selection step (see Note 24). Stable integrants will lead to colony formation. Each colony will typically grow from a single cell, and hence colonies are considered of clonal purity. Protein expression levels vary dramatically from colony to colony, as a function of the site(s) of integration and the copy number, just to mention two most important parameters. The investigator must therefore identify colonies for which expression of the two recombinant proteins is maximal. In this system, expression levels correlated with fluorescence levels (Figure 1B). The task is therefore to select colonies that exhibit high levels of both GFP and RFP derived fluorescence. This can be achieved by visual inspection and manual isolation of colonies (section 3.4.1) or, in an automated way, by FACS and cloning of individual cells (section 3.4.2) (see Note 25).
3.4.1 Manual picking of double-fluorescent colonies
Carefully scan the plate by visual inspection under the fluorescence microscope for bright, double fluorescent colonies (see Note 26).
Pick individual colonies by gentle aspiration with a 10 μL tip/pipette (see Note 27).
Transfer each aspirated colony to a well of a 96-well tissue culture plate containing 15 μL of Trypsin-EDTA (see Note 28).
Transfer the contents of each 96 well to a 24-well tissue culture plate containing 500 μL of fully supplemented DMEM media (with puromycin).
Leave in the incubator to grow to approximately 80% confluency.
Expand to 6-well format, allow growth to confluency and test for expression of the proteins of interest (see Note 29).
3.3.2 FACS Sorting
We present here a protocol describing how to prepare the cells for sorting, and the steps that should be followed for this experiment. However, operation of a cell sorter requires appropriately trained personnel. Details of how a cell sorter is operated depend on the make and model of the instrument, and are beyond the scope of this work.
Wait for the colony-containing plate to reach 50–80% confluency before scheduling the experiment.
The day before the experiment split the cells 1:3 into fresh medium (see Note 30).
Dislodge the cells by gentle trypsinization.
Resuspend the cells with 5–10 mL of fully-supplemented DMEM, transfer to a 15 mL tube and pellet by centrifugation for 5 minutes at 800 × g.
Remove the medium by aspiration and resuspend the pellet by pipetting up and down with 4 mL of serum-free, L15 medium (see Note 31).
Filter the cells through a nylon cell strainer (40–70 μM mesh size) (see Note 32).
Transfer cells to a 5 mL snap-cap tube.
In parallel, perform steps 2–6 for an un-transfected control and run these cells first through the cell sorter (see Note 33).
Determine the viable cell population using their forward and side scatter characteristics. Set the gates accordingly.
The fluorescent cells are excited with the 488 nm line of a krypton-argon laser. RFP emission is detected using a 590/20 nm band pass filter and GFP emission detected with a 525/30 nm band pass filter (see Note 34).
Select the top fluorescent cells of the double positive population (corresponding to 0.1% of the viable cell population) for cloning to single cell purity (Figure 1C).
Using the cell sorter’s Autoclone mode, deposit one selected cell in each well of a 96-well plate containing 100 μL of fully-supplemented DMEM media in each well (see Note 35).
Incubate the plate in a temperature controlled incubator at 37°C, in a humidified environment enriched with 5% CO2 for approximately 1 week.
Check the plate for bright, double fluorescent colonies under a fluorescence microscope. Expand a limited number of colonies and check for expression of proteins (see Note 36).
3.5 Large-scale protein expression
This protocol refers to how we scale up expression of recombinant Fabs.
Expand cells from 96 to 24 to 6 well plates.
Expand 6 well plates to 100 mm dishes to 150 cm2 flasks
Use 2–4 confluent 150 mm flasks of cells to set up each Hyper-Flask.
Fill the Hyper-Flask to the brim (approximately 500 mL of medium).
Leave for 7–14 days (until the medium turns orange/yellow).
Harvest the medium and proceed to purification of the recombinant protein(s).
3.6 His-Tag based protein purification
The recombinant Fab fragments carry a genetically-engineered hexa-histidine tag fused to the C-terminus of the heavy chain. We use this simple purification procedure both to screen colonies for the highest expressing ones and for milligram-scale protein purification (see Note 37).
Add 20 mM NaHepes pH 7.5, 400 mM NaCl and 10 mM Imidazole pH 7.5 to the harvested medium.
