Abstract
A main objective of synthetic biology is to make the process of designing genetically-encoded biological systems more systematic, predictable, robust, scalable, and efficient. The examples of genetic systems in the field vary widely in terms of operating hosts, compositional approaches, and network complexity, ranging from a simple genetic switch to search-and-destroy systems. While significant advances in synthesis capabilities support the potential for the implementation of pathway- and genome-scale programs, several design challenges currently restrict the scale of systems that can be reasonably designed and implemented. Synthetic biology offers much promise in developing systems to address challenges faced in manufacturing, the environment and sustainability, and health and medicine, but the realization of this potential is currently limited by the diversity of available parts and effective design frameworks. As researchers make progress in bridging this design gap, advances in the field hint at ever more diverse applications for biological systems.
Keywords: biological parts, evolutionary robustness, level-matching, genetically-encoded programs
INTRODUCTION
Synthetic biology aims to make cells programmable by designing genetic circuits to rewire endogenous gene regulatory systems, rescue cellular malfunctions, and control cellular morphology. The complexity of the functions of genetic circuits can range from a simple genetic toggle switch to a complex search-and-destroy system. In general, researchers begin with top-down approaches by constructing abstract work flow charts and decomposing the design goal into several executable instructions with concrete specifications. Then, each specification and consideration can be fulfilled with bottom-up approaches, by assembling smaller building blocks of biological ‘parts’ into larger ‘devices’ and finally into functional ‘systems’.
Although we can currently synthesize, assemble, and transfer artificial yeast chromosomes (1) and bacterial genomes (2) (equivalent to genetically-encoded programs on the order of a 1000 gene cassettes (3)), current examples of synthetic genetic systems generally consist of fewer than 10 genes. Two challenges that contribute to the disparity between our ability to design systems and our ability to synthesize systems include the lack of well characterized parts for desired functions and the lack of methods for reliably and robustly composing parts into devices. In this review, we will first summarize the availability of different classes of parts and methods for prospecting new parts. Then, we will discuss overall design strategies for genetic devices and systems, and propose methods for level matching between genetic components, and finally aspects of design beyond level matching. As the number of characterized parts and methods for reliable composition increase, so will the scalability and predictability of engineered genetic circuits, thus advancing the ability of synthetic biology programs to address needs in metabolic engineering, environmental remediation, and human health.
BIOLOGICAL PARTS
The building blocks in synthetic biology, commonly known as biological parts, are entities that can be genetically-encoded with distinct biological functions in vivo, such as a promoter sequence, an RNA aptamer, or a fluorescent protein. The composition, properties, and functions of biological parts are diverse and thus amenable to several types of categorization.
Categorizing Biological Parts
The number of biological parts exceeds the handling capacity of individual researchers, necessitating well-designed look-up tables. Different sorting architectures aimed to facilitate the bottom-up assembly have been proposed to organize biological parts. One standard approach is to categorize biological parts based on their underlying composition (DNA, RNA, or protein), which allows researchers to quickly identify the compositional characteristics and to determine a proper design scheme according to the genetic context (4). Another approach is to organize biological parts based on their operable host organisms. From this perspective, biological parts can be grouped into three main categories: host-specific, facultative, and universal biological parts (5). A host-specific part, which interacts specifically with the host-cell machinery, will only function in the one or a few closely related species. Facultative parts can usually be ported, but with varied regulatory roles across several organisms. This facultative activity results from diverse biological organisms using similar regulatory mechanisms for different purposes; for example, ribozyme cleavage of a transcript can either enhance or suppress gene expression depending on the RNA cleavage-associated degradation mechanism (6, 7). Universal parts, which exhibit similar function across different species, are usually less coupled to the gene expression process (e.g., RNA aptamers) or are the end product of gene expression (e.g., fluorescent reporters). Host-specificity can reduce the power of cross-platform engineering, by which biological parts can be functionally verified in a simpler model species and then applied to a more complicated system.
Another approach to categorize biological parts is by their functional roles, allowing researchers to quickly identify the biological parts needed to implement a genetic device (8). A typical genetic device can be decomposed into four major components, which either sense environmental signals (sensor), support calculations (regulator), generate outputs (actuator), or act to connect the individual components (adapter). Regulators play a central role in a genetic device by regulating gene expression at a constant level. Sensors and actuators integrate with device inputs and outputs, respectively, and provide the interfaces to regulators. Adaptors are supporting parts in a genetic device, which generally enable device fine-tuning for optimization and level matching without significantly changing the overall design architecture.
In this review, we provide a look-up table to demonstrate the organization of common biological parts previously used in engineered systems (Table 1). We first group biological parts by their functional roles, and then by their operating mechanisms, genetic material, and host-specificity, which allows researchers to quick identify the existing biological parts that can be used for their particular design needs. The table also serves as an overview of the current progress of prospecting biological parts and to highlight parts that are less well-understood and underpopulated, and where further engineering efforts may be beneficial.
Table 1.
Selected biological parts that have been characterized and commonly used in synthetic biological devices or systems.
| A) Regulators | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mechanism | Example | Material | Host-specificity | References | |||||
| pre-transcriptional | epigenetic regulation | synthetic TF recognizing DNA methylation | DNA & protein | facultative | (125) | ||||
| sequence orientation | integrase/excisionase (w/BP, LR sites) | DNA & protein | universal | (18) | |||||
| control | Cre recombinase (w/LoxP site) | DNA & protein | universal | (1) | |||||
| Hin recombinase and Fim recombinase | DNA & protein | universal | (16) | ||||||
| genome editing | synthetic retrotransposon | DNA | specific | (16, 17, 126) | |||||
| synthetic TALE nuclease | DNA & protein | can be universal | (127) | ||||||
| DNA replication | origin of replication | DNA | specific | (128, 129) | |||||
|
| |||||||||
| transcriptional | initiation | natural constitutive promoters | DNA | specific | (27, 62, 104) | ||||
| T7 promoter | DNA & protein | specific (bacteria) | (130) | ||||||
| synthetic promoter with zinc finger-based TF | DNA & protein | can be universal | (9, 131–133) | ||||||
| synthetic TALE-based TF | DNA & protein | can be universal | (134, 135) | ||||||
| termination | terminator | DNA | specific | (130) | |||||
|
| |||||||||
| co-transcriptional | exon rearrangement | alternative RNA splicing | RNA | specific (eukaryotes) | (93) | ||||
| synthetic intron | RNA | specific (eukaryotes) | (136, 137) | ||||||
|
| |||||||||
| post-transcriptional | mRNA stability | Rnt1p cleavage site | RNA | specific (yeast) | (110, 138) | ||||
| microRNA | RNA | specific (eukaryotes) | (56) | ||||||
| cis-regulating ribozyme | RNA | facultative | (6, 7, 139) | ||||||
| CRISPR-based RNA | RNA & protein | specific (bacteria) | (140) | ||||||
| mRNA availability | microRNA sponge as decoy | RNA | specific (eukaryotes) | (141) | |||||
|
| |||||||||
| co-translational | initiation | ribosome binding site | RNA | specific (bacteria) | (103) | ||||
| internal ribosome entry site | RNA | specific (eukaryotes) | (142, 143) | ||||||
| Kozac sequence | RNA | specific (eukaryotes) | (144) | ||||||
| termination | supD amber suppressor | RNA | specific (bacteria) | (111) | |||||
| ribosome specificity | 16S rRNA/SD sequence pair | RNA | specific (bacteria) | (114) | |||||
|
| |||||||||
| post-translational | protein stability | degradation tag | protein | specific | (11, 53) | ||||
| protein availability | peptide decoy | protein | can be universal | (12) | |||||
| extein rearrangement | protein splicing | protein | specific (eukaryotes) | (145, 146) | |||||
| ubiquitylation | ubiquitin ligase | protein | specific | (147, 148) | |||||
| B) Sensors | |||||||||
| Mechanism | Example | Material | Host-specificity | References | |||||
|
| |||||||||
| physical signals | light | phytochrome | protein | can be universal | (149, 150) | ||||
| histidine kinase | protein | specific | (151) | ||||||
| rhodopsins | protein | universal | (152) | ||||||
| temperature | thermosensor | RNA | universal | (153, 154) | |||||
| osmotic pressure | osmosensor | protein | specific | (155) | |||||
|
| |||||||||
| chemical signals | genetic signals | protein aptamer | RNA | universal | (156) | ||||
| antisense RNA | RNA | universal | (78, 157) | ||||||
| DNA damage sensor | protein | specific | (158) | ||||||
| physiological chemicals/secondary metabolites | nutrient sensor | protein | specific | (88, 159) | |||||
| hormone receptor | protein | universal | (160, 161) | ||||||
| small molecule aptamer | RNA | universal | (7, 97, 98, 162) | ||||||
| Ions | intracellular ion sensor | protein (163) or RNA (164) | can be universal | (163, 164) | |||||
| heavy metal sensor | protein | can be universal | (165) | ||||||
| pH (hydrogen) sensor | protein | can be universal | (163, 166) | ||||||
| environmental contaminants/toxins | arsenite sensor | protein | can be universal | (65) | |||||
| antibiotics sensor | protein | specific | (167) | ||||||
| PCB sensor | protein | can be universal | (168) | ||||||
| gas | H2O2 sensor | protein | can be universal | (65, 169) | |||||
| acetaldehyde sensor | protein | universal | (170) | ||||||
| C) Actuators | |||||||||
| Mechanism | Example | Material | Host-specificity | References | |||||
|
| |||||||||
| reporters | fluorescence | fluorescent protein | protein | universal | (171, 172) | ||||
| fluorescent RNA | RNA | universal | (173) | ||||||
| luminescence colorimetric | luciferase | protein | universal | (174) | |||||
| carotenoid synthesis enzyme | protein | can be universal | (175, 176) | ||||||
| β-galactosidase | protein | can be universal | (176) | ||||||
|
| |||||||||
| phenotypic | cell fate | apoptosis regulator | protein | specific | (157) | ||||
| actuators | MAPK (mating) | protein | specific | (155) | |||||
| CDK (cell cycle) | protein | specific | (177) | ||||||
| cell morphology | WASP (cell shape) | protein | specific | (63) | |||||
| CheZ (motility) | protein | specific | (67) | ||||||
| EPS-degrading enzyme (biofilm) | protein | specific | (178) | ||||||
| metabolism | secondary metabolite enzyme | protein | can be universal | (43) | |||||
| resistance or auxotrophic selection marker | protein | specific | (43) | ||||||
| transportation | nutrient importer | protein | specific | (35) | |||||
| D) Adapters | |||||||||
| Mechanism | Example | Material | Host-specificity | References | |||||
|
| |||||||||
| anchoring | membrane tags | ER tag | protein | mostly specific | (179) | ||||
| mitochondrion tag | protein | mostly specific | (180) | ||||||
| nuclear-localization signal | protein | mostly specific | (181) | ||||||
| vacuole tag | protein | mostly specific | (182) | ||||||
| secretion tags | secretory sequence motif | protein | mostly specific | (183) | |||||
|
| |||||||||
| molecular linker | static linker (spacer) | DNA insulator | DNA | mostly specific | (184) | ||||
| RNA spacer/transmitter | RNA | universal | (7, 185) | ||||||
| peptide linker | protein | universal | (23, 186) | ||||||
| scissile linker | mRNA cleavage signal | RNA | mostly specific | (105, 187) | |||||
| protein cleavage signal | protein | mostly specific | (188) | ||||||
Abbreviations: TALE: transcription activator–like effector. TF: transcription factor. ARS: autonomously replicating sequence. CRISPR: clustered regularly interspaced short palindromic repeat. SD sequence: Shine-Dalgarno sequence. PCB: polychlorinated biphenyls. ER: endoplasmic reticulum; EPS: extracellular polymeric substances; MAPK: mitogen-activated protein kinase; CDK: cyclin-dependent protein kinase; WASP: Wiskott–Aldrich syndrome protein
i) Regulators
Regulators are biological parts that function within a gene regulatory network by providing constitutive control over gene expression, which determines the maximal gene expression capacity of a biological device. Regulators are perhaps the best-understood biological parts and can be further categorized based on how they control gene expression (Table 1A). Transcriptional regulators comprise a large portion of biological parts in common use. Their regulatory functions are primarily encoded in a cis-acting DNA sequence, sometimes with the aid of a corresponding trans-acting protein factor (9). Another group of regulators controls gene expression through cotranscriptional, posttranscriptional, or co-translational mechanisms, with RNA as the major compositional material. Their target mechanisms include exon rearrangement, mRNA stability, translation efficiency, and ribosome specificity (10). Posttranslational-based regulators, which target protein degradation (11), phosphorylation (12), and ubiquitylation (13), are composed of protein. Recent progress in prospecting new classes of regulators is enabling targeting of pre-transcriptional regulatory mechanisms such as epigenetic methylation (14), direct genome editing (15), and DNA orientation control (16). These new biological parts are enabling researchers to manipulate gene expression at a more comprehensive level (17) and design more sophisticated genetic devices, such as those encoding memory (18).
