Abstract
Correctly calculating the structure of metal coordination sites in a protein during the process of nuclear magnetic resonance (NMR) structure determination and refinement continues to be a challenging task. In this study, we present an accurate and convenient means by which to include metal ions in the NMR structure determination process using molecular dynamics (MD) constrained by NMR-derived data to obtain a realistic and physically viable description of the metal binding site(s). This method provides the framework to accurately portray the metal ions and its binding residues in a pseudo-bond or dummy-cation like approach, and is validated by quantum mechanical/molecular mechanical (QM/MM) MD calculations constrained by NMR-derived data. To illustrate this approach, we refine the zinc coordination complex structure of the zinc sensing transcriptional repressor protein Staphylococcus aureus CzrA, generating over 130 ns of MD and QM/MM MD NMR-data compliant sampling. In addition to refining the first coordination shell structure of the Zn(II) ion, this protocol benefits from being performed in a periodically replicated solvation environment including long-range electrostatics. We determine that unrestrained (not based on NMR data) MD simulations correlated to the NMR data in a time-averaged ensemble. The accurate solution structure ensemble of the metal-bound protein accurately describes the role of conformational dynamics in allosteric regulation of DNA binding by zinc and serves to validate our previous unrestrained MD simulations of CzrA. This methodology has potentially broad applicability in the structure determination of metal ion bound proteins, protein folding and metal template protein-design studies.
Keywords: protein allostery, QM/MM MD, zinc sensor protein, metalloregulatory protein, transcriptional repressor, MRD-NMR refinement, MTK+, MCPB, metal ion based NMR refinement, CzrA, metal ion force field
Introduction
Metal ions are estimated to be present in 40% of all proteins and are involved in a wide range of cellular functions ranging from catalyzing enzymatic reactions to providing structural support in proteins (Waldron and Robinson 2009; Thomson and Gray 1998; Tus et al. 2012). Many proteins undergo a large change in structure and/or conformational dynamics upon binding metal ions (Ma et al. 2009; Mealman et al. 2012). While X-ray crystallographic methods provide a highresolution snapshot of a metal ion bound protein, solution-state NMR remains the experimental method of choice to obtain an accurate ensemble-guided view of the structure and associated conformational dynamics of the protein at an atomistic level (Snyder et al. 2005; Yang et al. 2007; Arunkumar et al. 2009; Brunger 1997; Montalvao et al. 2012; Boehr et al. 2009; Eisenmesser et al. 2005; Tolman et al. 1997; Henzler-Wildman et al. 2007; Mealman et al. 2012).
NMR structure determination efforts are, however, limited by a paucity of methods that accurately represent metal ion coordination complexes in a protein. As a result it is common practice to use experimentally derived distance and angle restraints with approximate force constants to constrain metal ions in place, as in the case when using modified amino acids with “dummy” coordination-covalent bonds to the metal with other coordination bonds constrained to X-ray absorption spectroscopy (XAS)-derived distances, or alternatively, to not include metal ions at all during the structure determination process (Banci et al. 2007; Arunkumar et al. 2009; Eustermann et al. 2010; Brockmann et al. 2012; Gao et al. 1998). The use of such approaches introduces possible errors in describing the metal-binding site as either excessively rigid or too flexible. To the best of our knowledge, CYANA (DYANA) (Mumenthaler et al. 1997; Guntert et al. 1997; Lopez-Mendez and Guntert 2006; Guntert et al. 1991; Herrmann et al. 2002), XPLOR-NIH (Schwieters et al. 2003; Clore and Schwieters 2006), and CNS (Brunger et al. 1998; Brunger 2007) protocols do not incorporate an accurate treatment for metal ions. While the Rosetta program (Rosato et al. 2012; Ramelot et al. 2009; Raman et al. 2010) provides for an explicit treatment of metal ions (Wang et al. 2010) based on observed coordination geometries, it is not applicable for novel metal-ligand complexes. In light of the large number of metal-bound proteins in nature, it is important that a broadly applicable general refinement protocol be developed that accurately represents metal ions in an NMR-derived protein structure.
In this report, we present an accurate and efficient means by which to include metal ion coordination structures in the NMR-structure determination procedure by effectively utilizing NMR-data biased all-atom explicit MD simulations coupled with QM/MM MD methods (Allen and Tildesley 1987; Case 2002; Senn and Thiel 2009; Hartsough and Merz 1995). While restrained dynamics methods that utilize NMR data (RD-NMR) have been successfully utilized to fold protein structures using MD platforms, QM/MM methods have been previously used to study metal ion coordination, and refine the metal binding site of zinc and copper bound metalloproteins (Case 2002; Chen et al. 2005; Robustelli et al. 2010; Chen et al. 2004; Bertini et al. 2011; Calhoun et al. 2008; Montalvao et al. 2012; Boehr et al. 2009; Showalter et al. 2007; Li et al. 2010; Markwick et al. 2010; Long and Bruschweiler 2011; Sgrignani and Pierattelli 2012). Using the approach described here, we implement a metal ion inclusive restrained dynamics NMR structure refinement method, denoted as MRD-NMR, in which the metal ions are modeled using specially developed force field parameters that accurately describe the metal ion coordination and electrostatics in NMR-data restrained MD simulations that sample on biologically relevant time-scales on the order of hundreds of nanoseconds (Chakravorty et al. 2012b; Pierce et al. 2012). QM/MM MD simulations complement and validate the classical force field models. They account for changes in metal ion coordination and water exchange while providing extensive sampling, which is not afforded by QM/MM calculations alone. The accurate treatment of metal ions will correctly account for first and second coordination-shell effects such as metal ion mediated hydrogen-bonding interactions and will help capture the polarizing influence of metal ion binding on the protein conformation, ultimately leading to a more precise ensemble of structures. In addition to the accurate treatment of metal ions, structure calculations using the MRD-NMR methodology benefit from the inclusion of explicit solvent models, periodic boundary conditions and an accurate calculation of long-range electrostatics using the particle mesh ewald (PME) approach (Bertini et al. 2011; York et al. 1993; Allen and Tildesley 1987; Jorgensen et al. 1983).
We demonstrate the applicability of our approach by modeling zinc ions in a previously determined NMR structure of the zinc-bound conformation of Staphylococcus aureus CzrA (Figures 1 and SI.1). CzrA is a zinc-sensing transcriptional repressor derived from the ArsR/SmtB family of transcriptional repressors (Ma et al. 2009). It is a two-fold symmetric homodimer (Figures 1, and SI.1) characterized by a winged-helix-turn-helix fold that mediates high affinity binding to the DNA operator (Arunkumar et al. 2009). Previous studies have determined that the transcriptional repressor functions by a mechanism of allosteric regulation in which Zn(II)-binding drives CzrA from a “closed” DNA-bound conformation to an “open” and “flat” conformation, thus reducing its DNA binding affinity by ∼6 kcal/mol (Arunkumar et al. 2009; Grossoehme and Giedroc 2009; Chakravorty et al. 2012b). The previous NMR structure of Zn(II)-CzrA reveals an open conformation, and was calculated using a total of 756 intra- and intersubunit NOE restraints and 112 backbone amide NH RDC restraints (Arunkumar et al. 2009; Tjandra and Bax 1997; Tolman et al. 1997). This structure was solved without the inclusion of zinc ions since the objective of that study was to document that the sparse nuclear Overhauser enhancements (NOE)/residual dipolar coupling (RDC)-based structure determination protocol used on highly deuterated CzrA was sufficiently sensitive to distinguish distinct allosteric states of CzrA, e.g., Zn(II)-bound vs. operator DNA-bound apo-CzrA (Arunkumar et al. 2009). The Zn(II)-CzrA NMR solution structure showed excellent overall agreement with the previously determined crystallographic structure (protein data bank (PDB) code: 1R1V) (Eicken et al. 2003), but appeared to adopt a “more open” conformation with an average backbone RMSD of 2.53 Å (Figure SI.2) and better agreement with the experimental RDC data (Figure 2). The question arises as to whether this difference reflects true differences in solution vs. crystallographically determined conformations, or arises as a result of a lack of structural constraints associated with structure determination of methyl-protiated, highly deuterated samples and/or a lack of constraints resulting from not considering the zinc coordination complex (Arunkumar et al. 2009). We therefore test our MRD-NMR methodology on this system, and also use it to validate our previous unrestrained MD and QM/MM MD simulations that found that the zinc-bound form of CzrA adopts an open conformation (Chakravorty et al. 2012b).
