Abstract
The proper choice of the CD4-helper or CD8-cytotoxic lineages by developing T cells is crucial for the generation of an antigen-responsive and functionally fit T cell repertoire. Here we present a brief overview of the transcriptional control of this process, with emphasis on two issues. The study of Cd4 expression, that had previously generated important paradigms for transcriptional regulation in eukaryotic cells, now brings new twists to the concept of ‘epigenetic memory’. On the other hand, connections are emerging between transcriptional regulators critical for commitment to either lineage. The present review attempts to integrate these findings and discusses the still elusive mechanisms that match CD4-CD8 lineage differentiation to MHC specificity.
Introduction
The decision by developing T cells to adopt either of the lineages defined by the expression of CD4 or CD8 glycoproteins has captured the attention of immunologists and developmental biologists for years, because of its importance for immune response and of its paradigmatic value [1]. In addition to the mutually exclusive expression of CD4 or CD8, each lineage is characterized by distinct antigen specificities and functions. CD4 T cells are MHC-II restricted and pre-programmed for helper functions, whereas CD8 T cells are MHC I-restricted and pre-programmed for cytotoxic functions. CD4 and CD8 subsets constitute the bulk of αβ T cells and are the main component of T-mediated immune responses. They differentiate in the thymus from CD4+CD8+ ‘double positive’ (DP) precursors [2], and a critical aspect of this process is the matching of CD4 or CD8 lineage differentiation (and of helper vs. cytotoxic functions) to MHC-II or MHC-I specificity, respectively (Fig. 1). This highlight of the recent literature is focused on the control of Cd4 expression and on the transcriptional mechanisms that underpin CD4-CD8 lineage differentiation in the thymus [3–5]. We refer the reader to recent reviews [1, 6] for a discussion of intrathymic signals that control lineage differentiation.
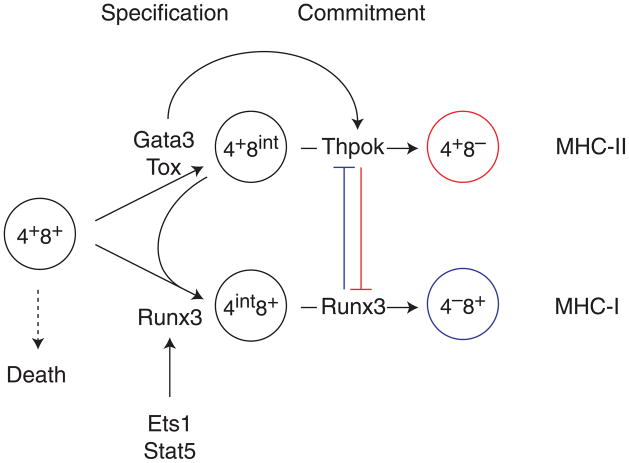
Figure 1. Specification and commitment to the CD4 and CD8 lineages.
DP thymocytes have rearranged genes encoding TCRα and TCRβ chains and express surface TCR complexes. These cells are programmed to undergo apoptotic cell death in the thymic cortex unless their TCR is productively engaged by MHC molecules expressed by the thymic epithelium, an event referred to as positive selection. Rescued thymocytes differentiate into CD4 or CD8 T cells, depending on whether they are MHC II- or MHC I-restricted, respectively. Lineage differentiation includes two conceptually distinct steps, specification and commitment. For the CD4 lineage, specification involves Gata3, Tox and E-proteins E2A and HEB (not shown), whereas commitment requires Thpok, which represses CD8-lineage genes including Runx3. For CD8 cells, specification involves Runx3, which also contributes to commitment by repressing Thpok and Cd4. Stat5 and Ets1 contribute to Runx3 expression, the latter binding the Runx3 locus. Note that the CD4+CD8int cells has precursor activity for both CD4 and CD8 lineages and is thought to include truly bi-potent cells [1]. In contrast the CD4intCD8+ subset only has CD8 precursor activity.