ii) Sensors
Sensors are biological parts that detect environmental or cellular signals and convert these signals to either gene-regulatory activities or conformational changes of other biological parts. Sensors are capable of adopting multiple conformations associated with different output signals depending on the level of their input signal(s) (19). Biological sensors can be further classified based on the nature of their input signals: physical and chemical (Table 1B). Physical signals typically achieve genetic regulation by changing the conformational state of a sensor in an energy-dependent way, such as the temperature-induced conformational changes (20). Chemical signals that can be either environmental (such as heavy metal ions or toxins) or cellular (such as nutrients and hormones) and act through direct binding to the sensors, leading to changes in the conformational distribution.
iii) Actuators
Actuators are biological parts that produce measurable outputs or phenotypic effects from genetic devices operated in the host cell. Actuators can be used to serve different design purposes. For example, reporters are commonly used to reveal the computational results of a synthetic device or system (21). Reporters can be based on the direct readout of fluorescence or through the indirect readout of a colorimetric/luminescent product from an enzymatic reaction. Most actuators directly act on the host cell by altering physiological or morphological phenotypes, such as changes in metabolism, cell shapes, or cell fates. These phenotypic actuators generally exhibit host specificity, which can lead to design restrictions in building broader sets of synthetic systems with a given part. However, strategies that incorporate bioinformatics and leverage available sequencing information can help to identify phenotypic actuator homologs across different species (22).
iv) Adapters
Adapter parts are usually composed of non-coding nucleotide sequences or unstructured peptide motifs that provide physical or functional insulation for connecting parts and play a critical role in optimizing the device or system performance. For example, peptide linkers are commonly used as adapter parts in designing chimeric proteins to physically prevent two protein domains from interfering with each other’s folding (23). Another functional role of adapters is to help establish synergistic effects of desired biological functions. For instance, adapter parts have been developed to support co-localization or anchoring of other biological parts (24). Adapters can also been used to fine-tune expression levels within operons by establishing a desired quantitative relationship between the linked genes (25).
Prospecting Biological Parts
The proposed look-up table for biological parts is particularly useful for bottom-up design approaches in synthetic biology, which focus on assembling individual parts into higher-level functional components such as genetic devices or systems (26). Many genetic devices and systems have been assembled as proof-of-concept models; however, current application of these model architectures to further address real world problems has been limited by the lack of biological parts that can execute the desired functions under a given in vivo cellular condition. This limitation significantly undermines the strength of modular design architectures and abstraction frameworks in synthetic biology. For instance, regulator parts are still the most available and well-characterized building blocks that have been widely used in synthetic biology, particularly those at the transcriptional level (Table 1). Yet, the design architecture can become unmanageable as more sophisticated design criteria are required if transcription-level architectures are the only approach considered (5). In cases where a spectrum of design variants with different characteristics is needed, it is more efficient to re-apply existing design frameworks while only replacing critical biological parts to achieve this diversity in characteristics. Thus, the engineering design process in synthetic biology will be greatly advanced by having a balanced reservoir of biological parts with diversity in their functional mechanisms, activities, and host-cell specificity (27, 28).
Evolutionary approaches for prospecting parts
Harvesting parts directly from the nature can be an effective approach, but is limited by our current understanding of biology. Computational strategies to parts prospecting require models with a certain level of accuracy to predict the function of biological parts, which are also limited by our current understanding (5). Thus, directed evolution, a parts prospecting approach that mimics the natural evolutionary process but applies artificial guidance, has become popular in the field of synthetic biology. Directed evolution consists of three major steps: identifying initial candidates, generating genetic diversity, and screening/selecting for parts with desired functions (29, 30). This approach has been successfully applied to different classes of biological parts in generating parts with novel functions (31), translating the functions of biological parts to a new host cell (32), and evolving part variants with different levels of activities (33).
Identifying candidates
The process of directed evolution begins with the identification of an initial biological part. If the desired function has been well-characterized, researchers can search for candidate “hits” through literature and databases (34, 35). However, in many instances the desired part might be uncharacterized or unidentified. Advances in sequencing and bioinformatics enable additional techniques for efficiently identifying putative candidates, such as functional genomics (36), comparative functional analysis (37), and bioinformatics analysis (38). Computationally-designed candidates can serve as a starting point for directed evolution when a biological part with a desired function has not yet been identified from a natural biological system. For example, recent studies have shown the feasibility of evolving a nonnatural enzymatic actuator from an in silico designed parent (31, 39).
Generating genetic diversity
After identifying candidate parts, the next step is to create genetic diversity, which determines the size and quality of sequence space that can be explored in the directed evolution process. Classic methods for generating diversity include DNA shuffling, random mutagenesis, and semi-random mutagenesis approaches, such as biased mutagenesis and site-directed mutagenesis (40).
Searching through sequence space for desired parts
A screen or selection is used to identify variants in the library with the desired activities. Screens and selections can be performed in vitro, such as through plate-based biochemical or photometric assays, affinity-based screens, and molecular display methods, or in vivo, such as through growth complementation assays (41) and fluorescence-activated cell sorting (40). The particular screen or selection used to search a library will often dictate the library size that can be effectively searched. Setting up high-throughput in vivo screens and selections can be challenging as many of biological activities of interest cannot be directly linked to a readily measurable output. In these cases, the desired biological activity may be coupled to a biosensor device, which can translate the desired activity into an easily detectable signal or cell survival (42). Synthetic biology has advanced the design of novel biosensor devices to address this significant need (43). For example, an RNA-based biosensor device was recently demonstrated to support a high-throughput, FACS-based screen for a P450 monooxygenase activity in vivo (32).
Challenges in Prospecting Parts
A number of recent technical advances are addressing challenges long associated with prospecting parts through evolutionary strategies. For example, one challenge has been limitations on the size and complexity of the library. However, recent advances in DNA synthesis and assembly technologies have substantially increased the length, accuracy, and complexity of DNA fragments that can be synthesized (44). Another challenge has been that only limited information about sequence-function relationships could be obtained across evolutionary trajectories. However, next generation sequencing technologies can now be used to gain a more complete understanding of the sequence-function landscape across evolutionary trajectories (45). A final challenge has been the limited accuracy and the time efficiency associated with multiple rounds of screens and selections. However, new technologies are increasing the accuracy and sensitivity of screens and selections for new biological parts (33, 46). In addition, researchers have recently developed strategies that dramatically speed up the process of directed evolution by taking advantage of continuous evolution systems that avoid the need for separate cloning and transformation steps. For example, phage-assisted continuous evolution (PACE) is a one-pot in vivo evolution system that utilizes bacteriophage as a library carrier accompanied with external mutagenesis forces (47).
GENETIC DEVICES
A genetic device is a functional assembly of biological parts that encodes and executes a human-defined function. Significant efforts in synthetic biology have been directed toward making genetic devices that establish a distinct input/output (I/O) relationship in vivo. A wide variety of controllable genetic devices have been implemented and analyzed based on different design and performance goals, such as genetic switches, logical gates, oscillators, arithmetic operators, and higher-order devices (Table 2). The sensors and actuators of the device interface with the inputs and outputs, respectively, and are generally linked by several regulator and adapter parts. The combined device components determine the device specifications such as device dynamics, temporal behaviors, and input sensitivity.
Table 2.
Examples of genetic devices.
| Genetic device | Key mechanism | Host organism (for the given examples) | Implemented functions | Addressed design considerations | References |
|---|---|---|---|---|---|
| switch/logic gates | transcription initiation with natural TF | bacteria | switch, logic gates, combinatorial logic gates (189), concentration band- pass filter (190) | device performance, noise propagation (70), device crosstalk | (70, 189–191) |
| transcription initiation with synthetic TF | yeast (9), higher eukaryotes (132) | switch, logic gates (132) | device performance, device crosstalk (9) | (9, 132) | |
| exon arrangement by RNA splicing | higher eukaryotes | switch | device performance | (93) | |
| mRNA stability mediated by cis- ribozyme | bacteria (6), yeast (7, 185), higher eukaryotes (139) | switch, logic gates (185), concentration band-pass filter (7) | device performance, device crosstalk | (6, 7, 139, 185) | |
| mRNA stability mediated by RNAi | higher eukaryotes | switch, combinatorial logic gates | device performance, device crosstalk, evolutionary robustness (56) | (56, 162, 192, 193) | |
| mRNA stability mediated by Rnt1p | yeast | switch | device performance | (57) | |
| mRNA availability mediated by antisense RNA | bacteria | switch, invertor & NOR gate, combinatorial logic gates | device performance, device crosstalk (78, 140) | (78, 108, 140) | |
| translation initiation by RBS availability | bacteria | switch | device performance | (97, 98) | |
| translation repression by RNA- binding protein | higher eukaryotes | switch | device performance | (156) | |
| translation termination by amber suppression | bacteria | switch, AND gate | device performance | (111) | |
|
| |||||
| cell-cell communication | chemical-based signaling | bacteria (194), yeast (161), higher eukaryotes (195) | single channel communication module | device performance, device crosstalk | (161, 194, 195) |
| phage-based signaling | bacteria | multiple genetic cassette communication module | device performance, device crosstalk | (196) | |
|
| |||||
| oscillator | ring oscillator | bacteria | static oscillator | device performance | (197) |
| relaxation oscillator | bacteria(198), higher eukaryotes(76, 77) | static and tunable oscillator | device performance | (76, 77, 198) | |
| metabolite-mediated oscillator | bacteria | static oscillator | device performance | (199) | |
|
| |||||
| memory | DNA orienting by recombinase | bacteria | SR latch (18), counter (200) | device performance, evolutionary robustness (18) | (16, 18, 200) |
Abbreviations: TF: transcription factor. RBS: ribosome binding site. RNAi: RNA interference.
Measurement Standards Driving Design
The first step in designing a genetic device is to determine the appropriate physical architectures and engineering strategies based on the design goals (48). In many engineering fields, the description of design constraints for devices has been standardized (e.g., “operating between -15 and 15 V with maximal current of 500 mA”) and can often be found in datasheets. Researchers in synthetic biology have proposed similar datasheets to describe such constraints for genetic devices with standardized performance measurements (49). However, significant variations in absolute expression levels when evaluating genetic device performance have been observed as a function of numerous experimental parameters, including host cell environments, culture media, and characterization methods. One proposed approach demonstrated to significantly increase consistency of device performance reported across different laboratories is to incorporate a normalizing construct and report relative device activity as a ratio of the activity of device of interest to that from the normalizing construct (50). Other classes of standard constructs have been proposed as a means of standardizing data reporting and accounting for gene expression noise, such as internal normalizing reporters (51).
