Abstract
Objective: There are many reports on associations between spermatogenesis and partial azoospermia factor c (AZFc) deletions as well as duplications; however, results are conflicting, possibly due to differences in methodology and ethnic background. The purpose of this study is to investigate the association of AZFc polymorphisms and male infertility in the Yi ethnic population, residents within Yunnan Province, China. Methods: A total of 224 infertile patients and 153 fertile subjects were selected in the Yi ethnic population. The study was performed by sequence-tagged site plus/minus (STS+/−) analysis followed by gene dosage and gene copy definition analysis. Y haplotypes of 215 cases and 115 controls were defined by 12 binary markers using single nucleotide polymorphism on Y chromosome (Y-SNP) multiplex assays based on single base primer extension technology. Results: The distribution of Y haplotypes was not significantly different between the case and control groups. The frequencies of both gr/gr (7.6% vs. 8.5%) and b2/b3 (6.3% vs. 8.5%) deletions do not show significant differences. Similarly, single nucleotide variant (SNV) analysis shows no significant difference of gene copy definition between the cases and controls. However, the frequency of partial duplications in the infertile group (4.0%) is significantly higher than that in the control group (0.7%). Further, we found a case with sY1206 deletion which had two CDY1 copies but removed half of DAZ genes. Conclusions: Our results show that male infertility is associated with partial AZFc duplications, but neither gr/gr nor b2/b3 deletions, suggesting that partial AZFc duplications rather than deletions are risk factors for male infertility in Chinese-Yi population.
Keywords: Azoospermia factor c (AZFc), AZFc polymorphism, b2/b3, gr/gr, Infertility
1. Introduction
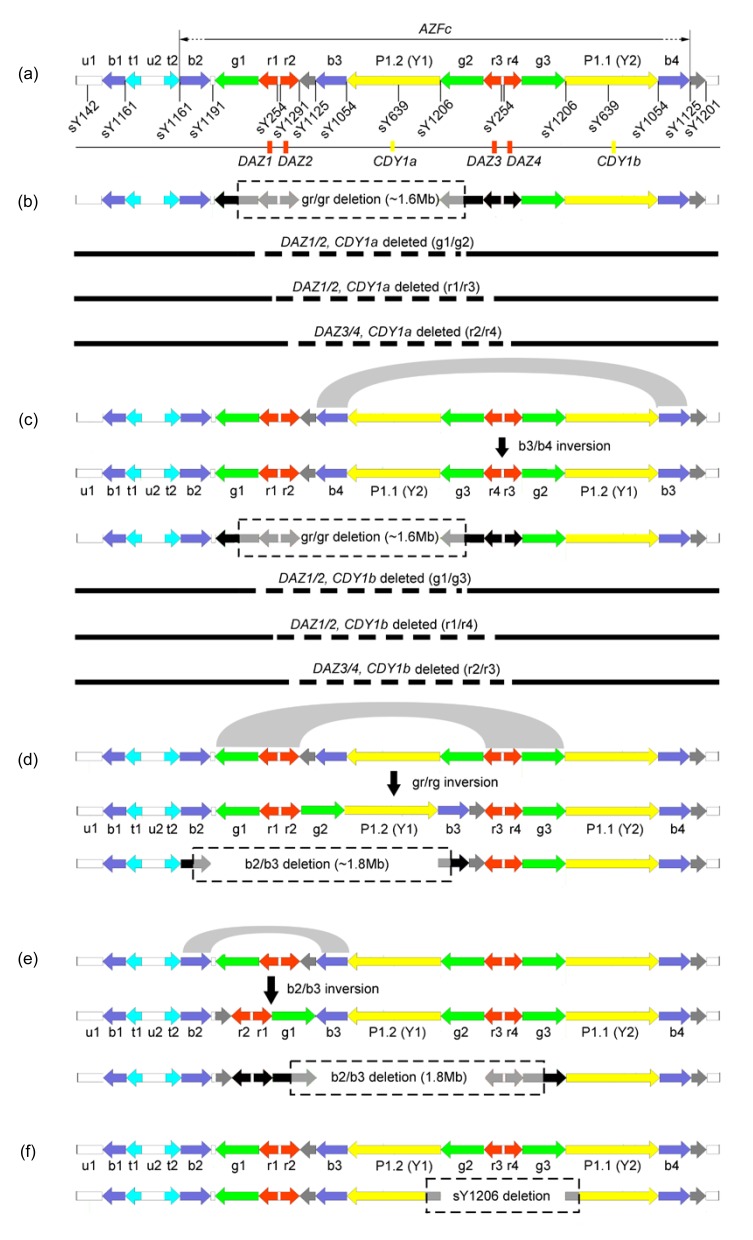
The azoospermia factor (AZF) region, mapped to Yq11, is associated with spermatogenesis (Bardoni et al., 1991; Vog et al., 1996; Pryor et al., 1997). There are three subregions within AZF, referred as AZFa, AZFb, and AZFc (Vog et al., 1996). Today, testing for microdeletions of AZF is routine in cases of male infertility. AZFc is comprised of long direct and inverted repetitive sequence blocks called “amplicons”, which make AZFc one of the most unstable regions in the human genome (Fig. 1a) (Kuroda-Kawaguchi et al., 2001; Skaletsky et al., 2003). The AZFc deletion, also known as b2/b4 deletion, leads to variable phenotypes ranging from azoospermia to oligozoospermia. It is the most commonly known genetic cause of spermatogenic failure (Ferlin et al., 2007; Simoni et al., 2008). In this region, three copies of basic protein Y2 (BPY2), two copies of chromodomain Y1 (CDY1), and four copies of deleted in azoospermia (DAZ) have