Cd4 gene expression
Previous studies of Cd4 gene expression had spawned insights critical for our understanding of gene silencing [7], and the last two years have brought new thought-provoking results. Two cis-regulatory elements involved in Cd4 expression had been identified earlier [7]: an upstream enhancer (‘proximal’, E4P) and an intronic silencer whose activity requires recruitment of repressor proteins Runx1 or Runx3 (Fig. 2). The conventional picture was that E4P is active throughout T cell development, whereas the silencer prevents Cd4 expression in CD8 cells and in CD4−CD8− (double negative, DN) thymocytes [8]. A first dent into this dichotomic view comes from the observation that E4P also contributes to Cd4 repression in CD4-negative cells, by recruiting the transcriptional repressor AP4 [9], suggesting an unsuspected inter-dependence of activation and repression functions within the Cd4 locus.
Figure 2. Cd4 cis-regulatory elements.
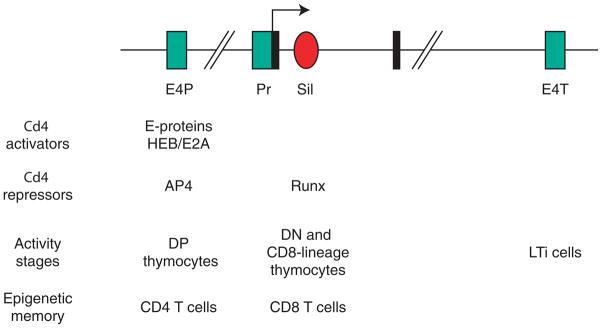
Schematic representation (not on scale) of the Cd4 locus, with exons 1 and 2 (black boxes), the silencer (red-filled oval) and positive regulatory elements (green-filled rectangles), including the proximal enhancer (E4P), promoter (Pr) and a downstream enhancer known as ‘thymic enhancer’ (E4T) even though it is now known to be active in LTi cells, not in thymocytes. Transcription factors important for the activity of each element are indicated, as are cell subsets in which each element is active, or determines ‘epigenetic memory’ despite having no intrinsic activity in the subset. Note that while AP4 does not bind the silencer, it interacts with Runx molecules and could therefore ‘bridge’ that element with E4P. Factors distinct from Runx proteins are thought to contribute to Cd4 silencing because the silencer contains functionally important motifs in addition to Runx binding sites [21].
A stronger challenge to the conventional view, together with clarifications of an old controversy, come from experiments ‘knocking-out’ E4P to explore its functions [10]. The first surprise is that E4P has stage-specific activity: germline deletion shows the enhancer to be required for Cd4 expression in DP thymocytes, but not in mature CD4 T cells or in CD4-differentiating thymocytes as CD4-expressing T cells develop despite germline E4P deletion. This was unexpected: if anything, the contrary could have been envisioned, because earlier experiments with transgenic reporters had suggested that an enhancer located downstream of the Cd4 gene was active in DP thymocytes but not mature T cells [11]. In fact, the new report shows that this element is dispensable for Cd4 expression at any stage of T cell development, but is active in lymphoid tissue inducer (LTi) cells, a subset of innate lymphoid cells that in mice express Cd4 [12].
These observations imply that another cis-regulatory element (unknown enhancer, or the Cd4 promoter itself) activates Cd4 in CD4-lineage cells. However, things are not that simple. Strikingly, disrupting E4P in thymocytes affects Cd4 expression in activated T cells [10]. Specifically, the CD4-expressing T cells that develop despite E4P deletion fail to sustain Cd4 expression after activation, whereas Cd4 expression is not affected, even in proliferating cells, by post-thymic deletion of a conditional (‘floxed’) E4P element [10]. These findings suggest that, in thymocytes, E4P ‘deposits’ activating epigenetic marks elsewhere on the Cd4 locus, and that such marks contribute to stable Cd4 expression in effector cells. What these marks are, and whether they target the promoter, a putative new enhancer or other components of the Cd4 locus remain to be determined, and is further discussed in a review accompanying the original study [13].
Lineage commitment
The rest of this review will address the transcriptional control of the CD4-CD8 differentiation decision. Our discussion will be based on the idea that two conceptually distinct, even if possibly overlapping, processes contribute to this event: specification, the initiation of lineage-specific gene expression, and commitment, a biological event defined by the loss of the bi-potency characteristic of the precursor stage and of alternate differentiation potential.