Device Composition
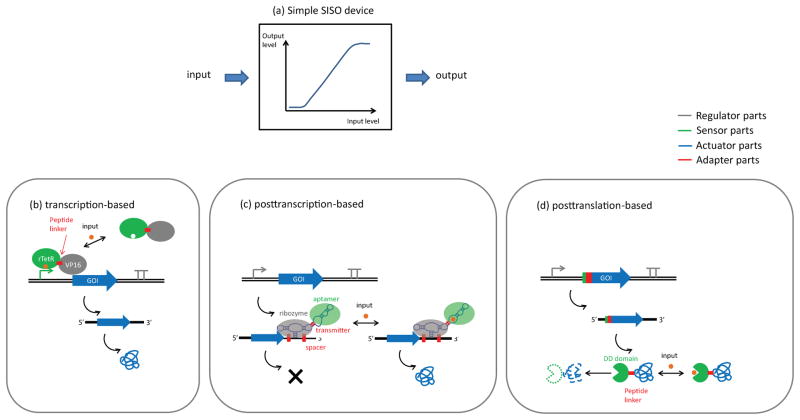
In many instances a given biological function can be achieved through different device compositions. For example, a genetic SISO (single-input, single output) ON switch (Figure 1a), in which small molecules are used as an input signal to turn on expression of a fluorescent reporter, can be implemented through the combination of a diverse set of parts.
Figure 1.
A single-input, single output (SISO) device. (a) The input/output curve of the SISO device. (b) A transcription-based scheme for implementing SISO. An inducible transcriptional activator is composed of a ligand-binding protein (sensor) fused with an RNA polymerase recruiting domain (regulator). Binding of the input ligand enables the sensor binding to its corresponding promoter region, which turns on gene expression. (c) A posttranscription-based scheme for implementing SISO. An RNA aptamer (sensor) is coupled with a ribozyme (regulator) through an RNA transmitter sequence (adapter). This device is turned on in the presence of input molecule, which binds to the sensor and disrupts the self-cleaving confirmation of the regulator. (d) A posttranslation-based scheme for implementing SISO. A protein destabilizing domain (sensor) is fused directly to the gene-of-interest (actuator) with a peptide linker (adapter). The input ligand can bind to the destabilizing domain; thus prevents the protein from degradation.
One implementation to achieve this design goal is a device that operates at the transcriptional level with an inducible promoter (Figure 1b). For example, a chimeric protein can be engineered in which a sensor part (rTetR) is fused with a regulator (VP16) that activates transcription (52). In the absence of the small molecule input, the chimeric rTetR-VP16 transcriptional activator fails to bind to its corresponding hybrid promoter region, resulting in low fluorescent output. In the presence of input, the conformation of the rTetR-VP16 activator changes, resulting in expression of the reporter gene. An alternative implementation for an SISO ON switch is a device that operates at the posttranscriptional level with an RNA switch placed downstream of the reporter gene (Figure 1c). For example, small molecule-responsive RNA switches can be engineered by coupling an aptamer sensor with a RNA regulator such as a ribozyme (7). In the absence of the small molecule input the RNA switch adopts a self-cleaving conformation, resulting in mRNA degradation and low reporter levels. Binding of the input to the aptamer sensor disrupts the self-cleaving conformation of the ribozyme, thus stabilizing the transcript and resulting in increased reporter levels. A third implementation for an SISO ON switch is a device that operates at a posttranslational level by controlling the stability of the reporter protein (Figure 1d). For example, the reporter protein can be fused to a small molecule-controlled destabilizing domain (DD) (53). In the absence of the small molecule input the DD adopts an active conformation and results in protein degradation, whereas in the presence of the input the conformation of the DD changes to one that does not induce degradation.
While the three genetic devices described above are functionally equivalent from an abstract device description perspective, they are implemented through distinct physical architectures. Devices implemented through different architectures may exhibit very different device characteristics. For example, transcription-based genetic devices typically exhibit relatively large dynamic ranges compared to other devices. Posttranslational genetic are typically characterized by a more rapid temporal performance by avoiding delays that can be introduced through the processes of transcription and translation (54). Posttranscriptional devices general exhibit relatively moderate performances in terms of temporal and dynamic properties, but have been shown to exhibit relatively high design modularity (5) and I/O programmability (19).
Design Considerations and Device Evaluations
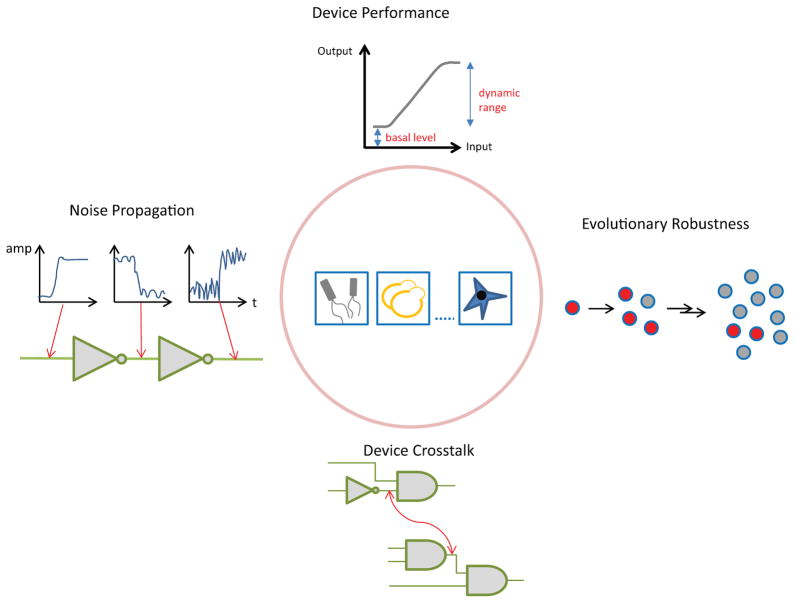
In Table 2, we summarize the current progress for designing genetic devices, with comments on their key mechanism, host organism, and addressed design considerations. In particular, we propose four major evaluation properties (Figure 3), which are critical in guiding the device design and ultimately the evaluation of device performance. These properties are often linked with trade-offs and thus must be considered simultaneously during the device design and evaluation process.
i) Device performance
The performance of a genetic device is typically the foremost property considered in device evaluation. Common performance measures when evaluating genetic I/O devices include basal activity, dynamic range, sensitivity to input, and temporal response (55). Specific tuning strategies can be established to predictably adjust specific performance measures. For example, the basal activity of an RNA-based I/O device can be efficiently and predictably reduced by implementing multiple copies of the device in tandem (56, 57). As another example, the input sensitivity of a genetic transcriptional device can be improved by introducing device cascading, which increases the degree of cooperativity of the combined device (58). However, optimization of one device performance measure can result in trade-offs with other performance measures. For example, researchers have demonstrated that while the degree of cooperativity can be increased by as much as 3-fold by cascading three transcriptional devices, this enhanced input sensitivity is earned at the cost of temporal delays and noise propagation (58).
Several studies have proposed that the introduction of feedback architectures can be a powerful way to optimize genetic device performance. In many cases, closed-loop control architectures can support improved device performance relative to open-loop architectures (59). For example, coherent feed-forward loop architectures can be used in genetic devices to accelerate the response time. In another example, researchers demonstrated that similar device performance can be achieved by changing ordering of feedbacks between two conceptually equivalent feedback configurations (60). They showed that two device parameters (response duration vs. magnitude variability) can be tuned by setting different feedback strengths, which is essentially a degree of freedom gained by using closed-loop configuration.
ii) Noise rejection
Noise is a disturbance that appears in any engineered system and can impact the desired performance of that system. One of the major noise sources in biological systems stems from plasmid-based expression vectors, which result in copy number variability and loosely regulated genetic behaviors (61). This type of noise can be minimized by expressing the genetic device from a less variable genetic context, such as the host genome or by selecting biological parts that are more resistant to extrinsic fluctuation (32, 62). Noise in the operation of genetic devices can have significant negative impacts on system performance, particularly when a cell population is designed to operate in a synchronized and coordinated fashion. Even if cells in a population are designed to launch a program simultaneously with the same expression capacity, as the cells grow and divide, stochastic interferences will accumulate and dephase the population. Synthetic cell-cell communication modules have been implemented to address this challenge, where each device-operating cell is capable of exchanging a genetically-encoded signal molecule to achieve collective behaviors such as temporal synchronization (63–65) and spatial pattern formation (66, 67). Thus, cell-cell communication modules can be used to share and confirm the “genetic state” of each individual in a population to minimize cell-to-cell desynchronization (68).
iii) Evolutionary robustness
The robustness of device performance over time is another important performance metric of genetic devices (49). One unique characteristic of biological systems is their capability to actively evolve toward optimal fitness behaviors for the set of conditions they are grown under (69). This evolvability has been used as a design strategy to optimize device performance (70), but can also negatively impact the robust activity observed from a genetic device over time in vivo (71). If a synthetic device introduces a significant metabolic burden on the host cell, loss-of-function becomes an evolutionarily favorable genotype, in which cells that acquire mutations that inactivate the device (and remove the associated load) can quickly overtake the population (72). The traditional approach used to address this evolutionary challenge is to couple the device operation to a separate selection pressure for its maintenance (43). Another approach is to reduce the evolutionary instability by removing unstable genetic elements from the host genome (73). However, both approaches can create new selection pressures by imposing additional stress on the host cells, potentially leading to undesired physiological responses (74).
As the complexity of the genetic device increases, more cellular resources may be needed for properly operating such devices, particularly when all the functions are implemented at transcriptional level. This overloading effect can reduce the evolutionary robustness by activating cellular stress-associated responses, which facilitate the loss-of-function of the genetic devices. Recent studies have begun to report and examine design principles for evolutionary robust genetic device function (18, 72, 75). Moreover, researchers have begun designing and implementing genetic functions using different expression mechanisms, such as antisense- or microRNA-based feedback (76, 77), which can increase the evolutionary robustness through alleviating the loading imposed by transcriptional devices.
iv) Device crosstalk
Crosstalk of a genetic device, or interactions of components in that device with other biological components in the system, can result in undesirable performance properties. One type of crosstalk, device-device crosstalk, generally arises from re-using biological parts in designing genetic devices. This type of crosstalk can become significant when designing MIMO or multi-layered genetic devices, as the device-device crosstalk can break the desired wiring network between devices. A recent study described a rational approach for designing orthogonal genetic devices based on the antisense binding specificity of RNA biological parts (78). The researchers screened repression of sense-antisense pairs using a library of 23 candidates and used this data to build a mathematical model to predict interaction of other candidates. The RNA sense-antisense pairs were sorted into distinct orthogonal groups by setting desired crosstalk levels. Such approaches may allow the crosstalk between devices to be minimized to desired levels, thus providing researchers with a sufficient number of orthogonal genetic devices to implement multi-layer or MIMO architectures.
SYNTHETIC BIOLOGY SYSTEMS
Synthetic biological systems combine one or more devices to execute a useful, human-defined function. In the past decade, synthetic biological systems ranging from microbes that incorporate information processing and communication capabilities to detect or form population-wide patterns (66, 79) to T cells re-wired with externally controllable switches to achieve safer and more effective cellular therapies (80) have been described. These systems are diverse in their goal, host, execution, and complexity of logic, but all attempt to systematically alter a natural system to exhibit a new, pre-defined function for applications in biosynthesis, environmental remediation, intracellular diagnostics, and therapeutics. Systems engineering has faced limitations in the availability of application-specific parts and frameworks supporting the functional assembly of large-scale genetic systems. Despite these and other challenges, synthetic parts and devices have been combined into systems exhibiting sophisticated behaviors that can be generally categorized as synthesizers, sensors, seekers, and strikers.
Synthetic Biology Synthesizer Systems
Synthesizers are synthetic biology systems engineered to produce a molecule of interest using an appropriate host organism. While engineering organisms as production hosts has long been of interest in biotechnology, synthetic biology introduces new tools and approaches that promise to systematically increase the achievable complexity, yields, and robustness of biosynthesis strategies. As one example, synthetic biology provides new tools for DNA synthesis and assembly, allowing for the combinatorial generation and testing of many pathway and enzyme variants (81, 82). As another example, synthetic biology also provides new tools for precise control of gene expression (7, 27) and enzyme assemblies (83–85), allowing for the implementation of control strategies to alter flux through pathways (86). As a final example, synthetic biology is resulting in the design of new biosensor platforms (87), providing new strategies for probing biosynthesis pathways (88).
