Abstract
Nontranslational functions of vertebrate aminoacyl tRNA synthetases (aaRSs), which catalyze the production of aminoacyl-tRNAs for protein synthesis, have recently been discovered. While these new functions were thought to be ‘moonlighting activities’, many are as critical for cellular homeostasis as the activity in translation. New roles have been associated with cytoplasmic forms as well as with nuclear and secreted extracellular forms that impact pathways for cardiovascular development, the immune response, and mTOR, IFN-γ and p53 signaling. The associations of aaRSs with autoimmune disorders, cancers and neurological disorders further highlight nontranslational functions of these proteins. Novel architecture elaborations of the aaRSs accompany their functional expansion in higher organisms and have been associated with the nontranslational functions for several aaRSs. While a general understanding of how these functions developed is limited, the expropriation of aaRSs for essential nontranslational functions may have been initiated by co-opting the amino acid binding site for another purpose.
Introduction
The aminoacylation reaction catalyzed by aminoacyl tRNA synthetases (aaRSs) fuses each amino acid to its cognate tRNA, in a reaction that requires amino acid activation through condensation of the amino acid with ATP to form an aminoacyl adenylate. The activated amino acid is then transferred to the 3′-end of the cognate tRNA1,2. Because tRNAs harbor the anticodon triplets of the genetic code, the specific aminoacylations catalyzed by the synthetases establish the rules of the universal code. Thus, aaRSs are essential components for protein synthesis in every living species (Box 1).
Box 1.
Canonical function of aminoacyl-tRNA synthetases: aaRSs activate amino acids by using ATP to catalyze formation of a high energy aminoacyl-adenylate (AA-AMP) intermediate. In a thermodynamically favorable reaction, the high energy mixed anhydride is then condensed with the 2′- or 3′-OH of the bound cognate tRNA to form an aminoacyl ester.
| (1) |
| (2) |
Given their central role in translation, it came as a surprise to learn that most of the synthetases have novel and critical functions in higher organisms. These functions are elicited not only in the cytoplasm, where translation occurs, but also in the nucleus and outside the cell. Moreover, some of these novel activities come from resected forms, where an alternative splice variant, or natural proteolytic fragment, harbors the non-translational activity (which is inactive in the native synthetase). The associations of synthetases with specific diseases has also stimulated investigations of disease mechanisms, and whether they arise from one of the non-translational functions. Here we summarize and highlight recent discoveries, discuss their conceptual implications, and consider their potential clinical utility. Taken together, these advances have established new paradigms that are likely to guide future research on aaRSs, as well as the development of new diagnostic and therapeutic strategies for treating human diseases.
Non-translational functions of aaRSs in lower organisms
The early work in lower organisms mostly illustrated how the connection of synthetases to nucleic acid binding was diversified to regulatory functions associated with control of translation and transcription. In both bacteria and eukaryotes, mRNAs of certain aaRSs encode short sequences that fold into the cloverleaf shape to mimic the cognate tRNAs3. When the aaRS protein is excess, binding of the “extra” aaRS to a tRNA-mimicking element blocks translation of the synthetase’s mRNA and thus keeps the specific aaRS at an optimal level. Similar regulation also occurs at the level of transcription. For example, E. coli AlaRS binds a DNA palindrome imbedded in the promoter of its gene and thereby represses transcription4. In a different vein, the broad connection of synthetases to nucleic acids is also seen in the examples of fungal mitochondrial aaRSs (S. cerevisiae mt-LeuRS and Neurospora crassa mt-TyrRS) participating in splicing of group I intron-containing pre-mRNAs5,6. These examples suggest that, in early evolution more than a billion years ago, selective pressures expropriated synthetases for new functions, perhaps leveraging off their original partnership with tRNAs. After establishing the specific precedents for repurposing synthetases, and as the Tree of Life was further ascended to engender metazoans, vertebrates, and mammals, the complexity associated with higher life forms was mirrored in the complexity and diversity of non-translational functions acquired by tRNA synthetases. This progressive accretion of new activities is suggestive of a key role of aaRSs in the etiology of the Tree of Life. Summarized below are specific examples of some of these novel functions of tRNA synthetases in vertebrates that include the fish and mammals.
The proliferation of functions of aaRSs in animals
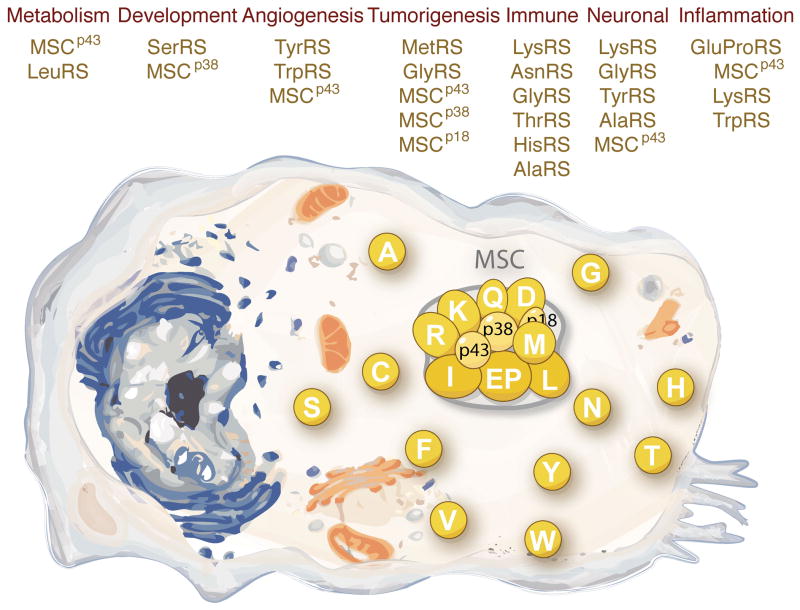
The full elaboration of non-translational functions of aaRSs is seen in the animal kingdom. These elaborations go far beyond a simple extension of the nucleic acid binding properties of synthetases, and typically require new sequences that have expanded the size and decorated the overall architecture of the proteins. In addition, the novel activities often are unmasked from a native molecule by the creation (such as through natural proteolysis or alternative splicing) of a fragment. These novel activities include but are not limited to: (1) mediation of glucose and amino acid metabolism; (2) regulation of the development of specific organs and tissues; (3) control of the ying-yang balance of angiogenesis for the vasculature; (4) triggering or silencing of inflammatory responses; (5) control of cell death and stress responses that may lead to tumorigenesis; and, (6) amplification or inhibition of the immune response (Figure 1).
Figure 1. The non-translational functions of aaRSs.
To a first approximation, aaRSs are organized in two groups in the cytoplasm of higher eukaryotes. Some are in a free form, while others are part of a high molecular weight multi-tRNA synthetase complex (MSC), which includes 3 scaffold proteins designated as MSCp43, MSCp38, and MSCp18. These proteins not only are an essential part of the translation apparatus, but also have a myriad of cytoplasmic, nuclear and extra-cellular functions. For simplicity, the various synthetases are designated by single letters, using the standard abbreviations for amino acids.
Amino acid binding sites link aaRSs to signaling pathways
In the development of the genetic code, during the transition from the RNA world to the theatre of proteins, the amino acid recognition properties of RNA aptamers were eventually assumed by the active sites of early tRNA synthetases. To acquire the needed specificity, aaRSs shaped their amino acid (AA) binding pockets to sterically fit with specific side chains and reinforced that fit with polar and apolar interactions. As perhaps the earliest highly specific AA binding sites, they were available for expropriation for other activities. The amino acid binding site also developed a crucial role in local signal transduction events within the aaRSs themselves, via reactions initiated by AA/ATP binding and activation, which lead to conformational changes for docking and dissociating tRNA7,8. Moreover, amino acids and related metabolites have been identified as central to cancer cell proliferation, metastasis, transformation and stem cell proliferation9–11. Thus, the recent discoveries of the role of the amino acid binding site in signal transduction events associated with non-translational functions of tRNA synthetases has a logical connection to the pre-existing properties of this amino acid detection and signaling system (Figure 2).