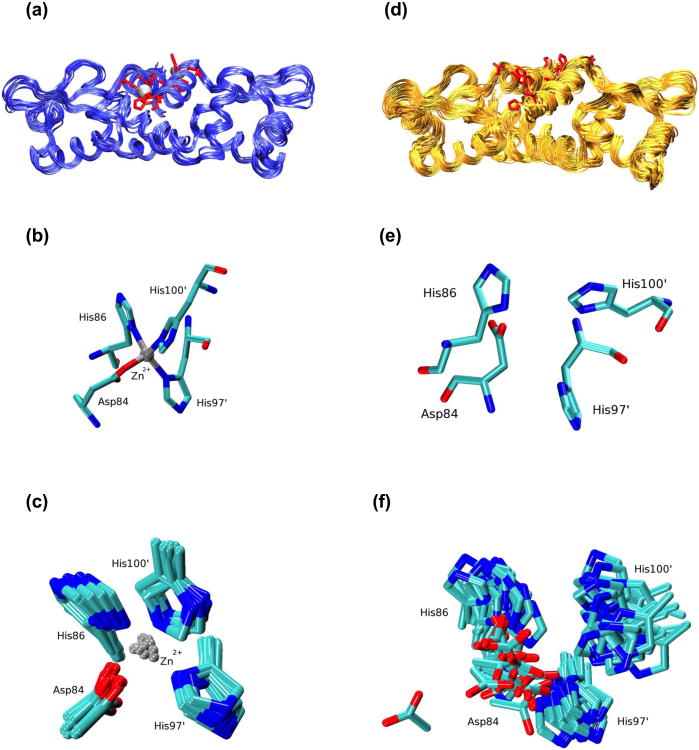
Fig. 1.
Calculated structural ensembles of Zn(II)-bound CzrA from NMR structure determination while including the metal ion (panels a-c) and without including the metal ion (panels d-f). (a) Ribbon representation of ten metal ion refined NMR structures of Zn(II)-CzrA from independent QM/MM MD calculations. Zn(II) ions are shown as silver spheres and the zinc binding residues (Asp84, His86, His97' and His100') are depicted in licorice notation. (b) The zinc ion coordination in a random model selected from the calculated QM/MM MD NMR ensemble of structures. (c) An overlay of metal-binding residue side-chain atoms at a metal binding site for the calculated QM/MM MD NMR ensemble of structures. In a similar manner, panel (d) shows a ribbon representation for the 2KJC ensemble of NMR structures. (e) Arrangement of zinc binding residues at a metal binding site for a random model for the 2KJC structural ensemble. (f) An overlay of zinc binding residues for the 2KJC structural ensemble. The zinc binding residues in panels (a) and (d) are presented from a different angle compared to panels (b), (c), (e) and (f). The backbone atoms of Asp84, His86, His97' and His100' were aligned to create figures (c) and (f)
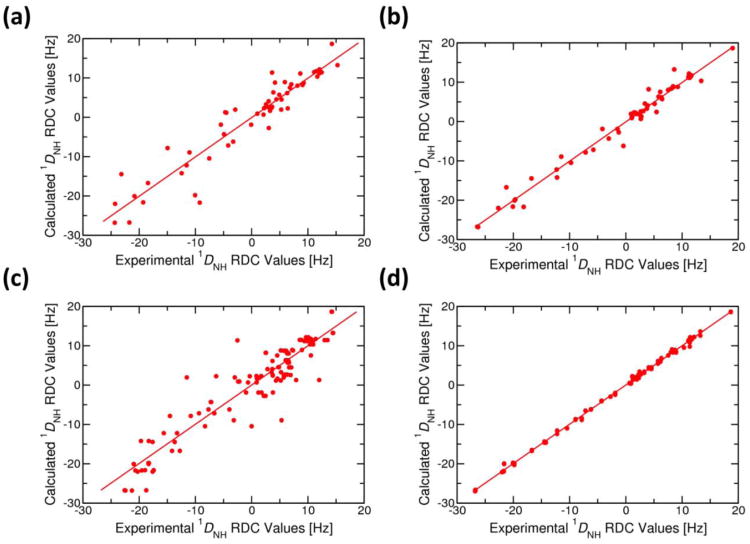
Fig. 2.
(a) Correlation of experimental backbone NH (1DNH) RDC constraints with those calculated from (a) the 1R1V crystal structure of Zn-bound CzrA (R = 0.944; y = 1.002x + 0.036), (b) the average NMR solution structure for the 2KJC ensemble (R = 0.988; y = 0.999x − 0.078), (c) model 1 of the 2KJC ensemble of NMR structures before (R = 0.934; y = 0.994x − 0.178) and (d) after the metal ion refinement MRD-NMR procedure described here (R = 0.999; y = 0.997x + 0.017). R is the correlation coefficient and the equation of the fitted line is in the form of y = mx +c where, ‘m’ defines the slope of the line and ‘c’ the value of x at y=0
Methods and Application
The assisted model building with energy refinement (AMBER) 11 suite of programs provides an integrated platform for MD, QM/MM, metal ion force-field generation and NMR-structure determination methods (Case et al. 2010; Case 2002; Bertini et al. 2011). All MD, QM/MM MD based NMR refinement calculations were performed on a potential energy surface described by the ff99sb force field (Hornak et al. 2006). The force field parameters required to represent the zinc ion and its coordinating ligands were calculated using the metal center parameter builder (MCPB) program that is available as part of the AmberTools package (Peters et al. 2010; Chakravorty et al. 2012b; Roberts et al. 2012; Chakravorty et al. 2011; Case et al. 2010). The Gaussian09 program was employed to perform electronic structure calculations that were needed to develop the metal ion force fields (Frisch et al. 2009). Structure-based and conformational analyses were performed using the ptraj module in AMBERTools Case et al. 2010), the PALES prediction of alignment of structure software (Zweckstetter and Bax 2000), the Maestro (Tools 2011) program and the visual molecular dynamics (VMD) (Humphrey et al. 1996) visualization program.
Simulations of metal ion bound proteins pose a unique set of challenges. A variety of bonded (Chakravorty et al. 2012b; Lee et al. 2012; Yang et al. 2010; Vedani and Huhta 1990; Hoops et al. 1991; Li et al. 2008; Lin and Wang 2010), semi-bonded (Pang 2001), non-bonded (Deeth et al. 2009; Stote and Standing 1995; Wu et al. 2011; Ponomarev et al. 2011), and polarizable force-field (Ponomarev et al. 2011; Wu et al. 2010) approaches have been proposed. The bonded model approach is an accurate and standardized implementation of the “pseudo-bond” or “dummy-cation” approach that is commonly used to model the metal ion binding sties in NMR structure determination calculations. Owing to its accuracy and comparative ease of generation, the bonded model approach has been widely used in MD simulations and is our method of choice to represent zinc and its coordinating ligands in these structure calculations (Lin and Wang 2010; Peters et al. 2010). In this approach, metal ion parameters are derived from geometry optimized structures of the metal-bound complex. Based on metal binding site geometry in the crystal structure of Zn(II)-CzrA, the zinc ion is bound to Asp84, His86, His97' and His100' residues (Figure 1) in a tetrahedral coordination environment (Eicken et al. 2003). We had previously determined the force field parameters for the Zn(II) coordination environment in CzrA from density functional theory (DFT) calculations performed at the TPSSKCIS/Zn=LANL2DZ/6-31G* level of theory (Toulouse et al. 2002; Rey and Savin 1998; Frisch et al. 2009; Cramer and Truhlar 2009; Ditchfie.R et al. 1971; Wadt and Hay 1985; Hay and Wadt 1985). We have since calculated metal ion parameters using the M052X, B3LYP, M06-L and M06-2X DFT functionals as well (Cramer and Truhlar 2009; Zhao and Truhlar 2008). Similar calculations may be performed for model metal ion binding sites for The charges for the metal ion and its surrounding ligands were calculated according to the RESP methodology (Hoops et al. 1991; Case et al. 2005; Cornell et al. 1993). In order to remain consistent with our earlier simulation studies of CzrA, we used our previously derived parameters to represent the zinc ion and its ligating ligands (Chakravorty et al. 2012b). The bonded model approach to representing metal ions, however, suffers from an inability to model changes in the metal coordination. In order to account for such effects while accurately sampling protein conformational ensembles, we employed semi-empirical QM/MM MD calculations performed at the SCC-DFTB3/ff99SB level of theory to supplement our classical simulations (Seabra et al. 2007; Gaus et al. 2011). Such QM/MM MD calculations have been extensively used to study zinc bound proteins, such as in our previous studies of CzrA and the enzymatic reactions in NaphB and protein farnesyl transferase (Chakravorty et al. 2012b; Yang et al. 2012a; Yang et al. 2012b).