Synthetic biology can be used to advance the engineering of microorganisms that produce valuable materials. As one example, the idea of feedback control is commonly employed in engineering fields and natural metabolic networks to improve the performance of a process. In engineered biosynthesis pathways, feedback control can improve efficiency and yield by reducing energy spent on the production of non-rate-limiting steps and decreasing the accumulation of toxic intermediates that impair the growth of the host (89). In one early demonstration of this idea, researchers improved lycopene production in E. coli by approximately 18-fold by increasing the production of the rate-limiting enzymes in the pathway in response to acetyl phosphate (ACP), a molecule that indicated excess flux was being diverted away from the desired final product (86). More recently, a similar dynamic sensor-regulator system (DSRS) was applied to improve production of fatty acid-based fuels in E. coli by 3-fold by engineering the later portions of the biosynthesis pathway to be expressed in response to the output of the earlier portions, to prevent over-production of toxic intermediates (ethanol and acyl-CoA) (88).
Synthetic biology can also be used to advance the engineering of cells that synthesize products in vivo. In one example, human embryonic kidney cells (HEK293) were engineered to secrete glucagon-like peptide-1 when stimulated by light and implanted subcutaneously to lower blood glucose levels in diabetic mice (90). In another example, human cervical cancer cells (HeLa) were engineered to secrete urate oxidase in response to excess uric acid (91). When implanted into urate oxidase-deficient mice, which develop acute hyperuricemia, the prosthetic gene-network device decreased urate levels to concentrations similar to animals given standard allopuirnol therapy (91). These in vivo systems demonstrate how synthetic biology can be applied to replace dysfunctional circuitry in human health, reducing problems with proper drug dosage and patient compliance.
The synthesizer systems highlighted above illustrate the utility of precisely regulating biosynthesis pathways for production of molecules, whether to improve yield or maintain homeostatic levels of a product. However, all of the examples relied on the identification and engineering of natural sensors to adapt them for use in a functional feedback controller. The ACP sensor used for improving lycopene production was only responsive to ACP after certain E. coli regulatory units were inactivated (86). The bacterial repressor and promoter used in uric acid homeostasis system required significant optimization to identify a device with suitable performance (91). Such engineering efforts are laborious and difficult to scale to other systems, highlighting the need for better parts.
Synthetic Biology Sensors Systems
Synthetic biological sensor systems utilize information processing and computation to transform inputs into useful genetically-encoded readouts. Synthetic biology enables advances in the design of sensing systems by supporting the development of new sensor parts and the circuitry for interchangeably linking diverse sensing modalities to information processing modules. The ability to modularly link an input to a genetic output with different information processing capabilities has three main consequences. First, it is possible to encode different modes of readout, such as linear, frequency, or threshold. Second, input detection can be readily linked to filters, logic, and communication devices for the detection of sophisticated conditions. And third, a genetic output permits noninvasive detection of conditions in live cells over time, which can be particularly powerful for diagnostic applications.
Synthetic sensor systems can be programmed to exhibit different modes of readout. To date, most readouts for biological sensors are linear (where the intensity of a photometric or colorimetric output signal is proportional to the input) and straightforward to detect by standard equipment. An alternative mode of detection is or frequency (where the frequency of an oscillating output signal is indicative of the input level), which can be easily digitized and for fluorescence measurements, less sensitive to technical factors such as excitation beam power and exposure time. By combining a microfluidic device with short- and long-distance quorum sensing circuits, researchers engineered E. coli-filled ‘biopixel’ arrays to report arsenic levels via the frequency of fluorescent blinking (65). This sensor system was built upon a previously developed synchronized oscillator network (64) and relied upon arsenic- and hydrogen peroxide-responsive promoters to support synchronization of the oscillations across the biopixels on a chip. The proper functioning of this frequency readout circuit required careful tuning of the concentrations and rates of diffusion of the communication molecules in order to sustain synchronized oscillations. A third mode of readout is thresholding, where the output is present only for levels of input past a given threshold value. This type of simple YES/NO output is useful for qualitative reporting and for integrating into digital logic. Researchers demonstrated that the arsenic sensor system could be rewired as a tunable threshold detector by placing a necessary component for cell-cell communication and synchronization under the control of an arsenic-responsive promoter (65).
Synthetic biology is advancing the integration of genetic filters and logic circuits into sensor systems to enable the detection of complex environmental conditions. In an early example, E. coli was engineered to act as a concentration band-pass filter, fluorescing only when exposed to intermediate ranges of a small molecule input produced by ‘sender’ cells (66). The ability to apply such detection filters to inputs communicated from other cells forms the basis of spatial patterning. The researchers demonstrated this idea by growing cells that fluoresce green in response to one range of input concentrations with a second population that fluoresce red in response to a different range of input concentrations on solid media surrounding a central population of sender cells to form a bullseye pattern (66).
By integrating communication with logic circuits, it is possible to program detection of complex patterns of input. For example, researchers demonstrated a synthetic system that could detect the edge between light and dark inputs projected onto E. coli cells grown on solid media by engineering cells with a genetic circuit that included a quorum-sensing component, a dark sensor, and a simple logic processor (79). To detect the boundary between light and dark, researchers relied on short-distance diffusion of a sugar molecule synthesized by cells receiving a dark input. Cells were programmed to express a black pigment only when they received both the light input (NOT dark) AND the sugar input (indicating their proximity to the dark region). The edge detection system relied on the development of a dark-sensing part, which consisted of a chimeric two-component system and the enzymes required to synthesize the associated chromophore.
Synthetic biology is advancing the frontier in intracellular diagnostics by enabling genetically-encoded noninvasive detection of combinations of small molecules, nucleic acids, and proteins in live cells over time. For example, a number of systems utilize RNA-based sensors to detect small molecules, proteins, or other RNAs inside live cells (92). In one example, RNA aptamers responsive to protein inputs were used to detect levels of endogenous proteins in the NF-κB and Wnt signaling pathways (93). The sensor system linked detection events to an altered splicing pattern, and thus expression, of an output gene. As RNA aptamers can be generated de novo to diverse targets through a variety of selection strategies (94, 95), this sensor system incorporates a sensing modality that can be potentially tailored to the detection of many cellular protein levels.
In a second example of intracellular diagnostics, a platform that integrates logic and sensing to detect a pattern of up to six endogenous miRNAs was recently described (96). This sensor platform was used to classify cancer cells (HeLa) in a mixed HeLa/HEK293 co-culture and can potentially classify any cell-type or phenotype associated with a unique miRNA signature. The parts needed for tailoring the sensor system to a specific application consist of sensors for the known signature miRNA sequences, which can be readily generated based on predicted hybridization interactions. While promising, this synthetic sensor system had to be extensively tuned to obtain a sufficiently large signal to noise ratio on the final output reporter gene for practical use.
Synthetic biology is advancing the design of genetically-encoded sensor systems by increasing the diversity of readout modes that can be implemented, the complexity of input patterns that can be processed, and the scope of molecular inputs that can be accessed. However, challenges remain in tailoring these designs to new inputs for broader applications. In particular, the development of new sensor systems is significantly limited by the number of available sensor parts with desired affinities. In addition, the ease with which new sensor parts can be coupled to information processing and reporting devices to achieve operational sensor systems with desired signal to noise properties remains a challenge in the field and highlights the continued need for improved composition frameworks.
Synthetic Biology Seeker Systems
Synthetic seeker systems are programmed to seek out particular environments in which to execute an action, such as processing an environmental compound. Synthetic biology contributes to the design of novel sensing and actuator components as well as the functional integration of sensing and actuation with movement control pathways for modular assembly of synthetic seeker systems. The ability to engineer robust ‘seeking’ systems that physically move toward particular stimuli may enable novel approaches for various applications.
As one example of a seeker system, E. coli was reprogrammed to chemotax toward the non-natural chemoattractant theophylline (97). This synthetic seeker activity was achieved by placing the expression of a key controller of E. coli motility under the control of an RNA aptamer-based switch that regulated translation by occluding the RBS in the absence of the molecular input (97). This seeker system can be tailored to ‘seek’ different molecules by replacing the RNA switch component with alternative switches designed to respond to different molecular inputs. Researchers demonstrated this capability by adapting this seeker system to engineer E. coli to seek out and destroy an environmental toxin (98). Specifically, E. coli was engineered to chemotax in response to the small molecule herbicide atrazine and simultaneously degrade the herbicide through the expression of an atrazine-degrading enzyme.
A second example of a seeker system demonstrated the potential to control mammalian lamellipodia formation in response to a non-natural input such as light. Specifically, mouse embryonic fibroblasts (NIH3T3) were engineered to produce lamellipodia at the direct location that a source of light hit the cells (63). By successively moving the location of the activating light source, cells were induced to extend lamellipodia in the direction of light movement. This light seeker system is based on an Arabidopsis thaliana phytochrome and phytochrome interacting factor (PIF), whose dimerization depends on red light and far-red light. When the phytochrome is membrane localized with PIF fused to a Rho-family GTPase that controls actin polymerization, activating light causes dimerization of the phytochrome and PIF, resulting in recruitment of GTPases to the membrane and activation of changes in cell shape.
Synthetic biology is advancing the design of novel component functions and their functional integration into genetically-encoded seeker systems. The seeker systems highlight several challenges in achieving a successful seeker design, including the need for application-specific sensor parts and methods for subsequently tuning the interaction between the parts of the system for proper overall function. For example, the atrazine seeker system required several rounds of in vitro and in vivo selection to generate a novel atrazine-responsive RNA switch that could properly match the atrazine input to chemotaxis-related gene expression output. Similarly, the final light seeker system in mammalian cells was achieved after careful tuning. Specifically, substantial optimization was required to identify a functional phytochrome and PIF pair with binding kinetics capable of driving the activation of the actin control circuit for lamellipodia extension.
Synthetic Biology Striker Systems
Synthetic striker systems program a host cell to specifically kill, re-program, or otherwise affect a target cell. The striker systems developed to date have been designed to address needs in human health. Synthetic biology provides new tools tailored to non-model organisms for integrating sensing, information processing, and control over therapeutics activities to achieve safer and more effective strategies based on mechanisms typically inaccessible by currently available treatments. For example, striker systems have been designed to invade target cells, deliver payloads into targets, and dynamically control therapeutic activities aimed at eliminating target cells in response to cellular and exogenous drug signals.
Genetically-encoded striker systems have been used to engineer viruses and bacteria to invade and deliver payloads to cells. In one example, bacteriophage were engineered to invade bacteria and execute genetic programs that circumvent antibiotic resistance (99), offering a novel approach to the growing clinical concern of antibiotic resistant infections. In particular, M13mp18 bacteriophage engineered to over-express proteins that disrupt the E. coli SOS network were shown to enhance killing of the bacteria by quinolones and survival of infected mice (99). This striker system was further engineered to enhance killing of the bacteria through over-expression of proteins that disrupt other essential E. coli functions, including superoxide stress response, biofilm formation, and antibiotic transport (99).
One limitation to engineering viruses as hosts for striker systems is that the size of the viral capsid restricts the amount of genetic material that can be incorporated into the program. Using bacteria as the host for the striker system can increase the size of synthetic programs that can be encoded. As one example, E. coli were programmed to specifically invade cancer cells by expressing the Yersinia pseudortuburculosis invasion gene under the control of a promoter responsive to hypoxic conditions (an indicator of a tumor microenvironment) (100). Bacterial invasion of mammalian cells can be used to deliver payloads, including RNA or proteins, to alter the function of a cell. In a separate example, bacteria were engineered to deliver RNAi substrates to human cells in vivo (101). In this system, E. coli expressing a combination of Inv and HlyA delivered shRNAs against catenin β-1, a cancer oncogene, to a human cancer xenograft in mice induced gene silencing of the target gene (101).