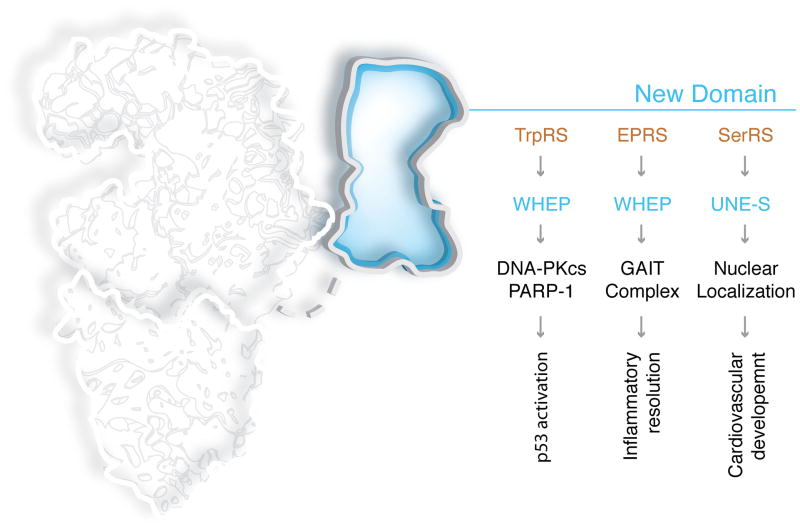
Figure 2. Amino acid binding pocket has a vital role in some of the non-translational functions.
Higher eukaryote tRNA synthetases are comprised of a catalytic domain (which is highly conserved through evolution), an RNA binding domain (for some synthetases, this domain recognizes the tRNA anticodon), and a new domain that is absent in lower organisms. The ancient amino acid-binding pocket of several aaRSs is essential for some of the non-translational functions. Ap4A: diadenosine tetraphosphate; VE-cad: vascular endothelial cadherin; RagD: small guanosine triphosphatase D; ASK1: apoptosis signal-regulating kinase 1.
Leucine-dependent TORC1 signaling through LeuRS
Emerging evidence indicates that the precisely controlled amino acid binding pockets of aaRSs also serve as intrinsic sensors of amino acid levels that are monitored to regulate homeostasis (a balanced and stable state of cellular and organismal functioning) associated with nutritional intake (Figure 2). The TOR pathway is among the most significant for relaying signals that are sensitive to nutritional status12. This pathway is regulated by two multi-protein signaling complexes known as TORC1 and TORC2. Under amino acids starvation, TORC1 becomes inactive and mRNA translation is blocked at the initiation step. Significantly, two recent studies showed that LeuRS is a leucine sensor that signals to TORC1 in both yeast and mammalian cells13,14.
Specifically, in human cells, cytoplasmic LeuRS promotes translocation of mammalian TORC1 (mTORC1) to the lysosomal membrane and activates it by binding to the RagD GTPase component of mTORC113. The association of LeuRS with the lysosomal membrane and its interaction with RagD is strictly regulated by levels of leucine. Disruption of the leucine-binding site abolishes the ability of LeuRS to bind RagD and renders the mTORC1 pathway insensitive to intracellular levels of leucine. Interestingly, LeuRS interacts with RagD through its C-terminal domain, which is far from its amino acid binding site in the 3-D structure. This observation suggests that a domain-domain conformational change is operating under the control of leucine. Although the exact mechanism of leucine activation of LeuRS to regulate the mTORC1 complex remains to be determined, it appears that the co-binding of ATP, and possibly the formation of the leucyl-AMP, is the critical step for triggering the conformational change required for the RagD interaction13.
Glutamine-regulated ASK signaling through GlnRS
Because AAs are not only nutrients for cell growth but also regulators of cell signaling and metabolism, control of cancer growth and metastasis is thought to require sensing of amino acid levels15,16. As an example, glutamine contributes both to building proteins in a dividing cell and to control of redox potentials through the synthesis of NADPH. It also serves as a major cancer cell anabolic substrate that regulates cell survival, signal transduction and autophagy17,18. The increased glutamine uptake and metabolism observed in many tumor cells leads to a glutamine-addictive state that is, at least in part, regulated by the oncogenic transcription factor Myc19. Interestingly, GlnRS suppresses the pro-apoptotic activity of the apoptosis signal-regulating kinase 1 (ASK1) that can control tumorigenesis and can facilitate stress responses triggered by TNFα, oxidative insult, or calcium influx20. This suppression requires the binding of glutamine to the catalytic site of GlnRS. Thus, GlnRS functions as a direct link between the anti-apoptotic role of glutamine and the control of the activity of ASK1. Consistently, deprivation of glutamine appears to activate ASK1-induced apoptosis, at least in HEK293 cells21.
The novel functions engendered by the AA sensor roles of GlnRS and LeuRS are logical expansions of the use of pre-existing AA bindings. However, the sensors do not act in isolation, but rather require the partial reshaping of the synthetase itself to connect with a new signaling pathway. Human LeuRS has added a sequence motif that is for interaction and signaling with the RagD GTPase. Thus, while the tRNA synthetases are a logical place in evolution to link signal transduction pathways of higher organisms to AA levels, they eventually need further modification to intervene in the signaling pathway that is specific to the AA (Figure 2).
LysRS as a regulator of the immune response
The activation of amino acids by condensation with ATP is the hallmark of aaRSs. While the activated amino acid can then be transferred to the cognate tRNA, it can also be released as a ‘leaving group’ when the bound AA-AMP reacts with a second molecule of ATP, to produce Ap4A. Although this Ap4A-producing reaction has long been associated with a subset of synthetases, it appears to be most robust with LysRS22. Characterized more than 30 years ago as an ‘alarmone’ in bacterial, yeast and mammalian cells23,24, detailed understanding of at least one of the functions of Ap4A emerged only recently. In mast cells, immune-stimulation results in a rise in the level of Ap4A which, in turn, promotes transcription of microphthalmia associated transcription factor (MITF) target genes that regulate the immune response25. This rise in Ap4A results from antigen activation that triggers the MAPK cascade to phosphorylate LysRS, which promotes its translocation to the nucleus26 and enhances its activity for Ap4A synthesis27. Interestingly, the phosphorylation event also inhibits the aminoacylation activity of LysRS. Thus, the synthetase is ‘switched’ from a translational to a transcriptional function27. Though the exact mechanism of how LysRS is so potent in Ap4A production remains to be determined, the AA binding site appears to be required to produce Ap4A28. Thus, LysRS is another example where the AA binding site of an aaRS serves a critical role in enabling a novel function.
Anti-angiogenesis function of TrpRS by inhibiting VE-cadherin
Extending from the utilization of the ancient AA binding sites for sensing amino acids, the amino acid-binding pockets can also play direct roles in docking target proteins. For example, the lysine-binding pocket of histone modification enzymes can interact with histone subunits and thus read and write the ‘histone code’29. In these cases, protein docking interactions can be mediated through the recognition of protruding side chains of target proteins that fit into the AA binding pockets of aaRSs. After the interaction with a potential partner is initiated in this way, during evolution it can be further anchored by making complementary surface adaptations in each partner.