Equilibrate Ni-NTA resin on a column, with 5 column-volumes of buffer containing 20 mM NaHepes pH 7.5, 400 mM NaCl and 10 mM Imidazole pH 7.5. Fit a 22 or 23 gauge needle to the column’s outlet (see Note 38).
Add recombinant-protein containing medium to the resin (see Note 39).
Wash the resin with 10 column volumes of 20 mM NaHepes pH 7.5, 400 mM NaCl, 25 mM Imidazole.
Elute the purified protein with 20 mM NaHepes pH 7.5, 400 mM NaCl, 200 mM Imidazole. Collect 0.5 column volume fractions and monitor for the presence of protein (see Note 40).
Pool the protein-containing fractions and dialyze overnight at 4 °C against a buffer of choice (see Note 41).
Footnotes
pFM1.2 (15) is an in-house generated vector to express proteins under the control of a strong constitutively active promoter (CMV), linking the expression of the gene of interest to that of GFP via an IRES element. Similar vectors are available commercially.
We took the RFP gene from this vector and cloned it into pFM1.2 to replace GFP and generate pFM1.2R (10). This parental vector could most likely serve the same purpose (untested in our hands) as pFM1.2R.
These plasmids were required to generate the heavy and light chains of the Fabs that were then cloned into pFM1.2 and pFM1.2R respectively. Fv regions were cloned from cDNA generated from RNA extracted from hybridoma cells using kits from Roche. The CH1 and CL regions of the Fab were taken from the anti-lysozyme D1.3 antibody (in pASK84), and fused to the respective variable regions as reported by Skerra (16).
pPURO is an in-house generated selection vector that confers puromycin resistance to eukaryotic cells. The expression of the puromycin-resitance gene is driven by the RSV promoter. Equivalent plasmids are readily available from various commercial sources (for explample, pPUR, Clontech, cat. no. 631601).
In our laboratory, we typically use the QuickChange site-directed mutagenesis kit (Stratagene, cat. no. 200515) to introduce mutations, mini-prep kit from Promega (cat. no.), restriction enzymes from New England Biolabs, DNA ligation kit from Roche (cat. no. 11 635 379 001), QIAquick gel extraction kit from Qiagen (cat. no. 28704).
Any other bacterial competent cell would work.
HEK293-T cells transfect well, are robust and relatively easy to handle. SV40 large T-antigen transformation (20) is supposed to boost expression levels of ectopically expressed proteins.
Whatever source for DMEM you choose to utilize, make sure it is supplied as high glucose in contents. We also recommend keeping cells in culture with DMEM from the same supplier.
We have also tested other transfection kits and obtained comparable results.
Our laboratory is equipped with a Nikon Diaphot 300 inverted fluorescence microscope.
This cell sorter has excitation capabilities at 488 nm, and emission detection is achieved using 590/20 nm (for RFP) and 525/30 nm (for GFP) band-pass filters.
As an alternative to Hyper-Flasks, we also recommend 2 layer cell stacks (Corning, cat. no.). Hyper-Flasks are excellent for secreted proteins.
Ingredients to make these solutions are from molecular biology grade chemicals from Sigma-Aldrich.
The approximate molecular weight of an Fab fragment is 50,000 Da. These dialyzing units with a 10,000 Da molecular weight cut-off are safe to use.
Sequencing and PCR amplification across IRES elements most often does not work due to their high GC contents. In this instance, the IRES element of pIRES2-DsRed-Express was removed by digestion with PstI, and re-inserted after the PCR reaction to introduce the 3′ SpeI site.
We recommend washing the DNA twice with 70% EtOH after precipitation with iso-propanol. This step improves the quality of the purified DNA. We typically resuspend the DNA pellet in 300 μL of ultrapure H20 to yield concentrations between 0.5 and 3 μg/μL as measured by absorbance at 280 nm.
This step increases the purity of the DNA to be used for transfection. Purity of the DNA is a very important parameter to maximize the efficiency of transfection. Also, it serves to minimize the risk of any residual bacteria contaminating the mammalian cells following transfection.
The cells should look healthy, not clumpy, perfectly distributed and at approximately 50–60 % confluency the following morning.
This and all other procedures to be carried out under a tissue culture hood following standard sterile techniques.