been identified (Fig. 1a) (Navarro-Costa et al., 2010b). These genes are considered as candidates important for spermatogenesis. In addition to the 3.5-Mb AZFc deletion, several partial deletions in this region have been reported, such as gr/gr and b2/b3 deletions, both of which remove almost half of the AZFc gene copies, including two copies of DAZ and one copy of CDY1 (Figs. 1b–1e) (Repping et al., 2003; 2004). However, the reports on associations between partial AZFc deletions and infertility have been conflicting (Kuroda-Kawaguchi et al., 2001; Giachini et al., 2008). Several studies about partial AZFc deletions and infertility have been performed in Chinese-Han population, but results have been conflicting and may be due to control selection, sample quantity, different ethnic backgrounds, or geographic areas (Zhang et al., 2006; Lin et al., 2007; Wu et al., 2007; Lu et al., 2009; Yang et al., 2010). Polymorphic expression among DAZ proteins has been reported, indicating unequal activities of different DAZ copies (Kim et al., 2009). Thus, analysis on deletion types of DAZ or other related genes is necessary to determine the relationship between partial AZFc polymorphisms and male infertility. In addition to the partial AZFc deletions, partial duplications in this region have been identified (Lin et al., 2005). Associations between partial duplications and male infertility have been found in the Chinese-Han population, but not in the Italian population (Lin et al., 2007; Giachini et al., 2008).
Fig. 1.
Representation of AZFc structure and arrangement
(a) The structures of amplicons, locations of sequence-tagged sites, and transcription units of DAZ and CDY1 gene families (Zhang et al., 2007); (b) The gr/gr deletion and sub-types (Krausz et al., 2009); (c) The gr/gr deletion following b3/b4 inversion and sub-types (Krausz et al., 2009); (d) The b2/b3 deletion following gr/rg inversion (Zhang et al., 2007); (e) The b2/b3 deletion following b2/b3 inversion (Zhang et al., 2007); (f) The sY1206 deletion
Though lots of studies have been carried out, the impact of partial AZFc deletions and duplications on spermatogenesis is inconclusive. In this paper, we report an association study on AZFc polymorphisms and male infertility in the Yi population, an ethnic minority in southwestern China. We carefully selected 224 infertile patients and 153 fertile controls. We did not find significant frequency differences between the case and control groups for partial AZFc deletions and subtypes analyzed by single nucleotide variant (SNV). However, the frequency of partial AZFc duplications in infertile patients was significantly higher than that in fertile men. Our results indicate that partial AZFc duplications, rather than deletions, are likely risk factors for male infertility in the Chinese-Yi population.