The current perspective is that commitment results from the opposing activity of two transcriptional repressors, Thpok and Runx3 (Fig. 1) [3]. The zinc finger protein Thpok is up-regulated in MHC II-restricted thymocytes as they undergo CD4-lineage differentiation and is needed for CD4 lineage commitment. In contrast, MHC I-restricted cells up-regulate Runx3, which promotes CD8-lineage commitment redundantly with the related protein Runx1 [5]. Thpok represses CD8-lineage genes, including Runx3 and Cd8, whereas Runx3 proteins repress Thpok and Cd4. Despite the apparent symmetry of this process, there is evidence for Thpok having a dominant effect on Runx3. Thpok antagonizes Runx3 functions in cells where both factors are expressed [14, 15]. In addition, transgenic Thpok expression prevents Runx3 expression and CD8 differentiation, whereas transgenic Runx3 expression is insufficient to repress Thpok and CD4 differentiation [Refs. 16, 17, 18, and our unpublished results]. Transgene dosage effects, a typical caveat of gain-of-function studies, do not seem to account for these differences [18–20]. Rather, one possible interpretation is that additional CD8 lineage-specific factors cooperate with Runx3 to seal commitment, a possibility in line with the idea that non-Runx factors, so far unidentified, contribute to repress Cd4 and Thpok in CD8 cells [21–23].
The repression of CD8-lineage genes by Thpok appears to operate, albeit with modifications, beyond lineage commitment in the thymus [24]. One notable variation is seen in IFNγ-producing (Th1) CD4 effectors, which express both Thpok and Runx3 [25–27], yet do not re-express Cd8 at least in part because of persistent Thpok expression [Ref. 25, and unpublished results from our laboratory]. Thus, the Thpok-Runx3 reciprocal repression has stage-specific characteristics that are not yet fully understood. In contrast, Cd8 repression seems a general effect of Thpok. That is illustrated in iNK T cells, an αβ T cell population recognizing lipids presented by the MHC-like molecule CD1d, and that are CD4+CD8− or CD4−CD8− [28]. Thpok is expressed in both subsets at similar levels, and represses Cd8 as in conventional T cells [29, 30]. Genome-wide analyses of Thpok binding will be helpful to determine the mechanistic bases of its distinct effects on Cd8 and Runx3 expression. Despite these remaining uncertainties, the Thpok-Runx3 mutual repression provides a robust mechanism explaining how cells embracing either lineage renounce the alternative fate.
Lineage specification
The question now hovering over the field is to decipher the initial lineage specification process: what sets this commitment machinery in motion by initiating expression of Thpok or Runx3, neither of which is expressed in DP thymocytes [16, 17, 31, 32]. Two factors potentially involved in Runx3 expression have been recently identified. Stat5, a target of IL-7 in the thymus, is critical for the generation of CD8 T cells; it acts redundantly with the related protein Stat6 and has been reported to promote Runx3 expression [33]. It is not clear yet whether this effect is direct or indirect and how it integrates with other effects of IL-7 during CD8 T cell differentiation [34]. The implication of these observations is that IL-7, possibly redundantly with other cytokines [34, 35], promotes CD8-lineage specification. This provocative idea is a key tenet of the ‘kinetic signalling’ model of CD4-CD8 differentiation, a discussion of which is beyond the scope of this overview; for more information, the reader is referred to recent reviews [1, 6] and original studies [33, 36, 37]. The second Runx3-promoting factor, Ets1, is expressed at all stages of T cell development. Ets1 binds the Runx3 locus and promotes Runx3 expression (and Cd4 silencing at least in part through its effect on Runx3) [20]. In addition, Ets1 contributes to IL-7 receptor expression [38]. While the latter finding provides another possible clue to its effect on Runx3, it also illustrates how arduous dissecting the pleiotropic functions of lineage specifying factors can become, an issue that we will encounter again with CD4 differentiation.