Model 1 of the 2KJC ensemble of NMR structures (Arunkumar et al. 2009) provided a starting point for our metal ion modeling approach. The structure was determined to be compliant with NOE restraints while not being over-fitted to a smaller number of RDC restraints (Figure 2). Long NMR-biased MD simulations on the time-scale of tens of nanoseconds allow the protein structure to sample conformations representative of the entire ensemble and as such remove dependencies associated with the starting structure. Charged amino acids were modeled in protonation states obtained from the H++ protonation state server (Gordon et al. 2005), while the metal-ligating residues were maintained in their metal-binding protonation states. The protein structure was maintained in its biologically active dimer state and was immersed in a periodically replicated rectangular box of SPCE water molecules allowing for an 8Å solvation shell around every protein atom (Jorgensen 2005). Explicit Cl- ions were added to neutralize the net charge on the solvated system (Joung and Cheatham 2008). Steered molecular dynamics and energy minimization methods were employed to bring the ligating residues into the zinc-bound coordination geometry obtained from DFT calculations performed at the TPSS/KCIS/LANL2DZ/6-31G(d) level of theory (Case et al. 2010). The calculated zinc-coordinating geometry was validated against the Zn(II) coordination in the 1R1V crystal structure of Zn(II)-CzrA and our previous QM/MM calculations (Eicken et al. 2003; Chakravorty et al. 2012b). A Zn(II) ion was then introduced into each metal binding site described by Asp84, His86, His97 and His100 residues (Eicken et al. 2003). In the absence of an experimentally determined structure, QM/MM methods may be used to introduce the metal ion in its coordination shell as demonstrated in our studies of Zn(II)-bound NmtR and Cu(I)-bound W44M CusF (Lee et al. 2012; Chakravorty et al. 2011).
Next, the solvated protein was energy minimized and equilibrated using a well-defined procedure (Chakravorty et al. 2009; Chakravorty et al. 2008; Chakravorty and Hammes-Schiffer 2010). NOE restraints with a force constant of 30 kcal/mol•Å2 were applied to the protein structure over the course of the equilibration process. Details of the equilibration protocol have been provided elsewhere (Chakravorty et al. 2012b; Chakravorty et al. 2009; Lee et al. 2012) and are included as part of supporting information. In brief, a five-step energy minimization protocol was implemented to gradually minimize the protein in its solvation environment followed by heating to 300 K over 200 ps of MD for a canonical ensemble (NVT) (Allen and Tildesley 1987). The protein was further equilibrated for an isobaric and isothermal ensemble (NPT) for 1 ns at 1 atm pressure (Allen and Tildesley 1987). Following the equilibration phase, a NOE restrained MD simulation (760 restraints) was propagated at 300K for 10 ns using a Langevin thermostat. A time step of 2 fs was employed during this simulation and frames were collected once for every 1000 steps of MD. Over the course of this simulation, long-range electrostatics were calculated using the particle mesh Ewald method (PME) and the system temperature was maintained using Langevin Dynamics (Allen and Tildesley 1987). The SHAKE algorithm was utilized to maintain heavy atom-hydrogen bond lengths and the translational center-of-mass motion was removed every 100 steps (Allen and Tildesley 1987). The protein backbone RMSD was determined to be stable over the course of this simulation. The protein was then gradually cooled to 0 K over 200 ps and further energy minimized to obtain energy minimized NOE-constrained protein structure. No significant NOE violations (>0.3 Å) were observed and the structure passed all structure checks performed using the Maestro program (Qsite; 5.7 ed.; Schrodinger).
The energy minimized NOE-restrained structure was then fitted to RDC information to obtain an NOE and RDC restraint compliant structure. 112 NH RDC restraints (1DHN) were applied to each protein substructure in addition to the NOE restraints at this stage (Arunkumar et al. 2009). Owing to the angular dependence of RDC restraints and their smaller numbers, specific care was taken to not over-fit to them. The RDC fitting cycle was performed in multiple stages starting with a force constant of 0.05 kcal mol-1 Å-2 applied to every RDC restraint until a correlation of ∼1 and an offset of ∼0 Hz were achieved between RDC values obtained from the calculated structure and the experimentally determined values. During this phase, the previously energy minimized NOE-constrained protein structure was run through an abridged version of the NMR refinement process while employing both NOE and RDC restraints to obtain an RDC and NOE refined structure. Details of the process are provided in supporting information. In the next stage of the refinement protocol, the metal ions were treated quantum mechanically to account for possible artifacts and deficiencies of the bonded model of Zn(II). For these QM/MM calculations the Zn(II)-binding residues (Asp84, His86, His97 and His100) and the zinc ions were included in the QM region. The metal binding residues were introduced into the QM regions by means of side-chain cuts between the α and β carbon atoms. The remaining simulated system was modeled using the ff99SB force field (Hornak et al. 2006). During this phase, the NOE and RDC fitted structure was further energy minimized at the SCC-DFTB3/ff99SB level of theory over two steps. In the first stage, the metal ion and its ligating residues were energy-minimized, while in the second stage the entire solvated system was energy minimized. Both minimizations steps were performed with NOE and RDC restraints to obtain the QM/MM energy-minimized NOE+RDC structures.
The RDC fitting cycle was performed with force constants of 0.05, 0.25 and 0.50 kcal mol-1 Å-2 applied to every RDC restraint. The resulting energy minimized RDC+NOE refined structures were checked for consistency with the AMBER and PALES programs. While a good correlation (∼1) was found between the structure-derived RDC values and experimental values, a large offset remained in the RDC refinement performed with a force constant of 0.05 kcal mol-1 Å-2. Energy minimization with a force constant of 0.25 kcal mol-1 Å-2 provided a structure with a correlation value of ∼1 and offset of 0.01 Hz, showing a marked improvement on the starting structure (Figure 2). Only two NOE violations in excess of 0.5 Å were observed in this structure. Furthermore, Ramachandran plots (Figure SI.3) and a comparison of protein backbone carbon Cα-Cα distances (Figure SI.4) for the Zn(II)-crystal structure (panel a), model 1 of the 2KJC NMR ensemble of structures (panel b), NOE-refined energy minimized structure (panel c) and the QM/MM energy minimized NOE+RDC structures (panel d) reveals that CzrA remains folded and maintains attributes that closely resembled the crystal structure. Thus, a force constant of 0.25 kcal mol-1 Å-2 was chosen for the RDC-fitting process.
In the final stage of the MRD-NMR structure refinement methodology, we generated an accurate ensemble of solution structures of Zn(II)-CzrA by collecting 120 ns of NOE and RDC restrained MD (NOE+RDC restrained-MD) data along with an additional 12 ns of QM/MM MD calculations. For this, ten independent simulations starting from the QM/MM energy minimized NOE+RDC refined structure of CzrA were equilibrated and propagated for 12 ns at 300K. Each MD simulation was followed by over 1 ns of QM/MM MD refinement in which the zinc ion and its ligating residues were treated at the SCC-DFTB3 level of theory while the water molecules and remaining protein atoms were modeled using the ff99SB force field (Hornak et al. 2006; Seabra et al. 2007; Gaus et al. 2011). In addition to these simulations, we performed 20 ns of NOE-restrained MD followed by 2 ns of QM/MM MD simulations on the NOE-refined energy minimized Zn(II)-CzrA structure, and 100 ns of unrestrained (not restrained by NMR data) MD starting from the 2KJC structure. The ensembles of structures derived from these simulations would serve to validate our previously obtained results from MD simulations of CzrA and gauge the performance of unconstrained MD simulations and NOE-constrained MD simulations in correlating to experimentally observed backbone 1DNH RDC values. A detailed description of the effort for a skilled user is provided as part of SI.