Alternatively, one can transfect later in the day, leave the cells incubated with only the transfection mix overnight, and replace this with fully-supplemented DMEM the next morning. Results are comparable.
Puromycin is added to select for stable integrants. At this point, RFP and GFP should be clearly visible under the fluorescent microscope. Fluorescence of the cells monitored at this stage is a useful indicator of the efficiency of transfection. Puromycin should be kept continuously present from this point on.
Un-transfected cells, and cells where integration has failed to occurred, should start to die a couple of days after the addition of puromycin. On occasion, the dead or dying cells do not lift off and stay clumped on the dish. Should this occurs, wash cells with PBS every other day.
Colonies should start to appear approximately a week after transfection. The number of colonies and their fluorescence profile varies according to several parameters including the efficiency of transfection and the toxicity of the expressed proteins to the cells. Do not expect all colonies to be double fluorescent.
Plasmids that are not integrated in the genome are rapidly eliminated from the cell within a few days following transfection. Recombinant protein expression typically peaks 48–72 hours after transfection, and declines thereafter.
The two methods have advantages and disadvantages. Manual picking saves times, as colonies as opposed to single cells that then need to grow into colonies are selected. However, timing is critical as colonies need to be picked when large enough to be aspirated, but small enough not to have grown into neighboring colonies. Also, only a limited number of colonies can be picked manually, and one always runs the risk of failing to identify the brightest, best expressing cells. FACS on the other hand is quicker to perform, and provides a more accurate readout of the fluorescence profile of the cells. Ultimately though, the procedure from transfection to a scaled-up cell line is lengthier as individual cells are selected and it takes time to expand these into a cell-line of clonal purity. FACS is the only feasible option when either colonies overgrow, or dead cells remain attached to the dish.
Care should be taken to ensure that selected colonies are not only bright for both GFP and RFP fluorescence, but also homogenous. Colonies that show non-fluorescent or partially-fluorescent patches should be discarded.
Ideally, the fluorescence microscope should be placed under a sterile hood. This is most likely not feasible. Care should be taken to minimize the chances of contamination. The time the lid is kept off the cell-containing dish should be kept to a minimum, and the microscope should not be located around potential sources of contamination such as A/C ventilation outlets.
This step is performed to disperse the colony into individual cells. Care should be taken not to leave the cells in Trypsin-EDTA more than a minute.
We recommend testing more than one double-fluorescent, bright colony, for protein expression. We typically screen 3–6. The degree of expansion of the cells required to test for expression depends on the proein levels and on the assay.
Make sure the cells have been well trypsinized, and that they are evenly dispersed.
L15, unlike DMEM, exhibits no [CO2]-dependent pH variability.
This step is necessary to remove clumps from the cells. Clumps often clog the cell sorter and/or negatively impact its accuracy.
Un-transfected cells are required to determine correct settings for the cell sorter. Cells exhibit background fluorescence. This can be set to zero by running cells expressing neither GFP nor RFP.
Settings depend on the emission spectra of the fluorescent proteins, and on the lasers and filters available with the instrument. If excitation is performed with one laser, the wavelength must be chosen carefully to compromise adequately between the two fluorofores. In our experiments, we were obliged to choose 488 nm for excitation. At this wavelength, GFP excitation is close to maximum, while RFP excitation is at approximately 40%. We could also have performed the experiment at 514 nm where RFP excitation would have been optimal, but GFP would have been below 10%. The reader is encouraged to consult www.metroflow.org under tools & tutorials for a better understanding on how to set up a two-color experiment.
To increase viability of the cloned cells, we recommend adding 30–50% of conditioned medium to the wells. To generate conditioned medium, split cells 1:5 into fresh medium, remove and sterile filter this medium after overnight incubation. Alternatively, consider using a higher percentage of serum (15–20%).
Do not expect to find a bright, double fluorescent colony in each well. Success rates depend primarily on cell viability (empty wells), and the accuracy of the instrument (empty wells and multiple colonies per well).
For small-scale (colony screening) purification we typically use 2 mL of medium from cells grown in 6-well format. For milligram-scale expression we purify medium from 1 Hyper-Flask (approximately 500 mL).
We use approximately 200 μL and 5 mL of Ni-NTA resin for screening and production purposes respectively. Column sizes are chosen accordingly.