The strategy is used by natural fragments of TrpRS, which are produced either by alternative splicing or natural proteolysis. These extracellular N-terminal fragments of TrpRS-- designated as TrpRSAct for simplicity –are potent anti-angiogenesis factors that act on VE-cadherin, an adhesion protein whose extracellular EC1 domains self-associate to form blood vessel tubes. The self-association is through a lock-and-key fitting of two protruding N-terminal Trp side chains of one EC1 with the Trp binding pockets of the adjacent EC130. During neovascularization, TrpRSAct interferes with the assembly of the network of EC1 interactions, by capturing the protruding Trp side chains of the EC1. One of the two Trp’s of an EC1 is docked into the Trp binding pocket of TrpRS, while the other is docked into the adenine-binding pocket of the adjacent ATP interaction site. Importantly, native TrpRS cannot interact with VE-cadherin, because the Trp binding pocket is partly covered by the segment that is removed by alternative splicing or natural proteolysis. Thus, the potent extracellular anti-angiogenic activity of TrpRS is masked in the native molecule.
New domains introduce more non-translational functions
Although the amino acid binding pocket can play a major role in endowing an aaRS with a new protein-protein interaction, the repertoire of interactions was expanded greatly through the progressive (in evolution) and accretive addition of new domains to aaRSs. Many of these domains are themselves dispensable for aminoacylation and capture partners by interactions that are outside of the AA binding pockets. When contrasted with other ancient protein families, such as ribosomal- or amino-acid binding-proteins, the progressive decoration of aaRSs with new domains (as the Tree of Life is ascended) appears unique. Interestingly, these new structural units-- such as leucine zippers and GST domains--are designed for protein-protein interactions. These domain additions typically correlate with major transitions in the Tree of Life, such as the invertebrate to vertebrate transition31.
WHEP domain of nuclear TrpRS links IFN-γ signaling
In addition to the AA binding pocket of TrpRSAct that was exploited for an anti-angiogenesis function, TrpRS also illustrates how a novel domain addition enables a new set of interactions that connect to a major signal transduction cascade. A helix-turn-helix WHEP domain (named after a subset of those synthetases that harbor this domain (TrpRS(W), HisRS(H) and GluProRS(EP)) was added to TrpRS at the time of insects and thereafter retained32. Deletion of the WHEP domain has little effect on the aminoacylation activity of human TrpRS33. Recent work showed that the mechanics of IFN-γ induced anti-proliferation is strongly correlated with an increase in nuclear TrpRS. Through the appended WHEP domains on the homodimeric enzyme, nuclear TrpRS bridges DNA-dependent protein kinase (DNA-PKcs) to poly(ADP-ribose) polymerase 1 (PARP-1) to activate p5334. Interestingly, the high-resolution crystal structure of human TrpRS showed that accessibility to the WHEP domain is regulated by the occupancy of the Trp-binding site. In fact, administration of a tight-binding non-hydrolysable Trp-AMP analog disrupted the ternary complex of TrpRS and DNA-PKcs and PARP-1, and thereby prevented IFN-γ triggered activation of p53. Therefore, through TrpRS, tryptophan may have a role in regulating p53 activation.
WHEP domains of EPRS enable translational silencing
EPRS (Glu-ProRS) is the only bifunctional aaRS that is encoded by a gene that fuses together the coding regions for two different tRNA synthetases. Three successive WHEP domains serve as the linker between GluRS and ProRS in human EPRS35. Though the reason for the fused synthetases is not known, the WHEP domain plays a critical role in recruiting partner proteins to associate with the synthetases. EPRS is a component of the higher eukaryote multi-tRNA synthetase complex (MSC), which is comprised of nine tRNA synthetases and three scaffold proteins36. In human monocytes, IFN-γ stimulation triggers phosphorylation on the WHEP domain linkers and releases EPRS from the MSC. Subsequently, EPRS binds to three other protein partners including NSAP1, L13a and GAPDH37, and the newly formed heterotetrameric GAIT (gamma-interferon activated inhibitor of translation) complex binds ‘GAIT elements’ in the 3′ UTRs of a specific set of mRNAs and thereby blocks their translation initiation38. Detailed studies showed that, in the linker, the first two WHEP domains direct high-affinity binding of EPRS to specific GAIT elements on the target mRNAs, while the second and third WHEP domains interact with the other members of the GAIT complex, and thereby implement GAIT element-specific translation suppression39. Here again, the WHEP domains appear to be versatile adaptors that have an essential role in enabling a non-translational function (Figure 3).
Figure 3. New domains that introduce and regulate non-translational functions.
aaRSs appear to be unique in the degree to which they acquired new domains that are not essential for catalytic activity. In addition to the common leucine zipper and GST domains, most of the novel domains are restricted to the aaRS family and are not found in other proteins. Shown here are two of the domains that are unique to vertebrate aaRSs and their related functions in inflammation, development and angiogenesis. DNA-PKcs: DNA-dependent protein kinase, catalytic subunit; PARP-1: poly [ADP-ribose] polymerase 1; GAIT complex: gamma-IFN-activated inhibitor of translation (GAIT) complex.
UNE-S domain of SerRS is essential for development of closed circulatory system
Early genetic screens in zebrafish embryos identified SerRS as essential for development of the closed circulatory system40,41. These studies also showed that this surprising role for SerRS was not associated with its aminoacylation activity. Interestingly, in the transition from the open circulatory system of invertebrates to the closed system of vertebrates, a unique 45 amino acid domain, annotated as UNE-S (unique to SerRS), was added to SerRS. UNE-S encodes a nuclear localization signal that is absent in invertebrate SerRSs42. Mutational analysis, and RNAi knockdown experiments, showed that UNE-S is dispensable for aminoacylation but is essential for vascular development. This role for UNE-S is associated with conferring nuclear localization of SerRS and with transcriptional regulation of the VEGF-A signaling pathway.
Additional novel domains added to higher eukaryote aaRSs
Including the UNE-S and WHEP domain, about 20 domains altogether have been joined to higher eukaryote tRNA synthetases. These include, among others, 8 UNE domains (unique to aaRSs but not found in other proteins) and the aforementioned leucine zipper and GST domains31. While they are believed to be largely dispensable for aminoacylation, most have not been assigned to specific functions. In some instances, the initial addition of domains to aaRSs may have resulted from their ability to enhance the canonical aminoacylation activity (such as the ‘N-helix’ of LysRS43 and of AspRS44), or to facilitate association and efficient transfer of charged tRNA (such as the GST domain of ValRS that binds to the elongation factor EF-1α45), or the extensions that associate with the MSC (such as in the GST domain of MetRS46). However, these sequence additions may have later evolved for broader functions. In several instances, such as the UNE-S or WHEP domain additions, there is no obvious connection to the enhancement of aminoacylation or to transfer of charged tRNA. The systematic survey of phenotypes in animal models with ablations of appended domains will greatly advance understanding of the non-translational function of aaRSs.
Multifaceted aaRSs scaffold proteins
Higher eukaryotes, starting from insects, evolved a large molecular weight multi-tRNA synthetase complex (MSC) comprised of nine tRNA synthetases and three scaffold proteins known as MSCp43, MSCp38, MSCp18 (also known as AIMP1, AIMP2, AIMP3)47,48. The three scaffold proteins facilitate the assembly of the MSC through interactions with each other and with specific aaRSs, and for this reason are irreplaceable for complex assembly and for the functions of the MSC49–51. The assembly of the MSC appears to facilitate protein synthesis through channeling of tRNAs between aaRSs, initiation/elongation factors, and the ribosome52,53. In addition, these aaRS-related scaffold proteins have broad non-translational functions (Figure 4).