Striker programs can also be encoded within specific human cells, such as T cells or stem cells, which can be isolated, engineered ex vivo, and re-implanted into the human patient. T cells are key cellular components of the immune response and therefore interesting targets to host striker programs for clearing virally infected and cancerous cells. An important feature to ensure the safety of adoptive T-cell therapies is to incorporate control strategies over the proliferation and survival of the engineered cells in the human patient to prevent uncontrolled proliferation. To address this challenge, researchers developed a striker system that achieved drug-responsive control over T-cell proliferation and survival in vivo (80). An RNA switch responsive to small molecule drugs was used to increase the expression level of IL-2 or IL-15, genes responsible for activating the T-cell proliferation pathway, in the presence of the drug. The striker system also included a kill switch that is activated by the presence of a second prodrug to ensure that the engineered system could be removed from the patient as desired. An alternative T-cell proliferation switch was described that utilizes bacterial proteins to repress T-cell activation through inactivation of components of the mitogen activated protein kinase (MAPK) pathway (102). Expression of these proteins from a doxycycline-inducible promoter inhibited T-cell activation in a ligand-dependent manner, while expression from a T cell receptor (TCR)-responsive promoter limited the amplitude of T-cell activation.
As the striker systems developed to date have been targeted to therapeutic applications they highlight several technical challenges within this application space that ongoing efforts in synthetic biology are working to address. In particular, the challenge of suitable parts for clinical applications presents a major limitation to current design. Specifically, limitations on delivering genetic programs into primary human cells creates addition considerations for system-level design, placing limits on the size of genetic programs that can be delivered to the cells. In addition, heterologous protein-based parts must be selected so as not to induce a nonspecific immune response and sensor components must respond to clinically-relevant inputs that will be nontoxic to patients. Finally, tools for tightly and robustly controlling activities encoded within striker systems are critical in therapeutics as many of the effectors exhibit potent activities and the consequences of mis-regulated activities can be severe.
LEVEL MATCHING BETWEEN PARTS AND DEVICES
It can be difficult to reliably compose different parts and devices together because the desired input and output levels of a module (in terms of concentration, gene expression levels, or enzyme activity at a biochemical level) are often unknown, difficult to measure quantitatively, or difficult to compare. Even when the quantitative I/O relationship for each device is available, it is possible that levels do not match or part/device performance is altered due to emergent properties of the combined system.
In many cases, proper level matching between parts and devices in a system can be critical to the proper function of the system. In one example, while both “set” and “reset” functions of a recombinase addressable data module worked separately in E. coli, these same functions failed when combined within the same cell (18). By developing computational models and testing variants of the device with different expression levels of the components, the researchers determined that the device required precise levels of the integrase and excisionase components to operate correctly. In general, mismatches in stoichiometric interactions (e.g., scaffolding components and other self-assembled structures) or enzymatic activities can lead to inefficiencies (energy directed to making unused RNAs, proteins, intermediates, etc), which aggravates the problems associated with heterologous gene expression, such as accumulation of toxic intermediates, bottlenecks, lower yields, mis-localization of proteins, instability of genetic programs, and growth inhibition (89).
A major goal to enable better design of genetically encoded systems is the development of methods for reliably combining genetic components in a manner that results in a properly functioning system. Several approaches to this goal have been taken in the field of synthetic biology, including the development of functional composition frameworks for predictable assembly of components into functional systems and evolutionary and screening strategies for case-by-case optimization or generation of a functional system. These strategies result in functional programs and can also provide a means to further optimize performance
Frameworks for Functional Composition
Frameworks for functional composition allow biological components to be systematically, reliably, and predictably assembled into a functional device or system. In general, the expression levels resulting from the integration of biological parts into a genetic device are often unreliable and unpredictable, as the function of a genetic component can be altered by its surrounding sequence context. Approaches to improve the predictability when composing biological components include designing parts such that they incorporate the influence of or are insulated from surrounding context.
As one example, manipulation of the ribosome binding site has been frequently used to tune the expression levels of protein components in genetic circuits in E. coli. However, the same RBS sequence results in different expression levels for different genes (103). The ‘RBS calculator’ is a computational design tool that uses an equilibrium statistical thermodynamic model to engineer RBS sequences with user-specified strengths within the context of a gene expression device (103). RBS sequences generated by the tool exhibit a 47% chance of producing expression levels within 2-fold of the desired level (103). Thus, the RBS calculator represents an early and useful first pass tool for engineering and optimizing functional synthetic gene circuits in E. coli.
Manipulation of promoter strength is another frequently used strategy used to tune protein expression in E. coli. One approach to insulating promoters is to expand the boundaries of a promoter sequence to including the flanking sequences from position -105 to +55 (104). Researchers demonstrated that the insulated promoters were less sensitive to stimulatory UP sequences introduced 5′ of the promoter and repressive ‘anti’ sequences introduced 3′ of the promoter compared to weak minimal promoters (104). A recent example demonstrated that using self-cleaving ribozymes as adapters to link an inducible promoter and the RBS of a gene significantly improves the predictability of expression levels of reporter genes with different N-terminal sequences (105). The researchers measured the ratio of CI-GFP versus GFP expression from the same inducible promoter, where perfect predictability was indicated by a ratio of 1. The introduction of a ribozyme adapter significantly improved the predictability of three promoters and the improved predictability is propagated through genetic logic gates (105).
Functional composition frameworks have also been proposed for the development of RNA devices exhibiting programmed information processing functions. These frameworks allow for diverse RNA components encoding sensor, information processing, and actuation functions to be modularly assembled into genetic devices that link the detection of molecular inputs to gene expression events (5, 10, 106, 107). In one early example, a RNA aptamer sensor was linked to a hammerhead ribozyme regulator through a distinct transmitter adapter (7). This transmitter adapter served to insulate the sensor and regulator parts, such that the sensor domain could be exchanged for various RNA aptamers to readily tailor the detection capability of the device. The transmitter adapter also encoded the information processing function of the device, such that altering the transmitter programmed the regulator activity in response to input (e.g., active versus inactive) in a predictable manner.
Finally, modular strategies have also been described for composing multiple RNA devices within a single transcript to support more complex cellular computation. In an early example, multiple ribozyme-based devices with different small molecule input responsiveness and information processing functions were placed in series within the 3′ UTR of a target gene to encode two-input logic gates (19). Similar composition strategies have been demonstrated with other classes of RNA devices, highlighting the generality of this strategy for expanding the scope of cellular computation functions in a predictable manner (93, 96, 108, 109). Finally, when linking RNA devices with similar sequences in series, misfolding of the RNA and thus loss of device activity can be a design concern. To support proper folding and predictable activity of RNA devices researchers have proposed a number of insulation strategies with RNA adapters, including unstructured spacer regions (7, 56), structured spacer regions (80), and device clamps (110).
Evolution for Optimization of Function
Another approach to matching the expression levels of components to achieve proper system function and optimal performance utilizes evolution and screening strategies. By creating libraries in which expression levels of components are diversified, it is possible to select for the desired functional output when the desired function can be linked to an assayable readout. In addition, recent development of powerful tools capable of optimizing the levels of a large number of targets in parallel allow level matching across entire pathways and genomes, thereby increasing the speed with which synthetic genetic circuits can be developed.
Directed evolution and screening strategies have been regularly applied to optimize the function of genetic circuits. In one early example, researchers applied directed evolution and screening to generate a functional inverter (70). Initial assembly of the inverter from its composite parts resulted in a nonfunctional device. By targeting mutations to the RBS and coding region of the repressor component and screening E. coli colonies for cells exhibiting the desired activity, the researchers identified functional system variants. As another example, the initial construction of a transcriptional AND gate that turns on gene expression only in the presence of two molecular inputs was nonfunctional (111). Specifically, the genetic device was activated in the presence of a single input, indicating a mismatch in the output levels of the components that respond to the different inputs. By mutating the RBS downstream of one of the component promoters and screening for the proper device response under all input combinations, the researchers isolated a functional AND gate.
Methods for tuning the expression of multiple genes simultaneously have also been described. In one example, PCR assembly strategies were used to assemble libraries of tunable intergenic regions (TIGRS), which combined RNA control elements, such as hairpins and RNase cleavage sites, to modulate the stability, translation initiation, and transcription termination of individual genes in an operon architecture in E. coli (25). Once assembled, the TIGR libraries were cloned between two target genes to alter their relative expression levels, and the resulting library was screened for the desired output. The strategy was applied to optimize a three gene biosynthetic pathway for mevalonate production. Notably, the researchers developed a mevalonate biosensor system to facilitate a medium throughput screen capable of identifying desirable variants in the libraries generated.
Recently, several methods for generating targeted diversity on a pathway or genome-wide scale have been described and applied to optimizing synthetic systems. As one example, multiplex automated genome engineering (MAGE) utilizes single-stranded oligonucleotides to target mismatches, insertions (~20 bp), and deletions (up to 100 kbp) to the E. coli genome (81). By automating the transformation process, the oligonucleotides can be sequentially introduced into the same population of E. coli, which allows the targeted introduction of discrete, relatively small modifications to accumulate across several different chromosomal locations. The utility of MAGE for tuning pathway expression levels was demonstrated by optimizing lycopene biosynthesis in E. coli. As another example, trackable multiplex recombineering (TRMR) uses barcoded synthetic DNA cassettes to introduce tagged mutations that can up-regulate or down-regulate expression of gene targets in the E. coli genome (112). The modified strains can be selected for desirable phenotypes, after which microarray analysis of the barcodes allow for easy identification of the genetic change responsible for the selected behavior. TRMR presents a strategy for identifying a single, rare target that can confer a desired function. This was demonstrated on identifying E. coli genes able to confer a growth advantage in media with acetate, low pH, and cellulosic hydrolysate (82).
The two major approaches to level matching parts and devices for a functional or optimized system are to use functional composition frameworks or evolutionary strategies. The functional composition framework approach provides tools for rational, predictable, and modular engineering; yet few such design tools have been developed. Therefore, directed evolution is currently often faster for composing a functional system from a specific set of components compared to the development and validation of a composition framework to support the predictable assembly of that system. While directed evolution methods have been commonly used in the design of synthetic systems, they require a screen or selection for the desired result, which is unavailable for many behaviors and phenotypes. In addition, directed evolution methods require screening for every possible combination of inputs to a device or system, and thus challenging to scale for systems intended to process a large number of inputs. As such, functional composition frameworks, once established, offer many advantages for the rapid and efficient design of devices and systems composed of diverse components (due to the time, availability, and scaling limitations associated with screening and selection strategies).
BEYOND LEVEL MATCHING
Having matched input and output levels between biological devices is necessary, but not always sufficient, to guarantee proper or optimal function of a synthetic biological system. It is also necessary to consider the physical interactions between components and how genetically encoded programs interact with the host organism. Spatial engineering strategies physically localize components of a pathway in a specific order, ratio, or orientation to better control the signaling or substrate shuttling through a pathway. In addition, engineering strategies that target the interactions between the synthetic program and host organism have generated orthogonal expression machinery and design principles for evolutionarily robust biological programs.
Matching Activity Across Space
Spatial engineering to physically localize components within a system while preventing undesired interactions can order the interactions between proteins in a signaling pathway, such as those within MAPK pathways, and scaffold biosynthesis proteins together for improved substrate shuttling. Synthetic scaffolds based on protein, RNA, and DNA have been successfully engineered to co-localize proteins to re-wire cellular signaling pathways or increase the yield of biosynthesis pathways.
A protein scaffold consists of a fusion of known protein binding domains. Protein components are recruited to the scaffold when tagged to complementary binding domains. The resulting scaffold organizes recruited components, is modular, and simple to design. Another advantage of protein scaffolds is the ease with which it is possible to modify an existing scaffold. In one example, researchers re-shaped the topology of a MAP kinase pathway in yeast to exhibit accelerated or delayed response times, ultrasensitivity, and adaptation by adding a leucine zipper heterodimerization domain to the Ste5 scaffold to recruit either positive or negative regulators (12). The re-shaped response dynamics could be tuned by varying the levels of the recruited regulators, adjusting the strength of the interaction between the regulators and scaffold, and altering levels of a ‘decoy’ leucine zipper that titrated regulators from the scaffold. Fully synthetic scaffolds have been demonstrated to improve the yield of biosynthetic pathways. As one example, a protein scaffold, comprised of a fusion of three well-characterized protein interaction domains, was used to optimize the stoichiometry of three mevalonate biosynthesis enzymes, each of which was fused to an appropriate corresponding protein interaction domain (83). The relative levels of scaffold to enzyme, as well as relative ratios of the three enzymes to each other, were shown to be important parameters for optimal mevalonate production in E. coli. Mevalonate production was increased 77-fold over levels from the system without the scaffold present. While useful, potential disadvantages of protein scaffolds include the limited ability to design the geometry of the scaffold and instability of the scaffold as a result of degradation, mis-folding, and aggregation.