The up-regulation of Runx3 is part of a broader CD8-lineage specification program in the thymus, that includes genes encoding cytotoxic proteins (such as perforin). New evidence documents the role of Runx3 itself in this process [39], in addition to it repressing Thpok and Cd4. The few CD8 cells that develop in the absence of Runx3 (and depend on the redundant function of Runx1 [31]) have impaired expression of perforin [22, 31, 39], and of Cd8 genes themselves. Even though the expression of CD8 is barely diminished in resting Runx3-deficient cells [32], it is strikingly reduced when these cells undergo effector differentiation and proliferation, consistent with the recruitment of Runx3 to Cd8 enhancers [40, 41]. It is interesting a that similar difference between resting and activated CD8 cells was found in their usage of Cd8 cis-regulatory elements [41], and future studies will determine if, as for Cd4 expression in CD4 cells [10], such differences result from epigenetic changes imprinted during intra-thymic differentiation.
The CD4-lineage specification problem has been even more difficult to tackle. In contrast to Runx3 for cytotoxic genes, it is not yet clear to which extent Thpok itself contributes to the expression of CD4-lineage genes, including its own [15, 25, 42, 43]. However, genetic studies have identified several transcription factors important for CD4 T cell generation, and thus candidates for a specification function; these include Gata3, the DNA-binding protein Tox, and E proteins E2A and HEB [44–46]. Both Gata3 and Tox are up-regulated by TCR signaling, albeit possibly through distinct intra-cellular signaling pathways [47, 48]; in addition, Gata3 but not Tox is expressed higher in MHC-II than MHC I-restricted thymocytes [49]. Both factors are required for Thpok expression [19, 45], and Gata3 actually binds a region of Thpok important for its transcription [19] (Fig. 3). However, ectopic Thpok expression in Gata3- or Tox-deficient thymocytes fails to restore CD4 T cell differentiation, suggesting that each factor has a broader range of targets in developing CD4 cells [25, 50].
Figure 3. Thpok cis-regulatory elements.
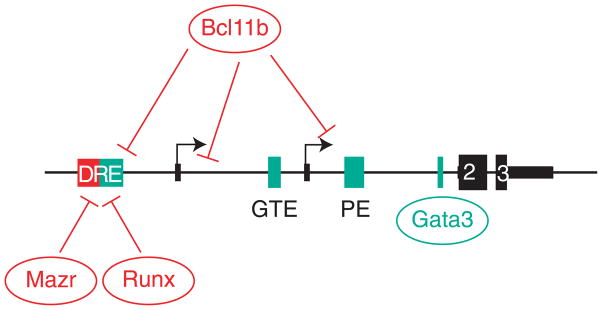
The Thpok locus is schematically represented (not on scale). The two transcription start sites, defined by alternate exons 1a and 1b, are shown as arrows and exons as black boxes; thick areas depict coding regions in exons 2 and 3. Boxes (top) indicate the distal regulatory element (DRE, including the silencer), the general T lymphoid element (GTE), the proximal enhancer (PE), and a Gata3 binding site, using the same color code as in Fig. 2. Binding areas of repressors (Runx molecules, Mazr and Bcl11b) and activators (Gata3) are schematically depicted. While the DRE is depicted as including distinct activating (green) and silencing (red) elements, it is not clear whether these activities are physically separated [19, 22, 23]. Note that Runx molecules also bind the proximal enhancer [22], although it is not know whether that binding serves an activating or repressive function. In CD4-lineage cells, which express Thpok, Thpok molecules bind the silencer [15].
What are these targets is an important but yet unanswered question. It has been proposed that Tox serves to sustain or increase Cd4 expression in MHC II-signaled cells [37, 50], an idea that would fit with the existence of a Cd4 cis-regulatory element specifically activated at this developmental stage [10]. However, this is unlikely to account for all Tox functions, as Tox inactivation severely disrupts the development of MHC II-restricted cells, whereas impaired CD4 expression redirects them to the CD8 lineage [36]. In addition, these analyses are complicated by the pleiotropic effects of these ‘CD4-specifying factors’: Tox affects late thymocyte maturation, and Gata3 TCR signal transduction [19, 50]. Such caveats apply to an even greater extent to E-proteins E2A and HEB: they are important for CD4-lineage differentiation and for the activity of the Cd4 E4P, but also serve as gate-keepers of the DP to SP transition, most spectacularly by enforcing the requirement for MHC-induced T cell receptor (TCR) signaling during positive selection [44].