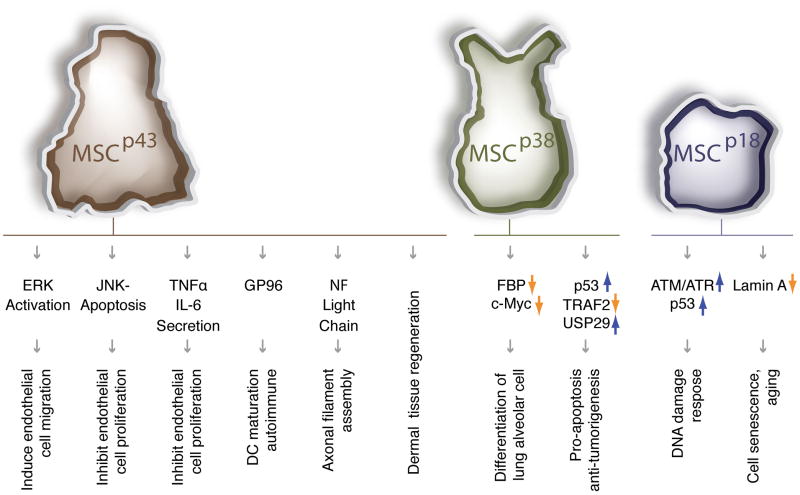
Figure 4. Multifaceted aaRSs scaffold proteins.
The three scaffold proteins that are required for the assembly of the MSC have diverse functions outside of the MSC.
TNFα: Tumor necrosis factor-alpha; IL-6: interleukin-6; GP96: 96-kDa glycoprotein; NF: neurofilament; FBP: fuse-binding protein; TRAF2: TNF receptor-associated factor 2; USP29: ubiquitin specific peptidase 29; ATM/ATR: ataxia telangiectasia mutated (ATM) and ATR (ATM and Rad3-related) protein kinases.
MSC component MSCp43 is also a secreted cytokine
Although MSCp43 interacts with ArgRS and MSCp38 in the MSC, it is also secreted as a cytokine controlling angiogenesis, immune responses, tissue regeneration and glucose homeostasis54. These functions of MSCp43 appear to vary depending on its cellular location. Among many experiments that define the roles of MSCp43, genetic depletion of MSCp43 showed prominent motor neuronal degeneration55. In addition, MSCp43 knockout mice also showed retardation of wound healing, and decreased fibroblast proliferation and collagen synthesis. These observations suggest that MSCp43 is critical for dermal tissue regeneration56.
MSCp38 is essential for lung development
MSCp38 is at the core for the MSC, where it interacts with LysRS, GlnRS, AspRS, EPRS, and IleRS, as well as with MSCp43 and MSCp18, 49,57. MSCp38 depletion leads to the complete disruption of the MSC, and destabilization of most of the MSC components57. Despite the essential role in assembly of the complex, MSCp38 also exists as a free form (including a nuclear distribution) having critical roles in development and tumorigenesis. MSCp38 mediates several anti-proliferative and pro-apoptotic signaling pathways related to FBP (FUSE-binding protein), Myc, TRAF2 and p53. For example, mice lacking MSCp38 are neo-natal lethal, mainly owing to severe over-proliferation of epithelial cells in the lung58. Free MSCp38 exerts its anti-proliferative activity by binding and promoting the ubiquitination of FBP, which down-regulates c-Myc and the differentiation of functional alveolar cells58. In addition, reduction of MSCp38 levels in heterozygous MSCp38 +/− mice elevated susceptibility to formation of various tumors in the lung, skin and colon59. These effects are, at least in part, associated with MSCp38′’s critical role as a regulator of ubiquitin delivery to its target proteins. Interestingly, MSCp38 itself is a substrate of the E3 ubiquitin ligase Parkin60.
MSCp18 is essential for DNA damage response
MSCp18 contains a sole GST domain and as such is the smallest component of the MSC. It associates with the similar GST domains of MetRS and MSCp38. UV irradiation-induced phosphorylation of MetRS causes release of MSCp18 from the MSC to the nucleus61. Other stresses—such as DNA damage—also induce nuclear translocation of MSCp18. In the nucleus, MSCp18 activates the ATM/ATR kinases and elevates the level of p53 so as to work against DNA damage62. In addition, depletion of MSCp18 blocked p53 induction and, as might be expected, low MSCp18 expression, or mutations in humans that ablate its interaction with ATM/ATR, are found in human cancers63.
Unanswered questions about the MSC scaffold proteins
The diverse functions of the three MSC scaffold proteins suggest that they were invented as critical hubs of protein-protein interactions, both within and beyond the MSC. One question is whether binding partners outside of the MSC evolved in a way that exploited the same protein-binding interfaces used in the MSC. Another question is how these scaffold proteins can be released from the MSC, under specific stress conditions, without causing disruption of the MSC. Possibly, only a sub-stoichiometric amount is released or a reshuffle of the MSC architecture occurs. A third issue is whether the three MSC scaffold proteins, which in addition to having contacts with specific synthetases and interact with each other, are designed to act cooperatively, such as in response to certain stress conditions like DNA damage. A high-resolution structure of the MSC, and more work on the side of functional and genetic analysis, would provide a foundation for developing answers to some of these questions.
Fragments of aaRSs introduce non-translational functions
Increasingly, examples are being uncovered of synthetases that have been reconfigured through natural proteolysis or alternative splicing. One of the most dramatic recent examples is HisRS, which is a prominent antigen in the chronic inflammatory conditions of polymyositis and interstitial lung disease, and which may be directly associated with the etiology of the disease64,65. An expressed alternative splice variant (HisRSδCD) ablates the entire catalytic domain, and links together N-terminal and C-terminal polypeptides to form a well organized 3-dimensional structure66. Lacking a catalytic site for aminoacylation, this splice variant is designed for repurposing HisRS from translation to a novel function. Because it harbors epitopes associated with polymyositis, HisRSδCD is being investigated for its mechanistic significance.
More generally, several examples of splice variants and products of natural proteolysis have been investigated for their functional significance. These examples probably represent a small part of the universe of alternative forms of tRNA synthetases that are active in non-translational pathways.
A natural fragment of TyrRS that is active in inflammatory pathways
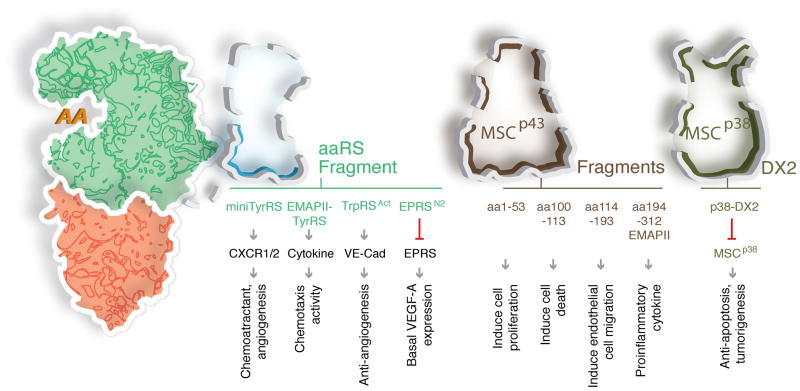
Endothelial monocyte-activating polypeptide II (EMAPII), the C-terminal portion of MSCp43, was initially discovered as a tumor-secreted proinflammatory cytokine and as an anti-angiogenic factor in tumor vascular development67. Later work showed that EMAPII is released from the MSC after cleavage of MSCp43 by proteases such as caspase 768. Interestingly, mammalian TyrRS contains a closely homologous EMAPII domain at the C-terminus. Under specific conditions, TyrRS is secreted and leukocyte elastase splits the protein into a free EMAPII domain and a second N-terminal part known as mini-TyrRS. Both released proteins are active in distinct cell-signaling pathways. For example, among other functions, mini-TyrRS induces the migration of polymorphonuclear leukocytes by binding to and activating CXCR1,2 receptors. Strikingly, the activities of its two imbedded cytokines are lacking in native full-length TyrRS (Figure 5).