An alternative scaffolding material is RNA, which has the design advantage of highly predictable local structure and thus offers more sophisticated control over scaffold geometry compared to proteins. RNA scaffolds rationally designed to self-assemble into discrete 1D and 2D patterns have been reported to improve hydrogen production from a two enzyme pathway in E. coli (84). RNA aptamers incorporated into the scaffolds recruited appropriately tagged enzymes to precise positions on the scaffold. By tuning the geometry of the scaffold, researchers increased hydrogen production by 48-fold over a system without the scaffold present. While RNA offers more control over the geometry of the scaffold than proteins, there are several disadvantages, including the relative difficulty in precisely designing RNA scaffolds and uncertainty as to whether similar systems would be functional in a eukaryotic host.
A third option for a scaffolding material is DNA, which has the design advantage of being less likely to mis-fold than either proteins or RNA. Plasmid DNA with zinc finger binding domains have been recently used as a scaffold to improve biosynthesis of resveratrol, 1,2-propanediol, and mevalonate in E. coli (85). Tuning the relative stoichiometries of enzymes in a pathway, the ratio of scaffold units to enzymes, ordering of enzymes along the DNA, and the spacer length between zinc finger binding sites were shown to improve biosynthesis yields. However, improvements in yield from protein and RNA scaffolds (77-fold and 48-fold, respectively) have been higher than those observed for DNA scaffolds (~5-fold). In addition, DNA scaffolds may not function in eukaryotic hosts as the DNA would likely be located in the nucleus, the repetitive scaffold sequences are sensitive to recombination and may be less stable, and the DNA binding tags used to recruit the pathway enzymes may unintentionally target the enzymes to parts of the genome.
These scaffolding examples highlight the need for parts and the importance of precise tuning for optimizing the function of synthetic biology programs. Thus far, scaffolding has required proteins to be tagged with peptide fusions in order to be recruited to the scaffolds. Relying on fusions limits the number of components that can be scaffolded to the number of available orthogonal binding pairs and results in impaired protein activity due to the addition of the peptide fusion. Therefore, an ideal scaffold architecture would not require modification of the recruited components. In this regard, RNA and protein scaffolds may be advantageous because it is possible to develop RNA- or peptide-based aptamers that bind directly to the desired protein components. Currently, such aptamers are typically unavailable and would need to be generated. In each system, tuning was critical for achieving improved yield with the scaffolded pathway. Thus, methods to carefully tune the interplay between relative pathway protein ratios, protein and scaffold ratios, and overall geometry are needed to fully take advantage of scaffolding for engineering synthetic systems.
Negotiation Between Synthetic Programs and Their Host
The predictability, performance, and robustness of engineered genetic systems are influenced by both the effect of the host on the synthetic gene circuit as well as the effect of the synthetic circuit on the host. One approach to improve the performance and predictability has been to limit the effect of host interference on engineered circuits by introducing orthogonal gene expression machinery such that the engineered circuits do not compete with the host for transcription and translation components. An approach to improve robustness of engineered genetic programs against elimination from the host due to evolutionary pressures is to incorporate design principles for evolutionarily stable genetic programs.
Designing beside and beyond the genetic code
The ability to execute genetic programs that run beside (i.e., orthogonal, parallel, and independent of) the host has been proposed as a strategy for making the design and execution of synthetic gene circuits more predictable, modular, amenable to abstraction, and thus easier to engineer. The synthetic program’s gene expression machinery is decoupled from the host machinery and may thus be less limited by the host’s growth and regulation requirements and can be used to program unnatural functions beyond the host cell’s original capabilities.
Orthogonal transcription has been achieved in bacteria by introducing a phage RNA polymerase, such as the commonly used T7 phage polymerase, and using its cognate promoter sequence to control the synthetic gene circuit (113). Orthogonal translation has been demonstrated in E. coli by screening for orthogonal mRNAs (O-mRNAs) that cannot be translated by the wild-type ribosomes followed by screening for orthogonal ribosomes (O-ribosomes) that can translate O-mRNA without disrupting cell function (114). These orthogonal transcription and translation schemes have been combined to introduce extended delays to gene expression and build transcription-translation feed-forward loops (115).
As orthogonal translation machinery is not essential for host survival, it can be engineered for new and unnatural functions beyond natural translation. One unnatural function of interest is the incorporation of unnatural amino acids into proteins. Researchers have examined the integration of diverse types of unnatural amino acids that support novel activities, including those that provide photocrosslinking capability for studying protein interactions and incorporate heavy atoms for studying protein structures (116–118). One in vivo approach for unnatural amino acid incorporation introduced the amber suppressor tyrosyl-tRNA synthetase (TyrRS)-tRNACUA pair from E. coli into S. cerevisiae (119). After selecting for TyrRS mutants that incorporate a unnatural amino acids but none of the endogenous amino acids in response to the nonsense codon TAG in yeast, researchers were able to achieve a minimum of 99.8% incorporation of a unnatural amino acid into the human superoxidase dismutase protein.
A limitation of the amber suppressor-based approach to synthesizing proteins with novel functionalities is that only one unnatural amino acid can be introduced into a protein at a time. One approach to permit the incorporation of multiple, distinct unnatural amino acids into proteins is to generate multiple, mutually orthogonal tRNA synthetase-tRNA pairs (120). Another limitation of the amber suppressor-based approach is that all TAG codons in cellular transcripts are affected. Potential approaches include incorporating unnatural tRNA synthetase-tRNA pairs with O-ribosomes could ensure only specific transcripts encode unnatural amino acids. An alternative potential approach for controlling unnatural amino acid incorporation in E. coli may be to use a strain where the genome has been re-coded to replace all TAG codons with TAA, for example a strain generated using CAGE (121), such that the codon is freed up for use only within the synthetic gene circuit.
Novel machinery capable of executing genetic programs in parallel with host programs has the potential to be leveraged to program unnatural functions beyond the host cell’s natural capabilities. Tools for genome-wide engineering can advance the creation of orthogonal machinery by providing a means to eliminate or alter genomic elements that interfere with the orthogonal machinery. Recent advances in the field have put in place the methods for large-scale engineering of organisms with useful unnatural abilities beyond those that can be encoded in the natural genetic code.
Designing for evolutionary stability
Evolutionary robustness is one aspect in the design of genetic programs that is important to address for long-term predictability and function. Unlike other engineered systems, biological systems have the potential to evolve over time and a key question in the field is whether it is possible to predict, control, and design how synthetic gene circuits evolve.
Host organisms are capable of altering the expression levels and activities of components in a synthetic biological program to maximize their fitness, even if the program harbors selection markers such as antibiotic resistance or auxotrophic genes (122). Un-programmed changes in expression levels or activities over time are generally undesirable to system performance, especially in gene circuits where level matching is important for proper system function. Another problem is the potential to lose complete circuit function over time. In one example, it was shown that a BioBrick receiver device consistently failed to function after 92 E. coli doublings when input level to the device was low and after 56 doublings when input level was high (49).
Studies have revealed three main strategies to design for genetic reliability in E. coli, which is defined as the number of culture doublings before a mutant device constitutes at least half of the population (49). The first design strategy is to avoid repeated sequences, as segments between repeated sequences are frequently of deleted due to recombination or replication slippage. Most of the studied device failures are due to repeated sequences, including direct and inverted repeats, repeated operator sites in a promoter, and even repeated sequences as short as 8 bp (49, 72, 75). The second design strategy is to decrease the expression level of system components as genetic reliability of gene circuits has been shown to exponentially decrease with increasing expression levels (and associated metabolic loads placed on the host) (72, 75). Finally, the third design strategy is to use inducible rather than constitutive promoters, which are hypothesized to increase genetic reliability by reducing metabolic load (72).
The examples highlighted above focus on the design of a genetic device or program that is more robust to undesired evolutionary changes over time. There are however, applications where the proposed design strategies for system robustness cannot be applied. For example, lentiviruses have potential as clinically usable vectors for gene therapy, but lentiviral expression vectors must harbor long terminal repeats, which make them challenging to propagate in E. coli. One approach that is promising for circumventing host evolutionary pressures has been to reduce the mutation rate of the host. Specifically, a reduced genome strain of E. coli in which mutation-generating insertion sequence (IS) elements were removed has been applied to propagate recombinant proteins and plasmids that are typically unstable (123). Specifically, the IS-free E. coli strain was able to propagate lentiviral expression clones more effectively than commercially available strains specifically developed for this application and a highly toxic synthetic vaccine protein (73).
While it is generally viewed as favorable in the field to increase the genetic reliability of a synthetic program, as our understanding of the conditions for genetic reliability improves, it will become possible to design new classes of devices that incorporate genetic instability as a parameter to program evolution or adaptation over time (75, 124). For example, researchers have suggested the possibility of designing an evolutionary timer in E. coli, where one color of a multi-colored device is lost after x generations, a second color is lost after x more generations, and so forth (75). The potential to evolve is a unique property of engineered biological systems and thus the ability to design systems with programmed evolutionary strategies will be a true frontier in biological engineering.
The examples highlighted above illustrate that the design of a genetically-encoded program can incorporate considerations of the physical interactions between biological components and the interactions of these components with the host organism, including selection pressures directed against the robust, long-term performance of the synthetic circuit. Spatial engineering strategies can improve the performance of synthetic biological systems, but are currently limited by the availability of parts for ready implementation in novel applications. In addition, research into orthogonal expression schemes and evolutionary robustness has revealed the potential to engineer non-natural functions that are inaccessible by natural biological systems and evolving functions inaccessible by other engineering fields.
CONCLUSIONS
Synthetic biology introduces new technologies, tools, and approaches that promise to systematically improve the design process and functional capacities of synthetic genetic programs. New tools include those for DNA synthesis and assembly, precise control of expression at the level of simple genetic circuits, pathways, and genomes, design of novel biological parts, and functional integration of parts into devices and systems. The diverse pool of recent work in synthetic biology highlights common challenges in the field, including the lack of well characterized and standardized part and the dearth of functional composition frameworks for reliable assembly of genetic components. In spite of these and other challenges, synthetic biology offers a powerful and promising novel approach to applications in diverse areas of interest.
Figure 2.
Design strategies for engineering genetic devices. Signal types and operating host organisms (indicated in the central circle with green and blue boxes, respectively) are usually determined by the design goal. When designing genetic devices, four major design considerations (the four surrounding sectors) are commonly taken into account.
KEY TERMS
- Biological parts
a discrete, genetically-encoded entity that exhibits a basic function, such as a promoter, an RNA aptamer, or a fluorescent reporter protein
- Genetic devices
a genetically-encoded assembly of one or more biological parts that executes a defined function, typically logical or information processing, such as genetic switches, logic gates, oscillators, or cell-cell communication modules
- Device specifications
an explicit set of performance requirements that need to be satisfied during the engineering process. For example, dynamic range, basal level activity, and response duration are commonly seen specifications in synthetic biology
- Design considerations
the collection of engineering principles and criteria that will affect performance and stability of devices. In synthetic biology, the design considerations that are frequently taken into account are: device performances, noise rejection, evolutionary robustness, and device crosstalk
- Synthetic biology systems
a genetically-encoded program consisting of one or more devices designed to execute a useful, human-defined function, such as synthesizing a molecule of interest, detecting and processing complex environmental inputs, or targeted delivery of a payload into a target cell
- Genetic reliability (or evolutionary robustness)
a measure of reliability for genetic devices. In E. coli, has been defined as the number of culture doublings before at least half of the population harbors a mutant device (49)
- Functional composition framework
a construction scheme that allows the systematic, reliable, and predictable assembly of a fully functional genetic device or synthetic biology program from their constituent components
- Level matching
the process of matching the input and output levels of the components in a genetic device or synthetic biology system (in terms of small molecule concentration, gene expression levels, or enzymatic activity) such that the output from upstream components is sufficient, but not excessive, as an input to downstream components
Contributor Information
Yen-Hsiang Wang, Email: wangyh@stanford.edu.