2. Materials and methods
2.1. Subjects
A total of 224 infertile patients and 153 control subjects were selected from the Population and Family Planning Institute in Yunnan Province, China. All the samples were from the Yi population in Yunnan Province with normal 46,XY karyotype. The men with Y-chromosome microdeletions were excluded. The infertile men had either non-obstructive azoospermia or severe oligozoospermia with a sperm count <5×106 spermatozoa/ml, on the basis of repeated semen analysis according to the World Health Organization (WHO) criteria. The subjects with history of orchitis and active orchitis, history of unilateral or bilateral cryptorchidism and varicocele, and hypogonadotrophic hypogonadism were also excluded from the infertile group. The fertile controls were healthy men who had given birth to at least one offspring without the help of assisted reproductive technology, which had been confirmed by paternity test.
2.2. Sequence tagged site plus/minus (STS+/−) analysis of AZFc
Overall, six AZFc specific STSs (sY1191, sY1291, sY1206, sY1201, sY142, sY1161) were screened for all the samples by polymerase chain reaction (PCR) analysis (Fig. 1a). The gr/gr deletion was represented by the absence of sY1291 with the presence of the other STSs. The b2/b3 was represented by the specific absence of sY1191. The primer sequences and PCR conditions were as described previously (Fernandes et al., 2002; Repping et al., 2003). For case 3, additional STSs (sY254, sY639, sY1054, sY1125) were screened (Fig. 1a).
2.3. SNV analyses of DAZ and CDY1 genes
The samples with the absence of sY1191 or sY1291 were carried out by SNV analysis using a PCR amplification-restriction digestion assay. SNV sY587 and CDY7750 were chosen for distinguishing DAZ1/2 from DAZ3/4, and CDY1a from CDY1b, respectively. The methods were according to Machev et al. (2004).
2.4. DAZ and CDY1 gene dosages
The dosage analyses of DAZ and CDY1 were performed as previously described (Machev et al., 2004; Yang et al., 2010). Primer pair o1130/o1313 was used to amplify DAZ (214 bp) and DAZL (217 bp), and oMY953a/o1023 was for amplification of CDY1 (134 bp) and CDY2 (137 bp). o1130 and oMY953a were labeled with 5′ FAM (Invitrogen). The PCR products were analyzed using ABI PRISM 3100 Avant genetic analyzer (Applied Biosystems).
2.5. Y chromosome haplogrouping
Y chromosome haplogroups were defined by 12 binary markers: M130, M174, M9, M175, M119, M95, M122, M324, P201, M7, M134, and M117. These single nucleotide polymorphisms (SNPs) were detected by Y-SNP multiplex assays based on single base primer extension technology using SNaPshot Multiplex kit (Applied Biosystems), which was performed as previously described (van Oven et al., 2011). Primers M174, M9, and M175 were designed as described by van Oven et al. (2011). The other primer sequences were kindly provided by Dr. Bo-wen CHENG (unpublished). These 12 markers defined 12 haplogroups, following the recommendation of Shi et al. (2005).
2.6. Statistical analysis
The frequencies of partial AZFc deletions, duplications, partial deletion sub-types, and the distribution of haplotypes between the cases and controls, were compared by Chi-squared test (P<0.05 was regarded as statistically significant) using the statistical package SPSS 10.0 (SPSS Inc.).
3. Results
3.1. Partial AZFc deletions
Based on the STS+/− analysis, gr/gr and b2/b3 deletions were detected in both infertile men and fertile control subjects, but no b1/b3 deletions were found (Figs. 1a‒1e and 2). We found 4 men in the azoospermia group and 13 in the severe oligozoospermia group with sY1291 absent (Table 1). Among the 17 gr/gr deletions in the infertile men, 15 were simple gr/gr deletions, 1 was gr/gr deletion with b2/b4 duplication (four DAZ copies and two CDY1 copies) and 1 was coupled with multiple b2/b4 duplications (six DAZ copies and three CDY1 copies) based on the DAZ and CDY copy analysis (data not shown). Thirteen fertile men were found with gr/gr deletions, all of which were simple gr/gr deletion (Table 1). The frequency of the gr/gr deletion did not show significant differences between the case and control groups.