Results and Discussion
S. aureus CzrA is an extensively studied member of the ArsR/SmtB family of metal ion sensing transcriptional repressor proteins (Chakravorty et al. 2012b; Pennella et al. 2006; Arunkumar et al. 2009; Eicken et al. 2003; Pennella et al. 2003; Grossoehme and Giedroc 2009) and serves as an excellent model system to expand on our understanding of the allosteric mechanism of regulation in metalloregulatory proteins (Giedroc and Arunkumar 2007). We successfully determined a structural ensemble for the zinc-bound form of CzrA (Figure 1(a)-(c)) by effectively using NOE and RDC data in combination with modern MD and QM/MM MD techniques to model metal ion coordination. 120,000 protein structures were collected from the NOE and RDC restrained MD and 12,000 protein structures were collected from the NOE and RDC restrained QM/MM MD simulations to form these ensembles. No significant differences were observed between the structural ensembles obtained from NOE and RDC restrained MD (NOE+RDC restrained-MD) and the QM/MM MD refinement calculations in terms of calculated RMSD values and radius of gyration values (Figure SI.4) suggesting that the classical force field accurately represents the metal-bound protein. Owing to these similarities, the two structural ensembles were merged to create the MRD-NMR structural ensemble whose atomic coordinates have been deposited in the protein data bank (PDB code 2M30). These simulations benefit from rapid convergence to an equilibrated state owing to the large number of restraints on the protein structure. A calculated ensemble comprising of 132,000 protein conformations was collected over the course of NMR-biased MD and QM/MM MD simulations and was found to correlate well with experimentally observed backbone 1DNH RDC values (Figure SI.5). The calculated MRD-NMR structural ensemble provides a detailed and accurate ensemble representation of Zn(II)-bound CzrA in solution and strongly benefits from correctly portraying the first-shell and second shell coordination effects of the metal ion binding to CzrA. In comparison to the NOE and RDC restrained MRD-NMR ensemble of refined structures (MRD-NMR structural ensemble), protein conformations visited over the course of our unconstrained MD, and NOE-restrained MD simulations were NOE compliant (all deviations <0.5 Å), but failed to match the experimentally obtained RDC values as well as the NOE+RDC-fitted ensemble of protein conformations (Figure 3(a)). Upon analyzing the distribution of deviations from the experimentally observed RDC values, we found that the structural ensembles from these simulations successfully matched the experimentally observed RDC data in a time-averaged manner as would be experienced in an experimental NMR setting (Figure 3(b)). These results are in agreement with previous studies (Azurmendi et al. 2002; Iwahar et al. 2006; Showalter et al. 2007; Marsh et al. 2009; Stelzer et al. 2009; Vendruscolo et al. 2011) that accurately calculated RDC data from MD simulations and suggest that MD simulations on biologically relevant time-scales can potentially capture the structural characteristics of the system under investigation.
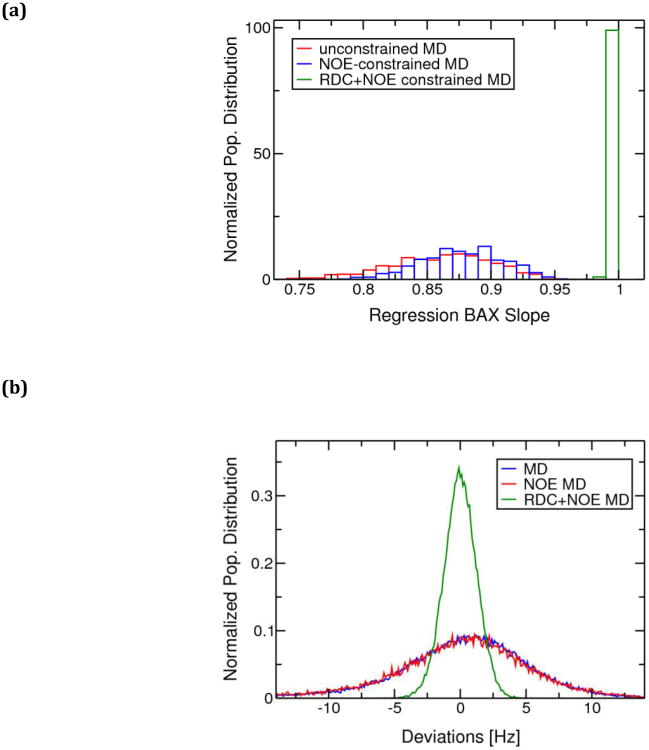
Fig. 3.
(a) Normalized population distribution of the Bax regression slopes(Zweckstetter and Bax 2000) indicating the quality of fitting between the experimental backbone NH(1DNH) RDC constraints with those calculated for ensembles of structures from NOE+RDC-restrained MD and QM/MD simulations (MRD-NMR structural ensemble), NOE-restrained MD simulations and unrestrained MD simulations of Zn(II)-CzrA. A higher regression slope indicates a better fit to the data. (b) Normalized population distributions of calculated NH(1DNH) RDC-deviations from experimentally determined values for the ensemble of structures from unrestrained MD simulations, NOE restrained MD simulations, and NOE + RDC restrained MD simulations of Zn(II)-CzrA
An aligned representation of the Zn(II) bound crystal structure, and a randomly selected model from the 2KJC NMR structural ensemble and the MRD-NMR structural ensemble (Figure SI.7) reveals that the fold is robustly maintained over the course of these simulations. In our refined model, the Zn(II) bound structure of CzrA maintained its zinc coordination with Asp84, His86, His97 and His100 residues in agreement with the geometry observed in the crystal structure (Figure 1(b)). The MRD-NMR ensemble of structures presents a significant improvement on the 2KJC structural ensemble in which the metal ion binding residues were not optimally arranged for metal ion binding as a result of not modeling the zinc ion during the structure determination process (Figure 1(d)-(f)). While the bonded model representation maintained a fixed coordination for the metal ion, the QM/MM MD simulations provided an opportunity to test the zinc coordination environment in CzrA. We found that the metal coordination bonds are maintained over the course of the QM/MM MD calculations, though the individual bond distances varied within a range of ± 0.1 Å from the observed distances in the crystal structure owing to inadequacies in the SCC-DFTB3 Hamiltonian (Figure 1(c)) (Settergren et al. 2008). While such errors in bond lengths may be considered reasonable, they may be resolved by utilizing DFT or ab initio based methods implemented in the AMBER suite of programs during the QM/MM MD phase of the metal ion structure refinement protocol (Case et al. 2010).