Matching lineage to MHC specificity
How does this specification machinery ensure expression of Thpok in CD4 cells and of Runx3 in CD8 cells? Because of the dominant effect of Thpok on Runx3 expression and function [14, 15, 25, 42], its up-regulation is thought to determine lineage choice, and there is strong evidence that it results primarily from the relief of repression [22, 23]. A cis-regulatory sequence called ‘distal regulatory element’ (DRE) or ‘silencer’ represses Thpok in DP thymocytes and CD8-lineage thymocytes (Fig. 3). Three transcriptional repressors bind the silencer and contribute to Thpok repression in DP thymocytes (and presumably in CD8-lineage cells) [22, 51, 52]: Runx proteins (Runx1 or Runx3), Mazr, a protein structurally related to Thpok, and Bcl11b, a molecule essential for early T lineage commitment [53]. For Runx proteins, there is evidence for direct repression through silencer binding [22]. While there is no such evidence for the other two factors, a similar mechanism is plausible for Mazr given its ability to associate with Runx1 [51]. The function of Bcl11b seems more complex as it may also contribute to repress Runx3 [52].
This does not mean that positive regulation does not contribute to Thpok expression, and positive cis-regulatory elements (enhancers) have been identified in the Thpok locus [22, 23] (Fig. 3). However, a ‘silencer-less’ Thpok allele or reporter transgene is expressed equally well in MHC I- and MHC II-restricted cells, indicating that these enhancers are not by themselves responsible for the lineage specificity of Thpok expression [22, 23]. Thus, the key question is to understand how the silencer is inactivated, and therefore Thpok expressed, in MHC II but not MHC I-signaled cells. This could involve up-regulating transcriptional activators that overcome Runx-mediated repression. Gata3 is a prime candidate for such a function, as in mature T cells it antagonizes the functions of Runx proteins [54] and its expression is higher in MHC-II than MHC I-restricted thymocytes [49 and our own observations]. Other possible mechanisms, at present speculative, include (i) lineage-specific down-regulation of silencer-binding repressors, although there is no evidence that this is the case for Runx1, Mazr or Bcl11b, or (ii) post-translational modifications of critical members of this circuitry.
In addition to their mechanistic interest, an important objective of these studies is to link silencer inactivation to the intrathymic signals that promote CD4-lineage differentiation and Thpok expression [1, 6, 36, 55, 56]. There is strong evidence, including from a recent ‘knock in’ study [36], that TCR signaling needs to persist longer in thymocytes for CD4 than for CD8 lineage choice, another tenet of the ‘kinetic signaling’ perspective [1]. One would then expect such ‘long’ signals to inactivate the Thpok silencer and promote Thpok expression, and there is indirect support for that possibility [23]. Conversely, it is possible that TCR signaling contributes to Runx3 repression: a recent study proposes that IRF4, a target of TCR signals in the thymus, could serve such a function by binding the Runx3 locus, among broader effects than cannot be attributed to Runx3 down-regulation [57]. However, additional analyses will be needed to fully appreciate the function of IRF4 in thymocytes, as IRF4 disruption did not affect CD4-CD8 differentiation [58].
Conclusions and perspectives
The last two years have seen connections emerging between transcription factors involved in the choice of the CD4 or CD8 lineage, bringing us a step closer to understanding the transcriptional circuits controlling that process. Important challenges lie ahead. The underpinnings of epigenetic control of gene expression and silencing will certainly be an important area of investigation for the coming years; clues may emerge from studies of non-coding RNAs and histone or DNA modifications [59]. Imaging studies are entering the field with provocative questions. Notably, the possibility that transcription at Cd4 and Cd8 loci are mechanistically coupled by Runx proteins has emerged from analyses showing close approximation of both loci in cells that express CD8 [60]. This effect has specific requirements, both in cis (Cd4 silencer and Cd8 enhancers) and in trans (Runx activity). The puzzling question is that, even when close, Cd4 and Cd8 loci remain separated by micrometer-range distances; elucidating the mechanistic bases of the association should provide insight into changes in subnuclear architecture that accompany and possibly affect lineage specific gene expression. Solving the maze of CD4-lineage differentiation is also a pressing challenge, notably in light of the clinical importance of CD4 cell deficiencies. An important objective of future research will be to determine whether this process results from the combined activities of multiple transcription factors, or whether a single factor, yet to be identified, serves as a major CD4-specifying activity.