Fig. 2.
Detection of the partial AZFc deletions with specific AZFc STSs
Marker: 100 bp ladder; Control: fertile man; Case 1: gr/gr deletion; Case 2: b2/b3 deletion; Case 3: sY1206 deletion
Table 1.
Frequencies of partial AZFc deletions
| Group | n | gr/gr deletion | b2/b3 deletion | sY1206 deletion |
| Fertile males | 153 | 13 (8.5%) | 13 (8.5%) | 0 (0.0%) |
| Azoospermia | 55 | 4 (7.3%) | 2 (3.6%) | 0 (0.0%) |
| Severe oligozoospermia | 169 | 13 (7.7%) | 12 (7.1%) | 1 (0.6%) |
| Infertile males | 224 | 17 (7.6%) | 14 (6.3%) | 1 (0.4%) |
Fourteen cases (2 in azoospermia and 12 in severe oligozoospermia), and 13 controls were found with b2/b3 deletion, among which 10 and 11 were simple deletions, respectively (Table 1). The others were coupled with b2/b4 duplications. No significant difference was found in this type of deletion between the infertile and fertile groups.
In addition, we found a new deletion type on a patient bearing severe oligozoospermia. In this case, sY1206 was negative while the other five sites were positive (Fig. 2, Table 1). To confirm the results and investigate the breakpoints, we tested other related STSs, including sY254, sY639, sY1054, and sY1125. All of these STSs were positive (data not shown), indicating that the breakpoints located between sY1206 and sY1054, namely between Y1 and Y2 (Fig. 1f) (Navarro-Costa et al., 2010a).
3.2. SNV analysis
We discriminated DAZ1/2 from DAZ3/4 and CDY1a from CDY1b by SNV analysis, which divided partial deletions into four sub-types: DAZ1/2+CDY1a, DAZ1/2+CDY1b, DAZ3/4+CDY1a, and DAZ3/4+CDY1b deletions (Figs. 1b‒1e). Among the 15 simple gr/gr deletions in cases, no DAZ3/4+CDY1a deletion was found, while 5 samples were found for the other three sub-groups, respectively (Table 2). The 13 gr/gr deletions in the control group were sub-grouped into DAZ1/2+CDY1a deletions (8) and DAZ3/4+CDY1b deletions (5) (Table 2). Five cases with the DAZ1/2+CDY1b deletion were identified, in contrast to none in the controls; however, the difference was not significant (P=0.064). All b2/b3 deletions in cases were grouped into DAZ3/4+CDY1b, while in the control group besides this type of deletion, one DAZ1/2+CDY1a deletion and one DAZ1/2+CDY1b deletion were found (Table 2). Case 3, with sY1206 missing, was sub-grouped into DAZ3/4+CDY1a deletion (data not shown).
Table 2.
Distribution of deletion types of DAZ and CDY1 in gr/gr and b2/b3 deletions
| Group | Simple gr/gr deletion |
Simple b2/b3 deletion |
||||||||
| n | DAZ1/2+CDY1a | DAZ1/2+CDY1b | DAZ3/4+CDY1a | DAZ3/4+CDY1b | n | DAZ1/2+CDY1a | DAZ1/2+CDY1b | DAZ3/4+CDY1a | DAZ3/4+CDY1b | |
| Fertile males | 13 | 8 | 0 | 0 | 5 | 11 | 1 | 1 | 0 | 9 |
| Azoospermia | 3 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 |
| Severe oligozoospermia | 12 | 4 | 4 | 0 | 4 | 9 | 0 | 0 | 0 | 9 |
| Infertile males | 15 | 5 | 5 | 0 | 5 | 10 | 0 | 0 | 0 | 10 |
3.3. DAZ and CDY1 gene dosages
Besides partial deletions, partial duplications are reported (Repping et al., 2003; Machev et al., 2004; Giachini et al., 2005). We analyzed the duplications by detecting the copy numbers of DAZ and CDY1 (Fig. 3). The partial duplications with six copies of DAZ and three copies of CDY1 were found in both infertile (9) and fertile (1) groups (Table 3), which showed significantly different frequencies (P<0.05). The frequency for this type of partial duplication is higher in the severe oligozoospermia group (4.7%) than in the azoospermia group (1.8%).