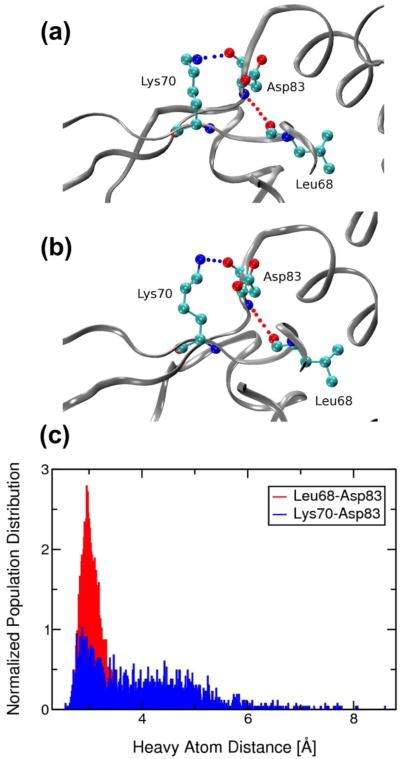
The MRD-NMR ensemble of structures sheds further light on the mechanism of allosteric regulation in CzrA. An interprotomer zinc-dependent second-coordination shell hydrogen bond between the conserved metal-binding ligand His97 and His67'of the αR helix extends to form a specific hydrogen-bonding pathway that connects the metal binding region to residues in the DNA-binding region (Figure 4) (Eicken et al. 2003). This hydrogen bond is only observed in the zinc-bound form of the protein, and as such has been implicated in playing a major role in the mechanism of allosteric regulation in CzrA (Eicken et al. 2003; Arunkumar et al. 2009; Chakravorty et al. 2012b). Computational studies suggest that this hydrogen bond is strengthened on the order of ∼10 kcal/mol on metal ion binding compared to an apo allosteric form of the protein (Chakravorty et al. 2012a). In excellent agreement with previous structural (Eicken et al. 2003), thermodynamic (Grossoehme and Giedroc 2009) and MD studies,(Chakravorty et al. 2012b) we find that this short hydrogen bond and the resulting hydrogen bonding pathway does indeed exist in the Zn(II) bound CzrA in solution (Figure 4 (a)). Upon comparing to the 2KJC ensemble of NMR structures, we find that the zinc-lacking model is unable to correctly describe the nature of this hydrogen bond (Figures 4(b) and SI.8(b)). It should be noted that while a hydrogen bond restraint between His97 and His67' was included in the 2KJC NMR structure calculations, no such constraints were used in these simulations (Arunkumar et al. 2009). In addition to this zinc-dependent hydrogen bond, we find that Asp83, a conserved residue that neighbours the invariant metal-binding residue Asp84, participates in an interaction observed in the crystal structure of Zn(II)-CzrA (Figure 5). This interaction was absent in the 2KJC ensemble of NMR structures (Arunkumar et al. 2009). The backbone carbonyl group of Asp83 now interacts with the positively charged side-chain of the β-wing residue, Lys70, while the backbone NH group of Asp83 interacts with the backbone carbonyl group of Leu68, possibly allowing Asp83 to communicate metal-binding to the β-wings. The energetic importance of Asp83 in mediating zinc-dependent allosteric switching in CzrA remains to be determined.
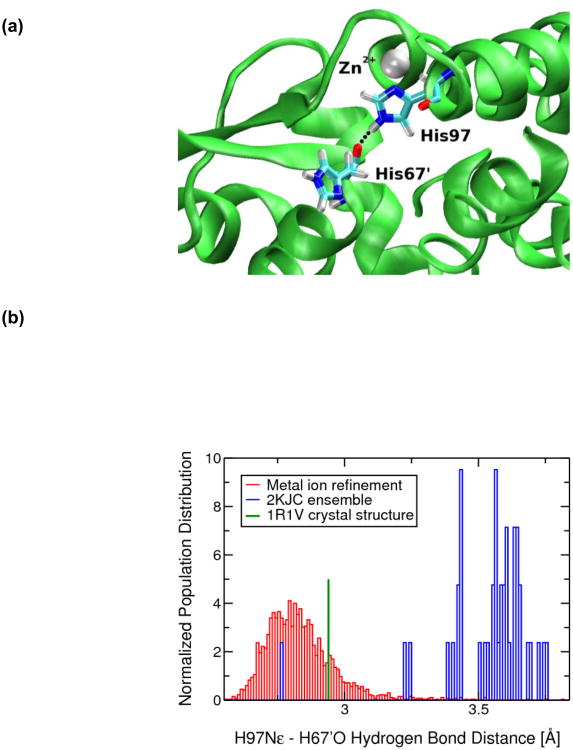
Fig. 4.
(a) The zinc mediated His97-His67' hydrogen bond that forms a part of the hydrogen-bonding pathway in Zn(II)-CzrA. The zinc ion is shown as a silver sphere while His97 and His67' residues are in licorice notation. The His97-His67' hydrogen bond is indicated with a dotted black line. (b) Normalized population distribution of heavy atom hydrogen bond distances (Nε-O) for the 2KJC ensemble of structures and the MRD-NMR ensemble of structures. The crystallographically determined His97-His67' distance is shown for comparison
Fig. 5.
Ribbon representation of (a) the 1R1V crystal structure of Zn(II)-CzrA and (b) a MRD-NMR refined structure of Zn(II)-CzrA near the metal binding site. Interactions between Asp83 and Lys70 (blue dotted line) and Asp83 and Leu68 (red dotted line) are indicated. (c) Normalized population distributions of the heavy atom hydrogen bond distances for the metal ion refined RDC+NOE constrained MD and QM/MM MD ensemble of structures
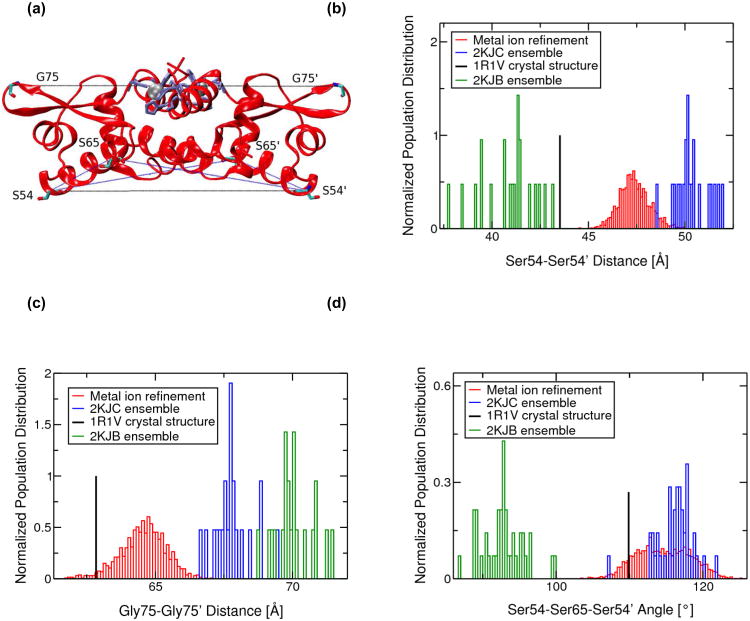
CzrA modulates its conformational dynamics to selectively sample functional allosteric ensembles that increase or decrease its binding affinity for the CzrO operator (DNA) or metal ion (Arunkumar et al. 2009; Chakravorty et al. 2012b). The 2KJC ensemble of structures and NMR data reveal that the zinc-bound protein adopts an open conformation that significantly reduces the DNA-binding affinity (Arunkumar et al. 2009). Incorporation of the metal ion chelate during the NMR structure refinement process yields more accurate statistical data. The interprotomer Ser54-Ser54' and Gly75-Gly75' Cα-distances provide useful metrics to measure the interprotomer distances between the αR helical DNA reading heads and the β-wing tips, respectively, and help distinguish between an “open” or “closed” conformation (Figures 6 and SI.9). In the open conformation, the DNA reading heads of the αR helices are widely separated while the β-wings lift upwards, bringing the wing tips closer. In addition, the Ser54-Ser65-Ser54' and Ser54-Ser65'-Ser54' Cα “torsion” angles describe the degree to which the αR helices are “torqued” relative to the core of the dimer. A “flatter” CzrA dimer (less torque) is less conducive to DNA binding and consequently corresponds to a lower DNA-binding affinity conformation. In agreement with our previous data, we find that the MRD-NMR refined ensemble of structures sample interprotomer Ser54-Ser54' and Gly75-Gly75' Cα-distances and Ser54-Ser65-Ser54' and Ser54-Ser65'-Ser54' Cα-angles that are indicative of the weakly DNA-binding “open” conformation (Figures 6 and SI.9) as compared to similar metrics obtained from the 2KJB ensemble of DNA-bound apo-CzrA that exists in the “closed” conformation (Arunkumar et al. 2009). In general agreement with 2KJC ensemble of NMR structures, we also find here that metal ion refined solution structures of CzrA adopt a more open conformation when compared to that of the 1R1V crystal structure. The spread of these distances and angles for the NOE+RDC restrained ensemble are in agreement with the range of distances sampled over our previous MD simulations of Zn(II)-CzrA, thus validating our previous study of the various allosteric forms of the transcriptional repressor involving unconstrained MD (Chakravorty et al. 2012b). A further analysis of the MRD-NMR Zn(II)-CzrA structural ensemble is provided in SI.
Fig. 6.