Kathy Y. Wei, Email: kywei@stanford.edu.
Christina D. Smolke, Email: csmolke@stanford.edu.
LITERATURE CITED
- 1.Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–6. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–6. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 3.Westberg J, Persson A, Holmberg A, Goesmann A, Lundeberg J, et al. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP) Genome Res. 2004;14:221–7. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slusarczyk AL, Lin A, Weiss R. Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet. 2012;13:406–20. doi: 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- 5.Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Mol Cell. 2011;43:915–26. doi: 10.1016/j.molcel.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–9. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 7.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci U S A. 2007;104:14283–8. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–79. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–58. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang AL, Wolf JJ, Smolke CD. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotechnol. 2012;23:679–88. doi: 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grilly C, Stricker J, Pang WL, Bennett MR, Hasty J. A synthetic gene network for tuning protein degradation in Saccharomyces cerevisiae. Mol Syst Biol. 2007;3:127. doi: 10.1038/msb4100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–43. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 13.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 14.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 15.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–85. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 16.Ham TS, Lee SK, Keasling JD, Arkin AP. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS One. 2008;3:e2815. doi: 10.1371/journal.pone.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–8. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 18.Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012;109:8884–9. doi: 10.1073/pnas.1202344109. Programmed 1 bit of re-writable memory in E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Win MN, Liang JC, Smolke CD. Frameworks for programming biological function through RNA parts and devices. Chem Biol. 2009;16:298–310. doi: 10.1016/j.chembiol.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldminghaus T, Kortmann J, Gesing S, Narberhaus F. Generation of synthetic RNA-based thermosensors. Biol Chem. 2008;389:1319–26. doi: 10.1515/BC.2008.150. [DOI] [PubMed] [Google Scholar]
- 21.van der Meer JR, Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat Rev Microbiol. 2010;8:511–22. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- 22.Petranovic D, Nielsen J. Can yeast systems biology contribute to the understanding of human disease? Trends Biotechnol. 2008;26:584–90. doi: 10.1016/j.tibtech.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Albertsen L, Chen Y, Bach LS, Rattleff S, Maury J, et al. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl Environ Microbiol. 2011;77:1033–40. doi: 10.1128/AEM.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen AH, Silver PA. Designing biological compartmentalization. Trends Cell Biol. 2012 doi: 10.1016/j.tcb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–32. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 26.Schwille P. Bottom-up synthetic biology: engineering in a tinkerer’s world. Science. 2011;333:1252–4. doi: 10.1126/science.1211701. [DOI] [PubMed] [Google Scholar]
- 27.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A. 2005;102:12678–83. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465–71. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyo K, Nevoigt E, Stephanopoulos G. Directed Evolution of Promoters and Tandem Gene Arrays for Customizing RNA Synthesis Rates and Regulation. Methods in Enzymology. 2011:497. doi: 10.1016/B978-0-12-385075-1.00006-8. [DOI] [PubMed] [Google Scholar]
- 30.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–76. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–5. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 32.Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab Eng. 2012;14:306–16. doi: 10.1016/j.ymben.2012.04.004. Genetic device-facilitated enzyme (P450) evolution. [DOI] [PubMed] [Google Scholar]
- 33.Liang JC, Chang AL, Kennedy AB, Smolke CD. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawkins KM, Smolke CD. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol. 2008;4:564–73. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CN, et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012;335:308–13. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 36.Hagel JM, Facchini PJ. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol. 2010;6:273–5. doi: 10.1038/nchembio.317. [DOI] [PubMed] [Google Scholar]
- 37.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–3. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 38.Bayer TS, Widmaier DM, Temme K, Mirsky EA, Santi DV, Voigt CA. Synthesis of methyl halides from biomass using engineered microbes. J Am Chem Soc. 2009;131:6508–15. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 39.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–13. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shivange AV, Marienhagen J, Mundhada H, Schenk A, Schwaneberg U. Advances in generating functional diversity for directed protein evolution. Curr Opin Chem Biol. 2009;13:19–25. doi: 10.1016/j.cbpa.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 41.Lin H, Tao H, Cornish VW. Directed evolution of a glycosynthase via chemical complementation. J Am Chem Soc. 2004;126:15051–9. doi: 10.1021/ja046238v. [DOI] [PubMed] [Google Scholar]
- 42.Dietrich JA, McKee AE, Keasling JD. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu Rev Biochem. 2010;79:563–90. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- 43.Siddiqui MS, Thodey K, Trenchard I, Smolke CD. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012;12:144–70. doi: 10.1111/j.1567-1364.2011.00774.x. [DOI] [PubMed] [Google Scholar]
- 44.Ma S, Tang N, Tian J. DNA synthesis, assembly and applications in synthetic biology. Curr Opin Chem Biol. 2012;16:260–7. doi: 10.1016/j.cbpa.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 46.Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci U S A. 2010;107:4004–9. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. A continuous, in vivo directed evolution system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng AA, Lu TK. Synthetic biology: an emerging engineering discipline. Annu Rev Biomed Eng. 2012;14:155–78. doi: 10.1146/annurev-bioeng-071811-150118. [DOI] [PubMed] [Google Scholar]
- 49.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–93. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 50.Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, et al. Measuring the activity of BioBrick promoters using an in vivo reference standard. J Biol Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cox RS, 3rd, Dunlop MJ, Elowitz MB. A synthetic three-color scaffold for monitoring genetic regulation and noise. J Biol Eng. 2010;4:10. doi: 10.1186/1754-1611-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–9. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 53.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–22. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 55.Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Comput Biol. 2009;5:e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beisel CL, Chen YY, Culler SJ, Hoff KG, Smolke CD. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2011;39:2981–94. doi: 10.1093/nar/gkq954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babiskin AH, Smolke CD. Engineering ligand-responsive RNA controllers in yeast through the assembly of RNase III tuning modules. Nucleic Acids Res. 2011;39:5299–311. doi: 10.1093/nar/gkr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci U S A. 2005;102:3581–6. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alon U. An introduction to systems biology : design principles of biological circuits. Boca Raton, FL: Chapman & Hall/CRC; 2007. [Google Scholar]
- 60.Cagatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Suel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–22. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 61.Bruggeman FJ, Bluthgen N, Westerhoff HV. Noise management by molecular networks. PLoS Comput Biol. 2009;5:e1000506. doi: 10.1371/journal.pcbi.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Partow S, Siewers V, Bjorn S, Nielsen J, Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27:955–64. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- 63.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–30. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J. A sensing array of radically coupled genetic ‘biopixels’. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–4. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 67.Liu C, Fu X, Liu L, Ren X, Chau CK, et al. Sequential establishment of stripe patterns in an expanding cell population. Science. 2011;334:238–41. doi: 10.1126/science.1209042. [DOI] [PubMed] [Google Scholar]
- 68.Tabareau N, Slotine JJ, Pham QC. How synchronization protects from noise. PLoS Comput Biol. 2010;6:e1000637. doi: 10.1371/journal.pcbi.1000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poelwijk FJ, de Vos MG, Tans SJ. Tradeoffs and optimality in the evolution of gene regulation. Cell. 2011;146:462–70. doi: 10.1016/j.cell.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 70.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–91. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.You L, Cox RS, 3rd, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–71. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 72.Sleight SC, Bartley BA, Lieviant JA, Sauro HM. Designing and engineering evolutionary robust genetic circuits. J Biol Eng. 2010;4:12. doi: 10.1186/1754-1611-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chakiath CS, Esposito D. Improved recombinational stability of lentiviral expression vectors using reduced-genome Escherichia coli. Biotechniques. 2007;43:466, 68, 70. doi: 10.2144/000112585. [DOI] [PubMed] [Google Scholar]
- 74.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–71. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 75.Sleight SC, Sauro HM. Design and construction of a prototype CMY (Cyan-Magenta-Yellow) genetic circuit as a mutational readout device to measure evolutionary stability dynamics and determine design principles for robust synthetic systems. Artificial Life. 2012:13. [Google Scholar]
- 76.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–12. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 77.Tigges M, Denervaud N, Greber D, Stelling J, Fussenegger M. A synthetic low-frequency mammalian oscillator. Nucleic Acids Res. 2010;38:2702–11. doi: 10.1093/nar/gkq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mutalik VK, Qi L, Guimaraes JC, Lucks JB, Arkin AP. Rationally designed families of orthogonal RNA regulators of translation. Nat Chem Biol. 2012;8:447–54. doi: 10.1038/nchembio.919. Rational designs of orthogonal, cascadable antisense RNA devices. [DOI] [PubMed] [Google Scholar]
- 79.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, et al. A synthetic genetic edge detection program. Cell. 2009;137:1272–81. doi: 10.1016/j.cell.2009.04.048. E. coli-based synthetic edge detection system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci U S A. 2010;107:8531–6. doi: 10.1073/pnas.1001721107. Synthetic biology for controlling the proliferation of cells for adopted T cell therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–8. doi: 10.1038/nature08187. Multiplex automated genomic engineering (MAGE) for fast, automatic, and cumulative genome engineering in E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sandoval NR, Kim JY, Glebes TY, Reeder PJ, Aucoin HR, et al. Strategy for directing combinatorial genome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:10540–5. doi: 10.1073/pnas.1206299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–9. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 84.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–4. doi: 10.1126/science.1206938. A RNA-based synthetic scaffold. [DOI] [PubMed] [Google Scholar]
- 85.Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 2012;40:1879–89. doi: 10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–7. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 87.Michener J, Thodey K, Liang J, Smolke C. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metabolic engineering. 2012;14:212–22. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–9. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 89.Glick BR. Metabolic load and heterologous gene expression. Biotechnol Adv. 1995;13:247–61. doi: 10.1016/0734-9750(95)00004-a. [DOI] [PubMed] [Google Scholar]
- 90.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–8. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 91.Kemmer C, Gitzinger M, Daoud-El Baba M, Djonov V, Stelling J, Fussenegger M. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat Biotechnol. 2010;28:355–60. doi: 10.1038/nbt.1617. [DOI] [PubMed] [Google Scholar]
- 92.Chang AL, Wolf JJ, Smolke CD. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr Opin Biotechnol. 2012 doi: 10.1016/j.copbio.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–5. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 95.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 96.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–11. doi: 10.1126/science.1205527. Xie et al. 2011: Generalizable RNAi-based classifier for identifying live mammalian cells. [DOI] [PubMed] [Google Scholar]
- 97.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J Am Chem Soc. 2007;129:6807–11. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6:464–70. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106:4629–34. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–27. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 101.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 102.Wei P, Wong WW, Park JS, Corcoran EE, Peisajovich SG, et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488:384–8. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–50. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davis JH, Rubin AJ, Sauer RT. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011;39:1131–41. doi: 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lou C, Stanton B, Chen YJ, Munsky B, Voigt CA. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat Biotechnol. 2012 doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–54. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 107.Wittmann A, Suess B. Engineered riboswitches: Expanding researchers’ toolbox with synthetic RNA regulators. FEBS Lett. 2012;586:2076–83. doi: 10.1016/j.febslet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 108.Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci U S A. 2011;108:8617–22. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sharma V, Nomura Y, Yokobayashi Y. Engineering complex riboswitch regulation by dual genetic selection. J Am Chem Soc. 2008;130:16310–5. doi: 10.1021/ja805203w. [DOI] [PubMed] [Google Scholar]