Fig. 3.
Examples of electrophoretograms showing different gene dosages for the DAZ/DAZL and CDY1/CDY2 genes
The peak areas reflect the dosage of PCR products of detected genes. Control: fertile man; Case 1: gr/gr deletion; Case 3: sY1206 deletion
Table 3.
Distribution of copy numbers of DAZ and CDY1
| Group | n | 6DAZ+3CDY1 | 4DAZ+2CDY1 | 4DAZ+1CDY1 | 2DAZ+2CDY1 | 2DAZ+1CDY1 |
| Fertile males | 153 | 1 (0.7%) | 127 (83.0%) | 1 (0.7%) | 0 | 24 (15.7%) |
| Azoospermia | 55 | 1 (1.8%) | 49 (89.1%) | 1 (1.8%) | 0 | 4 (7.3%) |
| Severe oligozoospermia | 169 | 8 (4.7%)* | 138 (81.7%) | 1 (0.6%) | 1 (0.6%) | 21 (12.4%) |
| Infertile males | 224 | 9 (4.0%)* | 187 (83.5%) | 2 (0.9%) | 1 (0.4%) | 25 (11.2%) |
P<0.05, compared to the “fertile males” group
In case 3, two copies of CDY1 were detected by dosage analysis, but only the CDY1b band was detected by SNV analysis, which was probably caused by gene conversion (Fig. 3). Only two copies of DAZ were detected, which turned out to be DAZ1/2 (Fig. 3).
3.4. Y chromosome haplogroups
It has been suggested that different haplogroups show diverse frequencies of partial AZFc deletions (Lin et al., 2007; Zhang et al., 2007). To investigate the haplogrouping of the samples in this study, 215 cases and 115 controls were tested and grouped by 12 Y chromosome markers (Fig. 4). The distribution of haplotypes was not significantly different between the cases and controls (Table 4). Partial AZFc deletions are obviously clustered in haplogroup K-R* (* indicates that the haplogroup may not be monophyletic). Eight cases out of 14 with simple gr/gr deletion and 9 cases out of 10 with b2/b3 deletion were found in 56 infertile members of haplogroup K-R*. Comparing to the control group, the 26 members of this group contributed 5 out of 10 gr/gr deletions and 9 out of 10 b2/3 deletions. Low frequencies of partial AZFc deletions were found in the other haplogroups. The distribution of partial AZFc duplications did not show significant differences between these haplogroups.
Fig. 4.
Phylogenetic tree of Y chromosome haplogroups
The asterisk (*) indicates that the haplogroup may not be monophyletic
Table 4.
Distribution of the subjects and partial AZFc deletions and duplications in Y chromosome haplogroups
| Group | n | C | D | K-R* | O* | O1a | O2a | O3* | O3a* | O3a3* | O3a3b | O3a3c* | O3a3c1 | Others |
| All | ||||||||||||||
| Fertile males | 115 | 12 | 2 | 26 | 4 | 6 | 16 | 3 | 17 | 5 | 3 | 8 | 8 | 5 |
| Infertile males | 215 | 19 | 6 | 52 | 7 | 10 | 22 | 5 | 30 | 11 | 6 | 16 | 19 | 12 |
| Simple gr/gr deletion | ||||||||||||||
| Fertile males | 10 | 0 | 1 | 5 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Infertile males | 14 | 1 | 1 | 8 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| Simple b2/b3 deletion | ||||||||||||||
| Fertile males | 10 | 0 | 0 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Infertile males | 10 | 1 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Partial duplication | ||||||||||||||
| Fertile males | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Infertile males | 9 | 2 | 0 | 2 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
The asterisk (*) indicates that the haplogroup may not be monophyletic
4. Discussion
Ethnic background is important for the association study between partial AZFc polymorphisms and male infertility. Different results have been observed in different ethnic populations (Navarro-Costa et al., 2010b). Yi ethnic minority, the 7th largest ethnic minority in China, has its own script and language which belongs to the Tibeto-Burman language group of the Chinese-Tibetan language family (Zhu et al., 2010). A total of 61% of Yi people reside in Yunnan Province (Zhu et al., 2010). In this study, we have focused on the Yi ethnic group by selecting cases as either non-obstructive azoospermia or severe oligozoospermia according to the WHO criteria. The controls are males who have given birth to at least one offspring without the help of assisted reproductive technology (confirmed by paternity test), but semen analysis