Acknowledgments
We thank A. Gégonne, P. Love and M. Vacchio for reading the manuscript and apologize to colleagues whose work could not be discussed because of space limitations. Research work in the authors’ laboratory is supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research.
References
- 1.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4− versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Bosselut R. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. J Immunol. 2009;183:2903–2910. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniuchi I. Transcriptional regulation in helper versus cytotoxic-lineage decision. Curr Opin Immunol. 2009;21:127–132. doi: 10.1016/j.coi.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annu Rev Immunol. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- 7.Taniuchi I, Ellmeier W. Transcriptional and epigenetic regulation of CD4/CD8 lineage choice. Adv Immunol. 2011;110:71–110. doi: 10.1016/B978-0-12-387663-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 8.Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 9.Egawa T, Littman DR. Transcription factor AP4 modulates reversible and epigenetic silencing of the Cd4 gene. Proc Natl Acad Sci U S A. 2011;108:14873–14878. doi: 10.1073/pnas.1112293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong MM, Simpson N, Ciofani M, Chen G, Collins A, Littman DR. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adlam M, Siu G. Hierarchical interactions control CD4 gene expression during thymocyte development. Immunity. 2003;18:173–184. doi: 10.1016/s1074-7613(03)00021-9. [DOI] [PubMed] [Google Scholar]
- 12.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 13.Sen R, Grosschedl R. Memories of lost enhancers. Genes Dev. 2010;24:973–979. doi: 10.1101/gad.1930610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, Bosselut R. The transcription factor zbtb7b promotes CD4 expression by antagonizing runx-mediated activation of the CD4 silencer. J Immunol. 2007;179:4405–4414. doi: 10.4049/jimmunol.179.7.4405. [DOI] [PubMed] [Google Scholar]
- 15.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 16.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 17.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 18.Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, et al. Runx3 Regulates Integrin {alpha}E/CD103 and CD4 Expression during Development of CD4−/CD8+ T Cells. J Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zamisch M, Tian L, Grenningloh R, Xiong Y, Wildt KF, Ehlers M, Ho IC, Bosselut R. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med. 2009;206:2685–2699. doi: 10.1084/jem.20092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniuchi I, Sunshine MJ, Festenstein R, Littman DR. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell. 2002;10:1083–1096. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- 22.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 23.He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Xiong Y, Bosselut R. Maintaining CD4-CD8 lineage integrity in T cells: Where plasticity serves versatility. Semin Immunol. 2011;23:360–367. doi: 10.1016/j.smim.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 2008;29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 27.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 29.Engel I, Hammond K, Sullivan BA, He X, Taniuchi I, Kappes D, Kronenberg M. Co-receptor choice by V{alpha}14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J Exp Med. 2010 doi: 10.1084/jem.20090557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Carr T, Xiong Y, Wildt KF, Zhu J, Feigenbaum L, Bendelac A, Bosselut R. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4(+) NKT cells. Eur J Immunol. 2010;40:2385–2390. doi: 10.1002/eji.201040534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woolf E, Xiao C, Fainaru O, Lotem J, Rosen D, Negreanu V, Bernstein Y, Goldenberg D, Brenner O, Berke G, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, Kimura MY, Cui Y, Lucas PJ, Gress RE, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Q, Erman B, Bhandoola A, Sharrow SO, Singer A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J Exp Med. 2003;197:475–487. doi: 10.1084/jem.20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinreich MA, Jameson SC, Hogquist KA. Postselection thymocyte maturation and emigration are independent of IL-7 and ERK5. J Immunol. 