- 110.Babiskin AH, Smolke CD. A synthetic library of RNA control modules for predictable tuning of gene expression in yeast. Mol Syst Biol. 2011;7:471. doi: 10.1038/msb.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol. 2010;28:856–62. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 113.Basu S, Maitra U. Specific binding of monomeric bacteriophage T3 and T7 RNA polymerases to their respective cognate promoters requires the initiating ribonucleoside triphosphate (GTP) J Mol Biol. 1986;190:425–37. doi: 10.1016/0022-2836(86)90013-6. [DOI] [PubMed] [Google Scholar]
- 114.Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol. 2005;1:159–66. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 115.An W, Chin JW. Synthesis of orthogonal transcription-translation networks. Proc Natl Acad Sci U S A. 2009;106:8477–82. doi: 10.1073/pnas.0900267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–9. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 117.Axup JY, Bajjuri KM, Ritland M, Hutchins BM, Kim CH, et al. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc Natl Acad Sci U S A. 2012;109:16101–6. doi: 10.1073/pnas.1211023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim CH, Axup JY, Dubrovska A, Kazane SA, Hutchins BA, et al. Synthesis of bispecific antibodies using genetically encoded unnatural amino acids. J Am Chem Soc. 2012;134:9918–21. doi: 10.1021/ja303904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–7. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 120.Chatterjee A, Xiao H, Schultz PG. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2012;109:14841–6. doi: 10.1073/pnas.1212454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–53. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–92. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 123.Posfai G, Plunkett G, 3rd, Feher T, Frisch D, Keil GM, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–6. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 124.Egbert RG, Klavins E. Fine-tuning gene networks using simple sequence repeats. Proc Natl Acad Sci U S A. 2012;109:16817–22. doi: 10.1073/pnas.1205693109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Haynes KA, Silver PA. Synthetic reversal of epigenetic silencing. J Biol Chem. 2011;286:27176–82. doi: 10.1074/jbc.C111.229567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 128.Tolmasky ME, Actis LA, Crosa JH. Encyclopedia of industrial biotechnology: bioprocess, bioseparation, and cell technology. John Wiley and Sons, Inc; New York, NY: 2010. Plasmid DNA replication; pp. 1–12. [Google Scholar]
- 129.Grabherr Reingard EE, Stefan Gross, Juergen Mairhofer. 2011 [Google Scholar]
- 130.Temme K, Hill R, Segall-Shapiro TH, Moser F, Voigt CA. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012;40:8773–81. doi: 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, et al. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21:2271–6. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–7. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gonzalez B, Schwimmer LJ, Fuller RP, Ye Y, Asawapornmongkol L, Barbas CF., 3rd Modular system for the construction of zinc-finger libraries and proteins. Nat Protoc. 2010;5:791–810. doi: 10.1038/nprot.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–53. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–6. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 136.Yoshimatsu T, Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science. 1989;244:1346–8. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]
- 137.Swinburne IA, Miguez DG, Landgraf D, Silver PA. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 2008;22:2342–6. doi: 10.1101/gad.1696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Babiskin AH, Smolke CD. Synthetic RNA modules for fine-tuning gene expression levels in yeast by modulating RNase III activity. Nucleic Acids Res. 2011;39:8651–64. doi: 10.1093/nar/gkr445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Auslander S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst. 2010;6:807–14. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 140.Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat Biotechnol. 2012;30:1002–6. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 141.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Venkatesan A, Dasgupta A. Novel fluorescence-based screen to identify small synthetic internal ribosome entry site elements. Mol Cell Biol. 2001;21:2826–37. doi: 10.1128/MCB.21.8.2826-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou W, Edelman GM, Mauro VP. Isolation and identification of short nucleotide sequences that affect translation initiation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:4457–62. doi: 10.1073/pnas.0437993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dai C, Cao Z, Wu Y, Yi H, Jiang D, Li W. Improved fusion protein expression of EGFP via the mutation of both Kozak and the initial ATG codon. Cell Mol Biol Lett. 2007;12:362–9. doi: 10.2478/s11658-007-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. Conditional protein splicing: a new tool to control protein structure and function in vitro and in vivo. J Am Chem Soc. 2003;125:10561–9. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 146.Schwartz EC, Saez L, Young MW, Muir TW. Post-translational enzyme activation in an animal via optimized conditional protein splicing. Nat Chem Biol. 2007;3:50–4. doi: 10.1038/nchembio832. [DOI] [PubMed] [Google Scholar]
- 147.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 148.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annual review of biochemistry. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 149.Shimizu-Sato S, Huq E, Tepperman J, Quail P. A light-switchable gene promoter system. Nature biotechnology. 2002;20:1041–44. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 150.Hughes R, Bolger S, Tapadia H, Tucker C. Light-mediated control of DNA transcription in yeast. Methods (San Diego, Calif) 2012 doi: 10.1016/j.ymeth.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tabor J, Levskaya A, Voigt C. Multichromatic control of gene expression in Escherichia coli. Journal of molecular biology. 2011;405:315–24. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Airan R, Thompson K, Fenno L, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–29. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 153.Waldminghaus T, Kortmann J, Gesing S, Narberhaus F. Generation of synthetic RNA-based thermosensors. Biological chemistry. 2008;389:1319–26. doi: 10.1515/BC.2008.150. [DOI] [PubMed] [Google Scholar]
- 154.Neupert J, Bock R. Designing and using synthetic RNA thermometers for temperature-controlled gene expression in bacteria. Nature protocols. 2009;4:1262–73. doi: 10.1038/nprot.2009.112. [DOI] [PubMed] [Google Scholar]
- 155.Park S-H, Zarrinpar A, Lim W. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science (New York, NY) 2003;299:1061–64. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 156.Saito H, Kobayashi T, Hara T, Fujita Y, Hayashi K, et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nature chemical biology. 2010;6:71–78. doi: 10.1038/nchembio.273. [DOI] [PubMed] [Google Scholar]
- 157.Deans T, Cantor C, Collins J. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–72. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 158.Kobayashi H, Kærn M, Araki… M. Programmable cells: interfacing natural and engineered gene networks. Proceedings of the …. 2004 doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Murphy KF, Balazsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci U S A. 2007;104:12726–31. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Collins C, Leadbetter J, Arnold F. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nature biotechnology. 2006;24:708–12. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- 161.Chen MT, Weiss R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat Biotechnol. 2005;23:1551–5. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- 162.Beisel C, Bayer T, Hoff K, Smolke C. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Molecular systems biology. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Arosio D, Ricci F, Marchetti L, Gualdani R, Albertazzi L, Beltram F. Simultaneous intracellular chloride and pH measurements using a GFP-based sensor. Nature methods. 2010;7:516–18. doi: 10.1038/nmeth.1471. [DOI] [PubMed] [Google Scholar]
- 164.Cromie M, Shi Y, Latifi T, Groisman E. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 165.Ivask A, Rõlova T, Kahru A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC biotechnology. 2009;9:41. doi: 10.1186/1472-6750-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Richter L, Tai W, Felton J, Saviola B. Determination of the minimal acid-inducible promoter region of the lipF gene from Mycobacterium tuberculosis. Gene. 2007;395:22–28. doi: 10.1016/j.gene.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 167.Urban A, Eckermann S, Fast B, Metzger S, Gehling M, et al. Novel whole-cell antibiotic biosensors for compound discovery. Applied and environmental microbiology. 2007;73:6436–43. doi: 10.1128/AEM.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Turner K, Xu S, Pasini P, Deo S, Bachas L, Daunert S. Hydroxylated polychlorinated biphenyl detection based on a genetically engineered bioluminescent whole-cell sensing system. Analytical chemistry. 2007;79:5740–45. doi: 10.1021/ac0705162. [DOI] [PubMed] [Google Scholar]
- 169.Belousov V, Fradkov A, Lukyanov K, Staroverov D, Shakhbazov K, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nature methods. 2006;3:281–86. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 170.Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, et al. Gas-inducible transgene expression in mammalian cells and mice. Nature biotechnology. 2004;22:1440–44. doi: 10.1038/nbt1021. [DOI] [PubMed] [Google Scholar]
- 171.Zhang J, Campbell R, Ting A, Tsien R. Creating new fluorescent probes for cell biology. Nature reviews. Molecular cell biology. 2002;3:906–18. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 172.Drepper T, Eggert T, Circolone F, Heck A, Krauss U, et al. Reporter proteins for in vivo fluorescence without oxygen. Nature biotechnology. 2007;25:443–45. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- 173.Paige J, Wu K, Jaffrey S. RNA mimics of green fluorescent protein. Science (New York, NY) 2011;333:642–46. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Binkowski B, Fan F, Wood K. Engineered luciferases for molecular sensing in living cells. Current opinion in biotechnology. 2009;20:14–18. doi: 10.1016/j.copbio.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 175.Farmer W, Liao J. Improving lycopene production in Escherichia coli by engineering metabolic control. Nature biotechnology. 2000;18:533–37. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 176.van der Meer J, Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nature reviews. Microbiology. 2010;8:511–22. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- 177.Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–79. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- 178.Lu T, Collins J. Dispersing biofilms with engineered enzymatic bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Seema N, Stanley W, Kevin T. Parts-Based Assembly of Synthetic Transmembrane Proteins in Mammalian Cells. ACS Synthetic Biology. 2012:1. doi: 10.1021/sb200007r. [DOI] [PubMed] [Google Scholar]
- 180.Farhi M, Marhevka E, Masci T, Marcos E, Eyal Y, et al. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metabolic engineering. 2011;13:474–81. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 181.Lange A, Mills R, Lange C, Stewart M, Devine S, Corbett A. Classical nuclear localization signals: definition, function, and interaction with importin alpha. The Journal of biological chemistry. 2007;282:5101–05. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bayer T, Widmaier D, Temme K, Mirsky E, Santi D, Voigt C. Synthesis of methyl halides from biomass using engineered microbes. Journal of the American Chemical Society. 2009;131:6508–15. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 183.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature protocols. 2007;2:953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 184.Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, et al. The insulation of genes from external enhancers and silencing chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(Suppl 4):16433–37. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–60. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen X, Zaro J, Shen W-C. Fusion Protein Linkers: Property, Design and Functionality. Advanced drug delivery reviews. 2012 doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pfleger B, Pitera D, Smolke C, Keasling J. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nature biotechnology. 2006;24:1027–32. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 188.Szymczak-Workman A, Vignali K, Vignali DA. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harbor protocols. 2012;2012:199–204. doi: 10.1101/pdb.ip067876. [DOI] [PubMed] [Google Scholar]
- 189.Moon T, Lou C, Tamsir A, Stanton B, Voigt C. Genetic programs constructed from layered logic gates in single cells. Nature. 2012 doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sohka T, Heins R, Phelan R, Greisler J, Townsend C, Ostermeier M. An externally tunable bacterial band-pass filter. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10135–40. doi: 10.1073/pnas.0901246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Guido N, Wang X, Adalsteinsson D, McMillen D, Hasty J, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–60. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 192.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature biotechnology. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 193.Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nature nanotechnology. 2010;5:666–70. doi: 10.1038/nnano.2010.135. Leisner et al, 2010: Synthetic MIMO devices implemented by microRNA-based, NOR logic gates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Basu S, Mehreja R, Thiberge S, Chen M-T, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6355–60. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bacchus W, Lang M, El-Baba M, Weber W, Stelling J, Fussenegger M. Synthetic two-way communication between mammalian cells. Nature biotechnology. 2012 doi: 10.1038/nbt.2351. [DOI] [PubMed] [Google Scholar]
- 196.Ortiz M, Endy D. Engineered cell-cell communication via DNA messaging. Journal of biological engineering. 2012;6:16. doi: 10.1186/1754-1611-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Elowitz M, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–38. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 198.Stricker J, Cookson S, Bennett M, Mather W, Tsimring L, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–19. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fung E, Wong W, Suen J, Bulter T, Lee S-g, Liao J. A synthetic gene-metabolic oscillator. Nature. 2005;435:118–22. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- 200.Friedland A, Lu T, Wang X, Shi D, Church G, Collins J. Synthetic gene networks that count. Science (New York, NY) 2009;324:1199–202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]