was not performed. Thus, the control subjects are fertile but were not confirmed for normozoospermia. Our results show that the frequency of gr/gr deletion is 7.6% in cases, in contrast to 8.5% in controls, and the frequencies of the b2/b3 deletion are 6.3% and 8.5% respectively in these two groups (Table 1). The frequencies of gr/gr and b2/b3 deletions were similar to those in previous studies, but they did not differ significantly between the case and control groups, which agrees with most studies in East Asian population (Zhang et al., 2006) but conflicts with results published by Giachini et al. (2008) which have demonstrated the gr/gr deletion to be a significant risk for normal spermatogenesis in the Italian population using a similar methodology. This difference may be due to different genetic backgrounds, indicating a diverse sensitivity to the gr/gr deletion depending on genetic background. The haplogrouping results show that frequencies of gr/gr and b2/b3 deletions are low in the common East Asian group O* but significantly high in the group K-R*, which is consistent with previous reports (Table 4) (Lin et al., 2007; Zhang et al., 2007). These data suggest that the frequencies of partial AZFc deletion are significantly influenced by genetic background. Thus, haplotype match of cases and controls is of great importance in the association study on partial AZFc deletions and male infertility.
Besides the deletions occurring via homologous recombination between ampliconic sequences, deletions via non-allelic homologous recombination (NAHR) have been reported, and are due to the inherently unstable palindromic structure (Noordam et al., 2011). In this study, we found a new deletion type with the absence of sY1206 on a patient bearing severe oligozoospermia (Table 1). A similar deletion, named b3/b4 deletion, has been reported by Ferlin et al. (2005) as well as Choi et al. (2012). They speculated that the breakpoints of this type of deletion are located between sY1125 and sY1054 for the absence of sY1054 and presence of sY1125, and that this deletion removes the block Y1-g2-r3-r4-g3-Y2 including two copies of CDY1 and DAZ3/4. The breakpoints of case 3 seem to locate between sY639 and sY1206. This deletion removes g2-r3-r4-g3, including DAZ3/4 (Fig. 1f), which is consistent with the copy numbers of DAZ (two copies) and CDY1 (two copies). Since the breakpoints locate at palindrome 1, deletion in case 3 and b3/b4 deletion seem to occur via NAHR mechanism (Navarro-Costa et al., 2010a).
We analyzed partial duplications by detecting copy numbers of DAZ and CDY1. We found more cases with partial duplications in infertile men (4.0%) than in controls (0.7%) (Table 3). Similar results have been reported by Lin et al. (2007) and replicated in the China-Han population (Zhang et al., 2007). When comparing the fertile subjects to azoospermic or severe oligozoospermic patients, a significantly higher occurrence of partial duplications is observed only in patients with oligozoospermia, similar with what occurs with gr/gr deletion (Stouffs et al., 2011). That the distribution of partial AZFc duplications does not show significant differences between these haplogroups reduces the influence of genetic background bias between cases and controls. Partial duplications increase the dosage of contained genes, such as DAZ, encoding RNA-binding proteins, and CDY1, encoding a histone acetyltransferase. These proteins seem to be involved in gene/protein synthesis regulation. Thus, increased dosage of these genes may disrupt the normal expression of targeted genes, which may affect normal spermatogenesis.
Acknowledgments
These authors would thank Dr. Shuai LI (Security Department of Yunnan Province, Kunming, China) for collecting information of the cases, Dr. Bo-wen CHENG (Security Department of Yunnan Province, Kunming, China) for providing primer information, and Dr. Gianpiero CAVALLERI (Molecular and Cellular Therapeutics Department, Royal College of Surgeons in Ireland, Dublin, Ireland) for revising the manuscript.