(a) Ribbon representation of the MRD-NMR structure of zinc bound CzrA indicating residues used to measure changes between the open and closed conformations. Zinc ions are shown as silver spheres and protein residues are shown in licorice depiction. Residues involved in metal ion binding are colored lilac. Normalized population distributions of (b) Ser54-Ser54' and (c) Gly75-Gly75' inter-subunit Cα-Cα distances and (d) Ser54-Ser65-Ser54' and Ser54'-Ser65'-Ser54 inter-subunit Cα-Cα-Cα angles for the 2KJC ensemble of NMR structures of Zn(II)-CzrA, the 2KJB ensemble of NMR structures of DNA-bound CzrA, and the metal ion refined RDC+NOE constrained MD and QM/MM MD ensemble of Zn(II)-CzrA structures. Distances from the 1R1V Zn(II)-bound crystal structure of CzrA are also indicated
Conclusions
In this study we show that the zinc coordination in NMR protein structures may be refined by coupling the NMR structure determination protocol with metal ion force field based MD simulations and QM/MM calculations (Case 2002; Peters et al. 2010; Bertini et al. 2011). Using the MRD-NMR methodology, we refined the zinc-ion coordination in the paradigm zinc sensor protein S. aureus CzrA to obtain an accurate solution structure representation of the allosterically inhibited zinc-bound state. We find that our classical bonded model approach accurately models the metal ion and its effect on protein structure in the NMR structure calculation process. Our ensemble of metal ion coordination refined structures allowed us to analyze the atomistic networks in greater detail and understand the mechanism of allosteric regulation in this system in solution not previously possible. NOE and RDC restraint-based simulations can identify multiple conformations or Markov states that may play a role in biological function (Chennubhotla and Bahar 2006). Indeed, this approach will aid in protein folding studies by presenting additional metal ion based restraints (Lange et al. 2012; Thompson et al. 2012) and help in the design and structure determination of novel metal-templated proteins (Brodin et al. 2010; Salgado et al. 2010; Salgado et al. 2011). QM/MM MD metal ion NMR-refinement calculations are particularly poised to tackle protein structures with previously undetermined metal-binding conformations (Chakravorty et al. 2012a; Lee et al. 2012). While the MCPB program (Peters et al. 2010) is available as part of the AMBERTools package (Case et al. 2010), this protocol could potentially be implemented to treat metal ions in the AMBER-based portal server for NMR structures (AMPS-NMR) as part of the WENMR portal (Bertini et al. 2011). The presented MRD-NMR methodology may be implemented during the NMR structure calculation or subsequent structure-refinement methods, and should be broadly applicable in the field of solution NMR structure determination of metal binding proteins.
Supplementary Material
Acknowledgments
We gratefully acknowledge the United States National Institutes of Health for support of this study (GM044974 to K.M.M. and GM042569 to D.P.G.) and high performance computing at the University of Florida (UFHPC) for providing and maintaining the computational resources used to perform this study. We also thank Sarah Gordon for helpful discussions.
List of abbreviations
- AMBER
assisted model building with energy refinement
- DFT
density functional theory
- MCPB
metal center parameter builder
- MD
molecular dynamics
- MRD-NMR
metal restrained dynamics nuclear magnetic resonance
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser enhancements
- QM/MM
quantum mechanical/molecular mechanical
- QM/MM MD
quantum mechanical/molecular mechanical molecular dynamics
- RDC
residual dipolar couplings
- RD-NMR
restrained dynamics nuclear magnetic resonance
- RMSD
root mean square deviation
- XAS
X-ray absorption spectroscopy
- PDB
protein data bank
References
- Allen MP, Tildesley DJ. Computer Simulations of Liquids. Clarendon Press; Oxford: 1987. [Google Scholar]
- Arunkumar AI, Campanello GC, Giedroc DP. Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(43):18177–18182. doi: 10.1073/Pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azurmendi HF, Bush CA. Tracking alignment from the moment of interal tensor (TRAMITE) of biomolecules in neutral dilute liquid crystal solutions. 2002;124(11):2426–2427. doi: 10.1031/ja017524z. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Cavet JS, Dennison C, Graham AI, Harvie DR, Robinson NJ. NMR structural analysis of cadmium sensing by winged helix repressor CmtR. The Journal of biological chemistry. 2007;282(41):30181–30188. doi: 10.1074/jbc.M701119200. [DOI] [PubMed] [Google Scholar]
- Bertini I, Case DA, Ferella L, Giachetti A, Rosato A. A Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics. 2011;27(17):2384–2390. doi: 10.1093/bioinformatics/btr415. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann C, Soucek S, Kuhlmann SI, Mills-Lujan K, Kelly SM, Yang JC, Iglesias N, Stutz F, Corbett AH, Neuhaus D, Stewart M. Structural basis for polyadenosine-RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export. Structure. 2012;20(6):1007–1018. doi: 10.1016/j.str.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin JD, Medina-Morales A, Ni T, Salgado EN, Ambroggio XI, Tezcan FA. Evolution of metal selectivity in templated protein interfaces. Journal of the American Chemical Society. 2010;132(25):8610–8617. doi: 10.1021/ja910844n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. X-ray crystallography and NMR reveal complementary views of structure and dynamics. Nature Structural Biology. 1997;4:862–865. [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2(11):2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Calhoun JR, Liu W, Spiegel K, Dal Peraro M, Klein ML, Valentine KG, Wand AJ, DeGrado WF. Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement. Structure. 2008;16(2):210–215. doi: 10.1016/j.str.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA. Molecular dynamics and NMR spin relaxation in proteins. Accounts of Chemical Research. 2002;35(6):325–331. doi: 10.1021/ar010020l. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham TE, I, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER. Vol. 11. University of California; San Francisco: 2010. [Google Scholar]
- Chakravorty DK, Hammes-Schiffer S. Impact of mutation on proton transfer reactions in ketosteroid isomerase: insights from molecular dynamics simulations. J Am Chem Soc. 2010;132(21):7549–7555. doi: 10.1021/ja102714u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty DK, Kumarasiri M, Soudackov AV, Hammes-Schiffer S. Implementation of umbrella integration within the framework of the empirical valence bond approach. Journal of Chemical Theory and Computation. 2008;4(11):1974–1980. doi: 10.1021/ct8003386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty DK, Parker TM, Guerra AJ, Sherrill CD, Giedroc DP, Merz KM., Jr . Journal of the American Chemical Society:ASAP article. 2012a. Energetics of zinc-mediated interactions in the allosteric pathways of metal sensor proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty DK, Soudackov AV, Hammes-Schiffer S. Hybrid quantum/classical molecular dynamics simulations of the proton transfer reactions catalyzed by ketosteroid isomerase: analysis of hydrogen bonding, conformational motions, and electrostatics. Biochemistry. 2009;48(44):10608–10619. doi: 10.1021/bi901353v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty DK, Wang B, Lee CW, Giedroc DP, Merz KM., Jr Simulations of allosteric motions in the zinc sensor CzrA. Journal of the American Chemical Society. 2012b;134(7):3367–3376. doi: 10.1021/ja208047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty DK, Wang B, Ucisik MN, Merz KM., Jr Insight into the cation-pi interaction at the metal binding site of the copper metallochaperone CusF. Journal of the American Chemical Society. 2011;133(48):19330–19333. doi: 10.1021/ja208662z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Im W, Brooks CL., 3rd Refinement of NMR structures using implicit solvent and advanced sampling techniques. Journal of the American Chemical Society. 2004;126(49):16038–16047. doi: 10.1021/ja047624f. [DOI] [PubMed] [Google Scholar]
- Chen J, Won HS, Im W, Dyson HJ, Brooks CL., 3rd Generation of native-like protein structures from limited NMR data, modern force fields and advanced conformational sampling. Journal of Biomolecular Nmr. 2005;31(1):59–64. doi: 10.1007/s10858-004-6056-z. [DOI] [PubMed] [Google Scholar]