Figure 5. New functions of aaRS fragments.
Several aaRSs and MSC scaffold proteins are resected by proteolysis, alternative splicing, or alternative polyadenylation. Depicted here are examples where fragmentation activates a novel non-translational function (TyrRS, TrpRS), and a case where multiple fragments are generated from a single protein, with each fragment having a distinct activity (MSCp43). In two examples, to achieve a homeostatic balance of downstream signaling, a novel activity associated with the full-length protein is inhibited by a fragment (EPRSN2, MSCp38-DX2) of the same protein (EPRS, MSCp38). CXCR1,2: C-X-C chemokine receptor type1, 2.
Splice variant of TrpRS is a potent angiostatic factor
As stated above, the splicing- or proteolytic-directed removal of the N-terminal WHEP domain unmasks the angiostatic activity of TrpRS, which is absent in the native enzyme. Extracellular TrpAct binds to protruding N-terminal Trp side chains on the extracellular domains of VE-cadherin and thereby blocks formation of nascent blood vessels. With native TrpRS, the WHEP domain blocks access of the Trp binding pocket to the protruding side chains of VE-cadherin69. In animal models for macular degeneration and cancer, TrpAct demonstrated potent activity in arresting these pathologies70,71.
Homeostatic mechanism of action for a C-terminal truncated EPRS
Recent work identified a new mechanism of homeostatic regulation by EPRS in conjunction with a specific natural fragment. IFN-γ treatment of myeloid cells releases EPRS from the MSC to form the GAIT complex for translational silencing of mRNAs encoding proinflammatory proteins including VEGF-A38. However, VEGF-A is essential for vessel maintenance and its deletion results in accelerated tumor growth. By a novel polyadenylation event that introduces a new stop codon, a C-terminal truncated version EPRS (EPRSN1) shields the mRNA of VEGF-A from the GAIT complex and a ‘homeostatic’ basal level of expression of VEGF-A can thus be maintained72.
Upregulation of a splice variant of MSCp38 in cancers
A similar example of a homeostatic role was also established for MSCp38. MSCp38 (AIMP2) showed a tumor suppressive function through its interactions with p53 and TRAF273,74. A splice variant known as DX2 lacks the segment needed for the interaction of MSCp38 with the MSC and thus exists as a free form. Through its GST domain, DX2 competes with native MSCp38 and abrogates its tumor suppressive interaction with TRAF2, thus compromising the TNFα-dependent pro-apoptotic activity of MSCp38 in ovarian cancer75. Similarly, through its GST domain, DX2 also inhibits the tumor suppressive activity of MSCp38 through competitive binding to p5376. Mice constitutively expressing this variant showed increased susceptibility to carcinogen-induced lung tumorigenesis, while suppression of this variant slowed tumor growth. Importantly, in non-small cell lung cancer patients, the higher expression of this variant is also correlated with a lower survival76.
Unknown functions of pathological aaRS mutants
Ataxia and neurodegeneration arise from a mild editing defect (2-fold reduction) in murine AlaRS77. The editing activity, which enhances the accuracy of aminoacylation so as to prevent mistranslation, is considered part of the canonical function of an aaRS in translation. (Stronger mutations in the site for editing are lethal, even for bacteria78.) However, a new class of mutations has been discovered in the human population that also leads to a neuropathy phenotype. Altogether 20 mutations in 4 human aaRSs (GlyRS (11), TyrRS (4), LysRS (3) and AlaRS (2)) have been causally associated with Charcot–Marie–Tooth (CMT), the most common heritable peripheral neuropathy79,80. Typically, the mutations are dominant, which suggests that the mutations confer a gain-of-function. Additionally, or alternatively, the mutations may enhance a pre-existing non-translational function, so that the enhanced function or interaction (such as a tighter binding (than normal) to a neuronal cell receptor) results in dis-regulation or enhancement of a normal non-translational role of the synthetase. Importantly, because some of the CMT-causing mutations do not affect the aminoacylation activity81,82, and because studies in the heterozygous GARS mouse showed that haplo-insufficiency in protein synthesis did not cause the CMT-like phenotype83,84, a defect in protein synthesis is less likely as the cause of CMT for many of the mutants. In addition, expression of a CMT-causing mutant GlyRS in motor neurons of a CMT-mouse did not promote formation of protein aggregates, such as aggregates of GlyRS or of other misfolded proteins84. Because of the differences of the mouse model, such as the lifespan and axon lengths, it remains to be determined if similar effects would occur with the analogous mutation in GlyRS in humans.
Other evidence showed that specific conformational change was a common feature of the tested CMT mutations in GlyRS85. This result suggests that a neomorphic form is responsible for the gain-of-function phenotype. These and other studies of the etiology of CMT that is caused by mutations in aaRSs offer promise for potentially linking aaRSs to neuronal development and homeostasis. In addition, by studying the mechanism of aaRS-associated CMT, new players and pathways in neurogenesis will likely be uncovered.
Emerging Clinical Opportunities
Prognostic Biomarkers
A number of biomarkers, such as used in the MammaPrint and Oncotype DX assays, have entered into clinical use for cancer treatment. These assays employ multigene expression signatures to estimate the likelihood of disease progression and recurrence, and provide guidance for treatment options86,87. However, because current assays are principally informative for the prognosis of patients within only certain subclasses of tumors, expansion of prognostic signatures to more diseases is needed. aaRSs and associated proteins appear to have critical mechanistic roles in a variety of cellular processes that are relevant for disease development/pathology, and these roles may be used as one possible avenue for improvement of diagnostics. For example, under conditions of hypoxia, TrpRS expression is down-regulated in cells from patients with colorectal cancer or metastatic pancreatic cancer, suggesting TrpRS as a useful marker for cancer88,89. Interestingly, a clinical study of patients with colon cancer showed a statistically significant correlation of levels of TrpRS with cancer progression, namely, a higher level of TrpRS was associated with a slower progression. As another example, GlyRS and MetRS are over-expressed in cells from patients with breast and colon cancers90. Because GlyRS is an active anti-tumor agent (see below), its overexpression in certain cancers may be part of an innate anti-tumor response.
In addition, the levels of several anti-aaRSs antibodies (including AlaRS, AsnRS, GlyRS, HisRS, IleRS and ThrRS) are correlated with 25% of the patients with polymyositis or dermatomyositis and, were identified in 38% of patients with type 1 diabetes mellitus91–93. In fact, these extracellular autoantigenic aaRSs possess chemoattractant activities towards several chemokine receptors and may lead to tissue specific immune-mediated pathologies75. Upon exposure to toxic chlorobenzene-like compounds, SerRS is one of 28 proteins whose expression is highly altered in apoptotic human lung epithelial cells94. Also, all three MSC scaffold proteins (MSCp43, MSCp38 and MSCp18) are down-regulated in gastric and colorectal cancer, and differential expression of MSCp43 has been suggested to be an injury-specific biomarker in the brain95,96. Collectively, these studies suggest the potential utility of monitoring aaRSs as biomarkers for different pathologies.