2011;186:1343–1347. doi: 10.4049/jimmunol.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adoro S, McCaughtry T, Erman B, Alag A, Van Laethem F, Park JH, Tai X, Kimura M, Wang L, Grinberg A, et al. Coreceptor gene imprinting governs thymocyte lineage fate. EMBO J. 2011 doi: 10.1038/emboj.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarafova SD, Van Laethem F, Adoro S, Guinter T, Sharrow SO, Feigenbaum L, Singer A. Upregulation of CD4 expression during MHC class II-specific positive selection is essential for error-free lineage choice. Immunity. 2009;31:480–490. doi: 10.1016/j.immuni.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grenningloh R, Tai TS, Frahm N, Hongo TC, Chicoine AT, Brander C, Kaufmann DE, Ho IC. Ets-1 maintains IL-7 receptor expression in peripheral T cells. J Immunol. 2011;186:969–976. doi: 10.4049/jimmunol.1002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato T, Ohno S, Hayashi T, Sato C, Kohu K, Satake M, Habu S. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–328. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Hassan H, Sakaguchi S, Tenno M, Kopf A, Boucheron N, Carpenter AC, Egawa T, Taniuchi I, Ellmeier W. Cd8 enhancer E8I and Runx factors regulate CD8alpha expression in activated CD8+ T cells. Proc Natl Acad Sci U S A. 2011;108:18330–18335. doi: 10.1073/pnas.1105835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong Y, Bosselut R. The enigma of CD4-lineage specification. Eur J Immunol. 2011;41:568–574. doi: 10.1002/eji.201041098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aliahmad P, O’Flaherty E, Han P, Goularte OD, Wilkinson B, Satake M, Molkentin JD, Kaye J. TOX Provides a Link Between Calcineurin Activation and CD8 Lineage Commitment. J Exp Med. 2004;199:1089–1099. doi: 10.1084/jem.20040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gimferrer I, Hu T, Simmons A, Wang C, Souabni A, Busslinger M, Bender TP, Hernandez-Hoyos G, Alberola-Ila J. Regulation of GATA-3 Expression during CD4 Lineage Differentiation. J Immunol. 2011;186:3892–3898. doi: 10.4049/jimmunol.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 50.Aliahmad P, Kadavallore A, de la Torre B, Kappes D, Kaye J. TOX Is Required for Development of the CD4 T Cell Lineage Gene Program. J Immunol. 2011;187:5931–5940. doi: 10.4049/jimmunol.1101474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakaguchi S, Hombauer M, Bilic I, Naoe Y, Schebesta A, Taniuchi I, Ellmeier W. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kastner P, Chan S, Vogel WK, Zhang LJ, Topark-Ngarm A, Golonzhka O, Jost B, Le Gras S, Gross MK, Leid M. Bcl11b represses a mature T-cell gene expression program in immature CD4(+)CD8(+) thymocytes. Eur J Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Santo JP. Immunology A guardian of T cell fate. Science. 2010;329:44–45. doi: 10.1126/science.1191664. [DOI] [PubMed] [Google Scholar]
- 54.Yagi R, Junttila IS, Wei G, Urban JFJ, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Taylor BJ, Sun G, Bosselut R. Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes. J Immunol. 2005;175:4465–4474. doi: 10.4049/jimmunol.175.7.4465. [DOI] [PubMed] [Google Scholar]
- 56.Park K, He X, Lee HO, Hua X, Li Y, Wiest D, Kappes DJ. TCR-mediated ThPOK induction promotes development of mature (CD24−) gammadelta thymocytes. EMBO J. 2010;29:2329–2341. doi: 10.1038/emboj.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao Y, Li H, Sun Y, Chen X, Liu H, Gao X, Liu X. Interferon regulatory factor 4 regulates thymocyte differentiation by repressing Runx3 expression. Eur J Immunol. 2010;40:3198–3209. doi: 10.1002/eji.201040570. [DOI] [PubMed] [Google Scholar]
- 58.Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 59.Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 60.Collins A, Hewitt SL, Chaumeil J, Sellars M, Micsinai M, Allinne J, Parisi F, Nora EP, Bolland DJ, Corcoran AE, et al. RUNX Transcription Factor-Mediated Association of Cd4 and Cd8 Enables Coordinate Gene Regulation. Immunity. 2011;34:303–314. doi: 10.1016/j.immuni.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]