For screening purposes we fit the column with a 23 gauge needle and allow the medium to slowly pass through the resin by gravity flow. For large-scale purification, we load the column overnight with a peristaltic pump.
Protein can be monitored by Bradford assay or by absorbance at 280 nm. Purified Fab typically elutes in fractions 2–4.
If the sample needs to be greater than 95% pure, we recommend concentrating the protein and loading it onto a size-exclusion chromatography column (for example a Superdex 200, GE Healthcare). This second purification step is very efficient at removing residual contaminants.
References
- 1.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 2.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O’Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 3.Romier C, Ben Jelloul M, Albeck S, Buchwald G, Busso D, Celie PH, Christodoulou E, De Marco V, van Gerwen S, Knipscheer P, Lebbink JH, Notenboom V, Poterszman A, Rochel N, Cohen SX, Unger T, Sussman JL, Moras D, Sixma TK, Perrakis A. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr D Biol Crystallogr. 2006;62:1232–1242. doi: 10.1107/S0907444906031003. [DOI] [PubMed] [Google Scholar]
- 4.Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 5.Yin J, Li G, Ren X, Herrler G. Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J Biotechnol. 2007;127:335–347. doi: 10.1016/j.jbiotec.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Geisse S, Gram H, Kleuser B, Kocher HP. Eukaryotic expression systems: a comparison. Protein Expr Purif. 1996;8:271–282. doi: 10.1006/prep.1996.0101. [DOI] [PubMed] [Google Scholar]
- 7.Eifler N, Duckely M, Sumanovski LT, Egan TM, Oksche A, Konopka JB, Luthi A, Engel A, Werten PJ. Functional expression of mammalian receptors and membrane channels in different cells. J Struct Biol. 2007;159:179–193. doi: 10.1016/j.jsb.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Lundstrom K. Semliki Forest virus vectors for rapid and high-level expression of integral membrane proteins. Biochim Biophys Acta. 2003;1610:90–96. doi: 10.1016/s0005-2736(02)00721-6. [DOI] [PubMed] [Google Scholar]
- 9.Birch JR, Racher AJ. Antibody production. Adv Drug Deliv Rev. 2006;58:671–685. doi: 10.1016/j.addr.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Assur Z, Schieren I, Hendrickson WA, Mancia F. Two-color selection for amplified co-production of proteins in mammalian cells. Protein Expr Purif. 2007;55:319–324. doi: 10.1016/j.pep.2007.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 13.Knop M, Barr F, Riedel CG, Heckel T, Reichel C. Improved version of the red fluorescent protein (drFP583/DsRed/RFP) Biotechniques. 2002;33:592–602. doi: 10.2144/02333rr02. [DOI] [PubMed] [Google Scholar]
- 14.Thomsen DR, Stenberg RM, Goins WF, Stinski MF. Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc Natl Acad Sci U S A. 1984;81:659–663. doi: 10.1073/pnas.81.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mancia F, Patel SD, Rajala MW, Scherer PE, Nemes A, Schieren I, Hendrickson WA, Shapiro L. Optimization of protein production in mammalian cells with a coexpressed fluorescent marker. Structure (Camb) 2004;12:1355–1360. doi: 10.1016/j.str.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Skerra A. A general vector, pASK84, for cloning, bacterial production, and single-step purification of antibody Fab fragments. Gene. 1994;141:79–84. doi: 10.1016/0378-1119(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 17.Julius D, MacDermott AB, Axel R, Jessell TM. Molecular characterization of a functional cDNA encoding the serotonin 1c receptor. Science. 1988;241:558–564. doi: 10.1126/science.3399891. [DOI] [PubMed] [Google Scholar]
- 18.Mancia F, Brenner-Morton S, Siegel R, Assur Z, Sun Y, Schieren I, Mendelsohn M, Axel R, Hendrickson WA. Production and characterization of monoclonal antibodies sensitive to conformation in the 5HT2c serotonin receptor. Proc Natl Acad Sci U S A. 2007;104:4303–4308. doi: 10.1073/pnas.0700301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amit AG, Mariuzza RA, Phillips SE, Poljak RJ. Three-dimensional structure of an antigen-antibody complex at 2.8 A resolution. Science. 1986;233:747–753. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- 20.Ali SH, DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11:15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]