Therapeutic Agents
As a result of encouraging animal studies, aaRSs are now considered as potent therapeutic agents for a variety of disease indications. For example, in addition to the aforementioned single-agent applications with TrpRSAct, the combination of TrpRSAct with a VEGF-directed aptamer completely and synergistically inhibited tumor angiogenesis in a brain tumor model70. Several additional studies also demonstrated the potential for aaRS-based therapeutic agents in cancer (Figure 6). Secreted GlyRS binds to K-cadherin on ras-activated tumor cells and promotes the de-phosphorylation and de-activation of ERK. IP administration of GlyRS protein to colon-cancer-bearing mice strongly suppressed tumor formation97. In addition, most amino acids are highly consumed in cancer cells, which may reflect a metabolic vulnerability that could in principle be targeted through aaRSs, especially in rapidly proliferating cells9,10. As another example, in various xenograft tumor models, including pancreatic tumors, gliomas and melanomas, EMAPII generated from MSCp43 suppressed primary and metastatic tumor growth67,98. Other fragments of MSCp43 that exhibit different activities are secreted under stress conditions and may be candidates for additional therapeutic interventions99.
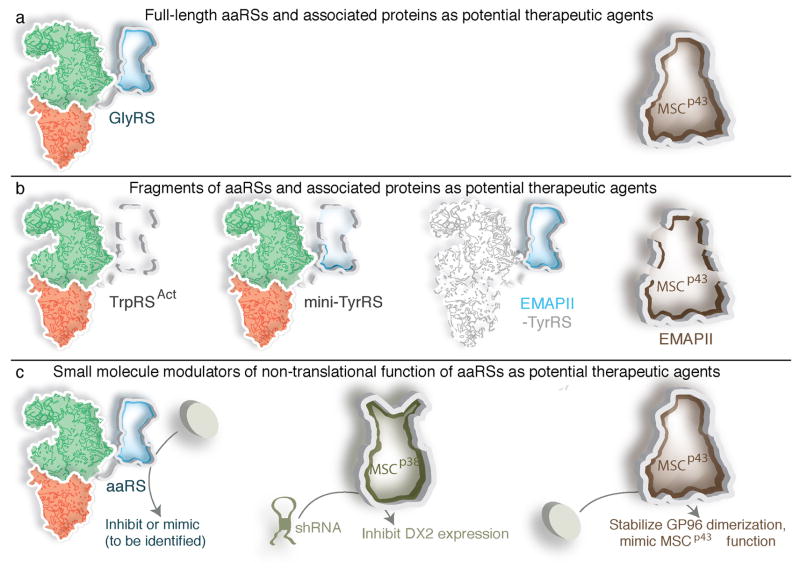
Figure 6. Potential therapeutic interventions derived from aaRS non-translational functions.
Animal models have established the potential therapeutic value of some of the aaRS -associated activities.
(a) Administration of native full-length GlyRS or of MSCp43 triggers a measurable response in tumor environments, where the proteins were initially discovered to be secreted as anti-tumor cytokines.
(b) Fragments of aaRS and MSC scaffold proteins that display non-translational functions are potent in various disease model settings. The parts deleted in fragmentation are shown as light shadows and outlined in dashes. MSCp43 has multiple fragments that each has a distinct activity.
(c) Small molecules that inhibit or mimic the non-translational functions of aaRSs and associated proteins may have clinical utility. Compounds displaying such efficacy are now being reported, namely mimetics of MSCp43, and MSCp38-DX2 shRNA.
Other studies on MSCp43 have identified a small molecule that is effective in treating a lupus-like autoimmune disease in the mouse. MSCp43 binds heat shock protein gp96 on the cell surface and stabilizes gp96 dimerization, which thereby enhances its transport to the endoplasmic reticulum (ER) and inhibits dendritic cell (DC) activation and the systemic lupus erythematosus (SLE)-like phenotype. Based on this observation, a small molecule (GPM1), specifically mimicking the non-translational function of MSCp43, has been developed100. GPM1 binds and suppresses gp96 by facilitating its dimerization and retrograde transport to the endoplasmic reticulum. Importantly, administration of GPM1 reduced maturation of DCs and the SLE-like symptoms in SLE-model mice. This result offers hope that small molecule modulators may also mimic non-translational functions of aaRSs in the future.
Concluding Remarks and Future Perspective
As an enzyme family, aaRSs appear by far to be the most robust for acquiring a large variety of non-translational functions in vertebrates. Specific features of aaRSs may provide a rationale for the prevalence of new functions. One consideration is that the synthetases are ubiquitous and are amongst the most ancient proteins and, therefore, have always been available for expropriation. Furthermore, because these enzymes are essential, the retention of newly acquired activities is greatly enhanced by linking the novel functions to aaRSs. Especially, by containing an amino acid binding site, the synthetases can be adapted to pathways that work through amino acid sensing mechanisms.
However, the fusing of novel structural units that are known to promote protein-protein interactions—such as leucine zippers and GST domains—suggests that nature used a broad strategy to promote interactions of synthetases with other proteins. Collectively, these features allow each aaRS to become a hub with spokes that connect to a variety of signaling pathways, which over evolutionary time scales made their non-translational functions integral to cellular homeostasis and indispensable for the creation of higher organisms. Possibly, the capacity of the genetic code to enable all forms of life was inherently linked to continuous Darwinian adaptations in the aaRSs, which themselves still retained their role of establishing the code through tRNA aminoacylations. These adaptations, in part, may be reflected in the aaRSs being able to generate discrete protein fragments that engender non-translational functions. These specific fragmentations, by natural proteolysis or alternative splicing, suggested an unexpected plasticity and functional heterogeneity within aaRSs and their associated proteins.