- Chennubhotla C, Bahar I. Markov propagation of allosteric effects in biomolecular systems: application to GroEL-GroES. Molecular Systems Biology. 2006;2:36. doi: 10.1038/msb4100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Schwieters CD. Concordance of residual dipolar couplings, backbone order parameters and crystallographic B-factors for a small alpha/beta protein: a unified picture of high probability, fast atomic motions in proteins. Journal of Molecular Biology. 2006;355(5):879–886. doi: 10.1016/j.jmb.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Kollman PA. Application of Resp Charges to Calculate Conformational Energies, Hydrogen-Bond Energies, and Free-Energies of Solvation. Journal of the American Chemical Society. 1993;115(21):9620–9631. [Google Scholar]
- Cramer CJ, Truhlar DG. Density functional theory for transition metals and transition metal chemistry. Phys Chem Chem Phys. 2009;11(46):10757–10816. doi: 10.1039/B907148b. [DOI] [PubMed] [Google Scholar]
- Crowhurst KA, Tollinger M, Forman-Kay JD. Cooperative interactions and a non-native buried trp in the unfolded state of an SH3 domain. Journal of Molecular Biology. 2002;322:163–178. doi: 10.1016/S0022-2836(02)00741-6. [DOI] [PubMed] [Google Scholar]
- Deeth RJ, Anastasi A, Diedrich C, Randell K. Molecular modelling for transition metal complexes: Dealing with d-electron effects. Coordin Chem Rev. 2009;253(5-6):795–816. doi: 10.1016/J.Ccr.2008.06.018. [DOI] [Google Scholar]
- Ditchfie R, Hehre WJ, Pople JA. Self-Consistent Molecular-Orbital Methods .9. Extended Gaussian-Type Basis for Molecular-Orbital Studies of Organic Molecules. Journal of Chemical Physics. 1971;54(2):724–728. [Google Scholar]
- Eicken C, Pennella MA, Chen XH, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. Journal of Molecular Biology. 2003;333(4):683–695. doi: 10.1016/J/Jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438(7064):117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nature Structural & Molecular Biology. 2010;17(2):241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JAJ, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamao C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople J. Gaussian 09 A.01 edn. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- Gao Y, Kaluarachchi K, Giedroc DP. Solution structure and backbone dynamics of Mason-Pfizer monkey virus (MPMV) nucleocapsid protein. Protein science: a publication of the Protein Society. 1998;7(11):2265–2280. doi: 10.1002/pro.5560071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus M, Cui QA, Elstner M. DFTB3: Extension of the Self-Consistent-Charge Density-Functional Tight-Binding Method (SCC-DFTB) Journal of Chemical Theory and Computation. 2011;7(4):931–948. doi: 10.1021/Ct100684s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc DP, Arunkumar AI. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Transactions. 2007;(29):3107–3120. doi: 10.1039/B706769k. [DOI] [PubMed] [Google Scholar]
- Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pK(a)s and adding missing hydrogens to macromolecules. Nucleic Acids Research. 2005;33:W368–W371. doi: 10.1093/Nar/Gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme NE, Giedroc DP. Energetics of Allosteric Negative Coupling in the Zinc Sensor S. aureus CzrA. Journal of the American Chemical Society. 2009;131(49):17860–17870. doi: 10.1021/Ja906131b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntert P, Braun W, Wuthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. Journal of Molecular Biology. 1991;217(3):517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. Journal of Molecular Biology. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Hartsough DS, Merz KM. Dynamic Force-Field Models - Molecular-Dynamics Simulations of Human Carbonic-Anhydrase-Ii Using a Quantum-Mechanical Molecular Mechanical Coupled Potential. Journal of Physical Chemistry. 1995;99(28):11266–11275. [Google Scholar]
- Hay PJ, Wadt WR. Abinitio Effective Core Potentials for Molecular Calculations - Potentials for K to Au Including the Outermost Core Orbitals. Journal of Chemical Physics. 1985;82(1):299–310. [Google Scholar]
- Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450(7171):838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. Journal of Molecular Biology. 2002;319(1):209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Hoops SC, Anderson KW, Merz KM. Force-Field Design for Metalloproteins. Journal of the American Chemical Society. 1991;113(22):8262–8270. [Google Scholar]
- Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14(1):33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(41):15062–15067. doi: 10.1073/Pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- Jorgensen WLTRJ. P Natl Acad Sci USA. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung IS, Cheatham TE., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. The journal of physical chemistry B. 2008;112(30):9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(27):10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Chakravorty DK, Chang FM, Reyes-Caballero H, Ye Y, Merz KM, Jr, Giedroc DP. Solution structure of Mycobacterium tuberculosis NmtR in the apo state: insights into Ni(II)-mediated allostery. Biochemistry. 2012;51(12):2619–2629. doi: 10.1021/bi3001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang J, Wang J, Wang W. Metal-coupled folding of Cys2His2 zinc-finger. J Am Chem Soc. 2008;130(3):892–900. doi: 10.1021/ja075302g. [DOI] [PubMed] [Google Scholar]
- Li X, Hayik SA, Merz KM., Jr QM/MM X-ray refinement of zinc metalloenzymes. J Inorg Biochem. 2010;104(5):512–522. doi: 10.1016/j.jinorgbio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Wang RX. Systematic Derivation of AMBER Force Field Parameters Applicable to Zinc-Containing Systems. Journal of Chemical Theory and Computation. 2010;6(6):1852–1870. doi: 10.1021/Ct900454q. [DOI] [PubMed] [Google Scholar]
- Long D, Bruschweiler R. In silico elucidation of the recognition dynamics of ubiquitin. PLoS computational biology. 2011;7(4):e1002035. doi: 10.1371/journal.pcbi.1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mendez B, Guntert P. Automated protein structure determination from NMR spectra. Journal of the American Chemical Society. 2006;128(40):13112–13122. doi: 10.1021/ja061136l. [DOI] [PubMed] [Google Scholar]
- Ma Z, Jacobsen FE, Giedroc DP. Coordination Chemistry of Bacterial Metal Transport and Sensing. Chemical Reviews. 2009;109(10):4644–4681. doi: 10.1021/Cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick PR, Cervantes CF, Abel BL, Komives EA, Blackledge M, McCammon JA. Enhanced conformational space sampling improves the prediction of chemical shifts in proteins. Journal of the American Chemical Society. 2010;132(4):1220–1221. doi: 10.1021/ja9093692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealman TD, Zhou MW, Affandi T, Chacon KN, Aranguren ME, Blackburn NJ, Wysocki VH, McEvoy MM. N-Terminal Region of CusB Is Sufficient for Metal Binding and Metal Transfer with the Metallochaperone CusF. Biochemistry. 2012;51(34):6767–6775. doi: 10.1021/Bi300596a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvao RW, De Simone A, Vendruscolo M. Determination of structural fluctuations of proteins from structure-based calculations of residual dipolar couplings. Journal of Biomolecular Nmr. 2012;53(4):281–292. doi: 10.1007/s10858-012-9644-3. [DOI] [PubMed] [Google Scholar]
- Mumenthaler C, Guntert P, Braun W, Wuthrich K. Automated combined assignment of NOESY spectra and three-dimensional protein structure determination. Journal of Biomolecular Nmr. 1997;10(4):351–362. doi: 10.1023/a:1018383106236. [DOI] [PubMed] [Google Scholar]
- Pang YP. Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method. Proteins-Structure Function and Genetics. 2001;45(3):183–189. doi: 10.1002/prot.1138. [DOI] [PubMed] [Google Scholar]
- Pennella MA, Arunkumar AI, Giedroc DP. Individual metal ligands play distinct functional roles in the zinc sensor Staphylococcus aureus CzrA. Journal of Molecular Biology. 2006;356(5):1124–1136. doi: 10.1016/J.Jmb.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Structural elements of metal selectivity in metal sensor proteins. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):3713–3718. doi: 10.1073/Pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MB, Yang Y, Wang B, Fusti-Molnar L, Weaver MN, Merz KM. Structural Survey of Zinc-Containing Proteins and Development of the Zinc AMBER Force Field (ZAFF) Journal of Chemical Theory and Computation. 2010;6(9):2935–2947. doi: 10.1021/Ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce LCT, Salomon-Ferrer R, de Oliveira CAF, McCammon JA, Walker RC. Routine Access to Millisecond Time Scale Events with Accelerated Molecular Dynamics. Journal of Chemical Theory and Computation. 2012;8(9):2997–3002. doi: 10.1021/Ct300284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev SY, Click TH, Kaminski GA. Electrostatic polarization is crucial in reproducing Cu(I) interaction energies and hydration. The journal of physical chemistry B. 2011;115(33):10079–10085. doi: 10.1021/jp2051933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qsite. 5.7. Schrodinger INY; NY: 2011. [Google Scholar]
- Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, Montelione GT, Baker D. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327(5968):1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA. Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins. 2009;75(1):147–167. doi: 10.1002/prot.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey J, Savin A. Virtual space level shifting and correlation energies. Int J Quantum Chem. 1998;69(4):581–590. [Google Scholar]
- Roberts BP, Miller BR, 3rd, Roitberg AE, Merz KM., Jr Wide-open flaps are key to urease activity. Journal of the American Chemical Society. 2012;134(24):9934–9937. doi: 10.1021/ja3043239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robustelli P, Kohlhoff K, Cavalli A, Vendruscolo M. Using NMR chemical shifts as structural restraints in molecular dynamics simulations of proteins. Structure. 2010;18(8):923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Rosato A, Aramini JM, Arrowsmith C, Bagaria A, Baker D, Cavalli A, Doreleijers JF, Eletsky A, Giachetti A, Guerry P, Gutmanas A, Guntert P, He Y, Herrmann T, Huang YJ, Jaravine V, Jonker HR, Kennedy MA, Lange OF, Liu G, Malliavin TE, Mani R, Mao B, Montelione GT, Nilges M, Rossi P, van der Schot G, Schwalbe H, Szyperski TA, Vendruscolo M, Vernon R, Vranken WF, Vries S, Vuister GW, Wu B, Yang Y, Bonvin AM. Blind testing of routine, fully automated determination of protein structures from NMR data. Structure. 2012;20(2):227–236. doi: 10.1016/j.str.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B, Tezcan FA. Metal templated design of protein interfaces. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(5):1827–1832. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado EN, Brodin JD, To MM, Tezcan FA. Templated construction of a Zn-selective protein dimerization motif. Inorg Chem. 2011;50(13):6323–6329. doi: 10.1021/ic200746m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Seabra GD, Walker RC, Elstner M, Case DA, Roitberg AE. Implementation of the SCC-DFTB method for hybrid QM/MM simulations within the amber molecular dynamics package. Journal of Physical Chemistry A. 2007;111(26):5655–5664. doi: 10.1021/Jp0700711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew Chem Int Ed Engl. 2009;48(7):1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- Settergren NM, Buhlmann P, Amin EA. Assessment of density functionals, semiempirical methods, and SCC-DFTB for protonated creatinine geometries. J Mol Struc-Theochem. 2008;861(1-3):68–73. doi: 10.1016/J.Theochem.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrignani J, Pierattelli R. Nuclear magnetic resonance signal chemical shifts and molecular simulations: a multidisciplinary approach to modeling copper protein structures. J Biol Inorg Chem. 2012;17(1):71–79. doi: 10.1007/s00775-011-0830-7. [DOI] [PubMed] [Google Scholar]
- Shao JY, Tanner SW, Thompson N, Cheatham TE. Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. Journal of Chemical Theory and Computation. 2007;3(6):2312–2334. doi: 10.1021/Ct700119m. [DOI] [PubMed] [Google Scholar]
- Showalter SA, Johnson E, Rance M, Bruschweiler R. Toward quantitative interpretation of methyl side-chain dynamics from NMR by molecular dynamics simulations. Journal of the American Chemical Society. 2007;129(46):14146–14147. doi: 10.1021/ja075976r. [DOI] [PubMed] [Google Scholar]
- Snyder DA, Bhattacharya A, Huang YJ, Montelione GT. Assessing precision and accuracy of protein structures derived from NMR data. Proteins. 2005;59(4):655–661. doi: 10.1002/prot.20499. [DOI] [PubMed] [Google Scholar]
- Stelzer AC, Frank AT, Bailor MH, Andricioaei I, Al-Hashimi HM. Constructing atomic-resolution RNA structural ensembles using MD and motionally decoupled NMR RDCs. Methods. 2009;49:167–173. doi: 10.1016/j.ymeth.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stote R, Standing L. Serial and Multiple Homicide - Is There an Epidemic. Soc Behav Personal. 1995;23(4):313–317. [Google Scholar]
- Thompson JM, Sgourakis NG, Liu G, Rossi P, Tang Y, Mills JL, Szyperski T, Montelione GT, Baker D. Accurate protein structure modeling using sparse NMR data and homologous structure information. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(25):9875–9880. doi: 10.1073/pnas.1202485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AJ, Gray HB. Bio-inorganic chemistry. Curr Opin Chem Biol. 1998;2(2):155–158. doi: 10.1016/s1367-5931(98)80056-2. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278(5340):1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR evidence for slow collective motions in cyanometmyoglobin. Nature Structural Biology. 1997;4(4):292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- Tools MDI. Maestro-Desmond Interoperability Tools Version 3.0 edn. Schrödinger; New York, NY: 2011. [Google Scholar]
- Toulouse J, Savin A, Adamo C. Validation and assessment of an accurate approach to the correlation problem in density functional theory: The Kriger-Chen-Iafrate-Savin model. Journal of Chemical Physics. 2002;117(23):10465–10473. doi: 10.1063/1.1521432. [DOI] [Google Scholar]
- Tus A, Rakipovic A, Peretin G, Tomic S, Sikic M. BioMe: biologically relevant metals. Nucleic Acids Research. 2012;40(Web Server issue):W352–357. doi: 10.1093/nar/gks514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedani A, Huhta DW. A New Force-Field for Modeling Metalloproteins. Journal of the American Chemical Society. 1990;112(12):4759–4767. [Google Scholar]
- Simone AD, Montalvao RW, Vendruscolo M. Determination of Conformational Equilibria in Proteins Using Residual Dipolar Couplings. Journal of Chemical Theory and Computation. 2011;7(12):4189–4195. doi: 10.1021/ct200361b. doi:101021/ct200361b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadt WR, Hay PJ. Abinitio Effective Core Potentials for Molecular Calculations - Potentials for Main Group Elements Na to Bi. Journal of Chemical Physics. 1985;82(1):284–298. [Google Scholar]
- Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7(1):25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- Wang C, Vernon R, Lange O, Tyka M, Baker D. Prediction of structures of zinc-binding proteins through explicit modeling of metal coordination geometry. Protein science: a publication of the Protein Society. 2010;19(3):494–506. doi: 10.1002/pro.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JC, Piquemal JP, Chaudret R, Reinhardt P, Ren P. Polarizable molecular dynamics simulation of Zn(II) in water using the AMOEBA force field. Journal of Chemical Theory and Computation. 2010;6(7):2059–2070. doi: 10.1021/ct100091j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RB, Lu ZY, Cao ZX, Zhang YK. A Transferable Nonbonded Pairwise Force Field to Model Zinc Interactions in Metalloproteins. Journal of Chemical Theory and Computation. 2011;7(2):433–443. doi: 10.1021/Ct100525r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LW, Eyal E, Chennubhotla C, Jee J, Gronenborn AM, Bahar I. Insights into equilibrium dynamics of proteins from comparison of NMR and X-ray data with computational predictions. Structure. 2007;15(6):741–749. doi: 10.1016/j.str.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chakravorty DK, Merz KM., Jr Finding a needle in the haystack: computational modeling of Mg2+ binding in the active site of protein farnesyltransferase. Biochemistry. 2010;49(44):9658–9666. doi: 10.1021/bi1008358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Miao Y, Wang B, Cui G, Merz KM., Jr Catalytic mechanism of aromatic prenylation by NphB. Biochemistry. 2012a;51(12):2606–2618. doi: 10.1021/bi201800m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang B, Ucisik MN, Cui G, Fierke CA, Merz KM., Jr Insights into the mechanistic dichotomy of the protein farnesyltransferase peptide substrates CVIM and CVLS. Journal of the American Chemical Society. 2012b;134(2):820–823. doi: 10.1021/ja209650h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York DM, Darden TA, Pedersen LG. The Effect of Long-Range Electrostatic Interactions in Simulations of Macromolecular Crystals - a Comparison of the Ewald and Truncated List Methods. Journal of Chemical Physics. 1993;99(10):8345–8348. [Google Scholar]
- Zhao Y, Truhlar DG. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor Chem Acc. 2008;120(1-3):215–241. doi: 10.1007/S00214-007-0310-X. [DOI] [Google Scholar]
- Zweckstetter M, Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: Aid to protein structure determination by NMR. Journal of the American Chemical Society. 2000;122(15):3791–3792. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.