Footnotes
Project (No. GREKF09-08) supported by the State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, China
Compliance with ethics guidelines: Jun-jie YE, Li MA, Li-juan YANG, Jin-huan WANG, Yue-li WANG, Hai GUO, Ning GONG, Wen-hui NIE, and Shu-hua ZHAO declare that they have no conflict of interest.
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000(5). Informed consent was obtained from all patients for being included in the study.
References
- 1.Bardoni B, Zuffardi O, Guioli S, Ballabio A, Simi P, Cavalli P, Grimoldi MG, Fraccaro M, Camerino G. A deletion map of the human Yq11 region: implications for the evolution of the Y chromosome and tentative mapping of a locus involved in spermatogenesis. Genomics. 1991;11(2):443–451. doi: 10.1016/0888-7543(91)90153-6. [DOI] [PubMed] [Google Scholar]
- 2.Choi J, Song SH, Bak CW, Sung SR, Yoon TK, Lee DR, Shim SH. Impaired spermatogenesis and gr/gr deletions related to Y chromosome haplogroups in Korean men. PLoS One. 2012;7(8):e43550. doi: 10.1371/journal.pone.0043550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, Garolla A, Engl B, Foresta C. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet. 2005;42(3):209–213. doi: 10.1136/jmg.2004.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, Garolla A, Lenzi A, Foresta C. Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab. 2007;92(3):762–770. doi: 10.1210/jc.2006-1981. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J, Rajpert de Meyts E, Skakkebaek NE, Habermann B, Krause W, Sousa M, et al. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod. 2002;8(3):286–298. doi: 10.1093/molehr/8.3.286. [DOI] [PubMed] [Google Scholar]
- 6.Giachini C, Guarducci E, Longepied G, Degl’Innocenti S, Becherini L, Forti G, Mitchell MJ, Krausz C. The gr/gr deletion(s): a new genetic test in male infertility? J Med Genet. 2005;42(6):497–502. doi: 10.1136/jmg.2004.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C. Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet. 2008;124(4):399–410. doi: 10.1007/s00439-008-0561-1. [DOI] [PubMed] [Google Scholar]
- 8.Kim B, Lee Y, Kim Y, Lee KH, Chun S, Rhee K, Seo JT, Kim SW, Paick JS. Polymorphic expression of DAZ proteins in the human testis. Hum Reprod. 2009;24(6):1507–1515. doi: 10.1093/humrep/dep032. [DOI] [PubMed] [Google Scholar]
- 9.Krausz C, Giachini C, Xue Y, O′Bryan MK, Gromoll J, Rajpert-de Meyts E, Oliva R, Aknin-Seifer I, Erdei E, Jorgensen N, et al. Phenotypic variation within European carriers of the Y-chromosomal gr/gr deletion is independent of Y-chromosomal background. J Med Genet. 2009;46:21–31. doi: 10.1136/jmg.2008.059915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29(3):279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 11.Lin YW, Thi DAD, Kuo PL, Hsu CC, Huang BD, Yu YH, Vogt PH, Krause W, Ferlin A, Foresta C, et al. Polymorphisms associated with the DAZ genes on the human Y chromosome. Genomics. 2005;86(4):431–438. doi: 10.1016/j.ygeno.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Lin YW, Hsu LCL, Kuo PL, Huang WJ, Chiang HS, Yeh SD, Hsu TY, Yu YH, Hsiao KN, Cantor RM, et al. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat. 2007;28(5):486–494. doi: 10.1002/humu.20473. [DOI] [PubMed] [Google Scholar]
- 13.Lu C, Zhang J, Li Y, Xia Y, Zhang F, Wu B, Wu W, Ji G, Gu A, Wang S, et al. The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population. Hum Mol Genet. 2009;18(6):1122–1130. doi: 10.1093/hmg/ddn427. [DOI] [PubMed] [Google Scholar]
- 14.Machev N, Saut N, Longepied G, Terriou P, Navarro A, Levy N, Guichaoua M, Metzler-Guillemain C, Collignon P, Frances A, et al. Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J Med Genet. 2004;41(11):814–825. doi: 10.1136/jmg.2004.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro-Costa P, Gonçalves J, Plancha CE. The AZFc region of the Y chromosome: at the crossroads between genetic diversity and male infertility. Hum Reprod Update. 2010;16(5):525–542. doi: 10.1093/humupd/dmq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro-Costa P, Plancha CE, Gonçalves J. Genetic dissection of the AZF regions of the human Y chromosome: thriller or filler for male (in)fertility? J Biomed Biotechnol. 2010;2010:936569. doi: 10.1155/2010/936569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noordam MJ, van Daalen SKM, Hovingh SE, Korver CM, van der Veen F, Repping S. A novel partial deletion of the Y chromosome azoospermia factor c region is caused by non-homologous recombination between palindromes and may be associated with increased sperm counts. Hum Reprod. 2011;26(3):713–723. doi: 10.1093/humrep/deq386. [DOI] [PubMed] [Google Scholar]