Acknowledgments
This work was supported by NIH grant GM 23562 and NCI grant CA92577, and by a fellowship from the National Foundation for Cancer Research (PS), by NIH grant GM 100136, a Kimmel Scholar Award for Cancer Research, and by funding from The State of Florida to Scripps Florida (MG).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–48. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Ryckelynck M, Giegé R, Frugier M. tRNAs and tRNA mimics as cornerstones of aminoacyl-tRNA synthetase regulations. Biochimie. 2005;87:835–45. doi: 10.1016/j.biochi.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Putney SD, Schimmel P. An aminoacyl tRNA synthetase binds to a specific DNA sequence and regulates its gene transcription. Nature. 1981;291:632–5. doi: 10.1038/291632a0. [DOI] [PubMed] [Google Scholar]
- 5.Sarkar J, Poruri K, Boniecki MT, McTavish KK, Martinis SA. Yeast mitochondrial leucyl-tRNA synthetase CP1 domain has functionally diverged to accommodate RNA splicing at expense of hydrolytic editing. J Biol Chem. 2012;287:14772–81. doi: 10.1074/jbc.M111.322412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–7. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
- 7.Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci. 1997;22:211–6. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 8.Torres-Larios A, Sankaranarayanan R, Rees B, Dock-Bregeon AC, Moras D. Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase. J Mol Biol. 2003;331:201–11. doi: 10.1016/s0022-2836(03)00719-8. [DOI] [PubMed] [Google Scholar]
- 9.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–4. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–9. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Guan KL. Amino acid signaling in TOR activation. Annu Rev Biochem. 2011;80:1001–32. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- 13.Han JM, et al. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell. 2012 doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 14.Segev N, Hay N. Hijacking Leucyl-tRNA Synthetase for Amino Acid-Dependent Regulation of TORC1. Mol Cell. 2012;46:4–6. doi: 10.1016/j.molcel.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman RM. Tumor-seeking Salmonella amino acid auxotrophs. Curr Opin Biotechnol. 2011;22:917–23. doi: 10.1016/j.copbio.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Ruggiero RA, et al. Concomitant tumor resistance: the role of tyrosine isomers in the mechanisms of metastases control. Cancer Res. 2012;72:1043–50. doi: 10.1158/0008-5472.CAN-11-2964. [DOI] [PubMed] [Google Scholar]
- 17.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–4. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 18.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hattori K, Naguro I, Runchel C, Ichijo H. The roles of ASK family proteins in stress responses and diseases. Cell Commun Signal. 2009;7:9. doi: 10.1186/1478-811X-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko YG, et al. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J Biol Chem. 2001;276:6030–6. doi: 10.1074/jbc.M006189200. [DOI] [PubMed] [Google Scholar]
- 22.Wahab SZ, Yang DC. Synthesis of diadenosine 5′,5′,‴-P1, P4-tetraphosphate by lysyl-tRNA synthetase and a multienzyme complex of aminoacyl-tRNA synthetases from rat liver. J Biol Chem. 1985;260:5286–9. [PubMed] [Google Scholar]
- 23.Zamecnik P. Diadenosine 5′,5‴-P1, P4-tetraphosphate (Ap4A): its role in cellular metabolism. Anal Biochem. 1983;134:1–10. doi: 10.1016/0003-2697(83)90255-5. [DOI] [PubMed] [Google Scholar]
- 24.Bochner BR, Lee PC, Wilson SW, Cutler CW, Ames BN. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell. 1984;37:225–32. doi: 10.1016/0092-8674(84)90318-0. [DOI] [PubMed] [Google Scholar]
- 25.Lee YN, Nechushtan H, Figov N, Razin E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–51. doi: 10.1016/s1074-7613(04)00020-2. [DOI] [PubMed] [Google Scholar]
- 26.Yannay-Cohen N, et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol Cell. 2009;34:603–11. doi: 10.1016/j.molcel.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Ofir-Birin Y, et al. Structural switch of lysyl-tRNA synthetases between translation and transcription. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.10.010. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanquet S, Plateau P, Brevet A. The role of zinc in 5′,5′-diadenosine tetraphosphate production by aminoacyl-transfer RNA synthetases. Mol Cell Biochem. 1983;52:3–11. doi: 10.1007/BF00230583. [DOI] [PubMed] [Google Scholar]
- 29.Justin N, De Marco V, Aasland R, Gamblin SJ. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr Opin Struct Biol. 2010;20:730–8. doi: 10.1016/j.sbi.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Tzima E, et al. VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J Biol Chem. 2005;280:2405–8. doi: 10.1074/jbc.C400431200. [DOI] [PubMed] [Google Scholar]
- 31.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–74. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiba K. Intron positions delineate the evolutionary path of a pervasively appended peptide in five human aminoacyl-tRNA synthetases. J Mol Evol. 2002;55:727–33. doi: 10.1007/s00239-002-2368-3. [DOI] [PubMed] [Google Scholar]
- 33.Wakasugi K, et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci U S A. 2002;99:173–7. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sajish M, et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-gamma and p53 signaling. Nature chemical biology. 2012 doi: 10.1038/nchembio.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray PS, et al. Evolution of function of a fused metazoan tRNA synthetase. Mol Biol Evol. 2011;28:437–47. doi: 10.1093/molbev/msq246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerini C, et al. A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 1991;10:4267–77. doi: 10.1002/j.1460-2075.1991.tb05005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arif A, Jia J, Moodt RA, DiCorleto PE, Fox PL. Phosphorylation of glutamyl-prolyl tRNA synthetase by cyclin-dependent kinase 5 dictates transcript-selective translational control. Proc Natl Acad Sci U S A. 2011;108:1415–20. doi: 10.1073/pnas.1011275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem Sci. 2009;34:324–31. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol Cell. 2008;29:679–90. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herzog W, Muller K, Huisken J, Stainier DY. Genetic evidence for a noncanonical function of seryl-tRNA synthetase in vascular development. Circ Res. 2009;104:1260–6. doi: 10.1161/CIRCRESAHA.108.191718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukui H, Hanaoka R, Kawahara A. Noncanonical activity of seryl-tRNA synthetase is involved in vascular development. Circ Res. 2009;104:1253–9. doi: 10.1161/CIRCRESAHA.108.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X, et al. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nature communications. 2012;3:681. doi: 10.1038/ncomms1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francin M, Kaminska M, Kerjan P, Mirande M. The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain. J Biol Chem. 2002;277:1762–9. doi: 10.1074/jbc.M109759200. [DOI] [PubMed] [Google Scholar]
- 44.Frugier M, Moulinier L, Giegé R. A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J. 2000;19:2371–80. doi: 10.1093/emboj/19.10.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Negrutskii BS, Shalak VF, Kerjan P, El’skaya AV, Mirande M. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J Biol Chem. 1999;274:4545–50. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 46.He R, Zu LD, Yao P, Chen X, Wang ED. Two non-redundant fragments in the N-terminal peptide of human cytosolic methionyl-tRNA synthetase were indispensable for the multi-synthetase complex incorporation and enzyme activity. Biochim Biophys Acta. 2009;1794:347–54. doi: 10.1016/j.bbapap.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 47.Kerjan P, Cerini C, Semeriva M, Mirande M. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim Biophys Acta. 1994;1199:293–7. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 48.Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: beyond translation. J Cell Sci. 2004;117:3725–34. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- 49.Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J Mol Biol. 2000;304:983–94. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 50.Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J Mol Biol. 1999;285:183–95. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 51.Han JM, et al. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem. 2006;281:38663–7. doi: 10.1074/jbc.M605211200. [DOI] [PubMed] [Google Scholar]
- 52.Kyriacou SV, Deutscher MP. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol Cell. 2008;29:419–27. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang T, et al. AIMP3/p18 Controls Translational Initiation by Mediating the Delivery of Charged Initiator tRNA to Initiation Complex. J Mol Biol. 2012;423:475–81. doi: 10.1016/j.jmb.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 54.Park SG, Choi EC, Kim S. Aminoacyl-tRNA synthetase-interacting multifunctional proteins (AIMPs): a triad for cellular homeostasis. IUBMB Life. 2010;62:296–302. doi: 10.1002/iub.324. [DOI] [PubMed] [Google Scholar]