- 18.Pryor JL, Kent-First M, Muallem A, van Bergen AH, Nolten WE, Meisner L, Roberts KP. Microdeletions in the Y chromosome of infertile men. N Engl J Med. 1997;336(8):534–539. doi: 10.1056/NEJM199702203360802. [DOI] [PubMed] [Google Scholar]
- 19.Repping S, Skaletsky H, Brown L, van Daalen SKM, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JWA, Oates RD, Silber S, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35(3):247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 20.Repping S, van Daalen SKM, Korver CM, Brown LG, Marszalek JD, Gianotten J, Oates RD, Silber S, van der Veen F, Page DC, et al. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83(6):1046–1052. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Shi H, Dong Y, Wen B, Xiao C, Underhill PA, Shen P, Chakraborty R, Jin L, Su B. Y-chromosome evidence of Southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet. 2005;77(3):408–419. doi: 10.1086/444436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simoni M, Tüttelmann F, Gromoll J, Nieschlag E. Clinical consequences of microdeletions of the Y chromosome: the extended Münster experience. Reprod Biomed Online. 2008;16(2):289–303. doi: 10.1016/S1472-6483(10)60588-3. [DOI] [PubMed] [Google Scholar]
- 23.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423(6942):825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 24.Stouffs K, Lissens W, Tournaye H, Haentjens P. What about gr/gr deletions and male infertility? Systematic review and meta-analysis. Hum Reprod Update. 2011;17(2):197–209. doi: 10.1093/humupd/dmq046. [DOI] [PubMed] [Google Scholar]
- 25.van Oven M, Ralf A, Kayser M. An efficient multiplex genotyping approach for detecting the major worldwide human Y-chromosome haplogroup. Int J Legal Med. 2011;125(6):879–885. doi: 10.1007/s00414-011-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vog PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Köhn FM, Schill WB, Farah S, Ramos C, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5(7):933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 27.Wu B, Lu NX, Xia YK, Gu AH, Lu CC, Wang W, Song L, Wang SL, Shen HB, Wang XR. A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han-Chinese population. Hum Reprod. 2007;22(4):1107–1113. doi: 10.1093/humrep/del499. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Ma M, Li L, Su D, Chen P, Ma Y, Liu Y, Tao D, Lin L, Zhang S. Differential effect of specific gr/gr deletion subtypes on spermatogenesis in the Chinese Han population. Int J Androl. 2010;33(5):745–754. doi: 10.1111/j.1365-2605.2009.01015.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang F, Li Z, Wen B, Jiang J, Shao M, Zhao Y, He Y, Song X, Qian J, Lu D, et al. A frequent partial AZFc deletion does not render an increased risk of spermatogenic impairment in East Asians. Ann Hum Genet. 2006;70(3):304–313. doi: 10.1111/j.1529-8817.2005.00231.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Lu C, Li Z, Xie P, Xia Y, Zhu X, Wu B, Cai X, Wang X, Qian J, et al. Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet. 2007;44(7):437–444. doi: 10.1136/jmg.2007.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu B, Yang G, Shen C, Qin H, Liu S, Deng Y, Fan S, Deng L, Chen F, Zhang P, et al. Distributions of HLA-A and -B alleles and haplotypes in the Yi ethnic minority of Yunnan, China: relationship to other populations. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2010;11(2):127–135. doi: 10.1631/jzus.B0900232. [DOI] [PMC free article] [PubMed] [Google Scholar]