- 55.Zhu X, et al. MSC p43 required for axonal development in motor neurons. Proc Natl Acad Sci U S A. 2009;106:15944–9. doi: 10.1073/pnas.0901872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park SG, et al. The novel cytokine p43 stimulates dermal fibroblast proliferation and wound repair. Am J Pathol. 2005;166:387–98. doi: 10.1016/S0002-9440(10)62262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JY, et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc Natl Acad Sci U S A. 2002;99:7912–6. doi: 10.1073/pnas.122110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim MJ, et al. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet. 2003;34:330–6. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 59.Choi JW, Um JY, Kundu JK, Surh YJ, Kim S. Multidirectional tumor-suppressive activity of AIMP2/p38 and the enhanced susceptibility of AIMP2 heterozygous mice to carcinogenesis. Carcinogenesis. 2009;30:1638–44. doi: 10.1093/carcin/bgp170. [DOI] [PubMed] [Google Scholar]
- 60.Ko HS, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–78. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon NH, et al. Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc Natl Acad Sci U S A. 2011;108:19635–40. doi: 10.1073/pnas.1103922108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park BJ, et al. The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell. 2005;120:209–21. doi: 10.1016/j.cell.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 63.Kim KJ, et al. Determination of three-dimensional structure and residues of the novel tumor suppressor AIMP3/p18 required for the interaction with ATM. J Biol Chem. 2008;283:14032–40. doi: 10.1074/jbc.M800859200. [DOI] [PubMed] [Google Scholar]
- 64.Howard OM, et al. Histidyl-tRNA synthetase and asparaginyl-tRNA synthetase, autoantigens in myositis, activate chemokine receptors on T lymphocytes and immature dendritic cells. J Exp Med. 2002;196:781–91. doi: 10.1084/jem.20020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamasaki Y, et al. Unusually high frequency of autoantibodies to PL-7 associated with milder muscle disease in Japanese patients with polymyositis/dermatomyositis. Arthritis Rheum. 2006;54:2004–9. doi: 10.1002/art.21883. [DOI] [PubMed] [Google Scholar]
- 66.Xu Z, et al. Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing. Structure. 2012;20:1470–7. doi: 10.1016/j.str.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu PC, et al. In vivo sensitivity of human melanoma to tumor necrosis factor (TNF)-alpha is determined by tumor production of the novel cytokine endothelial-monocyte activating polypeptide II (EMAPII) Cancer Res. 1999;59:205–12. [PubMed] [Google Scholar]
- 68.Shalak V, et al. The EMAPII cytokine is released from the mammalian multisynthetase complex after cleavage of its p43/proEMAPII component. J Biol Chem. 2001;276:23769–76. doi: 10.1074/jbc.M100489200. [DOI] [PubMed] [Google Scholar]
- 69.Zhou Q, et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat Struct Mol Biol. 2010;17:57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Banin E, et al. T2-TrpRS inhibits preretinal neovascularization and enhances physiological vascular regrowth in OIR as assessed by a new method of quantification. Invest Ophthalmol Vis Sci. 2006;47:2125–34. doi: 10.1167/iovs.05-1096. [DOI] [PubMed] [Google Scholar]
- 71.Tzima E, et al. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc Natl Acad Sci U S A. 2003;100:14903–7. doi: 10.1073/pnas.2436330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao P, et al. Coding Region Polyadenylation Generates a Truncated tRNA Synthetase that Counters Translation Repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han JM, et al. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc Natl Acad Sci U S A. 2008;105:11206–11. doi: 10.1073/pnas.0800297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi JW, et al. AIMP2 promotes TNFalpha-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J Cell Sci. 2009;122:2710–5. doi: 10.1242/jcs.049767. [DOI] [PubMed] [Google Scholar]
- 75.Choi JW, et al. Splicing variant of AIMP2 as an effective target against chemoresistant ovarian cancer. J Mol Cell Biol. 2012;4:164–73. doi: 10.1093/jmcb/mjs018. [DOI] [PubMed] [Google Scholar]
- 76.Choi JW, et al. Cancer-associated splicing variant of tumor suppressor AIMP2/p38: pathological implication in tumorigenesis. PLoS Genet. 2011;7:e1001351. doi: 10.1371/journal.pgen.1001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–5. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 78.Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22:668–75. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- 80.Zhao Z, et al. Alanyl-tRNA synthetase mutation in a family with dominant distal hereditary motor neuropathy. Neurology. 2012;78:1644–9. doi: 10.1212/WNL.0b013e3182574f8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Froelich CA, First EA. Dominant Intermediate Charcot-Marie-Tooth disorder is not due to a catalytic defect in tyrosyl-tRNA synthetase. Biochemistry (Mosc) 2011;50:7132–45. doi: 10.1021/bi200989h. [DOI] [PubMed] [Google Scholar]
- 82.Xie W, Nangle LA, Zhang W, Schimmel P, Yang XL. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2007;104:9976–81. doi: 10.1073/pnas.0703908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–26. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 84.Stum M, et al. An assessment of mechanisms underlying peripheral axonal degeneration caused by aminoacyl-tRNA synthetase mutations. Mol Cell Neurosci. 2011;46:432–43. doi: 10.1016/j.mcn.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He W, et al. Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc Natl Acad Sci U S A. 2011;108:12307–12. doi: 10.1073/pnas.1104293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 87.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 88.Paley EL, Paley DE, Merkulova-Rainon T, Subbarayan PR. Hypoxia signature of splice forms of tryptophanyl-tRNA synthetase marks pancreatic cancer cells with distinct metastatic abilities. Pancreas. 2011;40:1043–56. doi: 10.1097/MPA.0b013e318222e635. [DOI] [PubMed] [Google Scholar]
- 89.Ghanipour A, et al. The prognostic significance of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2949–56. doi: 10.1158/1055-9965.EPI-09-0456. [DOI] [PubMed] [Google Scholar]
- 90.Mun J, et al. A proteomic approach based on multiple parallel separation for the unambiguous identification of an antibody cognate antigen. Electrophoresis. 2010;31:3428–36. doi: 10.1002/elps.201000136. [DOI] [PubMed] [Google Scholar]
- 91.Kron MA, Petridis M, Haertlein M, Libranda-Ramirez B, Scaffidi LE. Do tissue levels of autoantigenic aminoacyl-tRNA synthetase predict clinical disease? Med Hypotheses. 2005;65:1124–7. doi: 10.1016/j.mehy.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 92.Levine SM, Rosen A, Casciola-Rosen LA. Anti-aminoacyl tRNA synthetase immune responses: insights into the pathogenesis of the idiopathic inflammatory myopathies. Curr Opin Rheumatol. 2003;15:708–13. doi: 10.1097/00002281-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 93.Park SG, et al. Autoantibodies against aminoacyl-tRNA synthetase: novel diagnostic marker for type 1 diabetes mellitus. Biomarkers. 2010;15:358–66. doi: 10.3109/13547501003777823. [DOI] [PubMed] [Google Scholar]
- 94.Morbt N, et al. Chlorinated benzenes cause concomitantly oxidative stress and induction of apoptotic markers in lung epithelial cells (A549) at nonacute toxic concentrations. J Proteome Res. 2011;10:363–78. doi: 10.1021/pr1005718. [DOI] [PubMed] [Google Scholar]
- 95.Kim SS, Hur SY, Kim YR, Yoo NJ, Lee SH. Expression of AIMP1, 2 and 3, the scaffolds for the multi-tRNA synthetase complex, is downregulated in gastric and colorectal cancer. Tumori. 2011;97:380–5. doi: 10.1177/030089161109700321. [DOI] [PubMed] [Google Scholar]
- 96.Yao C, et al. P43/pro-EMAPII: a potential biomarker for discriminating traumatic versus ischemic brain injury. J Neurotrauma. 2009;26:1295–305. doi: 10.1089/neu.2008.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park MC, et al. Secreted human glycyl-tRNA synthetase implicated in defense against ERK-activated tumorigenesis. Proc Natl Acad Sci U S A. 2012;109:E640–7. doi: 10.1073/pnas.1200194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schwarz RE, et al. Antitumor effects of EMAP II against pancreatic cancer through inhibition of fibronectin-dependent proliferation. Cancer Biol Ther. 2010;9:632–9. doi: 10.4161/cbt.9.8.11265. [DOI] [PubMed] [Google Scholar]
- 99.Han JM, Park SG, Lee Y, Kim S. Structural separation of different extracellular activities in aminoacyl-tRNA synthetase-interacting multi-functional protein, p43/AIMP1. Biochem Biophys Res Commun. 2006;342:113–8. doi: 10.1016/j.bbrc.2006.01.117. [DOI] [PubMed] [Google Scholar]
- 100.Han JM, et al. Identification of gp96 as a novel target for treatment of autoimmune disease in mice. PLoS ONE. 2010;5:e9792. doi: 10.1371/journal.pone.0009792. [DOI] [PMC free article] [PubMed] [Google Scholar]